Summary
The virus‐encoded viroporins are known to modify membrane permeability and play an essential role in virus budding. Here, a comparative analysis of the membrane permeabilization capacity of a number of viroporins was performed in baby hamster kidney cells. Synthesis of 6K protein from Sindbis virus, E from mouse hepatitis virus, M2 from influenza A virus, and 2B and 3A from poliovirus enhanced membrane permeability to different extents. We show that two proteins from hepatitis C virus, p7 and NS4A, also display viroporin activity to a level comparable to 6K protein. In addition to their capacity to disrupt ionic cellular homeostasis and promote bacterial cell lysis, the expressed viroporins were able to induce cell death. Degradation of internucleosomal DNA and generation of apoptotic bodies were observed upon viroporin expression. Consistently, cleavage of translation initiation factor 4GI and poly‐(ADP‐ribose) polymerase indicated activation of effector caspase‐3. We found that poliovirus 2B localizes partially in mitochondria and induces an anomalous perinuclear distribution of these organelles. Mitochondria morphology was also altered after expression of other viroporins. Finally, detection of cytochrome c release from mitochondria suggests involvement of the mitochondrial pathway in viroporin‐induced apoptosis. These findings suggest that viroporins induce caspase‐dependent programmed cell death.
Introduction
Alterations in cellular ion homeostasis are a common hallmark during infection of mammalian cells by animal viruses (Carrasco, 1995). The vast majority of RNA animal viruses encode cytotoxic pore‐forming proteins known as viroporins, which modify membrane function at late stages of infection and facilitate budding of virions from infected cells. Viroporins are small, non‐glycosylated, highly hydrophobic viral polypeptides which interact with cell membranes and increase their permeability to ions and other low‐molecular‐weight compounds (Carrasco, 1995; Gonzalez and Carrasco, 2003). They are usually integral proteins that possess at least one amphipathic α‐helix and, in some instances, a second hydrophobic domain. Upon membrane insertion, viroporins tend to oligomerize to assemble hydrophilic pores (Pinto et al., 1992; Grice et al., 1997; Agirre et al., 2002; Melton et al., 2002; Pavlovic et al., 2003; Wilson et al., 2004). Some viroporins are not required for production of viral progeny, although their expression significantly increases the formation of new virus particles (Klimkait et al., 1990; Watanabe et al., 2001; Kuo and Masters, 2003; DeDiego et al., 2007). However, these proteins are present in low amounts both in virions and at the plasma membrane (Klimkait et al., 1990; Yu et al., 1994; Loewy et al., 1995).
The 6K protein from Sindbis virus (SV; family Togaviridae), M2 from influenza A virus (IAV; family Orthomixoviridae), and 2B and 3A from poliovirus (PV; family Picornaviridae) represent examples of well‐known viroporins (Gonzalez and Carrasco, 2003). SV 6K, a palmitoylated protein of 55 amino acids, contains a conserved sequence motif, rich in aromatic residues, consisting of two interfacial domains at the N‐terminal region, which has been described to be important both for virus budding and to enhance membrane permeability (Sanz et al., 2003). IAV M2 (96 amino acids), one of the best characterized viroporins, forms a proton channel acting both during viral entry and virion budding (Ruigrok et al., 1991; Mould et al., 2000). PV 2B protein is associated with the viral replication complex and virus‐induced membranes (Bienz et al., 1992; Schlegel et al., 1996), and localizes to the ER and Golgi complex when expressed individually (Sandoval and Carrasco, 1997). Furthermore, the coxsackievirus (CV) 2B protein has been directly implicated in the disruption of Ca2+ homeostasis during infection (van Kuppeveld et al., 1997). PV 3A exhibits significant effects on membrane rearrangement, inhibits glycoprotein trafficking, and participates in initiation of viral RNA synthesis (Suhy et al., 2000; Choe et al., 2005). Recently, we described the viroporin activity of E protein from mouse hepatitis virus (MHV‐A59; family Coronaviridae) both in bacteria and in mammalian cells (Madan et al., 2005a). Moreover, MHV variants with mutations in the E gene present aberrant morphology of virus particles as also occurs when SV 6K gene is mutated (Fischer et al., 1998). These results provide evidence for a role of some viroporins in virion morphogenesis. The viroporin group also contains viral proteins, such as HIV‐1 Vpu, human respiratory syncitial virus SH, avian reovirus p10, Japanese encephalitis virus NS2B or SARS‐CoV E protein among others (Chang et al., 1999; Lombardo et al., 2000; Gonzalez and Carrasco, 2003; Liao et al., 2006). Additionally, a number of viral glycoproteins (e.g. NS3 of bluetongue virus or gp41 of HIV‐1) have also been described to possess viroporin‐like activity (Arroyo et al., 1995; Han and Harty, 2004).
Much research effort continues to be directed towards discovery of new viroporins and analysis of their function during infection. We found that both p7 and NS4A proteins from hepatitis C virus (HCV; family Flaviviridae) share typical features with the viroporin group. p7 is a small hydrophobic protein of 63 amino acids that is not required for RNA replication (Carrere‐Kremer et al., 2002) and forms oligomeric cation channels both in planar lipid bilayers and in cells (Griffin et al., 2003; Pavlovic et al., 2003; Clarke et al., 2006). On the other hand, HCV NS4A is a small non‐structural protein known to act as an essential cofactor for serine protease activity of HCV NS3 and is therefore required for the proteolytic processing of the viral polyprotein (Failla et al., 1994). NS4A localizes to ER membranes and is responsible for directing the NS3 protease to these membranes (Wolk et al., 2000).
An increasing number of animal viruses are known to induce a defensive apoptotic response in host cells at late stages of infection (O'Brien, 1998). Several picornaviruses, including PV and CV, are able to kill cells by apoptosis in a variety of situations (Tolskaya et al., 1995; Agol et al., 1998; Carthy et al., 1998). Many other enveloped RNA viruses, such as alphaviruses, coronaviruses, influenza viruses or HCV, also induce caspase‐3‐mediated apoptosis (Grandgirard et al., 1998; An et al., 1999; Kalkeri et al., 2001; Wurzer et al., 2003). Programmed cell death or apoptosis is involved in the organized elimination of cells during tissue development and somatic cell turnover, as well as during cell senescence and organism ageing (Kerr et al., 1972; Thompson, 1995). Several pathways can lead to execution of apoptosis by activating a family of proteases known as caspases (Cohen, 1997). The extrinsic or death‐receptor pathway requires binding of ligands such as CD95 to transmembrane death receptors, followed by activation of caspase‐8 (Ashkenazi and Dixit, 1998). The intrinsic or mitochondrial pathway, triggered by mitochondrial membrane depolarization, involves disruption of the mitochondrial membrane and subsequent release from the intermembrane space of cytochrome c, which participates in formation of the apoptosome together with apoptosis protease‐activating factor‐1 and pro‐caspase 9 (Zamzami and Kroemer, 2001). In addition, an ER‐specific apoptotic pathway is activated by perturbation of ER Ca(2+) homeostasis or under ER stress conditions, involving activation of caspase‐12 and caspase‐9 (Morishima et al., 2002). Although the upstream steps of these pathways are different, they converge with the activation of the effector caspase‐3, followed by extensive degradation of nuclear DNA and fragmentation of the nucleus of apoptotic cells.
In the present work, we analysed the permeabilization kinetics of SV 6K, MHV‐A59 E, IAV M2, PV 2B and PV 3A viroporins after expression in culture cells. Our findings reveal that both p7 and NS4A proteins from HCV have the capacity to permeabilize baby hamster kidney (BHK) cells. We also provide evidence that viroporins can induce an apoptotic response in these cells. Finally, we show that in addition to its localization to the ER and Golgi compartments, PV 2B is also associated with mitochondria and alters the normal morphology of these organelles, as occurs with HCV p7 and NS4A. Together, these data suggest a previously unrecognized role of viroporins in cytotoxicity and indicate that these proteins are able to induce apoptosis upon viral infection.
Results
Permeabilization of BHK cells by viroporins from different RNA animal viruses
A comparative analysis was performed of the capacity of different viroporins to permeabilize the plasma membrane of BHK cells. To this end, the following known viroporins were selected: 6K from SV, E protein from MHV‐A59, M2 from IAV, and 2B and 3A from PV. In addition, two small proteins from HCV, p7 and NS4A, were included in the study. These two proteins were selected as potential members of the viroporin group after examining their sequences, structural features, and their Kyte‐Doolittle hydrophobicity profiles (data not shown). To analyse membrane permeabilization to the translation inhibitor hygromycin B (HB), an expression system was developed based on a SV replicon. Seven replicons were obtained by cloning the viroporin sequence after the SV capsid (C) gene (Fig. 1). Upon translation of the subgenomic mRNA, the C protein is detached by its autoproteolytic activity and the remaining protein is synthesized at levels comparable to those obtained with late proteins in SV‐infected cells (Madan et al., 2005b). This system is suitable to examine the permeabilizing capacity of each protein at different times after transfection. In vitro‐transcribed RNAs from the different pT7SVrep C + Viroporin plasmids were electroporated in BHK cells and, at 5, 8 and 16 h post electroporation (hpe), protein synthesis was estimated in the absence or presence of HB (Fig. 2). C protein and viroporin synthesis were already detected by radiolabelling with [35S]Met/Cys at 5 hpe (data not shown). Figure 2A shows a representative PAGE analysis at 8 hpe, in which viroporin bands are clearly present and migrate according to their expected molecular weight. To detect NS4A expression at 8 hpe, Western blot analysis was carried out using a monoclonal anti‐NS4A antibody. The presence of non‐proteolysed C‐NS4A product was confirmed using a rabbit serum against the C protein (Fig. 2A, lower panels). Notably, the powerful permeabilization to HB induced by PV 2B protein from 5 hpe led to almost total inhibition of protein synthesis (Fig. 2B). MHV‐A59 E protein also promoted significant entry of HB at 5 hpe that was increased at 8 hpe, when translation was inhibited by HB to about 15% of control. The presence of small amounts of mature E protein alongside significant quantities of the non‐proteolysed product C‐E was sufficient to permeabilize BHK cells (Fig. 2A). The precursor C‐E itself is probably also capable of permeabilizing BHK cells. The other viral proteins analysed exhibited a delayed permeabilization activity compared with 2B or E proteins. For example, the well‐known viroporin 6K caused increasing levels of permeabilization beginning soon after electroporation, but membrane permeability to HB was not profoundly altered until 16 hpe. Finally, IAV M2, HCV p7 and NS4A proteins all caused entry of HB to a similar extent at 16 hpe, leading to 70–75% inhibition of protein synthesis. HCV p7 also has viroporin activity in bacterial cells. Specifically, expression of p7 in Escherichia coli resulted in membrane permeabilization to HB and [3H] choline, ultimately promoting cell lysis (data not shown). Despite the low expression of NS4A, inhibition of translation by HB was about 62% at 8 hpe (Fig. 2B). Previous works have reported that NS4A inhibits translation in mouse and Huh‐7 cells (Kato et al., 2002; Kou et al., 2006). Therefore, it is possible that NS4A interferes with translation of subgenomic mRNA in BHK cells. HB did not enter control BHK cells transfected with either transcription buffer alone or with RNA encoding only the C protein (Fig. 2A and B). These results indicate that p7 and NS4A from HCV are able to permeabilize mammalian cells. In contrast, PV 3A did not lead to significant cell permeabilization, as indicated by the 34% inhibition of protein synthesis due to HB at 16 hpe.
Figure 1.
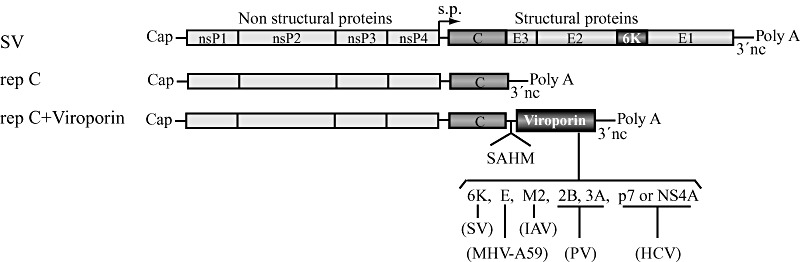
Schematic representation of the SV genome and SV‐derived replicons (RNA) with or without additional viroporin sequences. s.p., subgenomic promoter.
Figure 2.
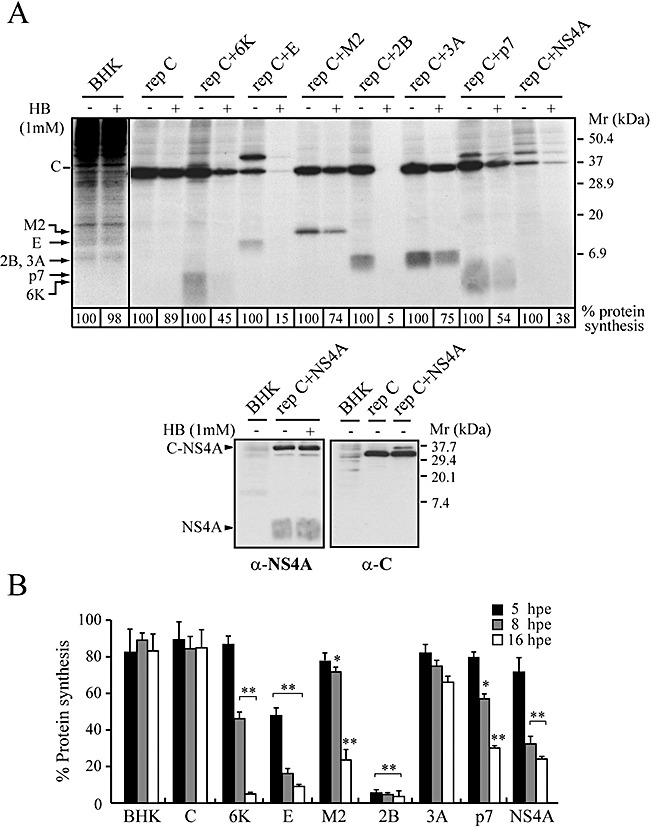
Membrane permeabilization induced by different viroporins in BHK cells. BHK cells were electroporated with in vitro‐synthesized RNA from the different constructs shown in Fig. 1. At 4, 7 or 15 hpe, proteins were labelled with [35S]Met/Cys in the absence (–) or presence (+) of 1 mM HB for 40 min. Samples were processed by SDS‐PAGE (17.5%), followed by fluorography and autoradiography. A. Membrane permeabilization assayed by the inhibition of translation as a result of HB entry induced by viroporins at 8 hpe. As negative controls, the cells were electroporated with transcription buffer alone (BHK) or with rep C. The numbers below each lane indicate the percentages of protein synthesis obtained by dividing the values obtained by densitometry in HB‐treated cells by the values obtained in untreated cells. Expression of NS4A and the non‐proteolysed product C‐NS4A at 8 hpe were analysed by Western blot using a monoclonal anti‐NS4A antibody and a rabbit polyclonal anti‐C antibody respectively (lower panels). B. Statistical analysis of membrane permeabilization caused by the indicated viroporins at different time points. Each bar represents the percentage of protein synthesis in HB‐treated cells compared with untreated cells. The SV C protein or cellular proteins in the case of control BHK cells were quantified by densitometry. All data are shown as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.005. Mr, molecular weight markers.
Viroporins promote apoptosis in BHK cells
An increasing number of viral proteins have been described to be endowed with apoptotic or anti‐apoptotic functions. HBx protein from hepatitis B virus (HBV), E1^E4 from human papillomavirus, and SV glycoproteins are examples of pro‐apoptotic proteins, whereas M11L from myxoma virus, F1L from vaccinia virus, and BHRF1 from Epstein‐Barr virus represent anti‐apoptotic proteins (Boya et al., 2004). Thus far, the viroporins MHV‐A59 E and Vpu from HIV‐1 have been reported to induce apoptosis (An et al., 1999; Akari et al., 2001). The aim of the present work was to assay the pro‐apoptotic activity of other proteins with pore‐forming properties. Chromatin condensation and nuclear DNA fragmentation with the appearance of apoptotic bodies were first analysed as typical cytological markers of programmed cell death. The nuclei of cells expressing the different viroporins selected in this study were stained with DAPI at 16 hpe and their morphology was examined. Control BHK cells and cells expressing C protein exhibited normal rounded nuclei. In contrast, all viroporins led to clear signs of apoptosis, as also occurred in positive‐control cells expressing PV 3Cpro (Barco et al., 2000) or MHV‐A59 E protein, as well as in cells treated with staurosporine (Fig. 3A). To further test whether viroporin‐mediated cell death was due to apoptosis, terminal deoxynucleotidyl transferase‐mediated dUTP nick end‐labelling (TUNEL) assay was carried out using fluorescence microscopy. An amount of TUNEL‐positive cells that was comparable to, but slightly lower than, the number with fragmented nuclei was detected at 16 hpe (Fig. 3A and B). The proportion of apoptotic cells was variable, depending on the viroporin analysed. Thus, the strongest apoptotic response was observed with PV 2B, 3A and HCV NS4A, whereas E, M2, 6K and p7 induced apoptosis in a lower proportion of cells (Fig. 3B). It should be noted that about 10% and 35% of the cells that expressed 2B and NS4A respectively were TUNEL‐positive at 8 hpe (data not shown). The proportion of apoptotic cells induced by the other viroporins was not significant at that time. Moreover, measurement of annexin V binding also revealed that the highest numbers of both annexin V‐positive cells and cells with fragmented and condensed nuclei were around 32% and 43% after 2B and NS4A expression respectively (data not shown). These results indicate that all viroporins tested can induce apoptosis of BHK cells, but 2B and NS4A are particularly powerful and rapid inducers of programmed cell death.
Figure 3.
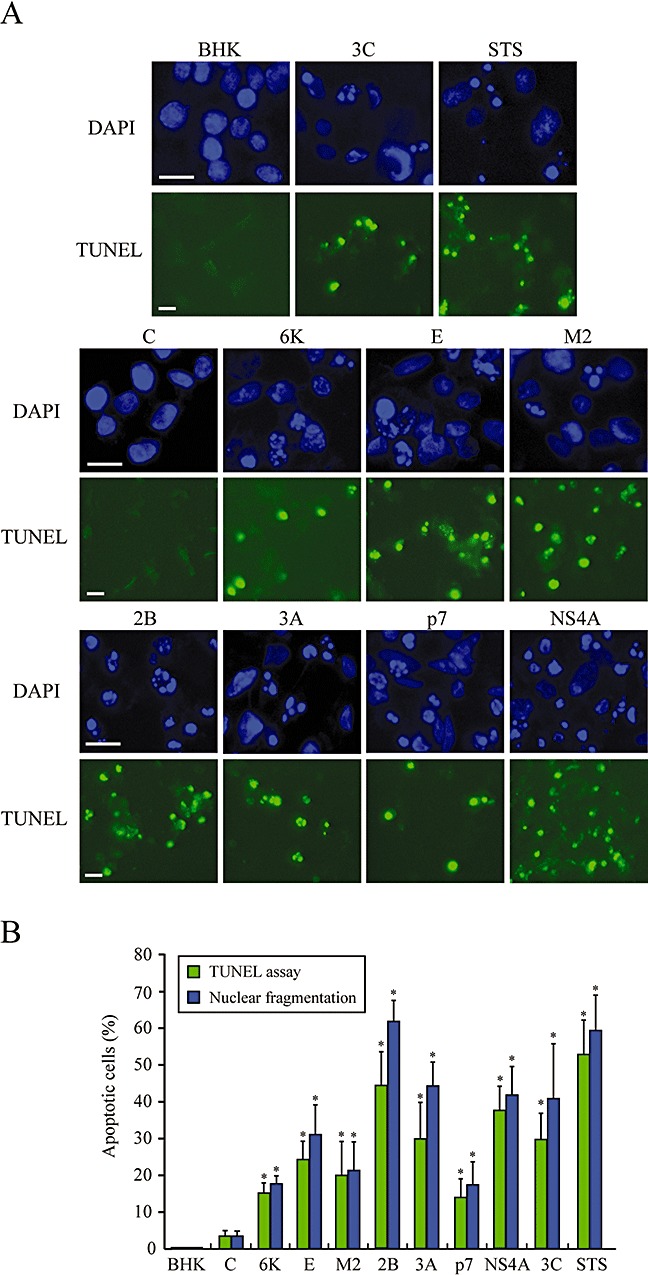
Nuclear fragmentation in BHK cells upon expression of different viroporins. A. Detection of apoptotic cells by DAPI staining and TUNEL assay. BHK cells were electroporated with the indicated viroporin replicons. At 16 hpe, the cells were fixed and permeabilized, and TUNEL assay was performed (see Experimental procedures). Cells were stained with 0.5 μg ml−1 DAPI. Cells expressing C protein or electroporated with transcription buffer alone (BHK) served as negative controls. Cells electroporated with rep C + 3Cpro, which express PV 3Cpro, and cells treated with 1 μM staurosporine (STS) for 23 h were used as positive controls. B. Percentage of TUNEL‐positive cells and cells showing nuclear fragmentation (mean ± SD). *P < 0.005.
Activation of caspase‐3 by viroporins
Activation of caspases, a family of cysteine proteases, plays a central role in the execution of apoptosis (Cohen, 1997). To assess the involvement of caspases in viroporin‐induced apoptosis, we examined activation of the effector caspase‐3. Hydrolysis of translation initiation factor 4GI (eIF4GI) is a typical phenomenon observed in apoptotic cells. Activated caspase‐3 cleaves eIF4GI at two different sites to generate characteristic fragments (Marissen and Lloyd, 1998). Thus, cleavage of eIF4GI was tested by Western blotting in cells expressing different viroporins (Fig. 4A). Previous analyses of eIF4GI using specific antibodies have revealed the existence of two proteins of ∼220 and ∼150 kDa in BHK cells. As described earlier, eIF4GI exhibits different mobility patterns in SDS‐PAGE of mammalian cells, possibly due to post‐translational modifications (Castello et al., 2006a). At 16 hpe, eIF4GI was cleaved in viroporin‐expressing cells to generate two cleavage products that migrate below the 50 kDa marker, as occurred in cells treated with inducers of apoptosis such as actinomicyn D (ActD) and cycloheximide (CHX) (Fig. 4A). Cleavage of eIF4GI also occurs in cells infected with picornaviruses or retroviruses through the action of different viral proteases (Etchison et al., 1982; Ventoso et al., 2001; Alvarez et al., 2003). Consequently, eIF4GI cleavage products observed in cells transfected with SV replicons that express either PV 2Apro or HIV‐1 PR differ from those detected in viroporin‐expressing cells (Fig. 4A, left lower panel) (Castello et al., 2006b). Notably, NS4A again acted as a potent inductor of apoptosis, such that apoptotic eIF4GI fragments could already be detected from 5 hpe and their presence was even clearer at 8 hpe. This result is consistent with the observation of a significant number of TUNEL‐positive cells at 8 hpe following transfection with rep C + NS4A (data not shown). However, at these early times, there was no evidence of eIF4GI proteolysis in cells transfected with other viroporins.
Figure 4.
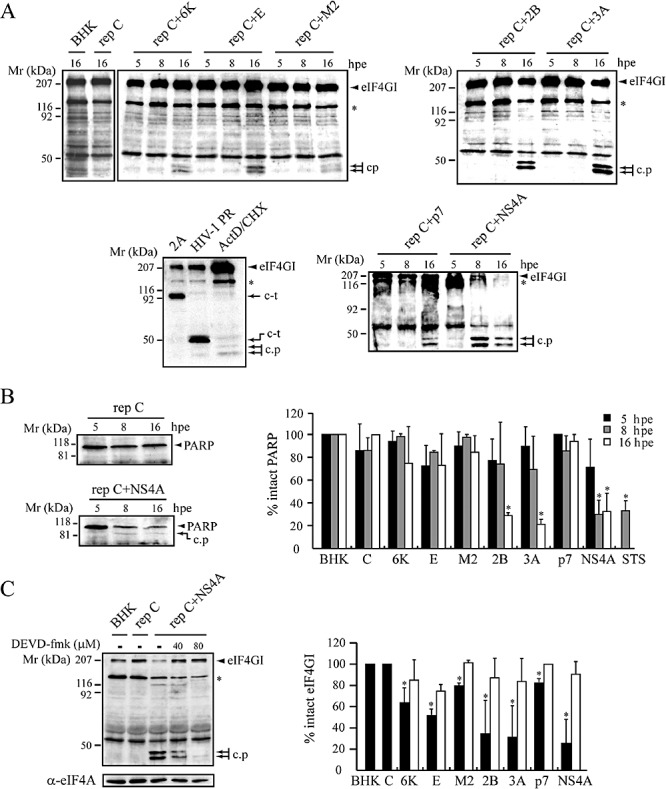
Activation of caspase‐3 induced by several viroporins. Whole‐cell lysates from BHK cells expressing viroporins at 5, 8 or 16 hpe were processed by SDS‐PAGE. Cells expressing C protein or electroporated with transcription buffer alone (BHK) served as negative controls. A. Cleavage of eIF4GI mediated by caspase‐3 activation. Two eIF4GI apoptotic cleavage products (c.p) were detected by Western blotting using a polyclonal anti‐eIF4GI antibody. In the lower left panel, the cleavage products of eIF4GI generated by viral proteases (PV 2 A and HIV‐1 PR) are shown. c‐t, eIF4GI carboxyl‐terminal fragment. As positive controls, cells were incubated with 5 μg ml−1 ActD and 25 μg ml−1 CHX for 16 h to induce apoptotic cleavage of eIF4GI by caspase‐3. The 150 kDa protein related to eIF4GI is indicated with an asterisk. B. Cleavage of PARP in BHK cells expressing viroporins. Proteolysis of PARP was analysed by Western blotting using a monoclonal anti‐PARP antibody. A representative experiment shows early PARP cleavage induced by NS4A expression (left panel). c.p, apoptotic PARP cleavage product. The percentage of intact PARP at 5, 8 and 16 hpe was calculated by densitometry of the PARP band (right diagram). Cells treated with 2 μM staurosporine (STS) for 8 h were used as positive controls (mean ± SD of three independent experiments). *P < 0.05. C. Inhibition of eIF4GI cleavage. Cells expressing the indicated viroporins were treated with or without DEVD‐fmk (40 μM) at 4 hpe and incubated for 12 h at 37°C. Cell lysates were analysed by Western blotting using anti‐eIF4GI antibodies. Inhibition of early eIF4GI cleavage in cells expressing NS4A at 8 hpe (left panel). To measure protein loading, eIF4A or α‐tubulin (not shown) were detected. Percentage of eIF4GI intact in untreated (solid bars) and DEVD‐fmk‐treated cells (grey bars) at 16 hpe was calculated by densitometry of the eIF4GI bands. Each bar shows the percentage of intact eIF4GI compared with control BHK cells (mean ± SD of three independent experiments). *P < 0.05.
Poly‐(ADP‐ribose) polymerase (PARP) is another well‐characterized caspase‐3 substrate. Although all the viroporins tested induced hydrolysis of eIF4GI, only expression of PV 2B, 3A and NS4A led to significant PARP cleavage at 16 hpe. NS4A was a powerful inducer of apoptosis, because a notable reduction in the level of intact PARP was already observed at 8 hpe (Fig. 4B). The ∼89 kDa cleavage product detected in these cells corresponds to that described for PARP proteolysis by caspase‐3 (Boulares et al., 1999). Taken together, these results indicate that eIF4GI is possibly a better substrate for caspase‐3 than PARP. Furthermore, caspase‐3 activation was indicated by the observation that DEVD‐fmk, an irreversible cell‐permeable inhibitor of caspase‐3, efficiently blocked eIF4GI cleavage induced by NS4A and the other viroporins at 8 and/or 16 hpe (Fig. 4C). Because transfection of rep C did not cause proteolysis of either eIF4GI or PARP, our findings indicate that viroporins activate caspase−3.
Association of viroporins with mitochondria
Most viroporins are described to be located mainly in the internal membrane systems, i.e. ER, ERGIC or Golgi apparatus, and to a lesser extent at the plasma membrane (Yu et al., 1994; Gonzalez and Carrasco, 2003). However, in the last few years, HCV p7 protein and several viral pro‐apoptotic proteins have been also found to be partially associated with mitochondria (Griffin et al., 2004; D'Agostino et al., 2005). Therefore, we decided to explore the involvement of the mitochondrial pathway in apoptosis induced by viroporins. First, we tested the possible colocalization of 2B and NS4A with Mitotracker and other organelle markers (Fig. 5A–F). Both proteins partially colocalized with Mitotracker in BHK (Fig. 5C and F) and Huh‐7 cells (data not shown). Interestingly, in both 2B‐ and NS4A‐expressing cells, mitochondria did not exhibit their typical long shape, appearing instead as punctate entities (Fig. 5, compare distribution of Mitotracker in C, F and G with control BHK cells in G, upper left panel). In addition, it should be noted that 2B and p7 expression also led to accumulation of mitochondria around the nucleus or at nuclear poles respectively (Fig. 5C and G). 2B was also present in the ER and predominantly in the Golgi, while NS4A colocalized with ER but appeared to be excluded from the Golgi (Fig. 5A, B, D and E). Mitochondrial localization of the other viroporins was also analysed. Unlike 2B and NS4A, none of the other viroporins analysed colocalized with mitochondria (data not shown). However, the expression of 6K, E and 3A altered the normal morphology of mitochondrial network (Fig. 5G, compare viroporin‐expressing cells with normal BHK or C‐expressing cells).
Figure 5.
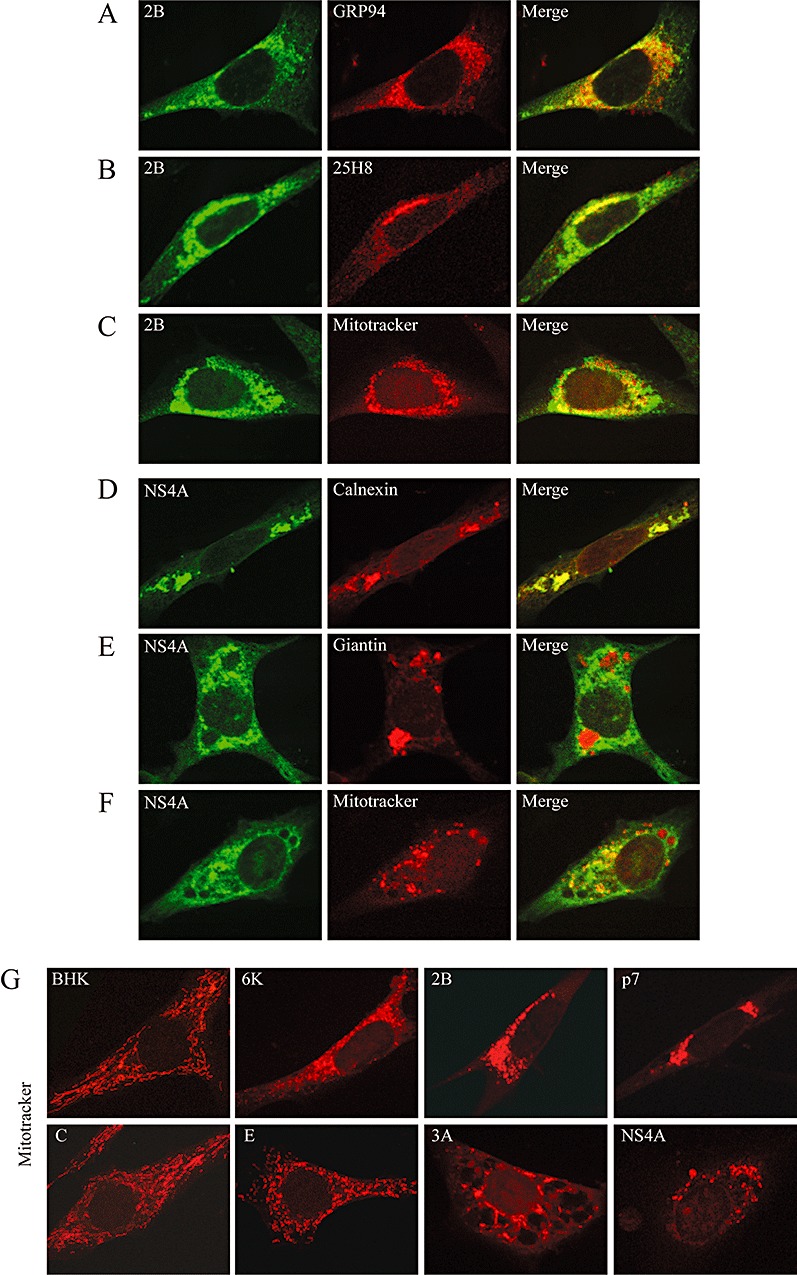
Colocalization of PV 2B and HCV NS4A proteins with mitochondrial and organelle markers. BHK cells expressing PV 2B (A–C) or HCV NS4A (D–F) were fixed at 8 hpe, permeabilized and double‐stained with anti‐2B and anti‐NS4A antibodies respectively, and different organelle markers. Antibodies against GRP94 and calnexin were used as ER markers (A, D). Antibody 25H8 and anti‐giantin antibodies were used as Golgi markers (B, E). Cells were incubated prior to fixation for 45 min with Mitotracker Red to stain mitochondria (C, F). Overlaid images are shown on the right. G. Mitochondria staining in cells transfected with viroporin replicons at 16 hpe.
Cytochrome c release from mitochondria in cells expressing viroporins
Disruption of the mitochondrial transmembrane potential and permeabilization of the outer mitochondrial membrane lead to the release of apoptogenic factors, such as cytochrome c, from the intermembrane space to the cytosol (Zamzami and Kroemer, 2001). Therefore, the distribution of cytochrome c in cells expressing viroporins was studied by immunostaining. In cells transfected with transcription buffer alone or which express C protein alone, cytochrome c is usually present in long structures corresponding to mitochondria dispersed throughout the cytoplasm (Fig. 6A). Interestingly, the distribution of cytochrome c changed drastically in viroporin‐transfected cells and was found to be spread throughout the entire cytoplasm, as observed in ActD–CHX‐positive control cells (Fig. 6A, white arrows). Notably, 16 h after expression of PV 2B, 3A and NS4A, nearly 60% of the cells had released cytochrome c, while in the case of NS4A, a similar proportion was observed as early as 7 hpe (Fig. 6B and data not shown). About 10% of cells that synthesized SV 6K and HCV p7 exhibited a diffuse staining pattern at 16 hpe, while pro‐apoptotic E protein and IAV M2 induced cytochrome c release in a slightly higher proportion (∼20%). These results are in accordance with our findings regarding the activation of caspase‐3. Therefore, mitochondrial alterations and cytochrome c efflux strongly suggest involvement of the mitochondrial pathway in apoptosis induced by viroporins.
Figure 6.
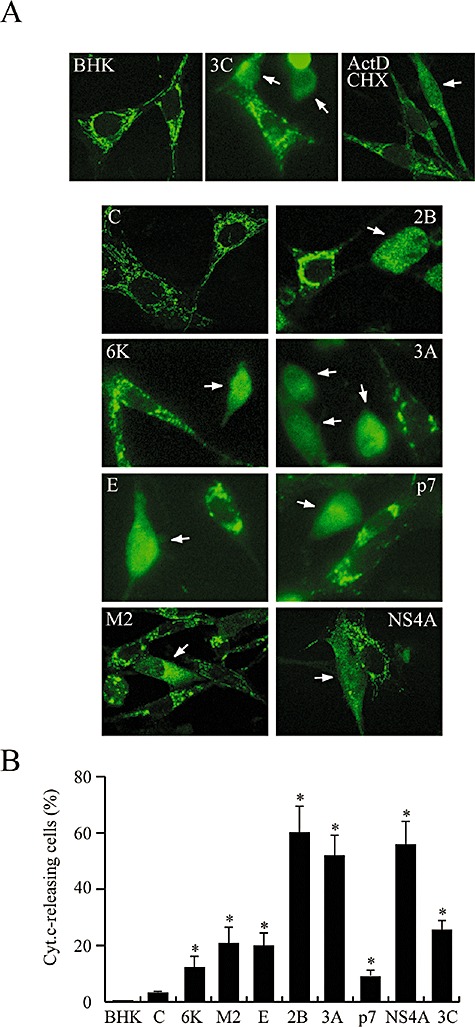
Cytochrome c release in cells expressing viroporins. A. BHK cells expressing the different viroporins as well as PV 3Cpro (upper middle panel) were fixed at 16 hpe, permeabilized and incubated with mouse anti‐cytochrome c antibodies and secondary Alexa Fluor 488‐conjugated anti‐mouse IgG. Cells treated with 5 μg ml−1 ActD and 100 μg ml−1 CHX, or 3C‐expressing cells were used as positive controls (upper right and middle panels). Cells expressing C protein or electroporated with transcription buffer alone (BHK) served as negative controls. Arrows indicate cells in which efflux of cytochrome c is clearly observed. B. Percentage of cells exhibiting cytochrome c release (mean ± SD). *P < 0.05.
Discussion
Recent studies have elucidated a novel role for changes in intracellular ion homeostasis as important modulators of the cell death programme (Franco et al., 2006). Our present results provide new evidence that viroporins induce apoptotic cell responses. Five viroporins (SV 6K, MHV‐A59 E, IAV M2, and PV 2B and 3A) encoded by different RNA viruses, as well as two potential viroporins, p7 and NS4A from HCV, were expressed in mammalian cells by means of a SV‐derived replicon. Our findings provide evidence that p7 and NS4A from HCV enhance membrane permeability in BHK cells. These results agree well with reports that p7 assembles cation‐selective ion channels in membranes, leading to increased membrane permeability of bacterial cells (Clarke et al., 2006; Madan, 2006). Although HCV NS4A fulfils many features of viroporins, its ability to assemble membrane pores still remains unexplored. The extent and time‐course of membrane permeabilization depend on the viroporin tested. Thus, the viroporins analysed can be classified in two groups: those that induce rapid permeabilization, and those in which permeabilization is induced more slowly. The first group includes PV 2B and MHV‐A59 E, because both polypeptides efficiently permeabilized the plasma membrane soon after their synthesis. In contrast, SV 6K, IAV M2, and HCV p7 and NS4A required a longer period of expression to exert extensive membrane permeabilization. PV 3A has the capacity to perturb bacterial membranes (Lama and Carrasco, 1992), but it does not lead to significant permeabilization of either BHK‐21 cells (this study) or HeLa cells (Aldabe et al., 1996). The lack of activity of PV 3A in mammalian cells, even after a long period of expression, may be due to its efficient inhibition of ER‐to‐Golgi transport, thereby interfering with the localization of 3A itself at the plasma membrane (Doedens et al., 1997). Alternatively, the delivery of 3A to the plasma membrane may require the presence of other PV proteins. Notably, PV 2B displayed the highest permeabilizing capacity. Picornavirus 2B also exhibits anti‐secretory activity, but in this case the inhibition is only partial (Cornell et al., 2006). 2B is endowed with a potent capacity to cause profound and rapid alterations in cellular ion homeostasis. Thus, CV 2B induces Ca2+ release from intracellular stores (ER and Golgi) and subsequent extracellular Ca2+ influx to the cytoplasm (van Kuppeveld et al., 1997). Therefore, a generalized permeabilization of both intracellular compartments and plasma membrane mediated by 2B can provoke an ionic imbalance, leading to disruption of the permeability barrier.
Several cellular ion channels participate in apoptotic cell death (Lang et al., 2003). Perturbation of ion homeostasis is a common hallmark of apoptosis, giving rise to depolarization of the plasma membrane associated with intracellular cation overload and cell volume decreases due to anion and H2O efflux (Waring, 2005; Burg et al., 2006; Franco et al., 2006). Therefore, deregulation of ion channels provokes various pathological conditions or channelopathies (Jentsch et al., 2004). Our data indicate that several viroporins trigger programmed cell death in BHK cells. Consistent with these findings, coronavirus E protein was described as an inducer of apoptosis before its viroporin activity was uncovered (An et al., 1999). The viroporins tested in this study produced both DNA fragmentation and generation of apoptotic bodies, as revealed by TUNEL assay and DAPI staining, after several hours of expression in BHK cells. Furthermore, cleavage of eIF4GI and PARP indicated that viroporin‐induced apoptosis was dependent on caspase activation. The fact that the inhibitor DEVD‐fmk abrogated eIF4GI cleavage provides further evidence supporting the involvement of caspase‐3 in this process. Notably, the most rapid induction of caspase‐3 activation was seen with HCV NS4A, followed by PV 2B and 3A. These results agree well with the recent finding that NS4A renders Huh‐7 cells prone to undergo apoptosis (Nomura‐Takigawa et al., 2006). It is possible that the ability of NS4A to inhibit host and viral translation (Kou et al., 2006), together with its permeabilizing capacity, accounts for the rapid development of apoptosis. Consistent with our findings, a number of viral pro‐apoptotic proteins contain amphipathic α‐helices that have potential pore‐forming properties, as occurs with both Vpr from HIV‐1 and HBx from HBV (Boya et al., 2004).
Our present findings indicate that membrane permeabilization takes place before the appearance of clear signs of apoptosis (i.e. caspase activation). The permeabilizing capacity of 2B, 6K, M2 and p7 was proportional to the degree of cell death observed in each case. However, a large number of cells expressing 3A underwent apoptosis, despite the fact that no significant plasma membrane permeabilization was detected. Most probably, inhibition of the vesicular transport system by PV 3A also activates alternative apoptotic pathways. Consistent with this possibility, a recent report indicated that mutations in GABA(A) receptors lead to retention of channel molecules in the ER, thereby causing ER stress followed by apoptosis (Hirose, 2006). On the other hand, coronavirus E induced efficient membrane permeabilization but did not provoke apoptosis in a high proportion of cells. These findings may suggest that besides induction of permeability changes, some viroporins could exert other effects on cellular functions to regulate the degree of apoptosis progression. Nevertheless, the pro‐apoptotic activity of each viroporin alone might be weakened or enhanced in the context of viral infection, where other pro‐apoptotic or anti‐apoptotic viral proteins can coexist.
The partial association of HCV p7 and NS4A with mitochondria has been reported previously (Griffin et al., 2004; Nomura‐Takigawa et al., 2006). Our study revealed that a low proportion of NS4A not only colocalized with mitochondria in BHK cells but also produced mitochondria fragmentation. NS4A was abundant in the ER but was excluded from the Golgi compartment. Notably, we found that 2B protein was also associated with mitochondria and induced a perinuclear redistribution of these organelles. A similar anomalous organization of the mitochondrial network was observed in cells expressing HCV p7, PV 3A or SV 6K, suggesting that viroporins may cause mitochondrial alterations. In accordance with these results, several pro‐apoptotic viral proteins, such as HBV HBx or E1^E4 protein from human papillomavirus, also induce dramatic alterations in mitochondrial morphology during the early stages of apoptosis (Takada et al., 1999; Karbowski and Youle, 2003; Raj et al., 2004). Furthermore, we found that 2B, 3A and NS4A triggered the release of cytochrome c in a high proportion of cells. The other viroporins also induced this mitochondrial response but affected a lower number of cells. Therefore, the present work suggests that viroporin expression induces mitochondria‐mediated apoptosis.
In contrast to our findings that PV viroporins induce apoptosis in BHK cells, an anti‐apoptotic effect mediated by picornavirus 2B and 3A has been reported in HeLa cells (Neznanov et al., 2001; Campanella et al., 2004). Several possibilities can be put forward to account for these apparent discrepancies. It is possible that a critical intracellular level of PV viroporins is required to trigger anti‐apoptotic or apoptotic pathways. Thus, transfection of plasmids or mRNAs renders a relatively low expression of 3A and 2B, which may result in their lack of effect or even inhibition of apoptosis (Neznanov et al., 2001; Campanella et al., 2004; Salako et al., 2006). However, in the present study the use of a SV‐derived replicon (rep C + 2B) guaranteed high expression of PV 2B protein, similar to that found during PV infection, leading to clear signs of apoptosis in transfected cells. Campanella and coworkers demonstrated that CV 2B expression decreases the Ca2+ content of both ER and the Golgi stores, and that this results in downregulation of Ca2+ signalling to mitochondria and prevents apoptosis (Campanella et al., 2004). Our findings provide evidence for a novel effect of 2B in mitochondria. The localization of a pool of 2B in mitochondria could directly affect their membrane permeability and disrupt Ca2+ regulation. Ca2+ plays an important role in the regulation of mitochondrial connectivity during apoptosis (Pinton et al., 2001; Rapizzi et al., 2002). Consistent with this possibility, 2B altered the normal mitochondrial network pattern. Pro‐apoptotic HBx protein from HBV also associates with mitochondria and mediates alteration of cytosolic calcium, which is a fundamental requirement for viral replication (Bouchard et al., 2001). A given viral protein may exhibit anti‐ or pro‐apoptotic activities depending on several factors, as is the case in NS1 from IAV and HIV‐1 Vpr (Lowy, 2003; Moon and Yang, 2006). This dual behaviour may have a physiological significance for viral replication. During the early phase of infection when the expression level of PV proteins is low, 2B and 3A may protect infected cells from apoptosis, thereby permitting RNA replication. However, at later stages, when high levels of viroporins are synthesized and permeabilization of cells is evident (Lopez‐Rivas et al., 1987; Carrasco, 1995), both 2B and 3A, together with 3Cpro and 2Apro, lead to induction of apoptosis in the infected cells (Barco et al., 2000; Calandria et al., 2004). Thus, apoptosis induced by 3Cpro, 2Apro, 2B and 3A could represent an efficient mechanism for the spread of PV virions, avoiding host immune inflammatory responses. The cell line used could also influence the development of apoptosis. Moreover, the reaction of the host apoptotic machinery to picornavirus infection has been reported to be generally cell‐dependent and affected by the cell's environment, the status of its differentiation, and the genotype (Romanova et al., 2005). In agreement with our results, the apoptotic programme induced by PV in murine cells involves damaged mitochondria, efflux of cytochrome c, and activation of caspase‐9, followed by activation of downstream effector caspases. The existence of possible interactions between viroporins and mitochondrial factors or some other endogenous proteins will open an interesting field of investigation that may lead us to a better understanding of virus biology and the role of ionic alterations during infection. Further studies of viroporins and their apoptotic effects could help to clarify the pathogenesis of different viral diseases and will lay the foundations for the development of novel antiviral compounds to block viroporin activity.
Experimental procedures
Cell culture and SV replicons
Baby hamster kidney (BHK)‐21 cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and non‐essential amino acids.
Five SV replicons containing the sequences coding for 2B and 3C proteins from PV (strain Mahoney‐1), M2 protein from IAV, and p7 and NS4A proteins from HCV type b (Con1 isolate, gift from Dr R. Bartenschlager, Johannes‐Gutenberg University, Mainz, Germany) were obtained by cloning the NdeI/BamHI‐digested PCR fragment, encoding each protein, after the sequence encoding the SV C protein in the plasmid pH 3′2 J‐C (Sanz and Carrasco, 2001), using the same restriction sites. In a second step, a fragment digested with AatII and XhoI was inserted into AatII/XhoI‐digested pT7SVwt vector (Sanz and Carrasco, 2001) to finally generate the plasmids pT7SVrep C + 2B, pT7SVrep C + 3C, pT7SVrep C + M2, pT7SVrep C + p7 and pT7SVrep C + NS4A. Replicons encoding 6K protein from SV, E protein from MHV‐A59 or only C protein have been described elsewhere (Madan et al., 2005a). Plasmid inserts from PCR amplifications were confirmed by sequencing with standard techniques.
Antibodies
Specific rabbit polyclonal antibodies against the PV protein 2B were obtained by inoculation of maltose binding protein−2B fusion proteins (Barco and Carrasco, 1995). Mouse monoclonal antibodies against HCV NS4A protein were generously provided by A.A. Kushch (Ivanovsky Institute of Virology, Moscow, Russia). Rat monoclonal antibodies against GRP94 (Affinity BioReagents) and rabbit polyclonal antibodies against calnexin (Stressgen) were used as organelle markers for ER. Mouse monoclonal antibodies 25H8 (a generous gift from I.V. Sandoval, Centro de Biología Molecular, Madrid, Spain) and rabbit polyclonal antibodies against giantin (Abcam) were used as Golgi markers. Alexa 488‐ or Alexa 555‐conjugated goat anti‐mouse, anti‐rabbit and anti‐rat IgG (Molecular Probes) were used as secondary antibodies.
Transfection of BHK‐21 cells
Baby hamster kidney (BHK)‐21 cells were electroporated with in vitro‐synthesized mRNAs. Transcription reactions were carried out with T7 RNA polymerase (Promega) and the corresponding plasmids as templates, according to the manufacturer's instructions. Subconfluent cells were harvested, washed with ice‐cold phosphate‐buffered saline (PBS), and resuspended in PBS at a density of approximately 2.5 × 106 cells ml−1. An aliquot (50 μl) of transcription mixture containing 15 μg RNA from each of the different DNA replicons was added to 0.4 ml of cell suspension. This mixture was transferred to a 2 mm electroporation cuvette (Bio‐Rad). Electroporation was performed at room temperature (RT) by generating two consecutive 1.5 kV, 25 μF pulses using a Gene Pulser apparatus (Bio‐Rad), as previously described (Liljestrom and Garoff, 1991). Finally, cells were diluted in DMEM supplemented with 10% FCS and seeded onto culture plates. Control BHK cells were electroporated with 50 μl transcription buffer in PBS.
Permeabilization assay
Baby hamster kidney (BHK) cells electroporated with the corresponding RNA synthesized in vitro from the different constructs or with transcription buffer alone were further seeded in wells of an L‐24 plate. At different time points, cells were pretreated with 1 mM HB (Clontech) for 20 min at 37°C, or left untreated. Next, proteins were radiolabelled for 40 min with 10 μCi [35S]Met/Cys (Promix; Amersham Pharmacia) in methionine/cysteine‐free DMEM in the presence or absence of 1 mM HB. Finally, cells were collected in sample buffer, boiled for 4 min and analysed by SDS‐PAGE (17.5%) and fluorography. Protein synthesis was quantified by densitometry using a GS‐710 calibrated Imaging Densitometer (Bio‐Rad) and calculated by dividing the values obtained for samples treated with HB by the corresponding values obtained from untreated cells. Protein synthesis was quantified by densitometry of either the C protein band or a cellular protein band.
Analysis of apoptosis
Changes in nuclear morphology and appearance of apoptotic bodies were assessed by DAPI staining of culture cells. BHK cells grown on coverslips were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min, permeabilized with 0.2% Triton X‐100 in PBS for 5 min and stained with DAPI (0.5 μg ml−1) at RT for 2 min. The morphology of the cell nuclei was examined with a fluorescence Axiovert 200 inverted microscope (Zeiss). The protein synthesis inhibitor CHX (100 μg ml−1) and the transcription inhibitor actinomycin D (5 μg ml−1), or staurosporine (1 μM), were used as non‐viral inducers of apoptosis.
Terminal deoxynucleotidyl transferase‐mediated dUTP nick end‐labelling (TUNEL) assay
Cells electroporated with the different SV replicons (RNAs synthesized in vitro) were seeded on coverslips and incubated at 37°C. For detection of apoptosis, TUNEL assay was performed using the Fluorescein FragELTM DNA Fragmentation Detection kit (Clontech) according to the manufacturer's instructions. Briefly, 18 h after electroporation, BHK cells were fixed with 4% PFA, permeabilized with 0.2% Triton X‐100 at RT and rinsed with Tris‐buffered saline (TBS; Tris 20 mM, pH 7.4 plus 140 mM NaCl). After equilibration for 20 min at RT, cells were overlaid with 60 μl of terminal deoxynucleotidyl transferase (TdT)‐labelling reaction mix containing 3 μl of TdT enzyme and incubated in a humidified chamber for 60 min at 37°C. Coverslips were incubated in TBS for 1 min at RT and washed again twice with TBS before DAPI staining (described above). Finally, coverslips were mounted in Moviol/DABCO (Calbiochem), and apoptotic nuclei were analysed by fluorescence microscopy. To obtain the percentages of TUNEL‐positive cells and DAPI‐positive cells (apoptotic cells), cells of 10 random fields (approximately 1000–1500 cells) were counted in each sample. Results were all expressed as means ± SD.
Immunofluorescence microscopy
Baby hamster kidney (BHK) cells electroporated with the SV replicons were seeded on coverslips, fixed in 4% PFA for 15 min, washed twice in PBS, and then permeabilized for 10 min with 0.2% Triton X‐100 in PBS. All antibody incubations were carried out for 1 h in PBS containing 0.1% FCS and 0.1% Triton X‐100. Coverslips were washed three times with PBS between primary and secondary antibody incubations. For mitochondria staining, cells were incubated with 2 μM Mitotracker Red CMH2Ros (Molecular Probes) for 45 min before fixation. Anti‐cytochrome c monoclonal antibodies (BD Pharmingen, clone 6H2.B4) were diluted 1:100 and incubated for 2 h. Coverslips were mounted in ProLong Gold anti‐fade reagent (Invitrogen) and examined with a Radiance 2000 (Bio‐Rad/Zeiss) confocal laser scanning microscope.
Western blotting
Electroporated cells expressing the different viral proteins were collected in sample buffer at the same times indicated for the permeabilization assay and the lysates were processed by SDS‐PAGE. After electrophoresis, proteins were transferred to a nitrocellulose membrane as described previously (Barco and Carrasco, 1995). The integrity of two caspase‐3 substrates, eIF4GI and PARP, was analysed using a rabbit anti‐eIF4GI serum raised against peptides derived from the N‐ and C‐terminal regions of human eIF4GI at a 1:1000 dilution and a mouse monoclonal anti‐PARP antibody (BD Pharmingen) at a 1:350 dilution respectively. Antibodies against eIF4A at a 1:50 dilution (a gift from Dr H. Trachsel, Institute for Biochemistry and Molecular Biology, University of Berne, Switzerland) were used to evaluate protein loading. To detect HCV NS4A protein and SV C protein, monoclonal anti‐NS4A antibodies (Masalova et al., 2002) at a 1:1000 dilution and a polyclonal rabbit serum against the SV C protein at a 1:10 000 dilution were used. Incubation with primary antibodies was performed for 2 h at RT, then the membrane was washed three times with PBS containing 0.2% Tween‐20 and incubated for 1 h with horseradish peroxidase‐conjugated anti‐mouse (Promega) or anti‐rabbit IgG antibodies (Amersham) at a 1:10 000 dilution. After washing three times, protein bands were visualized with the ECL detection system (Amersham).
Statistical analysis
Data are presented as mean values ± SD. Differences were tested for significance by means of the Student's t‐test. Viroporin effects on BHK cells were compared with respect to the control (transfection of rep C). A probability level P < 0.05 was considered significant.
Acknowledgements
This study was supported by Grant BFU2006‐02182/BMC from the Dirección General de Investigación Científica y Técnica, Spain, and an Institutional Grant awarded to the Centro de Biología Molecular Severo Ochoa by the Fundación Ramón Areces. A.C. is the holder of an FPI Fellowship.
References
- Agirre, A. , Barco, A. , Carrasco, L. , and Nieva, J.L. (2002) Viroporin‐mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J Biol Chem 277: 40434–40441. [DOI] [PubMed] [Google Scholar]
- Agol, V.I. , Belov, G.A. , Bienz, K. , Egger, D. , Kolesnikova, M.S. , Raikhlin, N.T. , et al. (1998) Two types of death of poliovirus‐infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252: 343–353. [DOI] [PubMed] [Google Scholar]
- Akari, H. , Bour, S. , Kao, S. , Adachi, A. , and Strebel, K. (2001) The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB‐dependent expression of antiapoptotic factors. J Exp Med 194: 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldabe, R. , Barco, A. , and Carrasco, L. (1996) Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem 271: 23134–23137. [DOI] [PubMed] [Google Scholar]
- Alvarez, E. , Menendez‐Arias, L. , and Carrasco, L. (2003) The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J Virol 77: 12392–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, S. , and Chen, C.J. , Yu, X. , Leibowitz, J.L. , and Makino, S. (1999) Induction of apoptosis in murine coronavirus‐infected cultured cells and demonstration of E protein as an apoptosis inducer. J Virol 73: 7853–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo, J. , Boceta, M. , Gonzalez, M.E. , Michel, M. , and Carrasco, L. (1995) Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Virol 69: 4095–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi, A. , and Dixit, V.M. (1998) Death receptors: signaling and modulation. Science 281: 1305–1308. [DOI] [PubMed] [Google Scholar]
- Barco, A. , and Carrasco, L. (1995) A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae . EMBO J 14: 3349–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco, A. , Feduchi, E. , and Carrasco, L. (2000) Poliovirus protease 3C (pro) kills cells by apoptosis. Virology 266: 352–360. [DOI] [PubMed] [Google Scholar]
- Bienz, K. , Egger, D. , Pfister, T. , and Troxler, M. (1992) Structural and functional characterization of the poliovirus replication complex. J Virol 66: 2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard, M.J. , Wang, L.H. , and Schneider, R.J. (2001) Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294: 2376–2378. [DOI] [PubMed] [Google Scholar]
- Boulares, A.H. , Yakovlev, A.G. , Ivanova, V. , Stoica, B.A. , Wang, G. , Iyer, S. , and Smulson, M. (1999) Role of poly (ADP‐ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3‐resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem 274: 22932–22940. [DOI] [PubMed] [Google Scholar]
- Boya, P. , Pauleau, A.L. , Poncet, D. , Gonzalez‐Polo, R.A. , Zamzami, N. , and Kroemer, G. (2004) Viral proteins targeting mitochondria: controlling cell death. Biochim Biophys Acta 1659: 178–189. [DOI] [PubMed] [Google Scholar]
- Burg, E.D. , Remillard, C.V. , and Yuan, J.X. (2006) K+ channels in apoptosis. J Membr Biol 209: 3–20. [DOI] [PubMed] [Google Scholar]
- Calandria, C. , Irurzun, A. , Barco, A. , and Carrasco, L. (2004) Individual expression of poliovirus 2Apro and 3Cpro induces activation of caspase‐3 and PARP cleavage in HeLa cells. Virus Res 104: 39–49. [DOI] [PubMed] [Google Scholar]
- Campanella, M. , De Jong, A.S. , Lanke, K.W. , Melchers, W.J. , Willems, P.H. , Pinton, P. , et al. (2004) The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J Biol Chem 279: 18440–18450. [DOI] [PubMed] [Google Scholar]
- Carrasco, L. (1995) Modification of membrane permeability by animal viruses. Adv Virus Res 45: 61–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrere‐Kremer, S. , Montpellier‐Pala, C. , Cocquerel, L. , Wychowski, C. , Penin, F. , and Dubuisson, J. (2002) Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J Virol 76: 3720–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthy, C.M. , Granville, D.J. , Watson, K.A. , Anderson, D.R. , Wilson, J.E. , Yang, D. , et al. (1998) Caspase activation and specific cleavage of substrates after coxsackievirus B3‐induced cytopathic effect in HeLa cells. J Virol 72: 7669–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A. , Alvarez, E. , and Carrasco, L. (2006a) Differential cleavage of eIF4GI and eIF4GII in mammalian cells. Effects on translation. J Biol Chem 281: 33206–33216. [DOI] [PubMed] [Google Scholar]
- Castello, A. , Sanz, M.A. , Molina, S. , and Carrasco, L. (2006b) Translation of Sindbis virus 26S mRNA does not require intact eukariotic initiation factor 4G. J Mol Biol 355: 942–956. [DOI] [PubMed] [Google Scholar]
- Chang, Y.S. , Liao, C.L. , Tsao, C.H. , Chen, M.C. , Liu, C.I. , Chen, L.K. , and Lin, Y.L. (1999) Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J Virol 73: 6257–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S.S. , Dodd, D.A. , and Kirkegaard, K. (2005) Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337: 18–29. [DOI] [PubMed] [Google Scholar]
- Clarke, D. , Griffin, S. , Beales, L. , Gelais, C.S. , Burgess, S. , Harris, M. , and Rowlands, D. (2006) Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro . J Biol Chem 281: 37057–37068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, G.M. (1997) Caspases: the executioners of apoptosis. Biochem J 326: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell, C.T. , Kiosses, W.B. , Harkins, S. , and Whitton, J.L. (2006) Inhibition of protein trafficking by coxsackievirus b3: multiple viral proteins target a single organelle. J Virol 80: 6637–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino, D.M. , Bernardi, P. , Chieco‐Bianchi, L. , and Ciminale, V. (2005) Mitochondria as functional targets of proteins coded by human tumor viruses. Adv Cancer Res 94: 87–142. [DOI] [PubMed] [Google Scholar]
- DeDiego, M.L. , Alvarez, E. , Almazan, F. , Rejas, M.T. , Lamirande, E. , Roberts, A. , et al. (2007) A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo . J Virol 81: 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens, J.R. , Giddings, T.H., Jr , and Kirkegaard, K. (1997) Inhibition of endoplasmic reticulum‐to‐Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol 71: 9054–9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison, D. , Milburn, S.C. , Edery, I. , Sonenberg, N. , and Hershey, J.W. (1982) Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220 000‐dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 257: 14806–14810. [PubMed] [Google Scholar]
- Failla, C. , Tomei, L. , and De Francesco, R. (1994) Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol 68: 3753–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, F. , Stegen, C.F. , Masters, P.S. , and Samsonoff, W.A. (1998) Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol 72: 7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, R. , Bortner, C.D. , and Cidlowski, J.A. (2006) Potential roles of electrogenic ion transport and plasma membrane depolarization in apoptosis. J Membr Biol 209: 43–58. [DOI] [PubMed] [Google Scholar]
- Gonzalez, M.E. , and Carrasco, L. (2003) Viroporins. FEBS Lett 552: 28–34. [DOI] [PubMed] [Google Scholar]
- Grandgirard, D. , Studer, E. , Monney, L. , Belser, T. , Fellay, I. , Borner, C. , and Michel, M.R. (1998) Alphaviruses induce apoptosis in Bcl‐2‐overexpressing cells: evidence for a caspase‐mediated, proteolytic inactivation of Bcl‐2. EMBO J 17: 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, A.L. , Kerr, I.D. , and Sansom, M.S. (1997) Ion channels formed by HIV‐1 Vpu: a modelling and simulation study. FEBS Lett 405: 299–304. [DOI] [PubMed] [Google Scholar]
- Griffin, S.D. , Beales, L.P. , Clarke, D.S. , Worsfold, O. , Evans, S.D. , Jaeger, J. , et al. (2003) The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett 535: 34–38. [DOI] [PubMed] [Google Scholar]
- Griffin, S.D. , Harvey, R. , Clarke, D.S. , Barclay, W.S. , Harris, M. , and Rowlands, D.J. (2004) A conserved basic loop in hepatitis C virus p7 protein is required for amantadine‐sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol 85: 451–461. [DOI] [PubMed] [Google Scholar]
- Han, Z. , and Harty, R.N. (2004) The NS3 protein of bluetongue virus exhibits viroporin‐like properties. J Biol Chem 279: 43092–43097. [DOI] [PubMed] [Google Scholar]
- Hirose, S. (2006) A new paradigm of channelopathy in epilepsy syndromes: intracellular trafficking abnormality of channel molecules. Epilepsy Res 70: S206–S217. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J. , Hubner, C.A. , and Fuhrmann, J.C. (2004) Ion channels: function unravelled by dysfunction. Nat Cell Biol 6: 1039–1047. [DOI] [PubMed] [Google Scholar]
- Kalkeri, G. , Khalap, N. , Garry, R.F. , Fermin, C.D. , and Dash, S. (2001) Hepatitis C virus protein expression induces apoptosis in HepG2 cells. Virology 282: 26–37. [DOI] [PubMed] [Google Scholar]
- Karbowski, M. , and Youle, R.J. (2003) Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ 10: 870–880. [DOI] [PubMed] [Google Scholar]
- Kato, J. , Kato, N. , Yoshida, H. , Ono‐Nita, S.K. , Shiratori, Y. , and Omata, M. (2002) Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo . J Med Virol 66: 187–199. [DOI] [PubMed] [Google Scholar]
- Kerr, J.F. , Wyllie, A.H. , and Currie, A.R. (1972) Apoptosis: a basic biological phenomenon with wide‐ranging implications in tissue kinetics. Br J Cancer 26: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait, T. , Strebel, K. , Hoggan, M.D. , Martin, M.A. , and Orenstein, J.M. (1990) The human immunodeficiency virus type 1‐specific protein vpu is required for efficient virus maturation and release. J Virol 64: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou, Y.H. , Chou, S.M. , Wang, Y.M. , Chang, Y.T. , Huang, S.Y. , Jung, M.Y. , et al. (2006) Hepatitis C virus NS4A inhibits cap‐dependent and the viral IRES‐mediated translation through interacting with eukaryotic elongation factor 1A. J Biomed Sci 13: 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, L. , and Masters, P.S. (2003) The small envelope protein E is not essential for murine coronavirus replication. J Virol 77: 4597–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kuppeveld, F.J. , Hoenderop, J.G. , Smeets, R.L. , Willems, P.H. , Dijkman, H.B. , Galama, J.M. , and Melchers, W.J. (1997) Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J 16: 3519–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama, J. , and Carrasco, L. (1992) Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J Biol Chem 267: 15932–15937. [PubMed] [Google Scholar]
- Lang, F. , Lang, K.S. , Wieder, T. , Myssina, S. , Birka, C. , Lang, P.A. , et al. (2003) Cation channels, cell Volume and the death of an erythrocyte. Pflugers Arch 447: 121–125. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Yuan, Q. , Torres, J. , Tam, J.P. , and Liu, D.X. (2006) Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349: 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljestrom, P. , and Garoff, H. (1991) Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol 65: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy, A. , Smyth, J. , Von Bonsdorff, C.H. , Liljestrom, P. , and Schlesinger, M.J. (1995) The 6‐kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol 69: 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo, E. , Maraver, A. , Espinosa, I. , Fernandez‐Arias, A. , and Rodriguez, J.F. (2000) VP5, the nonstructural polypeptide of infectious bursal disease virus, accumulates within the host plasma membrane and induces cell lysis. Virology 277: 345–357. [DOI] [PubMed] [Google Scholar]
- Lopez‐Rivas, A. , Castrillo, J.L. , and Carrasco, L. (1987) Cation content in poliovirus‐infected HeLa cells. J Gen Virol 68: 335–342. [DOI] [PubMed] [Google Scholar]
- Lowy, R.J. (2003) Influenza virus induction of apoptosis by intrinsic and extrinsic mechanisms. Int Rev Immunol 22: 425–449. [DOI] [PubMed] [Google Scholar]
- Madan, V. (2006) Viroporinas de virus animales con genoma RNA In Dpto. Biología Molecular. Madrid: Universidad Autónoma de Madrid. [Google Scholar]
- Madan, V. , Garcia Mde, J. , Sanz, M.A. , and Carrasco, L. (2005a) Viroporin activity of murine hepatitis virus E protein. FEBS Lett 579: 3607–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan, V. , Sanz, M.A. , and Carrasco, L. (2005b) Requirement of the vesicular system for membrane permeabilization by Sindbis virus. Virology 332: 307–315. [DOI] [PubMed] [Google Scholar]
- Marissen, W.E. , and Lloyd, R.E. (1998) Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol 18: 7565–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalova, O.V. , Lakina, E.I. , Abdulmedzhidova, A.G. , Atanadze, S.N. , Semiletov, Y.A. , Shkurko, T.V. , et al. (2002) Characterization of monoclonal antibodies and epitope mapping of the NS4 protein of hepatitis C virus. Immunol Lett 83: 187–196. [DOI] [PubMed] [Google Scholar]
- Melton, J.V. , Ewart, G.D. , Weir, R.C. , Board, P.G. , Lee, E. , and Gage, P.W. (2002) Alphavirus 6K proteins form ion channels. J Biol Chem 277: 46923–46931. [DOI] [PubMed] [Google Scholar]
- Moon, H.S. , and Yang, J.S. (2006) Role of HIV Vpr as a regulator of apoptosis and an effector on bystander cells. Mol Cells 21: 7–20. [PubMed] [Google Scholar]
- Morishima, N. , Nakanishi, K. , Takenouchi, H. , Shibata, T. , and Yasuhiko, Y. (2002) An endoplasmic reticulum stress‐specific caspase cascade in apoptosis. Cytochrome c‐independent activation of caspase‐9 by caspase‐12. J Biol Chem 277: 34287–34294. [DOI] [PubMed] [Google Scholar]
- Mould, J.A. , Drury, J.E. , Frings, S.M. , Kaupp, U.B. , Pekosz, A. , Lamb, R.A. , and Pinto, L.H. (2000) Permeation and activation of the M2 ion channel of influenza A virus. J Biol Chem 275: 31038–31050. [DOI] [PubMed] [Google Scholar]
- Neznanov, N. , Kondratova, A. , Chumakov, K.M. , Angres, B. , Zhumabayeva, B. , Agol, V.I. , and Gudkov, A.V. (2001) Poliovirus protein 3A inhibits tumor necrosis factor (TNF)‐induced apoptosis by eliminating the TNF receptor from the cell surface. J Virol 75: 10409–10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura‐Takigawa, Y. , Nagano‐Fujii, M. , Deng, L. , Kitazawa, S. , Ishido, S. , Sada, K. , and Hotta, H. (2006) Non‐structural protein 4A of Hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria‐mediated apoptosis. J Gen Virol 87: 1935–1945. [DOI] [PubMed] [Google Scholar]
- O'Brien, V. (1998) Viruses and apoptosis. J Gen Virol 79: 1833–1845. [DOI] [PubMed] [Google Scholar]
- Pavlovic, D. , Neville, D.C. , Argaud, O. , Blumberg, B. , Dwek, R.A. , Fischer, W.B. , and Zitzmann, N. (2003) The hepatitis C virus p7 protein forms an ion channel that is inhibited by long‐alkyl‐chain iminosugar derivatives. Proc Natl Acad Sci USA 100: 6104–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, L.H. , Holsinger, L.J. , and Lamb, R.A. (1992) Influenza virus M2 protein has ion channel activity. Cell 69: 517–528. [DOI] [PubMed] [Google Scholar]
- Pinton, P. , Ferrari, D. , Rapizzi, E. , Di Virgilio, F. , Pozzan, T. , and Rizzuto, R. (2001) The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide‐induced apoptosis: significance for the molecular mechanism of Bcl‐2 action. EMBO J 20: 2690–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, K. , Berguerand, S. , Southern, S. , Doorbar, J. , and Beard, P. (2004) E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J Virol 78: 7199–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapizzi, E. , Pinton, P. , Szabadkai, G. , Wieckowski, M.R. , Vandecasteele, G. , Baird, G. , et al. (2002) Recombinant expression of the voltage‐dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J Cell Biol 159: 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova, L.I. , Belov, G.A. , Lidsky, P.V. , Tolskaya, E.A. , Kolesnikova, M.S. , Evstafieva, A.G. , et al. (2005) Variability in apoptotic response to poliovirus infection. Virology 331: 292–306. [DOI] [PubMed] [Google Scholar]
- Ruigrok, R.W. , Hirst, E.M. , and Hay, A.J. (1991) The specific inhibition of influenza A virus maturation by amantadine: an electron microscopic examination. J Gen Virol 72: 191–194. [DOI] [PubMed] [Google Scholar]
- Salako, M.A. , Carter, M.J. , and Kass, G.E. (2006) Coxsackievirus protein 2BC blocks host cell apoptosis by inhibiting caspase‐3. J Biol Chem 281: 16296–16304. [DOI] [PubMed] [Google Scholar]
- Sandoval, I.V. , and Carrasco, L. (1997) Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro‐090179. J Virol 71: 4679–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, M.A. , and Carrasco, L. (2001) Sindbis virus variant with a deletion in the 6K gene shows defects in glycoprotein processing and trafficking: lack of complementation by a wild‐type 6K gene in trans. J Virol 75: 7778–7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, M.A. , Madan, V. , Carrasco, L. , and Nieva, J.L. (2003) Interfacial domains in Sindbis virus 6K protein. Detection and functional characterization. J Biol Chem 278: 2051–2057. [DOI] [PubMed] [Google Scholar]
- Schlegel, A. , Giddings, T.H. , Jr, Ladinsky, M.S. , and Kirkegaard, K. (1996) Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol 70: 6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy, D.A. , Giddings, T.H. , Jr, and Kirkegaard, K. (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy‐like origin for virus‐induced vesicles. J Virol 74: 8953–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S. , Shirakata, Y. , Kaneniwa, N. , and Koike, K. (1999) Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 18: 6965–6973. [DOI] [PubMed] [Google Scholar]
- Thompson, C.B. (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267: 1456–1462. [DOI] [PubMed] [Google Scholar]
- Tolskaya, E.A. , Romanova, L.I. , Kolesnikova, M.S. , Ivannikova, T.A. , Smirnova, E.A. , Raikhlin, N.T. , and Agol, V.I. (1995) Apoptosis‐inducing and apoptosis‐preventing functions of poliovirus. J Virol 69: 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso, I. , Blanco, R. , Perales, C. , and Carrasco, L. (2001) HIV‐1 protease cleaves eukaryotic initiation factor 4G and inhibits cap‐dependent translation. Proc Natl Acad Sci USA 98: 12966–12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring, P. (2005) Redox active calcium ion channels and cell death. Arch Biochem Biophys 434: 33–42. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. , Watanabe, S. , Ito, H. , Kida, H. , and Kawaoka, Y. (2001) Influenza A virus can undergo multiple cycles of replication without M2 ion channel activity. J Virol 75: 5656–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L. , McKinlay, C. , Gage, P. , and Ewart, G. (2004) SARS coronavirus E protein forms cation‐selective ion channels. Virology 330: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk, B. , Sansonno, D. , Krausslich, H.G. , Dammacco, F. , Rice, C.M. , Blum, H.E. , and Moradpour, D. (2000) Subcellular localization, stability, and trans‐cleavage competence of the hepatitis C virus NS3‐NS4A complex expressed in tetracycline‐regulated cell lines. J Virol 74: 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzer, W.J. , Planz, O. , Ehrhardt, C. , Giner, M. , Silberzahn, T. , Pleschka, S. , and Ludwig, S. (2003) Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J 22: 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Bi, W. , Weiss, S.R. , and Leibowitz, J.L. (1994) Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202: 1018–1023. [DOI] [PubMed] [Google Scholar]
- Zamzami, N. , and Kroemer, G. (2001) The mitochondrion in apoptosis: how Pandora's box opens. Nat Rev Mol Cell Biol 2: 67–71. [DOI] [PubMed] [Google Scholar]


