Summary
Interferon‐inducible transmembrane proteins 1, 2 and 3 (IFITM1, IFITM2 and IFITM3) have recently been identified as potent antiviral effectors that function to suppress the entry of a broad range of enveloped viruses and modulate cellular tropism independent of viral receptor expression. However, the antiviral effect and mechanisms of IFITMs in response to viral infections remain incompletely understood and characterized. In this work, we focused our investigation on the function of the extracellular IFITM3 protein. In cell models of DENV‐2 infection, we found that IFITM3 contributed to both the baseline and interferon‐induced inhibition of DENV entry. Most importantly, our study for the first time demonstrated the presence of IFITM‐containing exosome in the extracellular environment, and identified an ability of cellular exosome to intercellularly deliver IFITM3 and thus transmit its antiviral effect from infected to non‐infected cells. Thus, our findings provide new insights in the basic mechanisms underlying the actions of IFITM3, which might lead to future development of exosome‐mediated anti‐viral strategies using IFITM3 as a therapeutic agent. Conceivably, variations in the basal and inducible levels of IFITMs, as well as in intracellular and extracellular levels of IFITMs, might predict the severity of dengue virus infections among individuals or across species.
Background
Dengue virus is an enveloped, single‐stranded positive strand virus, belonging to the Flavivirus genus of the family Flaviviridae, with four related but distinct serotypes (DENV‐1 to 4) (Halstead, 2007; Simmons et al., 2012). DENV is the aetiologic agent of dengue fever (DF), the most prevalent arthropod‐borne viral disease in humans with more than 50–100 million cases worldwide annually. DENV is transmitted by mosquito Aedes aegypti or Aedes albopictus in the tropical and subtropical regions, where these mosquitoes are endemic, placing 2.5 billion people at risk of infection globally (Guzman et al., 2010; Simmons et al., 2012). Of note, 1% of infected individuals develop more severe, and often lethal, syndromes, designated dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS), with children bearing most of the disease burden (Halstead et al., 2007; Guzman et al., 2010). While genotypic differences of DENVs have been indicated to be associated with variations in viral virulence, heterotypic dengue virus antibodies are believed to be a risk factor for the development of DHF or DSS in secondary infections (Guzman et al., 2010). Despite the formidable toll imposed by DENV on world health, no approved vaccine or effective specific anti‐DENV therapies are currently available for DENV infection (Halstead, 2005; Guzman et al., 2010).
As a major host defence system against viral infection, the interferon (IFN) family, notably IFN‐alpha and IFN‐beta, initiates a potent antiviral response that activates the innate immunity mediated by induction of more than 300 IFN‐stimulated genes (ISGs), leading to the establishment of an anti‐viral state (Sadler and Williams, 2008). IFN inducible transmembrane proteins 1, 2 and 3 (IFITM1, 2 and 3) have recently been identified as antiviral mediators induced by IFN, to confer host cells resistance to a variety of pathogenic viruses such as influenza A virus (IAV) (Brass et al., 2009; Feeley et al., 2011; Huang et al., 2011; Everitt et al., 2012), West Nile virus (Brass et al., 2009; Jiang et al., 2010), DENV (Brass et al., 2009; Jiang et al., 2010; Chan et al., 2012), vesicular stomatitis virus (Weidner et al., 2010), Marburg virus (Huang et al., 2011), Ebola virus (Huang et al., 2011), SARS coronavirus (Huang et al., 2011), human immunodeficiency virus (Lu et al., 2011) and hepatitis C virus (Yao et al., 2012). Mechanistically, IFITMs are the only known ISG products that act to restrict the entry step of viral infection process (Feeley et al., 2011). In particular, Brass et al. demonstrated that the action of a single intrinsic immune effector, IFITM3, profoundly effected as an essential barrier to IAV infection in vitro, in a knockout mouse model and in humans, by blocking the virus‐cellular membrane fusion and thus preventing cytosolic entry of the virus (Brass et al., 2009; Feeley et al., 2011; Everitt et al., 2012). Interestingly, another study revealed that IFITM proteins could interfere with the antibody‐dependent enhancement (ADE) effect during secondary dengue virus infection, which bypassed the IFN‐mediated restriction (Chan et al., 2012). Taken together, these previous studies have defined IFITM3 as a mediator required for the anti‐viral action of IFN, representing a key component of human anti‐viral defence system, probably through targeting an early step of viral infection. Open questions, however, remain concerning the detailed biological functions of IFITM in IFN‐triggered cascades and the precise mechanisms how the host cellular defence system is involved in IFITM activation in response to viral infections. Such knowledge will provide insights in designing new antiviral therapeutics using IFITM proteins. Furthermore, it is worth noting that secreted or membrane‐bound proteins are of particular interest in drug development, because their extracellular nature renders them more accessible for therapeutic intervention.
As a novel alternative secretion system, exosomes are small vesicles (30–100 nm in diameter) of endocytic origin that are released from cell into the extracellular environment, under both normal and pathological conditions (Thery et al., 2002). Exosomes are formed through the inward budding of late endosomal membrane that gives rise to intracellular multivesicular bodies (MVBs), which involve their fusion with the plasma membrane and release of MVBs into the extracellular environment as exosome (Thery et al., 2002). Prior work has focused on exosome as a new family member of ‘bioactive vesicles’ that function to promote intercellular communication, by shuttling for proteins, lipids and RNAs of the cells, and participate in various biological processes including immunomodulatory events (Schorey and Bhatnagar, 2008). While shuttling for components of pathogenic microbes, exosome could stimulate immune responses. For instance, exosome isolated from cells infected with Mycobacterium tuberculosis, have been shown to contain bacterial components and promote antigen presentation and macrophage activation (Bhatnagar and Schorey, 2007a; Bhatnagar et al., 2007b). Moreover, exosome may also promote intercellular spreading of infectious cargo, such as the observed cell‐to‐cell transmission of HIV (Gould et al., 2003; Izquierdo‐Useros et al., 2009). On the other hand, despite these advances in our understanding of the functions of exosome, the physiological significance of exosome in shuttling bioactive molecules key to the host defence system remains enigmatic. More recently, Atanu et al. demonstrated that APOBEC3G, which belongs to an ISG family of cellular cytidine deaminases that effect to restrict replication of a variety of exogenous retroviruses, was exported by exosome and conferred antiviral phenotype to recipient cells (Khatua et al., 2009). Li et al. propose an antiviral mechanism of IFN‐α activity that involves the induction and intercellular transfer of antiviral molecules (such as LAMP‐2 protein, APOBEC3G proteins, IFI6 mRNA, DDIT3 mRNA, hsa‐miR‐638, hsa‐miR‐4284 and hsa‐miR‐1260 etc.) from liver non‐parenchymal cells to hepatocytes via exosomes, by using HBV infection as a model (Li et al., 2013). They also observed similar exosome‐mediated transfer of IFITM1 mRNA from macrophages to HepG2.2.15 cells, but there is not in‐depth study on whether the anti‐HBV activity is dependent or independent of exosome‐mediated transfer of IFITM1 mRNA (Li et al., 2013). It is therefore tempting to hypothesize that direct exchange of ISGs proteins transferred by exosome among host cells might contribute to the establishment of anti‐viral state in uninfected cells, in addition to the direct action of IFNs stimulation.
In this study, we focused our investigation on the function of the extracellular IFITM3 protein. In cell models of DENV‐2 infection, we found that endogenous basal protein levels of IFITMs inversely correlated to DENV‐2 infection, and induction of IFITMs in cells with low basal protein levels was sufficient to drive protection of host cells from DENV‐2 infection at entry. Conversely, loss of IFITM in host cell pronouncedly enhanced DENV‐2 infection. Most importantly, our study for the first time demonstrated the presence of IFITM‐containing exosome in the extracellular environment and the function of these exosome vehicles for inter‐cellular transmission of anti‐viral proteins.
Results
IFITM3 contributes to host cell resistance to DENV‐2 infection
In an effort to investigate whether endogenous IFITM3 in host cells plays a role in the interaction between DENV and the host cells, we first compared the expression of IFITM3 in DENV‐permissive cell lines with versus without DENV‐2 infection. Notably, following infection with DENV‐2 at multiplicity of infection (moi) of 2 in 9 human cell lines permissive for DENV‐2 replication, expression of IFITM3 was found to be inducible by DENV‐2 infection in various cell lines as demonstrated by Western blotting analysis (Fig. 1A). Next, we analysed the correlation of IFITM3 level in 8 cell lines with DENV‐2 infection. We found that while endogenous IFITM3 expression was varied in different cell lines, cells expressing low level of IFITM3 protein were more susceptible to DENV‐2 infection (Fig. 1B). Further exponential regression analysis showed that the level of IFITM3 protein in host cells inversely correlated with their susceptibility to DENV‐2 infection significantly (r 2 = 0.591, P = 0.026, n = 8, Fig. 1C), suggesting that IFITM3 might be an important cellular restriction factor for DENV infection.
Figure 1.
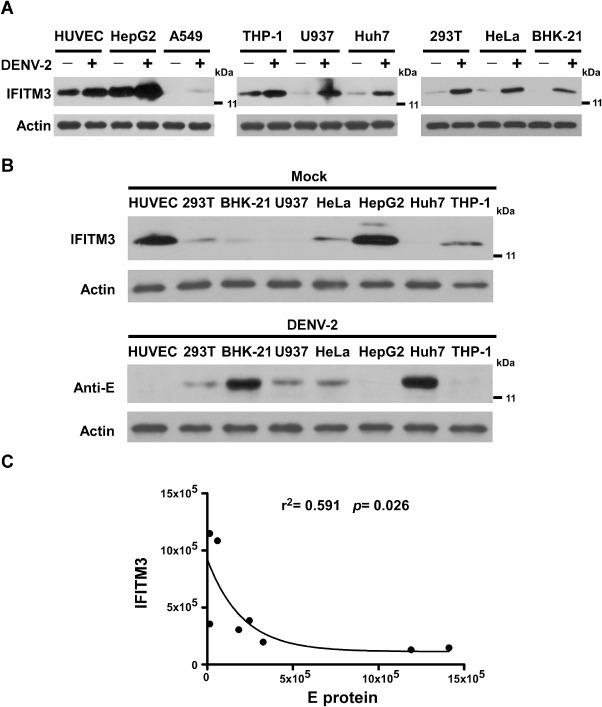
IFITM3 is required for basal levels of cellular resistance to DENV‐2 infection.
A. Western blotting analysis of IFITM3 expression induced by DENV‐2 infection in various permissive cell lines. Cells were infected with DENV‐2 at an moi of 2 and harvested at 24 h post infection. Actin was used as an internal control.
B. Baseline IFITM3 expression in various cell lines and DENV E protein in the post DENV‐2 infection cells, as indicated, were determined by Western blotting analysis.
C. Exponential regression analysis showing an inverse correlation between DENV‐2 infectivity and the level of IFITM3 protein in 8 permissive cell lines (r 2 = −0.591, P = 0.026, n = 8).
Expression of IFITMs elicits antiviral activity against DENV‐2 at entry
To evaluate the role of IFITM proteins in DENV infection, we generated permanent cell lines stably overexpressing human IFITM 1, 2 or 3, or control vector, respectively, with U937 and HeLa cells (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo). As show in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo, IFITM1, IFITM2 or IFITM3 overexpression drastically diminished the number of DENV‐2‐infected cells, as indicated by twofold to fourfold reduction of DENV‐2 viral E protein accumulation in virally infected U937 and HeLa cells. Similar profound restriction was also seen when IFITM3 was transiently transduced in 293T, despite a lesser extent of IFITM3 overexpression than that of IFITM1 or IFITM2 (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo), demonstrating the potency of IFITM3 in conferring host cells resistance to DENV infection. We next examined the effect of IFITM3 on the entry step of DENV‐2, and found that entry of DENV‐2 was reduced by IFITM3 by approximately twofold (Fig. 5A), as controlled with the 4G2 monoclonal neutralizing antibody to demonstrate a successful entry blockage, suggesting that the disruption of DENV replication by IFITM3 was associated with a targeted abrogation of viral entry.
Figure 5.
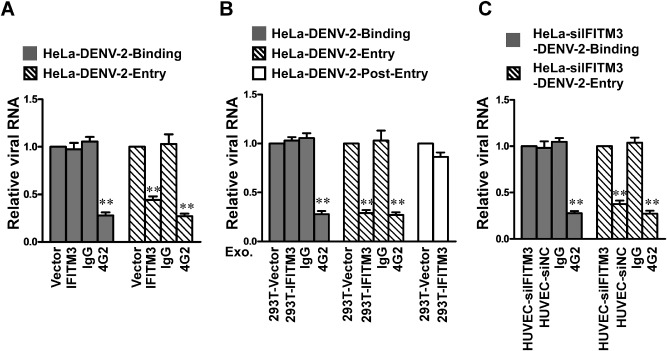
IFITM3 and IFITM3‐containing exosomes inhibit DENV‐2 entry. Virus binding and entry assays were performed with HeLa cells stably overexpressing IFITM3 (A), HeLa cells (B) or HeLa‐siIFITM3 cells (C) treated with exosome derived from each group of cells as indicated, using 4G2 neutralizing monoclonal antibodies as a positive control. Intracellular viral RNA was measured with real‐time RT‐PCR, and the results are expressed as relative folds of total cellular RNA. Error bars indicate standard deviations of the means (n = 3). Data points present means ± SD of triplicated experiments. Student's two‐tailed t test was performed, and statistical significance is shown with asterisks (**P < 0.01).
IFITM3 expression and inducibility contribute to the antiviral effect of interferon
Since IFN stimulated signalling plays a pivotal role in protecting cells from viral infection and damage, we investigated the functional significance of IFITM3 in IFN‐mediated anti‐DENV response. As shown in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo, compared with HUVEC cells transfected with control scramble siRNA, transfection of IFITM3‐siRNA‐1 and IFITM3‐siRNA‐2 efficiently depleted basal‐level IFITM3 expression, and increased the permissiveness of the transfected cells to DENV‐2 infection, as indicated by a > 2‐fold increase of infection, in HUVEC that constitutively express high basal level of endogenous IFITM3 expression. Similar effect of IFITM3 depletion was reproducible in another cell model, in which the basal IFITM3 level is low but greatly inducible by IFN‐α. siRNA silencing of IFITM3 markedly attenuated the protective effect of IFN‐α against DENV‐2 infection in HeLa cells, leading to profoundly increased infection by DENV‐2 in the presence of IFN‐α, by 5.9‐ or 7.2‐fold (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo), indicating that the depletion of IFITM3 could decrease the antiviral actions of IFN‐α. We also performed DENV binding/entry assay following knockdown of IFITM3 expression. Our results showed no effect of siRNA silencing of IFITM3 on the binding of DENV‐2 to HeLa cells, but the penetration of DENV‐2 into cells was significantly promoted by the depletion of IFITM3 (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo). Thus, our results suggest that IFITM3 is involved in mediating IFN‐induced cellular response against DENV infection.
IFITM3 can be exported from cells by exosome
In an attempt to analyse the biochemical properties of IFITM3, we found that the protein was present both intracelluarly and extracellularly. Specifically, cell culture supernatants were collected from parental HUVEC cells or HepG2 cells that express high basal level of endogenous IFITM3, from HUVEC or HepG2 cells transfected with IFITM3 siRNA‐1 (HUVEC‐siIFITM3 or HepG2‐siIFITM3), from HUVEC or HepG2 cells transfected with non‐targeting control siRNA (HUVEC‐siNC or HepG2‐siNC), from 293T cells transfected with pcDNA3 (293T‐Vector), and from 293T cells transfected with the pcDNA‐IFITM1, 2, 3‐Flag construct (293T‐IFITM1, 2, 3) for 48 h. Supernatants derived from each of the above groups with different treatments were collected and centrifuged at 1000 g for 10 min, and then 10 000 g for 10 min at 4°C to remove cells and cell debris, followed by filtration with 0.22 μm filters. Then the supernatants were concentrated by 10‐fold using an Amicon Ultra‐15 centrifugal filter unit with 10 kDa cut‐off value, and subjected to Western blotting analysis using an anti‐IFITM antibody. Our data showed that IFITM3 protein was present both in the cytosol and in the culture medium (Fig. 2A–D), suggesting the possibility that IFITM3 could be released to extracellular space. It was also noteworthy that the expression of IFITM3 in the supernatants collected from cultured HUVEC cells is lost when IFITM3 was knocked down or exosome secretion was inhibited by GW4869 (an exosome‐release inhibitor) (Fig. 2D).
Figure 2.
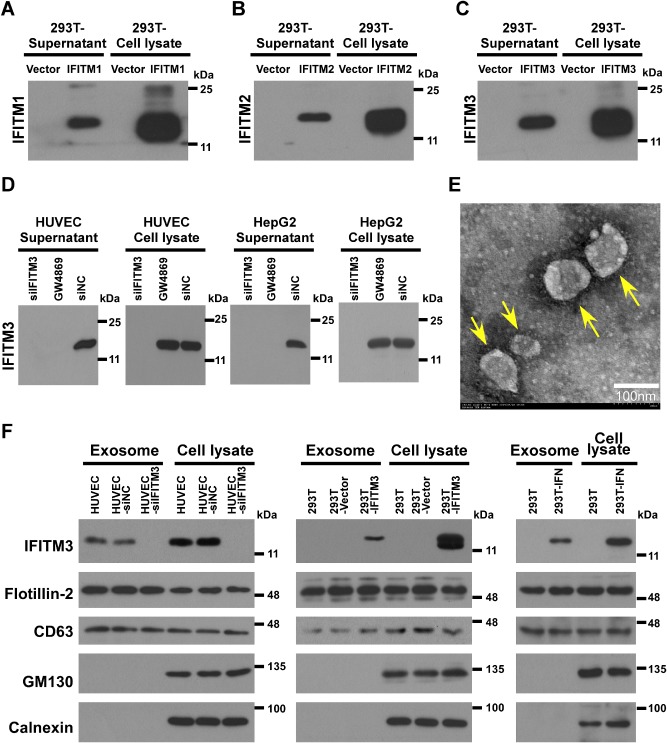
Identification of IFITM3‐containing exosomes derived from 293T and HUVEC cells.
A–D. Cell culture supernatants of HUVEC or HepG2 cells transfected with IFITM3 siRNA‐1 (HUVEC‐siIFITM3 or HepG2‐siIFITM3), from HUVEC or HepG2 transfected with non‐targeting control siRNA (HUVEC‐siNC or HepG2‐siNC), from293T cells transfected with pcDNA3 (293T‐Vector), or from 293T cells transfected with the pcDNA‐IFITM1, 2, 3‐Flag construct (293T‐IFITM1, 2, 3) for 48 h, were collected and concentrated for SDS‐PAGE and immunoblotting, using an anti‐IFITM3 antibody, with cell lysates as positive control.
E. Electron micrographs of crude exosomes negatively stained with uranyl acetate and examined at 80 kV are shown.
F. Purified IFITM3‐containing exosomes derived from each group of cells above described were analysed by immunoblotting with anti‐IFITM3, anti‐flotillin‐2, anti‐CD63, anti‐calnexin (endoplasmic reticulum, ER marker) and anti‐GM130 (Golgi marker) antibodies. IFITM3 is identified in the exosomes derived from each group of cells as indicated.
As the amino acid sequence of IFITM3 does not contain a putative signal peptide for protein secretion, we sought to explore the possibility that the exportation of IFITM3 could be through an exosome‐mediated mechanism. To achieve this, exosomes were prepared from the culture media of HUVEC, HUVEC‐siNC cells, HUVEC‐siIFITM3 cells, IFN‐α‐treated 293T cells (293T‐IFN), 293T‐IFITM3 cells, 293T‐Vector cells and 293T cells, and subjected to sucrose gradient ultracentrifugation. The electron microscopic examination of the exosomes revealed vesicles ranging in size from 30 nm to 100 nm (Fig. 2E). Interestingly, IFITM3 was detected in exosomes purified from the culture supernatant of HUVEC and HUVEC‐siNC cells, and was also abundant in those produced by 293T‐IFITM3 or 293T‐IFN cells. In contrary, however, exosomes isolated from the 293T cells, 293T‐vector cells and HUVEC‐siIFITM3 cells did not contain detectable IFITM3 (Fig. 2F). Purified exosomes were further characterized for the presence of conventional markers for exsosomes, namely, flotillin‐2 and CD63, and non‐exosomal markers calnexin (endoplasmic reticulum marker) and GM130 (Golgi matrix marker) using Western blotting analysis. As shown in Fig. 2F, exosomes collected from each group was shown to be positive for flotillin‐2 and CD63, but negative for calnexin and GM130, verifying that there is no contamination by other subcellular fractions in the purified exosome preparations. The IFITM3 protein contained in exosome composition was further verified by mass spectrometry (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo). Together, our data demonstrate that secretion of IFITM3 in exosome is not limited to HUVEC that express high level of endogenous IFITM3, but also by 293T cells undergoing IFN stimulation or transiently expressing exogenous IFITM3.
IFITM3‐laden exosome are internalizable by cells
The finding that IFITM3 is contained in exosome and exported from cells in which it is expressed prompted us to investigate whether the IFITM3‐containing exosomes are internalized by other cells in the system so that the anti‐DENV activity can be transferred to the recipient cells. In this study, we tested such a possibility by incubating HeLa cells, which express a low‐level of IFITM3, with exosomes collected from HUVEC, HUVEC‐siNC, HUVEC‐siIFITM3, 293T‐IFITM3 or 293T‐IFN cells. Indeed, as shown in Fig. 3A, an increased abundance of IFITM3 was detected in the recipient HeLa cells treated with HUVEC‐, 293T‐IFITM3 cell‐, or 293T‐IFN cell‐derived exosomes (60 μg ml−1), but not with those from 293T‐Vector cells. Moreover, detection of IFITM3 proteins in the lysates of recipient HeLa cells treated with IFITM3‐laden exosomes derived from 293T‐IFITM3 and HUVEC cells, as determined by immunoblotting, was dose‐ and time‐dependent (Fig. 3C and D). Notably, as it was reported that exosomes might be capable of delivering exogenous small RNA such as siRNA and microRNA in vitro and in vivo (van den Boorn et al., 2011; Pan et al., 2012; Shtam et al., 2013), in this experiment we preformed the protein transfer assay in recipient HeLa cells transfected with IFITM3‐siRNA (HeLa‐siIFITM3) to rule out the potential side‐effect of IFITM3‐siRNA possibly delivered by exosomes on the endogenous or exosome‐delivered IFITM mRNA in the recipient cells, Furthermore, as shown in Fig. 3B and D, increased abundance of IFITM3 was detected in the recipient HeLa‐siIFITM3 cells treated with HUVEC‐siNC‐derived exosomes in a time‐dependent manner, but not in those treated with HUVEC‐siIFITM3 cell‐derived exosomes.
Figure 3.
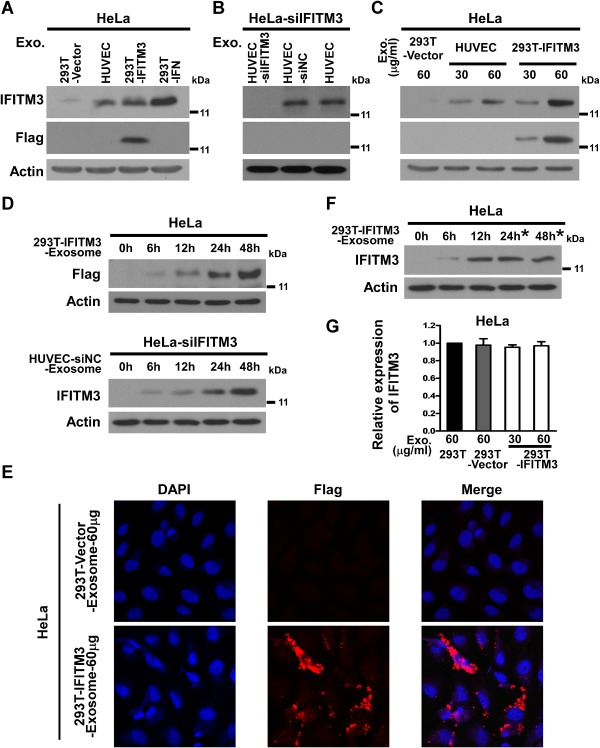
Inter‐cellular transfer of IFITM3‐containing exosome. HeLa cells were incubated with purified 60 μg ml−1 exosomes derived from supernatants of cultured HUVEC, HUVEC‐siNC, 293T‐IFITM3 or 293T‐IFN cells in serum‐free media for 24 h (A), incubated with increasing amounts of exosomes derived from 293T‐IFITM3 cells for 24 h (C), incubated with 60 μg ml−1 IFITM3‐exosomes derived from 293T‐IFITM3 cells or HUVEC for 6 h, 12 h, 24 h and 48 h (D), or incubated with 60 μg ml−1 IFITM3‐exosomes derived from 293T‐IFITM3 cells for 12 h, followed by washing exosomes after 12 h treatment (F). ‘*’ indicates that HeLa cells were maintained in DMEM after treatment with IFITM3‐exosomes for 12 h. HeLa cells with IFITM3 silenced (HeLa‐siIFITM3) were incubated with 60 μg ml−1 exosomes derived from HUVEC, HUVEC‐siNC and HUVEC‐siIFITM3 (B). Cell lysates were analysed by immunoblotting for IFITM3 or IFITM3‐Flag proteins. (E) Immunofluorescence staining and confocal microscopic analysis of IFITM3‐Flag in HeLa cells treated with 293T‐IFITM3‐exosome. (G) Expression of IFITM3 mRNA in HeLa cells from each group as indicated analysed by real‐time RT‐PCR. Expression levels were normalized with that of GAPDH. Exo = exosome.
Consistent with the Western blotting data, immunofluorescence staining and confocal microscopic analysis of IFITM3‐Flag showed that the localization in the cytoplasm in the recipient HeLa cells was significantly higher following treatment with exosomes derived from 293T‐IFITM3 cells (Fig. 3E). In contrast, our results showed that 293T‐IFITM3 cell‐derived exosomes did not change IFITM1 and IFITM2 expression in the recipient HeLa cells, when compared with treatments of 293T cell‐ or 293T‐Vector cells‐derived exosomes (http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo). Moreover, when the exosome treatments were terminated at 12 h, further incubation with blank culture media for a total length of 24 or 48 h did not increase the amount of endogenous IFITM3 within the recipient HeLa cells (Fig. 3F), suggesting that the exosome treatment procedure per se does not change endogenous IFITM3 expression in the recipient HeLa cells. In addition, in order to verify that the increased IFITM3 protein level in the transfer recipient cells was due to the exosomal transfer of IFITM3, rather than a stimulated expression of endogenous IFITM3, we measured IFITM3 mRNA levels in cells treated with IFITM3‐exosomes using quantitative real‐time RT‐PCR, and we found that IFITM3‐exosome treatment did not induce an increase of endogenous IFITM3 mRNA in HeLa cells (Fig. 3G). Furthermore, to evaluate the half‐life of IFITM3 in cell culture supernatant, we quantified IFITM3 protein in the supernatant of cultured HUVEC at various time points (0, 6, 12, 24, 36, 48, 60 and 72 h) using Western blotting. Specifically, supernatants derived from HUVEC cells were collected and centrifuged at 1000 g for 10 min, and then further centrifuged at 10 000 g for 10 min at 4°C to remove cells and cell debris, followed by filtration with 0.22 μm filters. The obtained supernatants were then concentrated by 10‐fold using an Amicon Ultra‐15 centrifugal filter unit with 10 kDa cut‐off value, divided into 8 groups, and incubated at 37°C in a humidified atmosphere of 5% CO2 for 0, 6, 12, 24, 36, 48, 60 and 72 h, respectively, before being subjected to Western blotting analysis using an anti‐IFITM3 antibody. As shown in the http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo, there was no or little change of the IFITM3 protein in the supernatant within 72 h, indicating a long half‐life (> 72 h) of IFITM3 protein in the supernatant.
The antiviral activity of internalized IFITM3‐laden exosome
To investigate whether that IFITM3‐laden exosome internalized into recipient cells could confer resistance to DENV‐2 infection, HeLa cells were cultured in serum‐free conditions and exposed to increasing amounts of purified IFITM3‐containing exosomes for 24 h. Purified IFITM3‐containing exosomes were used to treat HeLa cells that were simultaneously infected with DENV‐2 (moi = 2). At 24 h post infection when the infected cells were examined by real‐time RT‐PCR assay, strikingly, DENV‐2 infection was suppressed potently by the IFITM3‐containing exosomes in a dose‐dependent manner in parental HeLa cells (Fig. 4A) or in HeLa cells with endogenous IFITM3 knocked down (Fig. 4C), and the copy number of DENV‐2 viral RNA was reduced in the culture supernatant (Fig. 4B and D), indicating that the observed antiviral effect was mediated by the exosome‐transferred IFITM3. We also performed immunofluorescence analysis of infected HeLa cells treated with, or without, IFITM3‐exosomes, by staining for delivered IFITM3‐Flag and a DENV protein E. As shown in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo, our results showed that DENV protein E are present only in some cells that do not contain IFITM3‐Flag. The data from the CPE reduction assay demonstrated that IFITM3‐laden exosomes also attenuated CPE triggered by DENV‐2 infection (Fig. 4E). Moreover, our results showed no effect of IFITM3‐exosomes on the binding of DENV‐2 to host cells or post‐entry steps during DENV‐2 infection, but the penetration of DENV‐2 into cells was significantly inhibited by the IFITM3‐ containing exosomes, with a reduction of approximately fourfold (Fig. 5B). In contrast, the control exosomes prepared from 293T cells that contained only the vector plasmid without the IFITM3 expression cassette did not exhibit any anti‐DENV activity, suggesting that IFITM3 was the main exosomal component responsible for the detected antiviral activity of exosome.
Figure 4.
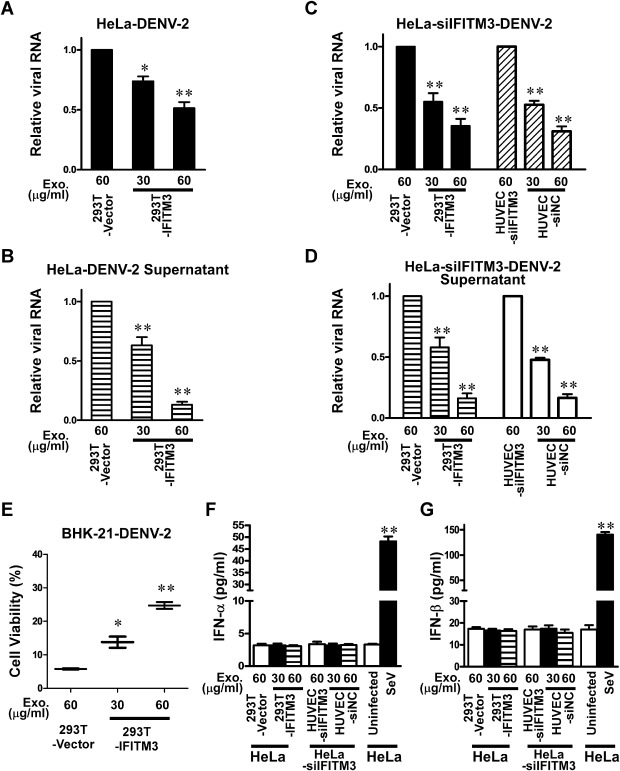
The antiviral activity of IFITM3‐containing exosome. After 24 h incubation with IFITM3‐containing exosomes, HeLa cells (parental) or HeLa cells with IFITM3 silenced were incubated with DENV‐2 virus for 24 h. Intracellular (A and C) and extracellular (in supernatant) (B and D) viral RNA was measured by real‐time RT‐PCR assay, and the results are presented as relative ratios of total cellular RNA. (E) IFITM3‐containing exosome suppressed CPE triggered by DENV‐2 infection. Intracellular viral RNA was measured in a real‐time RT‐PCR assay, and the results are expressed as relative fold of total cellular RNA. Conditioned media obtained from exosomes derived from each group of cells treated as indicated were quantitatively measured for contained IFN‐α (E) and IFN‐β (F) proteins by ELISA, using Sendai virus (SeV; 100 HAU ml−1) as a positive stimulus control. Data points are presented as means ± SD of triplicated experiments. Student's two‐tailed t test was performed, and statistical significance is shown with asterisks (*P < 0.05, **P < 0.01).
To verify that the enhanced antiviral effects caused by IFITM3‐exosome transfer was a result of IFITM3 action rather than a stimulated paracrine IFNs response, the concentration of human IFN‐α/β in culture supernatant of HeLa cells or HeLa‐siIFITM3 cells treated with IFITM3‐laden exosomes was measured with ELISA by using Sendai virus (SeV; 100 HAU ml−1) as a positive stimulus control. Our results showed that IFITM3‐exosomes treatment did not change IFN‐α and ‐β expression, as compared with the control‐exosomes (Fig. 4F and G), implicating that reduction of virus replication was not due to a non‐specific effect of IFITM3‐containing exosomes mediated by interferon.
Discussion
IFNs trigger a potent antiviral response that activates host innate immunity and leads to establishment of a cellular antiviral state, which is mediated by inducing expression of ISGs. Accordingly, ISGs represent a key line of defence against viral infection, and IFITMs are among the most potent ISG products with broad anti‐viral spectra. A key finding of our current study, namely, that IFITM3 can be transferred intercellularly via extracellular exosome, provides new insights in the functional significance of IFITM3 as an IFN‐induced anti‐viral protein. The discovery of the roles of free IFITM proteins and IFITM‐laden exosome as innate cellular defenders present an opportunity developing exosome‐based tools to actively combat existing or emerging pathogens. Conceivably, variations in the basal and inducible levels of IFITMs, as well as in intracellular and extracellular levels of IFITMs, might predict the severity of dengue infections among individuals or across species.
Exosome is secreted from many types of cells and remain stable in various body environments. By transferring proteins, mRNAs and microRNAs to neighbouring or distant cells, exosome contribute to modulation of important physiological or pathological processes such as immunity, angiogenesis, cell proliferation, cell migration and invasion, and cell‐to‐cell signalling (Schorey and Bhatnagar, 2008). Here, we propose a novel model for IFN‐inducible innate immune response against DENV‐2 infection mediated by IFITM‐containing exosome. Noteworthy in this context is that exosome released from IFN‐treated cells may remotely communicate with other cells through such a mechanism that the anti‐viral phenotype can be transferred to the exosome recipient cell that might not have the chance to be directly stimulated by IFN. Interestingly, this inter‐cellular communication model might also be applicable to explain a pioneering observation on the antiproliferative activity transmitted from IFN‐stimulated cells to non‐stimulated cells (Lloyd et al., 1983). At the subcellular level, following IFN induction of IFITM3 expression, IFITM3 is transferred to the multivesicular complexes and assembled into exosomal vesicles. Upon transport to the surface of a recipient cell, fusion of MVCs with the plasma membrane allows assembly of IFITM with a pre‐existing exosome receptor complex that is unknown thus far. Potentially, the released exosome particles containing IFITM protein can travel to non‐stimulated cells where receptor assembly also occurs. Such a model is consistent with the finding that the antiviral state can be transferred from IFNs induced cells to non‐treated cells in culture.
Hypothetically, this process might require IFITM proteins to be processed or relocalized in the host cells by an unknown mechanism so that it can be targeted for exosome secretion. Interestingly, it has been reported that cytosolic proteins present with exosome including members of the Rab family, are involved in the formation and secretion of exosome. Rab 1B, 5, 7 and GDI are found to promote exosome docking and the membrane fusion events (Thery et al., 2002). Interesting, Feeley et al. reported that IFITM3 primarily resides in the late endosomal compartment and partly colocalizes with Rab7 and LAMP1 (Feeley et al., 2011). In the context, therefore, it would be of great interest to further investigate the biogenesis of IFITM3 exosome as well as its regulation, which might provide important clues for a rationale development of optimal anti‐viral strategies.
While application of IFNs has been highly appreciated in the clinic, problems associated with the use of IFNs, such as suboptimal efficacies against many viruses and varied degrees of adverse effects caused by IFN treatment, remain unsolved (Borden et al., 2007). In the intensive efforts of identifying and developing new endogenous anti‐viral factors with potential clinical applicability, ISGs have been attractive candidate agents for therapeutic purposes. In such a context, exosome‐carried IFITM3, as discovered by our present study, appears to represent a novel, attractive potential anti‐viral strategy, as both natural and specially engineered exosome can serve as nano‐shuttle vehicles for drug delivery. Animal studies to test the feasibility and efficacy of applying exosome‐based delivery of IFITM3 for anti‐DENV purposes in vivo are therefore warranted.
Our findings provided the first set of evidence for the existence of an extracellular, exosome‐packaged form of IFITM3, which enables anti‐viral activities to transmit from one cell to another, leading to efficient establishment of an anti‐viral state. Further studies to decipher the structural and cell biological basis for the antiviral activities of IFITM‐exosome will be of theoretical as well as practical importance in this area of research.
Experimental procedures
Cell culture and virus
THP‐1, A549, Huh7, 293T, U937, HepG2, HeLa and BHK‐21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM L‐glutamine, 100 μg ml−1 streptomycin and 100 units ml−1 penicillin (Invitrogen). Human umbilical endothelial cells (HUVEC) were grown in human umbilical endothelial cell serum‐free medium (Invitrogen), supplemented with 15 g ml−1 endothelial cell growth supplements (Upstate, Billerica, MA, USA). The cultures were maintained at 37°C in a humidified atmosphere of 5% CO2. C6/36 Aedes albopictus cells were cultured at 28°C and 5% CO2 in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin.
The DENV‐2 strain NGC (New Guinea C, GenBank accession number M29095) was kindly provided by Guangzhou Centers for Disease Control and Prevention (CDC) (Tian et al., 2012; Wu et al., 2013) and was propagated in the C6/36 cell line. Briefly, a monolayer of C6/36 cells was infected with DENV‐2 at an moi of 1, and incubated at 28°C and 5% CO2 for 3 days. The supernatant was harvested and centrifuged for 5 min at 2000 g to remove pelleted cellular debris. DENV‐2 titres were determined by FACS assays in C6/36 cells, as previously described (Lambeth et al., 2005).
Western blotting analysis
Cells were harvested and lysed in 1× sampling buffer containing 50 mM Tris‐HCl (pH 7.4), 1 mM PMSF, 10% glycerol, 6% SDS, 5% mercaptoethanol and 0.1% bromophenol blue before sonication. The protein concentration of the lysate was determined using the Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's instructions with BSA as the standard. Protein molecular weight standards (GenStar Biosolutions Co. Ltd, Beijing, China) were used to determine molecular weights of sample proteins. An aliquot of the cell lysates containing 40 μg of protein was subjected to SDS‐PAGE, and then transferred to PVDF membranes. Following a blocking step using the blocking buffer (Tris‐buffered saline, TBS, containing 5% non‐fat milk) for 1 h at room temperature, the membranes were incubated overnight at 4°C with the following specific primary antibodies: monoclonal anti‐IFITM1, monoclonal anti‐IFITM2, polyclonal anti‐IFITM3 (Proteintech Group, Chicago, IL), monoclonal anti‐Flag (Sigma‐Aldrich, St Louis, MO), D1‐11 (anti‐DENV‐2 E mouse monoclonal antibody; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti‐Actin (Sigma‐Aldrich), polyclonal anti‐CD63 (H‐193; Santa Cruz Biotechnology), anti‐Flotillin 2 (C‐terminal; Sigma‐Aldrich), anti‐calnexin (Proteintech Group), and monoclonal anti‐GM130 (Cell Signaling, Danvers, MA) antibodies. Further incubation with appropriate horseradish peroxidase (HRP)‐conjugated secondary antibodies, depending on the primary antibody used, was performed for 1 h at room temperature. Membranes were washed three times in Tris‐buffered saline containing 0.1% Tween‐20 for 10 min after each incubation step. The bands were detected using enhanced chemiluminescence kit (Thermo Fisher Scientific) with a Kodak film. Actin was used as a loading control for quantification normalization. Intensities of the bands of interest on the PVDF membranes were quantitatively calculated with the Quantity One 4.6.3 measurement software (Bio‐Rad, Hercules, CA).
Overexpression and knockdown of IFITM in DENV‐permissive cells
Human IFITM1, 2, 3 cDNA with a C‐terminal HA epitope tag were amplified by RT‐PCR using primers shown in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo. The resultant PCR fragments were cloned into the pMSCV‐Puro vector (Clontech Laboratories, Inc., Mountain View, CA) at BglII and EcoRI sites to generate plasmids pMSCV‐IFITM1‐HA, pMSCV‐IFITM2‐HA and pMSCV‐IFITM3‐HA respectively. The cloned IFITM1, 2, 3 cDNAs were sequencing verified. A blank pMSCV‐Puro vector was generally used as a control. These constructs were used to establish stably transduced cell lines with U937 and HeLa cells to permanently express IFITMs. Briefly, retroviral particles were prepared by transfecting 20 μg of each of the pMSCV‐Puro‐IFITM1‐HA, pMSCV‐Puro‐IFITM2‐HA and pMSCV‐Puro‐IFITM3‐HA plasmid DNA into the packaging cells 293T, together with 20 μg of the pIK plasmid. Recombinant retroviral particles were used to infect U937 or HeLa cells, and stably transduced cell lines were selected in DMEM medium supplemented with puromycin (5 μg ml−1). To transiently express IFITMs protein, IFITMs genes were separately subcloned into the pcDNA vector (Invitrogen) at KpnI and BamHI sites and a Flag‐tag sequence was introduced into the coding sequence as the C‐terminus. 293T cells were transiently transfected with the resultant plasmid pcDNA‐IFITMs‐Flag by a standard calcium phosphate co‐precipitation method. HeLa cells were transiently transfected using Lipofectamine 2000 (Invitrogen) by following the manufacturer's instruction. The small interfering RNA (siRNA) duplexes against IFITM3, whose sequences are provided in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo) were published previously (Brass et al., 2009), and were purchased from Ribobio Inc. (Guangzhou, Guangdong, China). HUVEC and HeLa cells were transfected with siRNAs at indicated concentrations, using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions, followed by treatment with IFN‐α (0, 10, 100 U ml−1) and infection with DENV‐2 at an moi of 2. To assess the expression of IFITM proteins, cell lysates were harvested at 48 h after transfection and examined by Western blotting analysis with corresponding specific antibodies.
DENV infection assays
Dengue E protein was detected in cells infected with DENV‐2 by intracellular staining and flow cytometric (FCM) analysis, as previously described (Brass et al., 2009; Chan et al., 2012). Briefly, cells in growth DMEM medium were seeded and subsequently cultured for 24 h, followed by infection with DENV‐2 at an moi of 2. Cells harvested at indicated time points were washed with cold 1× phosphate‐buffered saline (PBS) and fixed with 4% paraformaldehyde at 4°C for 10 min. Fixed cells were scraped and then permeabilized in 90% methanol before being incubated with NYRDeng2 (anti‐DENV‐2 E mouse monoclonal antibody; Santa Cruz Biotechnology), and further with appropriate FITC‐conjugated goat anti‐mouse IgG secondary antibody (Santa Cruz Biotechnology). The FITC‐positive cells were acquired and scored by flow cytometry (Becton‐Dickinson Counter, San Jose, CA). The percentage of DENV‐infected cells was defined by FITC‐positive cells distribution in a fluorescence dot plot using the Cytometry List Mode Data Acquisition & Analysis software (Becton‐Dickinson Counter).
DENV in cell supernatants were quantified by determining the copy number of viral RNA using quantitative real‐time RT‐PCR as previously described (Mota and Rico‐Hesse, 2009). Cell supernatants were harvested at indicated time points, and total RNA was extracted using QIAamp MinElute Virus Spin kit by following the instruction of the manufacturer (Qiagen, Chatsworth, CA). cDNA was synthesized, and quantitative real‐time PCR was performed with standard curve analysis using a CFX96 Real‐Time PCR Detection System (Bio‐Rad). For relative quantification, results are presented as ratios relative to the quantification for GAPDH. Primer pairs used are shown in http://onlinelibrary.wiley.com/doi/10.1111/cmi.12339/suppinfo. To estimate DENV RNA copy number, a standard curve was generated using in vitro‐transcribed RNA standards, which were produced as follows. A 104 bp fragment of the DENV‐2 NGC strain was amplified by RT‐PCR and cloned into the pMD19‐T Simple Vector (TaKaRa, Dalian, China). The cloning plasmid was linearized with EcoRV, and RNA transcripts were generated with a T7 Megascript kit (Ambion) according to the manufacturer's instructions. Concentration of transcribed RNA was determined with the NanoDrop 2000c spectrophotometer (Thermo Scientific, Rockford, IL), and 10‐fold serial dilutions were prepared and used to construct a standard curve.
Virus binding and entry assays
The binding and entry experiments were performed as described previously (Kanlaya et al., 2010; Weidner et al., 2010). Briefly, for virus binding experiment, cells were seeded in 12‐well plates at a density of 5 × 105 cells per well and cultured for 24 h, followed by inoculation with DENV‐2 at moi of 2 and incubation on ice for 1 h to allow binding but impede cell entry. Such a multiplicity of infection ensures that at least 99% of cells were infected with a minimum of one infectious viral particle. Unbound virus was removed and cells were harvested to determine the amount of viral RNA accumulation by quantitative RT‐PCR 30 min later. To assess DENV‐2 virus entry into cells via endocytosis, after binding on ice for 1 h, virus inocula were removed after 1 h of binding on ice, and infected cells were washed with 1× PBS followed by incubation with pre‐warmed DMEM for 10 min at 37°C to initiate virus cell entry. Subsequently, cells were rinsed three times with 1× PBS and then treated with 0.25% Trypsin for 10 min, and again washed three times with 1× PBS to remove any cell‐associated virus that did not enter the cytoplasm. For assessments of post‐entry infection steps, after virus binding and entry procedures were finished, HeLa cells were subsequently treated with IFITM3‐containing or control exosomes. Total cellular RNA was extracted to measure the quantity of viral genomes that had entered cells by using a real‐time RT‐PCR assay.
Isolation and characterization of IFITM3‐containing exosome
Seven groups of cells, including HUVEC (with high basal‐level IFITM3 protein), non‐targeting control siRNA‐transfected HUVEC (HUVEC‐siNC), IFITM3 siRNA‐1‐transfected HUVEC (HUVEC‐siIFITM3), IFITM3‐transfected 293T cells (293T‐IFITM3), control vector‐transfected 293T cells (293T‐Vector), IFN‐α‐treated 293T cells (100 U ml−1; 293T‐IFN) and 293T cells, were cultured in medium supplemented with 10% exosome‐free FBS (overnight centrifugation, 120 000 g), and grown to monolayer with 80–90% confluence. Exosomes were isolated from cell supernatants by filtration steps and differential ultracentrifugation as previously described (Khatua et al., 2009). Briefly, supernatants derived from HUVEC and 293T cell culture with different treatments were collected and centrifuged at 1000 g for 10 min, and then 10 000 g for 10 min at 4°C to remove cells and cell debris, followed by filtration with 0.22 μm filters (Pall Life Sciences, Port Washington, NY) and ultracentrifugation at 150 000 g for 2 h at 4°C to pellet crude exosomes. For further purification, the crude exosomes were mixed with 2 ml of 2.5 M sucrose in PBS and placed on the bottom of a Optiseal centrifuge tube (Beckman Coulter Inc., Munich, Germany), overlaid with 6 ml 2 M sucrose and 3 ml 0.25 M sucrose, and ultracentrifuged for 16 h at 150 000 g. The purified exosomes accumulating at the 2 M/0.25 M interface were collected, washed twice by diluted in PBS, and pelleted by ultracentrifugation at 150 000 g for 90 min. Pelleted exosomes were re‐suspended in PBS and used immediately or kept at −80°C. GW4869 (Sigma‐Aldrich) was employed as an inhibitor of exosome secretion. Protein concentrations of exosome preparations were determined with the Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions, using BSA as the standard. Finally, the IFITM3‐laden exosome samples were characterized by Western blotting analysis, and the protein band was excised from the gel and subjected to mass spectrometry (MS) analysis according to a standard procedure (Khatua et al., 2009).
Transmission electron microscopy
Purified exosomes were processed by negative staining and analysed by transmission electron microscopy (TEM) as previously described (Khatua et al., 2009). Briefly, exosomes resuspended in 30 μl PBS were dropped onto a sheet of Parafilm, and a Formvar‐carbon coated nickel grid were floated for 5 min at room temperature to absorb exosomes, followed by wash with PBS twice and fixed with 2% paraformaldehyde After rinsing with PBS twice, the grids were negatively stained with 1% phosphotungstic acid for 2 min. Finally, dry samples were viewed using a Tecnai 10 transmission electron microscope at 80 kV or stored in a grid box for future work.
Exosome treatment and cytopathic effect (CPE) reduction assay
Cells were seeded in 24‐well plates at 1 × 105 cells per well 24 h prior to viral infection or treatments described below. To evaluate the anti‐DENV ability of purified IFITM3‐laden exosomes, 30 and 60 μg ml−1 of IFITM3‐exosomes, respectively, was added to cultured cells and incubated for 12 h. Subsequently, mixture of NGC DENV‐2 (moi = 2) and 30 or 60 μg ml−1 of IFITM3‐exosomes, respectively, which had been pre‐incubated for 1 h at 4°C, was added to cells. After three washes with PBS to remove the unbound virus, IFITM3‐exosomes was added to cells and incubated for the indicated lengths of time (6, 12, 24 or 48 h). The control‐exosomes was prepared from 293T cells that contained only the vector plasmid without the IFITM3 expression cassette. IFN‐α and IFN‐β was quantified in the supernatants derived from cells treated with IFITM3‐exosomes, by using Enzyme Linked Immuno‐sorbent Assay (ELISA) kits, according to the manufacturer's instruction (Keygen Biotech, China; PBL Interferon Source, Piscataway, NJ) respectively. Following a pre‐lysis treatment with trypsin to remove adherent exosomes, infected cells and culture medium were harvested at various times after infection and subjected to RNA extraction. The amounts of dengue viral RNA were measured by using a quantitative or relative real‐time RT‐PCR assay.
For CPE reduction assay, at 48 h post infection, microscopic examination was performed to determine the antiviral effect, and the data were confirmed using a cell viability assay. The viability of BHK‐21 cells was measured by using the MTS [3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐2‐(4‐sulfophenyl)‐2H‐tetrazolium] assay according to the manufacturer's instructions. Briefly, 20 μl MTS solution (CellTiter 96Aqueous One Solution reagent, Promega, Madison, WI) was added to each well and incubated for an additional 4 h at 37°C, and the absorbance was subsequently measured at 490 nm using a microplate reader (Bio‐Tek, Winooski, VT). Cells treated with control‐exosome were used as the control group. Cell growth inhibition rates were determined using the following formula according to a previously published method (Muhamad et al., 2010): Cell viability (%) = (OD490 of treated cells/OD490 of control cells) × 100%. The experiment was repeated at least three times from which mean and standard deviation (SD) values were calculated.
Statistical analysis
The data given in the text were presented as means ± SD. Comparison between two groups was evaluated by the 2‐tailed Student's t test, using 0.05 as the cut‐off P‐value. Correlations were determined with the exponential regression analysis using the IBM SPSS Statistics 19 software.
Supporting information
Fig. S1. The IFITM protein family restricts DENV‐2 infection at entry. U937 (A) or HeLa (C) cells were transduced to express IFITM1, 2 or 3 protein or control vector alone, as verified by Western blotting analysis. IFITM protein expression in 293T cells was measured by Western blotting analysis, using aliquots of the same cells assayed in (E) respectively. Actin was included as a loading control. DENV‐2‐infected cells were detected using a monoclonal antibody (NYRDeng2) directed to the envelope (E) glycoprotein of the virus, and the numbers of DENV‐2‐infected U937 (B), HeLa (D) or 293T (F) cells were determined by flow cytometry.
Fig. S2. IFITM3 is required for the antiviral effects of interferons and silencing IFITM3 enhances DENV‐2 infection. (A) HUVEC cells transfected with indicated siRNAs, or a non‐targeting control siRNA (siNC), were assessed for IFITM3 levels by Western blotting analysis. At 36 h post transfection with indicated siRNA, the percentage of infected cells was assessed 24 h after virus addition by flow cytometry for E protein (B). HeLa cells were transfected with the indicated siRNAs, or a control, non‐targeting siRNA. At 36 h post transfection, cells were incubated with 0, 10 or 100 IU ml−1 IFN‐α for 24 h, and the levels of IFITM3 were examined by Western blotting (C). (D) HeLa cells transfected with indicated IFITM3‐trageting siRNAs were incubated with the indicated concentrations of IFN‐α for 24 h at 6 h post transfection, followed by infected with DENV‐2 at an moi of 2. The percentages of infected cells were assessed 24 h after virus addition by flow cytometry for the DENV E protein. E, Effect of siRNA silencing of IFITM3 on the binding and entry of DENV‐2 to HeLa cells. Data points are presented as means ± SD of triplicated experiments. Student's two‐tailed t test was performed, and statistical differences are shown with asterisks (**P < 0.01).
Fig. S3. Mass spectrometric analysis of IFITM3 peptides. Results automatically generated by the mass spectrometer. Briefly, purified IFITM3‐containing exosomes derived from 293T cells overexpressing IFITM3 were assayed by mass‐spectrometric analysis of proteolytic peptides. The red vertical lines represent peptide ion intensities, and the horizontal axis shows the mass‐to‐charge (m/z) values around the peptide ions of interest. The green dash lines were automatically given by the spectrometer for pointing the labels to the red peaks.
Fig. S4. 293T‐IFITM3 cell‐derived exosomes do not change the quantities of IFITM1 and IFITM2 in the recipient HeLa cells, as compared with the 293T cell‐ and 293T‐Vector cell‐derived exosomes.
Fig. S5. IFITM3 half‐life in the cell culture supernatant. The levels of IFITM3 protein in cell culture supernatant of HUVEC at various times (0, 6, 12, 24, 36, 48, 60 and 72 h) was assayed with Western blotting. Briefly, supernatants derived from HUVEC cells were collected and centrifuged at 1000 g for 10 min, and then further centrifuged at 10 000 g for 10 min at 4°C to remove cells and cell debris, followed by filtration with 0.22 μm filters. The obtained supernatants were then concentrated by 10‐fold using an Amicon Ultra‐15 centrifugal filter unit with 10 kDa cut‐off value, divided into 8 groups, and incubated at 37°C in a humidified atmosphere of 5% CO2 for 0, 6, 12, 24, 36, 48, 60 and 72 h, respectively, before being subjected to Western blotting analysis using an anti‐IFITM3 antibody. Intensities of the bands of interest on the PVDF membranes were quantitatively calculated with the Quantity One 4.6.3 measurement software. The results are presented as relative ratios of intensity of the band for the 0 h treatment group.
Fig. S6. Immunofluorescence staining and confocal microscopic analysis of IFITM3‐Flag and DENV‐2 viral E protein in HeLa cells treated with 293T‐IFITM3‐exosome followed by infection with DENV‐2.
Table S1. Primer sequences and siRNA oligonucleatides.
Acknowledgements
We owe our special thanks to Professor Xi Huang of Sun Yat‐sen University for providing materials essential for the study. This work was supported by the Key (Key grant) Project of Chinese Ministry of Education (No. 311030); Natural Science Foundation of China (No. 81330058, No. 81272417, No. 81102370); the National Science and Technique Major Project (No. 2012ZX10004‐213, No. 201305017, No. 2012ZX09102101‐017); Guangdong Recruitment Program of Creative Research Groups (No. 2009010058).
References
- Bhatnagar, S. , and Schorey, J.S. (2007a) Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem 282: 25779–25789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, S. , Shinagawa, K. , Castellino, F.J. , and Schorey, J.R.S. (2007b) Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo . Blood 110: 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boorn, J.G. , Schlee, M. , Coch, C. , and Hartmann, G. (2011) SiRNA delivery with exosome nanoparticles. Nat Biotechnol 29: 325–326. [DOI] [PubMed] [Google Scholar]
- Borden, E.C. , Sen, G.C. , Uze, G. , Silverman, R.H. , Ransohoff, R.M. , Foster, G.R. , and Stark, G.R. (2007) Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6: 975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass, A.L. , Huang, I.C. , Benita, Y. , John, S.P. , Krishnan, M.N. , Feeley, E.M. , et al. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y.K. , Huang, I.C. , and Farzan, M. (2012) IFITM proteins restrict antibody‐dependent enhancement of dengue virus infection. PLoS ONE 7: e34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt, A.R. , Clare, S. , Pertel, T. , John, S.P. , Wash, R.S. , Smith, S.E. , et al. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley, E.M. , Sims, J.S. , John, S.P. , Chin, C.R. , Pertel, T. , Chen, L.M. , et al. (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7: e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S.J. , Booth, A.M. , and Hildreth, J.E. (2003) The Trojan exosome hypothesis. Proc Natl Acad Sci USA 100: 10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, M.G. , Halstead, S.B. , Artsob, H. , Buchy, P. , Farrar, J. , Gubler, D.J. , et al. (2010) Dengue: a continuing global threat. Nat Rev Microbiol 8: S7–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead, S.B. (2005) More dengue, more questions. Emerg Infect Dis 11: 740–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead, S.B. (2007) Dengue. Lancet 370: 1644–1652. [DOI] [PubMed] [Google Scholar]
- Halstead, S.B. , Suaya, J.A. , and Shepard, D.S. (2007) The burden of dengue infection. Lancet 369: 1410–1411. [DOI] [PubMed] [Google Scholar]
- Huang, I.C. , Bailey, C.C. , Weyer, J.L. , Radoshitzky, S.R. , Becker, M.M. , Chiang, J.J. , et al. (2011) Distinct patterns of IFITM‐mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7: e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo‐Useros, N. , Naranjo‐Gomez, M. , Archer, J. , Hatch, S.C. , Erkizia, I. , Blanco, J. , et al. (2009) Capture and transfer of HIV‐1 particles by mature dendritic cells converges with the exosome‐dissemination pathway. Blood 113: 2732–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, D. , Weidner, J.M. , Qing, M. , Pan, X.B. , Guo, H. , Xu, C. , et al. (2010) Identification of five interferon‐induced cellular proteins that inhibit west Nile virus and dengue virus infections. J Virol 84: 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanlaya, R. , Pattanakitsakul, S.N. , Sinchaikul, S. , Chen, S.T. , and Thongboonkerd, V. (2010) Vimentin interacts with heterogeneous nuclear ribonucleoproteins and dengue nonstructural protein 1 and is important for viral replication and release. Mol Biosyst 6: 795–806. [DOI] [PubMed] [Google Scholar]
- Khatua, A.K. , Taylor, H.E. , Hildreth, J.E. , and Popik, W. (2009) Exosomes packaging APOBEC3G confer human immunodeficiency virus resistance to recipient cells. J Virol 83: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth, C.R. , White, L.J. , Johnston, R.E. , and de Silva, A.M. (2005) Flow cytometry‐based assay for titrating dengue virus. J Clin Microbiol 43: 3267–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Liu, K. , Liu, Y. , Xu, Y. , Zhang, F. , Yang, H. , et al. (2013) Exosomes mediate the cell‐to‐cell transmission of IFN‐alpha‐induced antiviral activity. Nat Immunol 14: 793–803. [DOI] [PubMed] [Google Scholar]
- Lloyd, R.E. , Blalock, J.E. , and Stanton, G.J. (1983) Cell‐to‐cell transfer of interferon‐induced antiproliferative activity. Science 221: 953–955. [DOI] [PubMed] [Google Scholar]
- Lu, J. , Pan, Q. , Rong, L. , He, W. , Liu, S.L. , and Liang, C. (2011) The IFITM proteins inhibit HIV‐1 infection. J Virol 85: 2126–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota, J. , and Rico‐Hesse, R. (2009) Humanized mice show clinical signs of dengue fever according to infecting virus genotype. J Virol 83: 8638–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhamad, M. , Kee, L.Y. , Rahman, N.A. , and Yusof, R. (2010) Antiviral actions of flavanoid‐derived compounds on dengue virus type‐2. Int J Biol Sci 6: 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q. , Ramakrishnaiah, V. , Henry, S. , Fouraschen, S. , de Ruiter, P.E. , Kwekkeboom, J. , et al. (2012) Hepatic cell‐to‐cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi). Gut 61: 1330–1339. [DOI] [PubMed] [Google Scholar]
- Sadler, A.J. , and Williams, B.R. (2008) Interferon‐inducible antiviral effectors. Nat Rev Immunol 8: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey, J.S. , and Bhatnagar, S. (2008) Exosome function: from tumor immunology to pathogen biology. Traffic 9: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtam, T.A. , Kovalev, R.A. , Varfolomeeva, E.Y. , Makarov, E.M. , Kil, Y.V. , and Filatov, M.V. (2013) Exosomes are natural carriers of exogenous siRNA to human cells in vitro . Cell Commun Signal 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, C.P. , Farrar, J.J. , Nguyen, V.V. , and Wills, B. (2012) Dengue. N Engl J Med 366: 1423–1432. [DOI] [PubMed] [Google Scholar]
- Thery, C. , Zitvogel, L. , and Amigorena, S. (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2: 569–579. [DOI] [PubMed] [Google Scholar]
- Tian, J. , Zeng, G. , Pang, X. , Liang, M. , Zhou, J. , Fang, D. , et al. (2012) Identification and immunogenicity of two new HLA‐A*0201‐restricted CD8+ T‐cell epitopes on dengue NS1 protein. Int Immunol 24: 207–218. [DOI] [PubMed] [Google Scholar]
- Weidner, J.M. , Jiang, D. , Pan, X.B. , Chang, J. , Block, T.M. , and Guo, J.T. (2010) Interferon‐induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol 84: 12646–12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , He, L. , Li, Y. , Wang, T. , Feng, L. , Jiang, L. , et al. (2013) miR‐146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J Infect 67: 329–341. [DOI] [PubMed] [Google Scholar]
- Yao, L. , Dong, H. , Zhu, H. , Nelson, D. , Liu, C. , Lambiase, L. , and Li, X. (2012) Identification of the IFITM3 gene as an inhibitor of hepatitis C viral translation in a stable STAT1 cell line. J Viral Hepat 18: e523–e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The IFITM protein family restricts DENV‐2 infection at entry. U937 (A) or HeLa (C) cells were transduced to express IFITM1, 2 or 3 protein or control vector alone, as verified by Western blotting analysis. IFITM protein expression in 293T cells was measured by Western blotting analysis, using aliquots of the same cells assayed in (E) respectively. Actin was included as a loading control. DENV‐2‐infected cells were detected using a monoclonal antibody (NYRDeng2) directed to the envelope (E) glycoprotein of the virus, and the numbers of DENV‐2‐infected U937 (B), HeLa (D) or 293T (F) cells were determined by flow cytometry.
Fig. S2. IFITM3 is required for the antiviral effects of interferons and silencing IFITM3 enhances DENV‐2 infection. (A) HUVEC cells transfected with indicated siRNAs, or a non‐targeting control siRNA (siNC), were assessed for IFITM3 levels by Western blotting analysis. At 36 h post transfection with indicated siRNA, the percentage of infected cells was assessed 24 h after virus addition by flow cytometry for E protein (B). HeLa cells were transfected with the indicated siRNAs, or a control, non‐targeting siRNA. At 36 h post transfection, cells were incubated with 0, 10 or 100 IU ml−1 IFN‐α for 24 h, and the levels of IFITM3 were examined by Western blotting (C). (D) HeLa cells transfected with indicated IFITM3‐trageting siRNAs were incubated with the indicated concentrations of IFN‐α for 24 h at 6 h post transfection, followed by infected with DENV‐2 at an moi of 2. The percentages of infected cells were assessed 24 h after virus addition by flow cytometry for the DENV E protein. E, Effect of siRNA silencing of IFITM3 on the binding and entry of DENV‐2 to HeLa cells. Data points are presented as means ± SD of triplicated experiments. Student's two‐tailed t test was performed, and statistical differences are shown with asterisks (**P < 0.01).
Fig. S3. Mass spectrometric analysis of IFITM3 peptides. Results automatically generated by the mass spectrometer. Briefly, purified IFITM3‐containing exosomes derived from 293T cells overexpressing IFITM3 were assayed by mass‐spectrometric analysis of proteolytic peptides. The red vertical lines represent peptide ion intensities, and the horizontal axis shows the mass‐to‐charge (m/z) values around the peptide ions of interest. The green dash lines were automatically given by the spectrometer for pointing the labels to the red peaks.
Fig. S4. 293T‐IFITM3 cell‐derived exosomes do not change the quantities of IFITM1 and IFITM2 in the recipient HeLa cells, as compared with the 293T cell‐ and 293T‐Vector cell‐derived exosomes.
Fig. S5. IFITM3 half‐life in the cell culture supernatant. The levels of IFITM3 protein in cell culture supernatant of HUVEC at various times (0, 6, 12, 24, 36, 48, 60 and 72 h) was assayed with Western blotting. Briefly, supernatants derived from HUVEC cells were collected and centrifuged at 1000 g for 10 min, and then further centrifuged at 10 000 g for 10 min at 4°C to remove cells and cell debris, followed by filtration with 0.22 μm filters. The obtained supernatants were then concentrated by 10‐fold using an Amicon Ultra‐15 centrifugal filter unit with 10 kDa cut‐off value, divided into 8 groups, and incubated at 37°C in a humidified atmosphere of 5% CO2 for 0, 6, 12, 24, 36, 48, 60 and 72 h, respectively, before being subjected to Western blotting analysis using an anti‐IFITM3 antibody. Intensities of the bands of interest on the PVDF membranes were quantitatively calculated with the Quantity One 4.6.3 measurement software. The results are presented as relative ratios of intensity of the band for the 0 h treatment group.
Fig. S6. Immunofluorescence staining and confocal microscopic analysis of IFITM3‐Flag and DENV‐2 viral E protein in HeLa cells treated with 293T‐IFITM3‐exosome followed by infection with DENV‐2.
Table S1. Primer sequences and siRNA oligonucleatides.


