Summary
CD81 is a major receptor for Hepatitis C Virus (HCV). It belongs to the tetraspanin family whose members form dynamic clusters with numerous partner proteins and with one another, forming tetraspanin‐enriched areas in the plasma membrane. In our study, we combined single‐molecule microscopy and biochemistry experiments to investigate the clustering and membrane behaviour of CD81 in the context of cells expressing EWI‐2wint, a natural inhibitor of HCV entry. Interestingly, we found that EWI‐2wint reduces the global diffusion of CD81 molecules due to a decrease of the diffusion rate of mobile CD81molecules and an increase in the proportion of confined molecules. Indeed, we demonstrated that EWI‐2wint promotes CD81 clustering and confinement in CD81‐enriched areas. In addition, we showed that EWI‐2wint influences the colocalization of CD81 with Claudin‐1 – a co‐receptor required for HCV entry. Together, our results indicate that a change in membrane partitioning of CD81 occurs in the presence of EWI‐2wint. This study gives new insights on the mechanism by which HCV enters into its target cells, namely by exploiting the dynamic properties of CD81.
Introduction
Viral entry relies on a fine interplay between a viral particle and a host cell. This process is initiated by binding to an attachment factor present at the plasma membrane of target cells, which helps to concentrate virions on the cell surface. After this initial step, the viral particle interacts with one or several specific receptor(s), which actively promote virus entry by inducing conformational changes of the viral particle and/or by activating signalling pathways or internalization of the virion. A number of cell surface components exploited by viruses have now been identified (reviewed in Grove and Marsh, 2011). Although many viruses use a single specific receptor (i.e. Poliovirus and human Rhinovirus 2), several viruses exploit more than one molecular species as receptors, each with equivalent roles [i.e. angiotensin‐converting enzyme (ACE) or liver‐SIGN (L‐SIGN) for SARS coronavirus]. However, other viruses require multiple cell surface components, each of which is essential. One of the best examples of this kind of viruses is the Hepatitis C Virus (HCV).
HCV entry into hepatocytes is a multistep process involving viral envelope glycoproteins as well as several cellular attachment and entry factors. After attachment to the host cell, a complex process occurs in which the virion interacts with a series of cellular entry factors, including the tetraspanin CD81 (Pileri et al., 1998), the scavenger receptor type B class I (SRB1) (Scarselli et al., 2002), two tight junction proteins, claudin‐1 (CLDN‐1) (Evans et al., 2007) and occludin (Ploss et al., 2009) and additional host factors (Lupberger et al., 2011; Sainz et al., 2012). HCV entry is regulated by receptor tyrosine kinases including EGFR (Lupberger et al., 2011). Among these entry factors, CD81 plays a major role in HCV entry (reviewed in Dubuisson et al., 2008; Farquhar et al., 2011). Indeed, through its large extracellular loop (LEL), CD81 is involved in a direct interaction with the E2 envelope glycoprotein exposed at the surface of HCV virion. Numerous studies have shown that cell susceptibility to HCV infection is closely related to the CD81 expression level. In addition, CD81 associates with CLDN‐1 to form co‐receptor complexes that are critical for HCV entry (Harris et al., 2008, 2010; Krieger et al., 2010). It is believed that the engagement of CD81 activates Rho GTPase and MAPK signalling cascades (Brazzoli et al., 2008). Although MAPK pathway is likely important for as yet unidentified post‐entry events, CD81‐triggered Rho GTPase signalling probably induces actin remodelling, allowing lateral movement of CD81 necessary for HCV entry (Brazzoli et al., 2008).
CD81 belongs to the tetraspanin family whose members have a strong propensity to multimerize and interact with numerous partner proteins forming scaffolds in membranes, recruiting or excluding specific proteins involved in particular cellular processes (reviewed in Charrin et al., 2009a). The precise mechanisms of interaction within and outside these clusters called tetraspanin‐enriched microdomains (TEMs) or tetraspanin webs is still poorly understood. However, from high‐resolution fluorescence microscopy techniques, we now know that tetraspanins are distributed along the whole cell surface with a higher concentration in dot‐like areas called tetraspanin‐enriched areas (TEAs). These TEAs are stable platforms in position and shape but in permanent exchange with the rest of the membrane, indicating that inside the tetraspanin web, protein–protein interactions are transient and highly dynamic (Barreiro et al., 2008; Espenel et al., 2008). In addition, cellular lipids such as cholesterol, glycosphingolipids and palmitic acid seem to play an important role in interactions between tetraspanins and their partners and therefore in building the tetraspanin web (Charrin et al., 2009a).
The direct interaction between tetraspanins and their partner proteins can result in the modulation of their functions. For instance, CD81 functions in cellular processes and infectious diseases can be affected by the association with proteins of the EWI family whose members have a single transmembrane domain and several extracellular Ig‐domains with a conserved EWI motif, as well as a very short cytosolic tail (Stipp et al., 2001). Indeed, EWI‐2 modulates cellular migration (Stipp et al., 2003; Zhang et al., 2003; Sala‐Valdés et al., 2006) and EWI‐F/CD9P‐1 inhibits Plasmodium infection (Charrin et al., 2009b). A few years ago, we have identified EWI‐2wint, a cleavage product of EWI‐2 in which the first of the four extracellular Ig‐domains is cleaved off (Rocha‐Perugini et al., 2008). This shorter protein forming heterodimers with EWI‐2 (Montpellier et al., 2011) still interacts with CD81 and, can be found in most cell lines expressing EWI‐2 but not in hepatocytes. Importantly, in contrast to full‐length EWI‐2, EWI‐2wint inhibits HCV infection by inhibiting viral entry (Rocha‐Perugini et al., 2008). In addition to the presence of specific entry factors in the hepatocytes, the lack of this specific inhibitor may contribute to the hepatotropism of HCV. Although determinants for the interaction of EWI‐2/EWI‐2wint and CD81 have been recently identified (Montpellier et al., 2011), the mechanism by which EWI‐2wint blocks HCV entry has remained elusive.
Limited information is available on the exact role of each receptor in HCV entry. However, HCV entry is believed to be mediated through the formation of a tightly orchestrated HCV‐entry factor complex at the plasma membrane, but for which interaction kinetics need to be defined. Since the mobility is a critical determinant for the interaction capabilities and for the functions of membrane proteins, we sought to determine the impact of EWI‐2wint on CD81 membrane dynamics. By combining biochemistry experiments and single‐molecule microscopy experiments, we demonstrated that a change in membrane dynamics and partitioning of CD81 occurs in the presence of EWI‐2wint, which affects cell‐free transmission of HCV. Together, our results indicate that HCV likely exploits the dynamic properties of CD81 to enter into its target cells.
Results
EWI‐2wint promotes a change in CD81 organization at the plasma membrane
In a previous work, we described a human hepatoma Huh‐7 cell line (Huh‐7w7) which has lost CD81 expression and can be infected by HCV when mouse CD81 (mCD81) is ectopically expressed (Rocha‐Perugini et al., 2009). We took advantage of these permissive cells expressing mCD81 and the previously described MT81/MT81w monoclonal antibodies (mAbs) (Silvie et al., 2006) to analyse the role of tetraspanin web‐associated CD81 in HCV infection. Indeed, whereas MT81 mAb recognizes all CD81 molecules, MT81w antibody only recognizes a fraction of mCD81 associated with the tetraspanin web or at least CD81 engaged in protein clusters enriched in cholesterol. Although MT81 was fully neutralizing, HCV infection was poorly inhibited by MT81w, suggesting that only a fraction of CD81 is preferentially used for the virus entry (Rocha‐Perugini et al., 2009). Based on these data, we made the hypothesis that EWI‐2wint might inhibit HCV entry by modifying the organization of CD81 at the plasma membrane.
In order to test this hypothesis, we generated Huh‐7w7 clones expressing mCD81 in combination with EWI‐2 or EWI‐2wint and used the MT81w mAb to probe modifications of CD81 organization. EWI‐2 is endogenously expressed in hepatoma cells but its cleavage does not naturally occur preventing EWI‐2wint production, as previously described (Rocha‐Perugini et al., 2008). In addition, direct expression of EWI‐2wint sequence leads to the production of an unstable protein in these cells. To circumvent these technical difficulties, we used a modified EWI‐2 protein that contains a furin cleavage site (EWI‐2Fur), directly upstream of the first EWI‐2wint residue, leading to the production of EWI‐2wint in virtually all cell lines (Fig. 1A) (Rocha‐Perugini et al., 2008). We generated Huh‐7w7 cells stably expressing mCD81 in combination with either the parental EWI‐2 protein (Huh‐7w7/mCD81/EWI‐2), or the cleavage‐competent EWI‐2Fur construct (Huh‐7w7/mCD81/EWI‐2wint) or the empty vector (Huh‐7w7/mCD81/pcDNA3.1) (Fig. 1B). Protein expression in each cell line was controlled by flow cytometry (data not shown) and immunoprecipitation (IP) of cell surface biotinylated lysates (Fig. 1B). Each of the ectopic EWI proteins was correctly expressed, as controlled by direct IP with a mAb directed against a C‐terminally expressed FLAG tag. As shown by co‐IP with mCD81, in addition to endogenous EWI‐2 protein (EWI‐2 endo), ectopically expressed EWI‐2 and EWI‐2wint proteins interacted with mCD81, in MT81 and MT81w IPs. The conservation of an interaction between human EWI‐2/EWI‐2wint proteins and murine CD81 was expected since the regions involved in protein–protein contact between these two proteins are well conserved (Montpellier et al., 2011). The cell lines were next used to generate cell clones for which expression of ectopic EWI proteins was controlled (data not shown). These cell clones were infected with infectious HCV particles produced in cell culture (HCVcc) (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005), which represent the most relevant tool to study HCV life cycle. We used JFH1‐based Renilla luciferase (R‐Luc) reporter HCVcc (JFH1/CSN6A4/5′C19Rluc2AUbi), as described previously (Delgrange et al., 2007; Rocha‐Perugini et al., 2008; Goueslain et al., 2010). Interestingly, HCV infection levels were significantly reduced in cell clones producing EWI‐2wint (Fig. 1C), as compared with cells expressing the empty vector (pcDNA3.1) or overexpressing EWI‐2 (EWI‐2). Altogether these results strongly suggest that EWI‐2wint interacts with mCD81 and inhibits HCV infection mediated by the murine receptor. It should be noted that EWI‐2wint expression does not affect expression levels of mCD81 and other entry factors (data not shown).
Figure 1.
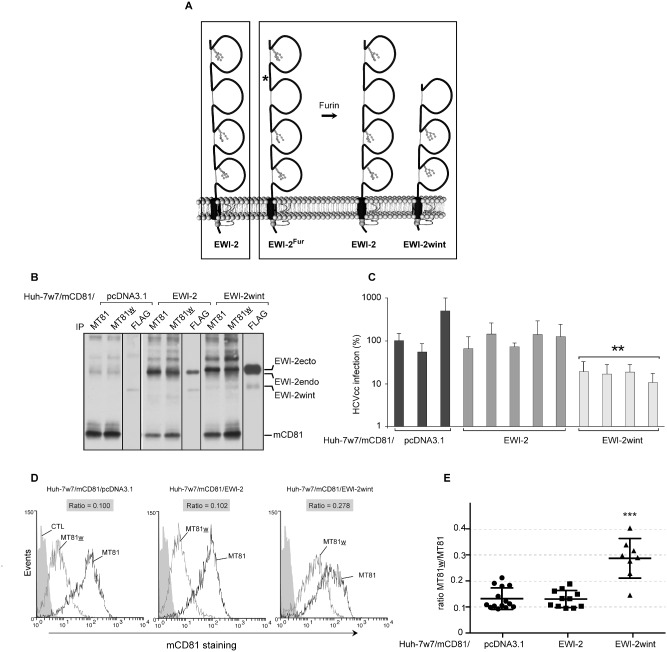
EWI‐2wint favours CD81 association with other tetraspanins.
A. Schematic representation of EWI‐2 and EWI‐2 Fur proteins. The EWI‐2 Fur construct consists in the insertion of a cleavage sequence for furin (RGRR, see asterisk) between the first and the second immunoglobulin (Ig) domains of EWI‐2, so that cells containing this construct overexpress EWI‐2 and express EWI‐2wint.
B. Huh‐7w7 cells were stably transfected with mCD81 and either the empty vector (pcDNA3.1), EWI‐2 or EWI‐2 Fur construct (EWI‐2wint). Cell surface biotinylation followed by immunoprecipitation assays were performed to control the expression of ectopic proteins. MT81 and MT81w mAbs immunoprecipitate mCD81. A FLAG tag inserted at the C‐terminus of the EWI‐2 Fur construct allows to immunoprecipitate EWI‐2 and EWI‐2wint. Proteins were revealed by Western blotting with HRP‐conjugated streptavidin.
C. Several Huh‐7w7/mCD81 clones were tested in HCVcc infection. Results are presented as related percentages to the first control clone (pcDNA3.1) and reported as the mean ± SD of three independent experiments. ** means a P value below 0.01, as determined by unpaired t‐test.
D. Huh‐7w7/mCD81 clones expressing pcDNA3.1, EWI‐2 or EWI‐2wint were stained with MT81 and MT81w mAbs followed by PE‐labelled secondary antibody and analysed by flow cytometry. Cells stained with only the secondary antibody were used as controls (CTL). Stainings of three representative clones are shown.
E. MT81w/MT81 ratios for 14 clones expressing pcDNA3.1, 12 clones expressing EWI‐2 and 8 clones expressing EWI‐2wint (one representative flow cytometry experiment). Black bars correspond to mean ± SD for each group and *** means a P value below 0.001, as determined by the Mann–Whitney U ‐test.
Using the MT81w anti‐mCD81 mAb, we next characterized the influence of EWI‐2wint on the cell surface organization of mCD81. As shown by flow cytometry analysis of Huh‐7w7/mCD81/pcDNA3.1 and Huh‐7w7/mCD81/EWI‐2 cells, the intensity of MT81w labelling was 15 times weaker than that of MT81 (Fig. 1D), confirming that only a fraction of CD81 molecules is recognized by MT81w. Interestingly, as shown for one representative cell clone, cells expressing EWI‐2wint exhibited a stronger staining with MT81w, indicating that EWI‐2wint modifies CD81 membrane organization, probably by increasing the association of CD81 with the tetraspanin web. In order to carry out statistical analyses of EWI‐2wint effect, we stained 14 clones expressing pcDNA3.1, 12 clones expressing EWI‐2 and 8 clones expressing EWI‐2wint with MT81 and MT81w antibodies. For each clone, we calculated the ratio MT81w/MT81. As shown in Fig. 1E, the ratio MT81w/MT81 was significantly increased in cell clones expressing EWI‐2wint indicating that EWI‐2wint likely promotes a change of CD81 organization within the tetraspanin web.
EWI‐2wint significantly restricts the diffusion of CD81 molecules
To strengthen the hypothesis that EWI‐2wint induces a change in CD81 organization at the plasma membrane, we next analysed the membrane dynamics and partitioning of human CD81 (hCD81) molecules in Huh‐7 cells expressing or lacking EWI‐2wint by single‐molecule fluorescence microscopy, which is a method of choice to characterize the diffusion of proteins and lipids in different regions of the plasma membrane (Owen et al., 2009). This emerging technique has been used previously to address the dynamics of the tetraspanin CD9 which has been proposed to diffuse in nanoclusters of tetraspanin/partner, patrolling in the plasma membrane in permanent exchange with TEAs (Espenel et al., 2008).
Before carrying out single‐molecule experiments, we first generated Huh‐7 cell clones expressing EWI‐2wint. For this purpose, we used the modified EWI‐2 protein that contains a furin cleavage site (EWI‐2Fur), directly upstream of the first amino acid of EWI‐2wint, as described in the previous section. Recently, we found that the mutation (LAL) of a glycine‐zipper motif in the transmembrane domain of EWI‐2/EWI‐2wint abolishes their interaction with CD81 and the inhibitory effect of EWI‐2wint on HCV infection (Montpellier et al., 2011). Moreover, we demonstrated that, if palmitoylation of two juxtamembranous cysteines is conserved, the cytosolic tail of EWI‐2/EWI‐2wint, which is involved in interactions with the actin cytoskeleton (Sala‐Valdés et al., 2006), can be replaced by that of MHC II without affecting their biological function (Qcc) (Montpellier et al., 2011). Thus, in our study, we generated Huh‐7 cell clones stably expressing either the parental EWI‐2 protein (Huh‐7/EWI‐2), or the cleavage‐competent EWI‐2Fur construct (Huh‐7/EWI‐2wint clones 1, 2 and 3), or the EWI‐2Fur protein in which residues that are essential for the interaction with CD81 were mutated (Huh‐7/LAL) (Montpellier et al., 2011), or the EWI‐2Fur protein in which the cytosolic tail was replaced but the two palmitoylatable cysteines conserved (Huh‐7/Qcc) or cells expressing the empty vector (Huh‐7/pcDNA3.1) (Fig. 2A). Protein expression in each clone was controlled by flow cytometry (data not shown) and immunoprecipitation of cell surface biotinylated lysates (Fig. 2B). Each of the ectopic EWI‐2 proteins was correctly expressed, as controlled by direct immunoprecipitation with a mAb directed against a C‐terminally expressed FLAG tag (Fig. 2B, left panel) or a mAb directed against an N‐terminally expressed HA tag (Fig. 2B, right panel). As shown by co‐immunoprecipitation with CD81, in addition to endogenous EWI‐2 protein (EWI‐2 endo), ectopically expressed EWI‐2 and EWI‐2wint proteins interacted with CD81, whereas LAL related proteins did not. It should be noted that, although Qcc related proteins barely co‐precipitate with CD81, the interaction occurs between these molecules, as previously demonstrated (Montpellier et al., 2011). Cells were next infected with infectious HCVcc particles (JFH‐1/CSN6A4) and infection levels were evaluated by flow cytometry using a mAb directed against the non‐structural protein 5 (NS5) (Fig. 2C). The proportion of infected cells was reduced in cells producing EWI‐2wint (EWI‐2wint 1, 2 and 3) whereas it remained unmodified in cells overexpressing EWI‐2 (EWI‐2) or EWI‐2 proteins that no longer interact with CD81 (LAL), as previously described (Montpellier et al., 2011). The proportion of infected cells was also reduced in cells expressing the Qcc protein, which still interacts with CD81 (Qcc). It should be noted that EWI‐2wint expression does not affect expression levels of CD81 and other entry factors (Rocha‐Perugini et al., 2008; Montpellier et al., 2011). In addition, levels of CD81 expression at the cell surface were equivalent for all the clones (data not shown).
Figure 2.
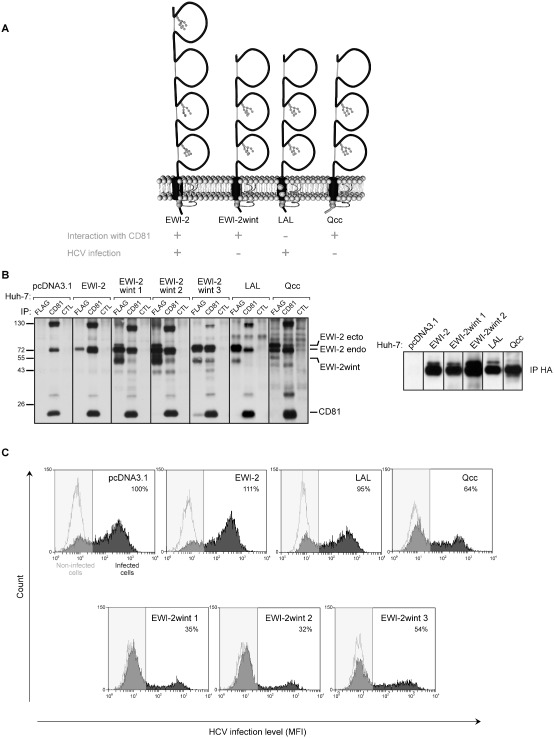
EWI‐2wint needs to interact with CD81 to inhibit HCV infection.
A. Schematic representation of EWI‐2 and mutated EWI‐2 Fur proteins. EWI‐2wint corresponds to EWI‐2 without its first Ig domain. In the LAL mutant, mutation of a glycine zipper motif in the transmembrane domain of EWI‐2 Fur abolishes the interaction of EWI‐2/EWI‐2wint with CD81. In the Qcc mutant, the cytosolic tail of EWI‐2 Fur was replaced by that of MHC II but which still contains the two juxtamembranous palmitoylatable cysteines.
B. Huh‐7 clones expressing the different FLAG and HA‐tagged EWI constructs were cell surface biotinylated and lysed in PBS/BrijO10/EDTA. Lysates were used to carry out immunoprecipation assays with the TS81 anti‐CD81 mAb, the M2 anti‐FLAG mAb or the HA11 anti‐HA mAb. The control lines (CTL) correspond to lysates incubated with no mAb. Proteins are detected using HRP‐Streptavidin.
C. Each Huh‐7 clone was infected with HCVcc and stained using anti‐NS5 mAb (2F6/G11) and a secondary antibody conjugated with PE. Non‐infected cells served as negative controls. A representative histogram of every clone is shown. Percentages corresponding to infected cells were calculated using Weasel software.
Before carrying out single‐molecule experiments on generated cell clones, the distribution of CD81 molecules at the basal surface of cells plated on collagen I was analysed by immunofluorescence on living cells using total internal reflection fluorescence (TIRF) microscopy. As controls, we also analysed the distribution of the tetraspanin CD9 and of CD46, a type I membrane protein excluded from rafts and TEMs (Espenel et al., 2008). CD81, CD9 and CD46 molecules were labelled with Atto647N‐labelled Fab fragments of TS81, SYB‐1 and 11C5 mAbs, respectively, as previously described (Espenel et al., 2008). CD81 molecules, as well as their counterparts CD9, were distributed along the whole cell surface, with a higher concentration in dot‐like areas (Fig. 3 and Fig. S1). These structures have been described as TEAs in which tetraspanins are confined but from which they can also escape and freely diffuse in the plasma membrane (Espenel et al., 2008). As shown in Fig. 3 and Fig. S1, no major change in the distribution of CD81, as well as that of CD9 and CD46, was observed in cell clones expressing or lacking EWI‐2wint, indicating that this protein does not affect the global distribution of membrane proteins expressed at the plasma membrane.
Figure 3.
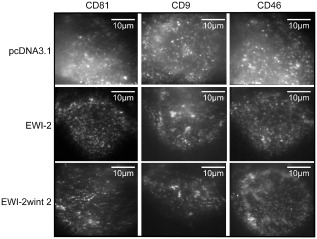
Cell surface distribution of CD81 and control proteins. Immunofluorescence images showing the basal membrane of living Huh‐7 clones expressing pcDNA3.1 control vector, EWI‐2 or EWI‐2wint 2. Experiments were performed at 37°C using TIRF microscopy. Cells were stained with Atto647N‐labelled Fab fragments of TS81, SYB‐1 and 11C5 mAbs to observe CD81, CD9 and CD46 respectively.
Dynamics and partitioning of CD81 were next investigated using single‐molecule tracking (SMT), a technique based on the labelling of a low number of molecules allowing individual molecules to be optically isolated and their position accurately determined. With this technique, the position of proteins was determined frame by frame and their trajectories reconstructed. For this purpose, Huh‐7 cell clones were plated on collagen I‐coated glass coverslips to allow attachment and cell spreading. Single CD81 molecules were tracked using Atto647N‐labelled Fab fragments of TS81 mAb. For each cell clone, the apparent diffusion coefficient (ADC) of at least 500 individual CD81 molecules (10–20 trajectories per cell) was calculated using a linear fit to the mean squared displacement (MSD) versus time plots for the first three points of the curve (Michalet, 2010) and their distribution displayed with a scatter plot. We first analysed the dynamics of CD81 in Huh‐7/pcDNA3.1 and Huh‐7/EWI‐2 cells for which we found no difference (data not shown). Since expressing EWI‐2wint in Huh‐7 cells required a mutated form of EWI‐2 to be overexpressed, we considered that cells overexpressing the parental EWI‐2 protein (Huh‐7/EWI‐2) were a better control than unmodified Huh‐7 cells (Huh‐7/pcDNA3.1). As shown in the scatter plot in Fig. 4A in which each trajectory is represented by a dot, CD81 molecules are distributed in three subpopulations (EWI‐2): (i) a subpopulation characterized by the highest ADC, (ii) a subpopulation characterized by the lowest ADC and (iii) a major subpopulation with intermediate ADC. Strikingly, in cell clones expressing EWI‐2wint (EWI‐2wint 1 and EWI‐2wint 2), a highly significant global shift of CD81 trajectories was observed with an increase of the subpopulation 2 with a low ADC at the expense of the subpopulation 1 with a high ADC. This effect was characterized by a decrease of the mean value of CD81 ADC in EWI‐2wint 1 (1 × 10−2 μm2 s−1) and EWI‐2wint 2 (1.1 × 10−2 μm2 s−1) cells, as compared with EWI‐2 cells (3.3 × 10−2 μm2 s−1) (Table 1). These results indicate that EWI‐2wint induces a global change of the behaviour of CD81 molecules in the plasma membrane. Interestingly, CD81 trajectories in LAL cells (in which EWI‐2wint no longer interacts with CD81) were distributed similarly to those of EWI‐2 cells demonstrating the specificity of the observed effect and that EWI‐2wint needs to interact with CD81 to alter its diffusion. We next analysed the distribution of CD81 trajectories in cells expressing the mutant Qcc which still interacts with CD81 but in which the cytosolic tail of EWI‐2/EWI‐2wint was replaced by that of MHC II. As in EWI‐2wint 1 and EWI‐2wint 2 cells, however at a lesser extent, a global shift of CD81 trajectories towards slower trajectories was seen in Qcc cells. Since the cytosolic tail of EWI proteins interacts with the actin cytoskeleton (Sala‐Valdés et al., 2006), our results indicate that differences in the diffusion of CD81 in cells expressing EWI‐2wint cannot be attributed to its differential anchorage to the actin cytoskeleton.
Figure 4.
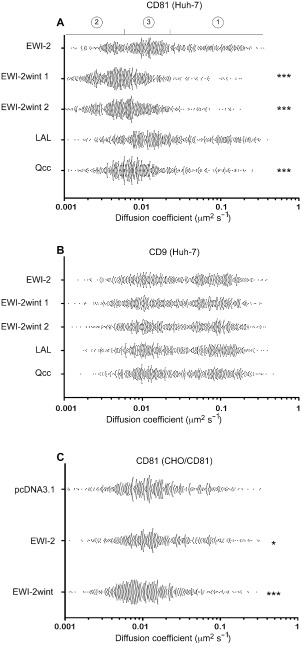
EWI‐2wint affects CD81 dynamics.
A and B. Panel (A) and panel (B) represent the distribution of all the apparent diffusion coefficients (ADC) calculated for CD81 and CD9 respectively, in Huh‐7 clones expressing EWI‐2, EWI‐2wint (two clones), LAL or Qcc. To stain proteins of interest, cells were labelled with Atto647N‐labelled Fab fragments of TS81 and SYB‐1 mAbs respectively. Each dot represents one trajectory, 500 trajectories are represented for each cell clone and *** indicates a P value below 0.0001 in comparison with the ADC in the Huh‐7/EWI‐2 clone, as determined by the Mann–Whitney U ‐test.
C. CHO cells were transfected with human CD81 in combination with either the empty vector (pcDNA3.1), a non‐cleavable EWI‐2 construct (EWI‐2) or EWI‐2, which is in these cells cleaved to produce EWI‐2wint (EWI‐2wint). Cells were used in single‐molecule tracking experiments and labelled with Atto647N‐labelled Fab fragments of TS81. * and *** indicate a P value below 0.01 and 0.0001, respectively, in comparison with the ADC in CHO/CD81/pcDNA3.1 cells, as determined by the Mann–Whitney U ‐test.
Table 1.
ADC of CD81, CD9 and CD46 molecules
| Cells | Proteins | D (·10−2 μm2 s−1)a |
|---|---|---|
| EWI‐2 | CD81 | 3.3 |
| CD9 | 4.8 | |
| CD46 | 3.6 | |
| EWI‐2wint 1 | CD81 | 1.0*** |
| CD9 | 5.0 | |
| CD46 | 2.7 | |
| EWI‐2wint 2 | CD81 | 1.1*** |
| CD9 | 5.3 | |
| CD46 | 2.3 | |
| LAL | CD81 | 4.3 |
| CD9 | 5.9 | |
| CD46 | 3.2 | |
| Qcc | CD81 | 1.9*** |
| CD9 | 5.9 | |
| CD46 | 2.8 | |
| CHO/CD81/pcDNA3.1 | CD81 | 2.6 |
| CHO/CD81/EWI‐2 | CD81 | 3.0* |
| CHO/CD81/EWI‐2wint | CD81 | 1.9*** |
ADC mean value of all trajectories (corresponding to the three modes of diffusion).
* and *** indicate a P value below 0.01 and 0.0001, respectively, in comparison with the ADC values of CD81, CD9 or CD46 in EWI‐2‐expressing Huh‐7 cells as determined by the Mann–Whitney U‐test. In CHO cells, statistics were done in comparison with the CHO/CD81/pcDNA3.1 cells.
To determine whether EWI‐2wint has a general effect on diffusion of membrane proteins, we also analysed the membrane behaviour of CD9 and CD46 molecules in cell clones expressing or lacking EWI‐2wint. As shown in Fig. 4B and Fig. S2, no significative difference was found in the distribution of CD9 and CD46 ADC values, indicating that EWI‐2wint does not affect their membrane diffusion. Together these results demonstrate that EWI‐2wint specifically affects the dynamics of CD81 molecules.
Since previous experiments were performed in Huh‐7 cell clones, we next investigated whether similar results could be obtained in another cell line. For this purpose, we generated CHO cell lines expressing the human CD81 alone (CHO/CD81/pcDNA3.1), or in combination with either a non‐cleavable EWI‐2 construct (CHO/CD81/EWI‐2) or a wild‐type EWI‐2 construct that leads to the production of EWI‐2 and EWI‐2wint proteins in CHO cells, which both interact with CD81 (CHO/CD81/EWI‐2wint) (Rocha‐Perugini et al., 2008; Montpellier et al., 2011), as shown in Fig. S3. Although the distribution of CD81 trajectories in CHO cells was rather different from that in Huh‐7 cells, we found again a statistically significant shift of CD81 trajectories towards slower trajectories (Fig. 4C) in presence of EWI‐2wint. This effect was characterized by a decrease of the ADC mean value of CD81 in EWI‐2wint‐expressing cells (1.9 × 10−2 μm2 s−1), as compared with control cells (3.1 × 10−2 μm2 s−1) (Table 1). Altogether, our results indicate that EWI‐2wint significantly restricts the diffusion of CD81 molecules at the plasma membrane.
EWI‐2wint restricts the diffusion of CD81 and confines it in plasma membrane areas
The different modes of diffusion of CD81 molecules were next evaluated by the analysis of MSD versus time plots of the trajectories of individual molecules (for details see the data analysis section in Experimental procedures) in each cell clone. As described for CD9 (Espenel et al., 2008) three different diffusion modes were observed for CD81 trajectories in EWI‐2‐expressing cells (Fig. 5A and B and Table 2): (i) pure Brownian diffusion (40% of total trajectories) characterized by an ADC mean value of 6.49 × 10−2 μm2 s−1, (ii) pure confined or restricted diffusion (50% of total trajectories) characterized by a lower ADC mean value of 0.94 × 10−2 μm2 s−1, and (iii) diffusion with different combinations of Brownian and confined modes referred to as ‘Mixed trajectories’ (9% of total trajectories). Importantly, the proportion of Brownian trajectories was reduced from 40% to 23–26% of the total trajectories in EWI‐2wint 1 and EWI‐2wint 2 cells (Fig. 5B). This effect was characterized by an increase of confined and mixed trajectories. In addition to the increase in confinement, the ADC mean value of Brownian trajectories was 2.4‐ and 3.1‐fold reduced in EWI‐2wint 1 and EWI‐2wint 2 cells as compared with EWI‐2 cells respectively (Table 2 and Fig. 5C). The effect of EWI‐2wint on CD81 diffusion can be easily observed by the eye on the overlapping of all the tracked molecules with CD81 ensemble labelling as shown in Fig. 5A. In addition, the analysis of the distribution of Brownian and confined CD81 trajectories (Fig. 5C and D) both showed a highly significant shift of trajectories towards slower diffusion. Indeed, a majority of Brownian proteins diffused more slowly than 0.1 μm2 s−1 at the expense of the highly mobile fraction in EWI‐2wint‐expressing cells. Moreover, the emergence of a subpopulation with a very low ADC (approximately 0.2 × 10−2 μm2 s−1) was observed in confined trajectories in EWI‐2wint cells (Fig. 5D, arrows). As shown in Fig. 5B, CD81 molecules could also be transiently confined (mixed trajectories) and, in order to determine whether EWI‐2wint affects the escape of CD81 molecules from confined areas, we calculated the dwell time of CD81 molecules in confined areas for each cell clone. Interestingly, CD81 molecules were trapped significantly longer in confined areas in cells expressing EWI‐2wint than in control cells with a dwell time of 4.8 s (EWI‐2wint 1)/4.6 s (EWI‐2wint 2) versus 3.4 s (EWI‐2) (Table 2). Interestingly, the effect of EWI‐2wint mainly depends on its interaction with CD81 since the dynamic behaviour in terms of ADC mean values, diffusion modes and dwell times of CD81 was not significantly modulated in cells expressing LAL as compared with EWI‐2 cells (Fig. 5). Although less pronounced, the behaviour of CD81 trajectories in Qcc‐expressing cells was similar to that in EWI‐2wint cells (Fig. 5), indicating that the observed effects on the diffusion of CD81 in cells expressing EWI‐2wint cannot be attributed to a difference in its anchorage to the actin cytoskeleton by its cytosolic tail.
Figure 5.
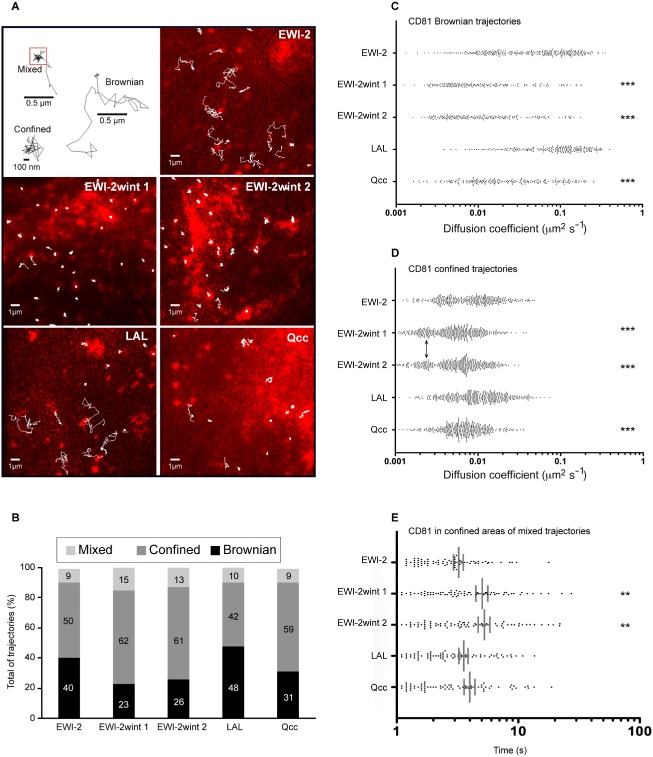
EWI‐2wint restricts the diffusion of CD81 at the plasma membrane.
A. In the first box are represented the different diffusion modes for CD81 molecules. The red square in the mixed trajectory corresponds to transient confinement. The five other images represent superposition of ensemble CD81 labelling (in red) and several CD81 single‐molecule trajectories obtained after tracking Atto647N‐labelled Fab fragments of TS81 (white thin lines) for Huh‐7 clones expressing EWI‐2, EWI‐2wint (two clones), LAL or Qcc. The ensemble labelling was obtained by stacking several frames of tracking experiments.
B. Histogram representing the percentage of each type of trajectory relative to the total number of trajectories for each clone. The black, dark grey and light grey boxes correspond to Brownian, confined and mixed trajectories respectively.
C–E. Distribution of Brownian (C) and confined (D) trajectories was analysed separately. Regarding the analysis of mixed trajectories, time spent in transient confined zones are reported for every clone in histogram (E). ** and *** correspond to P values below 0.001 and 0.0001, respectively, in comparison with cells expressing EWI‐2, as determined by the Mann–Whitney U ‐test.
Table 2.
ADC and dwell time of CD81
| Huh‐7 cells | D Browniana (·10−2 μm2 s−1) | D confinedb (·10−2 μm2 s−1) | T mixed (s)c |
|---|---|---|---|
| EWI‐2 | 6.5 | 0.9 | 3.4 |
| EWI‐2wint 1 | 2.7 | 0.5 | 4.8 |
| EWI‐2wint 2 | 2.1 | 0.7 | 4.6 |
| LAL | 7.4 | 1.0 | 3.5 |
| Qcc | 4.4 | 0.8 | 4 |
ADC mean value of pure Brownian trajectories.
ADC mean value of pure confined trajectories.
Dwell time of CD81 molecules in confined areas of mixed trajectories.
Together, our results demonstrate that EWI‐2wint restricts the diffusion of CD81 (i) by decreasing the diffusion properties of the mobile Brownian fraction and (ii) by increasing the subpopulation of CD81 molecules confined in plasma membrane areas.
EWI‐2wint promotes confinement of CD81 in CD81‐enriched areas
A previous study on the CD9 tetraspanin (Espenel et al., 2008) has shown that a fraction of molecules can be purely confined in areas that are mainly enriched in CD9 and CD81 or more generally in tetraspanins. No exchange with the rest of the membrane was observed for these confinement areas. We therefore compared CD81 trajectories with the ensemble distribution of CD9 and CD46 (Fig. 6A and B). To do so, overlapping of CD81 trajectories with ensemble CD9 and CD46 labelling was performed. In all cell clones, about 40% of CD81 confinement areas colocalized with CD9‐enriched areas whereas less than 5% were colocalized with CD46. Although somewhat higher, we did not find any significant increase of colocalization of CD81 with CD9 in cells expressing EWI‐2wint. In contrast, the proportion of CD81 trajectories overlapping CD81‐enriched areas was significantly increased in cells expressing EWI‐2wint, as compared with EWI‐2 cells (Fig. 6D). No significant change was observed for cells expressing the LAL mutant whereas an intermediate effect was observed for the Qcc mutant. We also analysed the colocalization of CD81 with EWI‐2/EWI‐2wint proteins (Fig. 6C). Interestingly, we found an increase of the colocalization of CD81 with EWI proteins in cells expressing EWI‐2wint, indicating that EWI‐2wint or at least EWI‐2/EWI‐2wint heterodimers probably interact more strongly with CD81 than parental EWI‐2 proteins.
Figure 6.
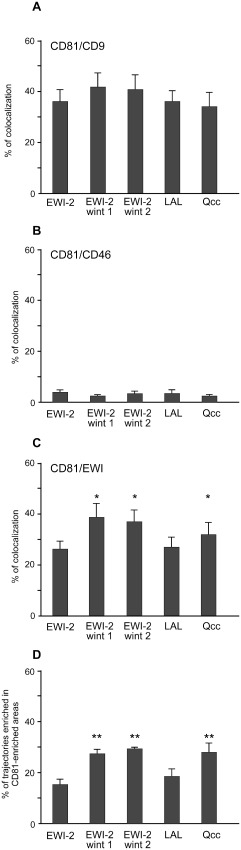
Colocalization of CD81 with its partners.
A–C. Histograms represent the percentage of colocalization between CD81 and CD9 (A), CD81 and CD46 (B), and CD81 and EWI proteins (C). Percentages of colocalization of CD81 (red) with its partners (green) were defined as colocalized pixels (yellow) divided by the number of yellow + red pixels, using images from single‐molecule tracking experiments.
D. The percentage of trajectories that coincided with CD81‐enriched areas. * and ** correspond to P values below 0.01 and 0.001, respectively, in comparison with the EWI‐2‐expressing cells, as determined by the Mann–Whitney U ‐test.
Taken together, our results indicate that EWI‐2wint restricts the global diffusion of CD81 and promotes its confinement in some areas of the plasma membrane that are enriched in CD81.
Effect of EWI‐2wint on colocalization of CD81 with CLDN1
It has been shown that CD81 and CLDN1 colocalize and interact in areas of the plasma membrane (Harris et al., 2008, 2010), suggesting that CLDN1 is likely a partner of CD81 in a HCV receptor complex. Concordant with this hypothesis, mutations in the first extracellular loop of CLDN1 or the use of antibodies directed against CLDN1 or CD81 inhibit HCV entry by disrupting CLDN1‐CD81 interaction (Evans et al., 2007; Cukierman et al., 2009; Harris et al., 2010; Krieger et al., 2010). Since we demonstrated that EWI‐2wint modifies the partitioning of CD81, we therefore analysed whether EWI‐2wint has an impact on the colocalization of CD81 with CLDN1. Cell surface expression of CD81 and CLDN1 on fixed pcDNA3.1, EWI‐2 and EWI‐2wint cells was studied by confocal microscopy (Fig. 7A), and colocalization levels were quantified using the CoLocalizer Pro software. Interestingly, we found a significant increase of colocalization between CD81 and CLDN1 in cells expressing EWI‐2wint (Fig. 7B), indicating that in addition to the trapping of CD81, EWI‐2wint likely constraints CLDN1 in areas of the plasma membrane.
Figure 7.
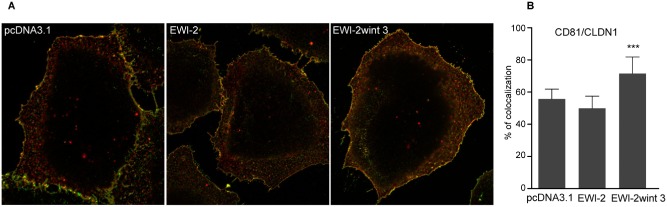
EWI‐2wint increases CD81–CLDN1 colocalization.
A. Representative images from confocal microscopy experiments. Fixed Huh‐7 cells expressing pcDNA3.1 (left) EWI‐2 (middle) or EWI‐2wint 3 (right) were labelled with TS81 anti‐CD81 and anti‐CLDN1 mAbs followed by secondary antibodies conjugated with Alexa488 (green staining) and Alexa555 (red staining) respectively.
B. Colocalization was calculated as the number of yellow pixels (green and red pixels) over the number of yellow + red pixels, using CoLocalizer Pro software. *** means a P value below 0.001 as determined by the Mann–Whitney U ‐test.
The MT81w antibody recognizes CD81‐enriched structures
In the first part of our results, we showed that EWI‐2wint induces an increase of CD81 recognition by the MT81w antibody. Since, as mentioned above, MT81w antibody only recognizes a fraction of CD81 associated with the tetraspanin web or at least CD81 engaged in protein clusters, we speculated that EWI‐2wint increased the association of CD81 within membrane areas such as TEAs that can favour its clustering. Therefore, we next tried to investigate the oligomerization of mCD81 and analyse whether its change has an influence on the recognition by the MT81w mAb.
In a previous study, we showed that CD81/CD82 chimeras in which the third (TM3) or the fourth (TM4) transmembrane domain of CD81 was replaced by the corresponding domain of CD82, have a tendency to oligomerize (Montpellier et al., 2011). Based on this, we constructed mCD81/CD82 chimeras in which the TM3 (Fig. 8A; mTM3) or the TM4 (Fig. 8A; mTM4) of mCD81 was replaced by the corresponding domain of human CD82. Huh‐7w7 cells that no longer express hCD81 were transfected with plasmids encoding mCD81, mTM3, mTM4 or the empty vector. The oligomerization status of mCD81 was next analysed by Western blotting with the MT81 antibody. In addition to the ≈ 25 kDa major band corresponding to the monomeric form of CD81, an additional band of ≈ 40 kDa was seen in cells expressing the mTM3 chimera (Fig. 8B). In cells expressing the mTM4 chimera, several other additional bands (≈ 60/70 kDa and ≈ 90 kDa) were detected (Fig. 8B). The presence of all these additional bands, which likely correspond to dimeric, trimeric and tetrameric forms of CD81 (Yang et al., 2006) indicate that the replacement in mCD81 of TM3 or TM4 by the corresponding region of CD82 increases the homo‐oligomerization of CD81. It has to be noted that similar results were obtained in CHO cells (data not shown). We then took advantage of these chimeras to analyse whether the increase of CD81 oligomers modulates the recognition by the MT81w mAb. Huh‐7w7 cells expressing mCD81, mTM3 or mTM4 were stained with MT81 and MT81w mAbs and analysed by flow cytometry (Fig. 8C). It has to be noted that cells expressing mCD81, mTM3 or mTM4 were equally stained with MT81 mAb, indicating transmembrane domain exchanges in chimeras did not affect their cell surface expression, as compared with unmodified mCD81. However, to avoid any bias of the CD81 expression level, the modulation of the recognition by MT81w was evaluated by the MT81w/MT81 ratio. Interestingly, the recognition by MT81w was twice more efficient in cells expressing the mTM4 chimera for which CD81 homo‐oligomerization is the strongest.
Figure 8.
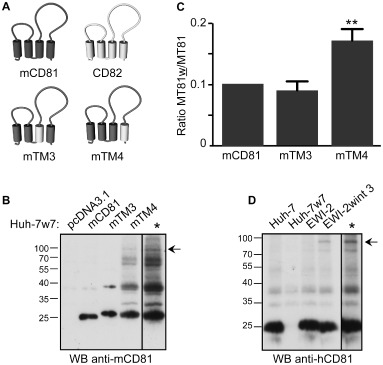
MT81w recognizes clusters of mCD81.
A. Schematic representation of mTM3 and mTM4 chimeras. These chimeras were generated by replacing the third (mTM3) or the fourth (mTM4) transmembrane domain of mCD81 by the corresponding domain of CD82.
B. Huh‐7w7 cells were transiently transfected with the empty vector (pcDNA3.1), mCD81, mTM3 or mTM4, lysed in PBS/BrijO10/EDTA and analysed by Western blot with MT81 mAb.
C. Huh‐7w7 cells transiently expressing pcDNA3.1, mCD81, mTM3 or mTM4 were labelled with MT81 and MT81w mAbs followed by PE‐conjugated secondary antibody and analysed by flow cytometry. MT81w/MT81 ratios are presented for each cell line and reported as the mean ± SD of three independent experiments. ** corresponds to a P value below 0.001, in comparison with cells expressing mCD81, as determined by the Mann–Whitney U ‐test.
D. Huh‐7 clones expressing EWI‐2 or EWI‐2wint were used in cross‐link experiments. Huh‐7 and Huh‐7w7 cells were used as controls. Cells were treated with 2‐BP to expose membrane proximal cysteines and cross‐linked with the DTME reagent. After lysis in PBS/BrijO10/Cysteine, 5A6 anti‐CD81 mAb was used to immunoprecipitate CD81 complexes. Proteins were revealed using biotinylated 1.3.3.22 anti‐CD81 mAb followed by HRP‐Streptavidin. The molecular weights of the prestained molecular ladder are indicated in kDa. Black arrows indicate a ≈ 90 kDa band probably corresponding to CD81 tetramers.
In (B) and (D) asterisk indicates a longer exposure of mTM4 and EWI‐2wint 3 respectively.
We next analysed the oligomerization status of CD81 in Huh‐7 cell lines expressing EWI‐2 or EWI‐2wint 3 (see Fig. 2). We treated cells with 2‐bromopalmitate (2‐BP) to expose membrane proximal cysteine residues and then cross‐linked tetraspanin multimers using the homobifunctional cysteine‐reactive reagent DTME. This method has previously been used to detect homo‐oligomerization of tetraspanins (Kovalenko et al., 2004). The oligomerization status of CD81 in cells expressing EWI‐2 or EWI‐2wint 3 was compared with that of Huh‐7 cells and Huh‐7w7 cells, which no longer express CD81. Although in our experimental conditions we did not detect any dimers of CD81, we found a specific additional band of ≈ 90 kDa in cells expressing EWI‐2wint (Fig. 8D, arrow). This band was seen in four independent experiments. Interestingly, a band with a similar migration pattern was detected in Huh‐7w7 cells (Fig. 8B, arrow) and CHO cells (data not shown) expressing mTM4.
Taken together, our results indicate that CD81 tends to form CD81‐enriched structures upon EWI‐2wint expression. In addition, these structures are preferentially recognized by the MT81w antibody.
Effect of EWI‐2wint on HCV cell‐to‐cell transmission
In addition to cell‐free infection, HCV can also be transmitted via cell‐to‐cell contact for which the mechanism needs to be elucidated (Timpe et al., 2008; Witteveldt et al., 2009; Brimacombe et al., 2011). Indeed, HCV is transmitted in the presence of mAbs or patient‐derived antibodies that are able to neutralize virus‐free infectivity (Timpe et al., 2008; Brimacombe et al., 2011). Since cell‐to‐cell transmission has been suggested to be a major route of transmission for HCV (Brimacombe et al., 2011), we next analysed the effect of EWI‐2wint on this process. Briefly, HCV‐infected Huh‐7 cells were labelled with 5‐chloromethylfluorescein diacetate (CMFDA) and co‐cultured with the naïve target cells expressing or lacking EWI‐2/EWI‐2wint proteins. Co‐cultures were performed in the presence and in absence of neutralizing antibodies (3‐11 anti‐E2 mAb) to analyse cell‐to‐cell and total transmission respectively. After 24 h of co‐culture, de novo transmission events were determined by staining for HCV NS5 and they were quantified by flow cytometry. It is worth noting that no HCV transmission occurred in Huh‐7w7 cells that do not express CD81, in agreement with this entry factor being essential to both cell‐free and cell‐to‐cell transmission. As demonstrated by our data in Fig. 9A, EWI‐2wint expression has no significant effect on HCV cell‐to‐cell transmission. To support this claim, we performed another assay in which the cell seeding density was lowered or increased to reduce or increase cell–cell contacts, as compared with standard cell seeding density (medium). Cells seeded at different densities were infected with HCVcc and infection levels were evaluated by flow cytometry at 48 h post infection (Fig. 9B). The more the cells were confluent at the time of infection, the less they were infected. In contrast, subconfluent cells were better infected. This is probably due to the differences in multiplicities of infection for each condition. Interestingly, this effect was not observed in cells expressing EWI‐2wint, it was even somewhat the opposite. When compared with control cell lines, the effect of EWI‐2wint on HCV infection was less pronounced in cells seeded at high density, indicating that HCV infection was less inhibited by EWI‐2wint in the presence of numerous cell‐to‐cell contacts. In contrast, EWI‐2wint highly reduced HCV infection in subconfluent cells, as compared with control cells. Together, these results demonstrate that EWI‐2wint does not inhibit cell‐to‐cell transmission of HCV.
Figure 9.
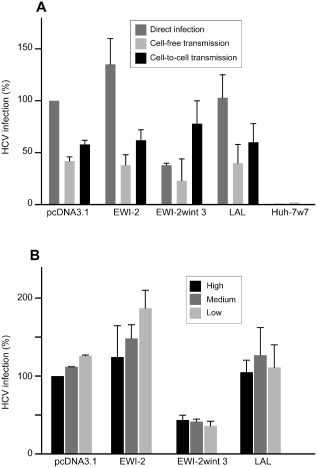
EWI‐2wint does not inhibit cell‐to‐cell transmission.
A. Huh‐7 donor cells were infected with HCVcc and stained with CMFDA. Acceptor cells are Huh‐7 clones expressing EWI‐2, EWI‐2wint, LAL or Qcc. Huh‐7w7 cells serve as negative control. Co‐culture of donor and acceptor cells with or without neutralizing 3‐11 anti‐E2 mAb allowed to monitor either cell‐to‐cell or total (cell‐to‐cell and cell‐free) transmission of HCV. Cells were labelled with anti‐NS5 mAb followed by PE‐conjugated secondary antibody and analysed by flow cytometry. In these conditions, newly infected cells are negative for CMFDA staining and positive for PE staining. For cell‐free (light grey) and cell‐to‐cell transmission (black), results are presented as percentages relative to the total transmission. Levels of infectivity of each clone are shown in dark grey.
B. Three conditions of cell density (high, medium and low) were used for HCVcc infection, for each Huh‐7 clone. After 48 h, cells were labelled with anti‐NS5 mAb followed by PE‐labelled secondary antibody and analysed by flow cytometry. Results are presented as related percentages to the infection of pcDNA3.1 control cells in high‐confluency condition.
Results are reported as the mean ± SD of three independent experiments (A and B).
Discussion
In our study, we combined biochemistry experiments and single‐molecule experiments to investigate the role of EWI‐2wint in the clustering and membrane behaviour of CD81 in the context of HCV infection. We found that a change in membrane partitioning of CD81 occurs in the presence of EWI‐2wint, which inhibits cell‐free infection of HCV.
The dynamics and partitioning of CD81 were probed using SMT, a technique based on the labelling of a low number of molecules allowing individual molecules to be optically isolated and their position accurately determined. Here, we especially focused on CD81 behaviour in cells expressing EWI‐2, one of the primary partners of CD81 (Stipp et al., 2001; Charrin et al., 2003a), or EWI‐2wint, a truncated form of EWI‐2 that inhibits HCV entry (Rocha‐Perugini et al., 2008). Interestingly, membrane dynamics of CD81 was largely reduced upon EWI‐2wint expression. We demonstrated that the lower ADC mean value of CD81 is explained by two principal effects of EWI‐2wint expression. First, the number of pure confinement largely increased, suggesting that CD81 molecules are trapped within membrane domains. Ensemble labelling revealed that these domains are often enriched in CD81 and in EWI molecules, suggesting that these domains likely correspond to TEAs. Second, the diffusion rate of CD81 Brownian molecules significantly decreased when EWI‐2wint was expressed. This result can be explained by the fact that CD81 diffuses in larger complexes that can be formed thanks to EWI‐2wint interacting more strongly with CD81. Indeed, although the diffusion of membrane proteins can be modulated by the lipidic composition of plasma membrane or the interaction with the membrane cytoskeleton, it is currently accepted that the diffusion coefficient of a molecule is dependent on its molecular weight (Saffman and Delbruck, 1975; Gambin et al., 2006). In our study, although we did not analyse the effect of EWI‐2wint on the membrane lipid composition, our data suggest that CD81 diffusion is modulated by protein–protein interactions. Indeed, we showed that the observed effects on the diffusion of CD81 molecules in cells expressing EWI‐2wint were not related to anchorage to the actin cytoskeleton by its cytosolic tail. Our colocalization studies indicated that EWI‐2wint or at least EWI‐2/EWI‐2wint heterodimers probably interact more strongly with CD81. In addition, high‐molecular‐weight complexes enriched in CD81 were detected in cells expressing EWI‐2wint. These complexes likely correspond to the subpopulation of CD81 molecules with a very low diffusion coefficient, which have been identified in cells expressing EWI‐2wint.
We demonstrated that CD81 molecules display similar diffusion modes as compared with the other tetraspanin CD9, namely Brownian, confined and mixed, a combination of the two first modes. However, as compared with CD9, the percentage of CD81 confined trajectories is more important, which is characterized by a low number of mixed trajectories (see Espenel et al., 2008; Krementsov et al., 2010). Such difference in CD81 and CD9 behaviour has also been observed in different cell lines such as CHO, PC3 and HeLa cells (P. Rassam et al., in preparation).
It has been largely demonstrated that CD81 associates with the tight junction protein CLDN1 to form complexes that are essential to HCV entry (Evans et al., 2007; Cukierman et al., 2009; Harris et al., 2010; Krieger et al., 2010). Although we did not perform quantitative analyses of the interaction between CD81 and CLDN1, we quantified the level of colocalization of these two HCV entry factors and found a significant increase in the presence of EWI‐2wint. This suggests that EWI‐2wint, by sequestering CD81 in membrane areas, might also sequester CLDN1 and promote CD81–CLDN1 interactions. This also suggests that CD81 and CLDN1 likely interact transiently with each other in TEAs, then dissociate and escape from these areas. In cells expressing EWI‐2wint, CD81–CLDN1 complexes might be sequestered in TEAs. Single‐molecule microscopy experiments will thus be interesting to investigate the dynamics and partitioning of CLDN‐1 molecules in cells expressing EWI‐2wint.
CD81 forms homodimers (Kitadokoro et al., 2001; Harris et al., 2008), which may promote higher‐order oligomeric structures. In 2006, in order to characterize tetraspanin microdomains, Silvie and colleagues generated new anti‐mouse CD81 antibodies among them the MT81w mAb (Silvie et al., 2006). They demonstrated that MT81w immunoprecipitates CD81 only from lysates made under detergent conditions preserving tetraspanin–tetraspanin interactions. Moreover, CD9 depletion completely removed the pool of CD81 molecules recognized by MT81w. Based on this, it has been suggested that MT81w specifically recognizes CD81 associated with other tetraspanins (including other CD81 molecules) or at least CD81 proteins interacting with cholesterol that directly interact with tetraspanins (Charrin et al., 2003b). We next took advantage of this tool to analyse the role in HCV infection of CD81 molecules recognized by the MT81w antibody (Rocha‐Perugini et al., 2009). We showed that MT81w did not strongly affect HCV infection. Furthermore, cholesterol depletion, which inhibits HCV infection and reduces total cell surface expression of CD81, did not affect recognition by MT81w. In addition, sphingomyelinase treatment, which also reduces HCV infection and cell surface expression of total CD81, raised the number of CD81 molecules recognized by MT81w. We therefore concluded that HCV does not use preferentially CD81 associated with other tetraspanins. In the present study, we used again this antibody in order to define whether EWI‐2wint promotes a change in CD81 partitioning and dynamics. We found that the proportion of CD81 molecules recognized by MT81w increased upon EWI‐2wint expression, suggesting that EWI‐2wint promotes the association of CD81 with other tetraspanins. However, our colocalization studies showed that CD81 was trapped in areas particularly enriched in CD81 and not in CD9, in cells expressing EWI‐2wint. We also demonstrated that MT81w preferentially stains cells expressing oligomeric forms of CD81 indicating that MT81w likely does not recognize CD81 associated with other tetraspanins but it is rather a low‐affinity antibody that better recognizes clusters of CD81. Antibodies with low monovalent affinity are indeed particularly dependent on bivalent binding, and consequently are particularly sensitive to antigen clustering. We can speculate that in depletion experiments made by Silvie and co‐workers, depletion of CD9, which shares the same partners as CD81, liberates partner proteins that can then interact with CD81 and thereby impede CD81 homo‐oligomerization. Under these conditions, the MT81w will no longer recognize CD81. Our results therefore suggest that MT81w is somehow similar to C9BB, a low‐affinity antibody that preferentially recognizes multimerized CD9 (Yang et al., 2006). In accordance with the claim that MT81w is a low‐affinity antibody that better recognizes clusters of CD81, we showed that recognition by MT81w was increased following ceramide enrichment (Rocha‐Perugini et al., 2009), which is known to induce receptor clustering for many receptor molecules (reviewed in Zhang et al., 2009) essential to initiation and amplification of receptor and stress‐mediated signalling in almost all cell types. Since receptor aggregation and trapping in ceramide‐enriched microdomains may limit lateral diffusion of membrane proteins, it will be interesting to study in single‐molecule microscopy whether CD81 molecules in sphingomyelinase‐treated cells behave similarly to those in cells expressing EWI‐2wint. Thus, our data suggest that MT81w antibody is likely a low‐affinity antibody that specifically recognizes CD81‐enriched structures, which might be identified as TEAs. Taken together, our results indicate that EWI‐2wint increased association of CD81 within membrane areas such as TEAs that favours CD81 clustering and which are specifically recognized by the MT81w antibody.
EWI family members have a propensity for oligomerization. For example, it has been demonstrated that EWI‐F/CD9P‐1 forms oligomers at the cell surface that are directly associated with the tetraspanins CD9 and CD81 (André et al., 2009). EWI‐2 has also been observed to form protein complexes containing at least two EWI‐2 molecules (André et al., 2009). In addition, EWI‐2wint is associated with EWI‐2 in heterodimers that are associated with CD81 and CD9 (Montpellier et al., 2011). We therefore cannot exclude the possibility that EWI‐2wint needs EWI‐2 to interact with CD81 and consequently to modulate its partitioning and membrane diffusion.
We found that the replacement of the TM3 and TM4 of CD81 by the corresponding domains of hCD82 increases the capacity of human and murine CD81 to homo‐oligomerize (the present work and Montpellier et al., 2011). Particularly, in the context of the mTM4 chimera, CD81 formed high‐molecular‐weight complexes that may correspond to dimers, trimers and tetramers. Interestingly, a high‐molecular‐weight band was seen in cells expressing EWI‐2wint, suggesting that CD81 forms high‐order complexes upon EWI‐2wint expression. This result was in accordance with our microscopy experiments demonstrating that CD81 molecules were confined in CD81‐enriched areas of the plasma membrane. Our previous (Rocha‐Perugini et al., 2008; 2009; Montpellier et al., 2011) and present results support the model in which CD81 clusters are deleterious for HCV entry. First, TM3 and TM4 chimeras, which form CD81 oligomers, are not functional in HCV entry. Second, EWI‐2wint, which promotes CD81 clustering, inhibits HCV infection. Third, the MT81w mAb, for which we showed a stronger affinity for cells expressing EWI‐2wint or mTM4 chimera, poorly neutralizes HCV infection.
In a previous work, we showed that EWI‐2wint restricts HCV entry by impeding E2–CD81 binding (Rocha‐Perugini et al., 2008). In the present study, we demonstrated that the expression of EWI‐2wint promotes the formation of CD81 oligomers that diffuse more slowly in the plasma membrane and, which are recognized by the MT81w mAb. Since this antibody poorly neutralizes HCV infection, we assume that the CD81 oligomers are not used during HCV entry. From all these data, we can therefore conclude that, by reorganizing CD81 molecules on the cell surface, EWI‐2wint impedes the interaction between CD81 and E2 envelope glycoproteins. Although additional experiments will be necessary to strengthen this hypothesis, we can assume that the effects of EWI‐2wint on CD81 membrane diffusion and E2–CD81 binding are probably two inseparable functions.
Enveloped viruses can spread via two distinct routes, either through the cell‐free aqueous environment or by remaining cell associated and being passed on by direct cell–cell contact. This latter mode of spread designated as cell‐to‐cell transmission not only facilitates rapid viral dissemination, but may also promote immune evasion and influence disease (reviewed in Sattentau, 2008). Increasing evidences suggest that cell‐to‐cell transmission is a major route of transmission for HCV. Indeed, HCV is transmitted in the presence of mAbs and patient‐derived antibodies that are able to neutralize virus‐free infectivity (Timpe et al., 2008; Brimacombe et al., 2011). In addition, cell‐to‐cell transfer of virions has been shown to be a feature common to chimeric viruses expressing the structural proteins representing the seven major HCV genotypes (Brimacombe et al., 2011). Although HCV may exploit a contact structure similar to the virological synapse of HIV (Brimacombe et al., 2011) or may use membrane nanotubes, the precise mechanism by which HCV is transmitted from a cell to another cell needs to be elucidated. It has been established that SRB1 and the tight‐junction components CLDN1 and occludin are involved in HCV cell‐to‐cell transmission. So far, the role played by CD81 remains controversial. Indeed, several studies reported HCV cell‐to‐cell transmission as a CD81‐dependent pathway (Russell et al., 2008; Brimacombe et al., 2011), whereas others demonstrated a CD81‐independent transmission (Timpe et al., 2008; Witteveldt et al., 2009; Jones et al., 2010). In our study, we found that Huh‐7w7 cells, which do not express CD81, were resistant to both cell‐free and cell‐to‐cell transmission indicating that CD81 is necessary for both cell‐free and cell‐to‐cell spread of HCV. We showed that EWI‐2wint inhibits cell‐free transmission but not cell‐to‐cell transmission of HCV. This finding demonstrates that cell‐free and cell‐to‐cell transmissions are driven by different mechanisms. According to the SMT experiments, we could speculate that EWI‐2wint inhibits cell‐free transmission by modulating CD81 membrane diffusion and its clustering and that these processes are not critically involved in cell‐to‐cell transmission.
In conclusion, by using EWI‐2wint, a natural inhibitor of HCV, we defined the dynamic properties and partitioning of CD81 molecules that are necessary for productive infection. Our results open the way to the understanding of the dynamics by which HCV enters into its target cells.
Experimental procedures
Antibodies
Anti‐human CD81 (5A6) was kindly provided by S. Levy. Anti‐human CD81 (TS81) and anti‐mouse CD81 (MT81 and MT81w) mAbs have been described previously (Charrin et al., 2001; Silvie et al., 2006). 3‐11 (anti‐HCV E2) hybridoma was kindly provided by J. McKeating. M2 anti‐FLAG mAb was from Sigma. Anti‐NS5 (2F6/G11) was from AUSTRAL Biologicals. HA11 anti‐HA mAb was from Covance. PE‐labelled goat anti‐mouse and anti‐rat were from BD Pharmingen. Peroxidase‐conjugated goat anti‐mouse and anti‐rat were from Sigma and Jackson ImmunoResearch respectively. Fab fragments of mAbs (anti‐CD81 TS81, anti‐CD9 SYB‐1, anti‐CD46 11C5) labelled with Atto647N or Cy3B have been described previously (Espenel et al., 2008). Fab fragments of 8A12 mAb [anti‐EWI‐2 (Charrin et al., 2003a)] were prepared and labelled with Cy3B as previously described (Espenel et al., 2008). Anti‐CLDN1 mAb has been previously described (Fofana et al., 2010).
Plasmids
pcDNA3.1 plasmids expressing EWI‐2, EWI‐2Fur, LAL, Qcc, EWI‐2AGA, human CD81, murine CD81 have been described previously (Charrin et al., 2003a; Rocha‐Perugini et al., 2008; Montpellier et al., 2011). The mTM3 and mTM4 chimeras were constructed by a PCR‐based method. In mTM3 chimera, the amino acid (aa) sequence corresponding to the third transmembrane domain of mCD81 (aa 87–113) was replaced by the aa sequence corresponding to the third transmembrane domain of hCD82 (aa 81–108). In mTM4 chimera, the aa sequence corresponding to the fourth transmembrane domain and cytosolic tail of mCD81 (aa 204–236) was replaced by the aa sequence corresponding to the fourth transmembrane domain and cytosolic tail of hCD82 (aa 228–267). Cloning details and oligonucleotide sequences are available upon request.
Cell lines and transfection
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 nM non‐essential amino acids. Huh‐7 and CHO cells were from ATCC. Huh‐7w7 (CD81−) cells were described previously (Rocha‐Perugini et al., 2009).
Cell transfections and generation of cell lines/clones were performed as described previously (Rocha‐Perugini et al., 2008). Since the cleavage of EWI‐2 generating EWI‐2wint naturally occurs in CHO cells, the mutation of two arginine residues abolishing the cleavage of EWI‐2 (Montpellier et al., 2011) was introduced in the coding sequence of the plasmid (pcDNA3.1/EWI‐2AGA; Montpellier et al., 2011) used to generate CHO/EWI‐2 cells.
Detection of cell surface biotinylated proteins
Twenty‐four hours post transfection, cells were surface biotinylated as described in Ref (Rocha‐Perugini et al., 2008) and lysed in PBS 1% Brij‐O10 2 mM EDTA (PBS/Brij/EDTA) and protease inhibitors (Complete, Roche Applied Science). Lysates were pre‐cleared for 2 h at 4°C with protein A‐Sepharose (GE Healthcare), then incubated for 2 h at 4°C with specific mAbs immobilized onto protein A‐Sepharose beads. Complexes were eluted with non‐reducing Laemmli buffer, resolved by SDS‐PAGE, transferred to a nitrocellulose membrane (GE Healthcare), and immunoblotted with peroxidase‐conjugated streptavidin (Vector).
HCVcc infection assays
HCVcc used in this study were based on the JFH1 strain (Wakita et al., 2005) and contained cell culture‐adaptive CS, N6 and A4 mutations (Delgrange et al., 2007; Goueslain et al., 2010).
HCVcc (JFH1/CSN6A4/5′C19Rluc2AUbi) expressing Renilla luciferase were produced as described (Delgrange et al., 2007; Rocha‐Perugini et al., 2008). HCVcc were added to Huh‐7 cells [multiplicity of infection (moi) = 1] seeded the day before in 24‐well plates and incubated for 2 h at 37°C. The supernatants were then removed and the cells were incubated in DMEM 10% FBS at 37°C. At 40–48 h post infection, Renilla luciferase assays were performed as indicated by the manufacturer (Promega).
HCVcc without reporter gene (JFH‐1/CSN6A4) stocks were produced as previously reported (Delgrange et al., 2007; Goueslain et al., 2010). For the HCVcc infection assays, cells grown in 24‐well plates were incubated with HCVcc (moi = 1) for 2 h at 37°C, washed, and incubated for an additional 48 h at 37°C. HCVcc infections were scored by staining for HCV non‐structural protein NS5 followed by flow cytometry analyses.
Flow cytometry
For NS5 staining, infected cells were permeabilized with PBS 2% BSA 0.05% saponin and washed with PBS 2% BSA. For MT81/MT81w stainings, cells grown in 24‐well plates were directly washed with PBS 2% BSA. Cells were then incubated 1 h at 4°C with anti‐NS5 or anti‐mCD81 (MT81 and MT81w) mAb. After rinsing, cells were incubated with PE‐labelled goat anti‐mouse or anti‐rat, respectively, for 45 min at 4°C, washed, detached with PBS 2 mM EDTA and fixed with formalin solution (formaldehyde 4%, Sigma). Labelled cells were analysed using a FACSCalibur (Becton Dickinson).
SMT experiments
SMT experiments were carried out as previously described (Espenel et al., 2008). Briefly, cells plated on collagen‐1 were incubated in DMEM at 37°C for 10 min with Atto647N‐labelled Fab fragments of mAbs raised against CD81 (TS81), CD9 (SYB‐1) and CD46 (11C5). A homemade objective‐type TIRF set‐up allowing multicolour single‐molecule imaging and equipped with a Plan Fluor 100×/1.45 NA objective (Zeiss, Le Peck, France Brattleboro, VT) was used. All the experiments were performed with a 100 ms integration time. For some experiments, to achieve a better specificity in the detection of the two fluorescent signals, alternating‐laser excitation (ALEX) was performed using an acousto‐optical tunable filter and controller (AOTF; AA Optoelectronics) (Margeat et al., 2006).
All the movies were analysed using a homemade software (named ‘PaTrack’) implemented in visual C++. Trajectories were constructed using the individual diffraction limited signal of each molecule. The centre of each fluorescence peak was determined with subpixel resolution by fitting a two‐dimensional elliptical Gaussian function. The two‐dimensional trajectories of single molecules were constructed frame per frame. Only trajectories containing at least 40 points were retained. Diffusion coefficient values were determined from a linear fit to the MSD (mean square displacement)‐τ plots between the first and the fourth points (D1–4) according to the equation MSD(t) = 4Dt.
The determination of the motional modes was performed using a homemade algorithm based on a neural network that has been trained using synthetic trajectories to detect pure Brownian, confined and directed motion modes (P. Dosset et al., in preparation). Due to a sliding window, the trajectory is analysed and the different modes detected within a trajectory for segments larger than 10 frames. Once the motion mode is identified, the different segments are analysed by plotting the MSD versus time lag. The MSD curve is linearly fitted (Brownian) or adjusted with a quadratic curve (4Dt + v2t2) (directed diffusion) or exponential curve L2/3[1 − exp(−12Dt/L2)] (confined diffusion), where L is the side of a square domain, the confinement diameter being related to L by dconf = (2/√L). The algorithm has been tested with simulated trajectories displaying pure Brownian, confined or directed behaviour or a combination of this three modes and successfully applied to a set of single‐molecule experiments previously recorded for tetraspanins diffusing into plasma membrane (Espenel et al., 2008; Krementsov et al., 2010).
Colocalization analyses
Colocalization analyses were carried out using the CoLocalizer Pro software. Briefly, images representing a single optical Z‐section of cells were converted to tiffs and imported into CoLocalizer Pro software. Background corrections were applied and the total number of pixels overlapping between the two different channels was calculated. Colocalized pixels were divided by the total number of pixels for a chosen channel (as indicated in figure legends), yielding per cent colocalization. Colocalization values from at least 10 cells were averaged to give mean per cent colocalization.
Cross‐linking
Cells were incubated with PBS containing 2% fatty acid‐free bovine serum albumin (Sigma) and 50 μM 2‐bromopalmitate (2‐BP, Sigma) for 18 h at 37°C. Then, cells were washed with PBS containing 2 mM MgCl2 (PBS/MgCl2) and treated with cysteine cross‐linker DTME (0.2 mg ml−1 dithiobismaleimidoethane, Pierce) in PBS/MgCl2 for 2 h at 4°C. Cells were washed again with PBS/MgCl2 containing 5 mM cysteine and lysed with PBS containing 1% BrijO10 (Sigma), 2 mM cysteine and protease inhibitors. Lysates were used for immunoprecipitation assays.
Cell‐to‐cell transmission assay
Huh‐7 cells were infected with HCVcc (moi = 2) 24 h prior to their use in the assay. Infected cells were then labelled with 5‐chloromethylfluorescein diacetate (CMFDA) (Molecular Probes, Invitrogen) by incubation at 37°C during 30 min with 10 μM CMFDA in medium without FBS. Cells were trypsinized, washed and incubated 15 min with 3‐11 mAb (50 μg ml−1) to neutralize any remaining viral particle at the cell surface. After washing, CMFDA‐labelled donor cells were mixed with naïve target cell lines (ratio 1:4), seeded in 24‐well plates in presence (cell‐to‐cell transmission) or in absence (cell‐free and cell‐to‐cell transmission) of neutralizing mAb (3‐11, 50 μg ml−1). After 24 h, de novo transmission events were determined by staining for HCV NS5 and were quantified by flow cytometry, as described above. Cell‐to‐cell transmission levels were defined as newly infected cells in presence of 3‐11 mAb. Cell‐free transmission levels were evaluated using the following calculation: (de novo infected cells in absence of 3‐11) − (de novo infected cells in presence of 3‐11).
For infection with different seeding densities, 2 × 105, 1 × 105 and 5 × 104 cells per well of a 24‐well plate were infected with HCVcc (moi = 1). After 48 h, HCV infection levels were determined by staining for HCV NS5 followed by flow cytometry analyses.
Supporting information
Fig. S1. Cell surface distribution of CD81 and control proteins. Immunofluorescence images showing the basal membrane of living Huh‐7 clones expressing EWI‐2wint 1, LAL or Qcc. Experiments were performed at 37°C using TIRF microscopy. Cells were stained with Atto647N‐labelled Fab fragments of TS81, SYB‐1 and 11C5 mAbs to observe CD81, CD9 and CD46 respectively.
Fig. S2. EWI‐2wint has no effect on CD46 dynamics. The distribution of all the apparent diffusion coefficients (ADC) was calculated for CD46 in Huh‐7 clones expressing EWI‐2, EWI‐2wint (two clones), LAL or Qcc. Each dot represents one trajectory and 500 trajectories are represented for each cell clone. CD46 molecules were stained usingAtto647N‐labelled Fab fragments of 11C5 mAb.
Fig. S3. EWI‐2/EWI‐2wint expression in CHO cells. CHO cells were stably transfected with hCD81 and either the empty vector (pcDNA3.1), the uncleavable EWI‐2 (EWI‐2, see Experimental procedures) or wild‐type EWI‐2 construct (EWI‐2wint). It has to be noted that the cleavage of EWI‐2 generating EWI‐2wint naturally occurs in CHO cells. Cell surface biotinylation followed by immunoprecipitation assays were performed to control the expression of proteins. FLAG‐tagged EWI‐2 constructs and CD81 were immunoprecipitated with M2 and 5A6 mAbs respectively. Proteins were revealed by Western blotting with HRP‐conjugated streptavidin. Asterisks indicate additional cleavage products of EWI‐2wint, as previously described (Rocha‐Perugini et al., 2008). Triangles indicate an unidentified partner of CD81, which is not related to EWI‐2/EWI‐2wint proteins.
Acknowledgements
We thank Sophana Ung and André Pillez for their technical assistance. We thank Yves Rouillé for critical reading of the manuscript. We are grateful to S. Levy, J. McKeating, T. Wakita, for providing us with reagents. We thank Antonino Bongiovanni and Frank Lafont from the BioImaging Center Lille‐Nord de France for access to the instruments and technical advice. We thank Nicole Lautrédou for her technical advice. This work was supported by the ‘Institut Fédératif de Recherche–142’ (IFR142) and by a grant from the ‘Agence Nationale de Recherches sur le Sida et les hépatites virales’ ANRS. P.R. was supported by a fellowship from the ANRS. B.A.T. was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme. C.‐I.P. was supported by the postdoctoral programme POSDRU/89/1.5/S/60746 from European Social Fund.
During the revision of the manuscript, another study showing that HCV exploits the dynamic properties of CD81 for its entry has been published (Harris et al., 2012), which is in accordance with our conclusions.
References
- André, M. , Chambrion, C. , Charrin, S. , Soave, S. , Chaker, J. , Boucheix, C. , et al. (2009) In situ chemical cross‐linking on living cells reveals CD9P‐1 cis‐oligomer at cell surface. J Proteomics 73: 93–102. [DOI] [PubMed] [Google Scholar]
- Barreiro, O. , Zamai, M. , Yáñez‐Mó, M. , Tejera, E. , López‐Romero, P. , Monk, P.N. , et al. (2008) Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol 183: 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazzoli, M. , Bianchi, A. , Filippini, S. , Weiner, A. , Zhu, Q. , Pizza, M. , and Crotta, S. (2008) CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol 82: 8316–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe, C.L. , Grove, J. , Meredith, L.W. , Hu, K. , Syder, A.J. , Flores, M.V. , et al. (2011) Neutralizing antibody‐resistant hepatitis C virus cell‐to‐cell transmission. J Virol 85: 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S. , Le Naour, F. , Oualid, M. , Billard, M. , Faure, G. , Hanash, S.M. , et al. (2001) The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J Biol Chem 276: 14329–14337. [DOI] [PubMed] [Google Scholar]
- Charrin, S. , Le Naour, F. , Labas, V. , Billard, M. , Le Caer, J.‐P. , Emile, J.‐F. , et al. (2003a) EWI‐2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem J 373: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S. , Manié, S. , Thiele, C. , Billard, M. , Gerlier, D. , Boucheix, C. , and Rubinstein, E. (2003b) A physical and functional link between cholesterol and tetraspanins. Eur J Immunol 33: 2479–2489. [DOI] [PubMed] [Google Scholar]
- Charrin, S. , Le Naour, F. , Silvie, O. , Milhiet, P.‐E. , Boucheix, C. , and Rubinstein, E. (2009a) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J 420: 133–154. [DOI] [PubMed] [Google Scholar]
- Charrin, S. , Yalaoui, S. , Bartosch, B. , Cocquerel, L. , Franetich, J.‐F. , Boucheix, C. , et al. (2009b) The Ig domain protein CD9P‐1 down‐regulates CD81 ability to support Plasmodium yoelii infection. J Biol Chem 284: 31572–31578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman, L. , Meertens, L. , Bertaux, C. , Kajumo, F. , and Dragic, T. (2009) Residues in a highly conserved claudin‐1 motif are required for hepatitis C virus entry and mediate the formation of cell–cell contacts. J Virol 83: 5477–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgrange, D. , Pillez, A. , Castelain, S. , Cocquerel, L. , Rouille, Y. , Dubuisson, J. , et al. (2007) Robust production of infectious viral particles in Huh‐7 cells by introducing mutations in hepatitis C virus structural proteins. J Gen Virol 88: 2495–2503. [DOI] [PubMed] [Google Scholar]
- Dubuisson, J. , Helle, F. , and Cocquerel, L. (2008) Early steps of the hepatitis C virus life cycle. Cell Microbiol 10: 821–827. [DOI] [PubMed] [Google Scholar]
- Espenel, C. , Margeat, E. , Dosset, P. , Arduise, C. , Le Grimellec, C. , Royer, C.A. , et al. (2008) Single‐molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol 182: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M.J. , Hahn, T. , Tscherne, D.M. , Syder, A.J. , Panis, M. , Wölk, B. , et al. (2007) Claudin‐1 is a hepatitis C virus co‐receptor required for a late step in entry. Nature 446: 801–805. [DOI] [PubMed] [Google Scholar]
- Farquhar, M.J. , Harris, H.J. , and McKeating, J.A. (2011) Hepatitis C virus entry and the tetraspanin CD81. Biochem Soc Trans 39: 532–536. [DOI] [PubMed] [Google Scholar]
- Fofana, I. , Krieger, S.E. , Grunert, F. , Glauben, S. , Xiao, F. , Fafi‐Kremer, S. , et al. (2010) Monoclonal anti‐claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139: 953–964, 964.e1–.e4. [DOI] [PubMed] [Google Scholar]
- Gambin, Y. , Lopez‐Esparza, R. , Reffay, M. , Sierecki, E. , Gov, N.S. , Genest, M. , et al. (2006) Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA 103: 2098–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goueslain, L. , Alsaleh, K. , Horellou, P. , Roingeard, P. , Descamps, V. , Duverlie, G. , et al. (2010) Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84: 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove, J. , and Marsh, M. (2011) The cell biology of receptor‐mediated virus entry. J Cell Biol 195: 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, H.J. , Farquhar, M.J. , Mee, C.J. , Davis, C. , Reynolds, G.M. , Jennings, A. , et al. (2008) CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol 82: 5007–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, H.J. , Davis, C. , Mullins, J.G.L. , Hu, K. , Goodall, M. , Farquhar, M.J. , et al. (2010) Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285: 21092–21102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, H.J. , Clerte, C. , Farquhar, M.J. , Goodall, M. , Hu, K. , Rassam, P. , et al. (2012) Hepatoma polarization limits CD81 and hepatitis C virus dynamics. Cell Microbiol doi: 10.1111/cmi.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.T. , Catanese, M.T. , Law, L.M.J. , Khetani, S.R. , Syder, A.J. , Ploss, A. , et al. (2010) Real‐time imaging of hepatitis C virus infection using a fluorescent cell‐based reporter system. Nat Biotechnol 28: 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadokoro, K. , Bordo, D. , Galli, G. , Petracca, R. , Falugi, F. , Abrignani, S. , et al. (2001) CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J 20: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko, O.V. , Yang, X. , Kolesnikova, T.V. , and Hemler, M.E. (2004) Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross‐linking. Biochem J 377: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsov, D.N. , Rassam, P. , Margeat, E. , Roy, N.H. , Schneider‐Schaulies, J. , Milhiet, P.‐E. , and Thali, M. (2010) HIV‐1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, S.E. , Zeisel, M.‐B. , Davis, C. , Thumann, C. , Harris, H.J. , Schnober, E.K. , et al. (2010) Inhibition of hepatitis C virus infection by anti‐claudin‐1 antibodies is mediated by neutralization of E2–CD81–claudin‐1 associations. Hepatology 51: 1144–1157. [DOI] [PubMed] [Google Scholar]
- Lindenbach, B.D. , Evans, M.J. , Syder, A.J. , Wölk, B. , Tellinghuisen, T.L. , Liu, C.C. , et al. (2005) Complete replication of hepatitis C virus in cell culture. Science 309: 623–626. [DOI] [PubMed] [Google Scholar]
- Lupberger, J. , Zeisel, M.‐B. , Xiao, F. , Thumann, C. , Fofana, I. , Zona, L. , et al. (2011) EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeat, E. , Kapanidis, A.N. , Tinnefeld, P. , Wang, Y. , Mukhopadhyay, J. , Ebright, R.H. , and Weiss, S. (2006) Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J 90: 1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet, X. (2010) Mean square displacement analysis of single‐particle trajectories with localization error: Brownian motion in an isotropic medium. Phys Rev E Stat Nonlin Soft Matter Phys 82: 041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpellier, C. , Tews, B.A. , Poitrimole, J. , Rocha‐Perugini, V. , D'Arienzo, V. , Potel, J. , et al. (2011) Interacting regions of CD81 and two of its partners, EWI‐2 and EWI‐2wint, and their effect on hepatitis C virus infection. J Biol Chem 286: 13954–13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, D.M. , Williamson, D. , Rentero, C. , and Gaus, K. (2009) Quantitative microscopy: protein dynamics and membrane organisation. Traffic 10: 962–971. [DOI] [PubMed] [Google Scholar]
- Pileri, P. , Uematsu, Y. , Campagnoli, S. , Galli, G. , Falugi, F. , Petracca, R. , et al. (1998) Binding of hepatitis C virus to CD81. Science 282: 938–941. [DOI] [PubMed] [Google Scholar]
- Ploss, A. , Evans, M.J. , Gaysinskaya, V.A. , Panis, M. , You, H. , De Jong, Y.P. , and Rice, C.M. (2009) Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457: 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha‐Perugini, V. , Montpellier, C. , Delgrange, D. , Wychowski, C. , Helle, F. , Pillez, A. , et al. (2008) The CD81 partner EWI‐2wint inhibits hepatitis C virus entry. PLoS ONE 3: e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha‐Perugini, V. , Lavie, M. , Delgrange, D. , Canton, J. , Pillez, A. , Potel, J. , et al. (2009) The association of CD81 with tetraspanin‐enriched microdomains is not essential for Hepatitis C virus entry. BMC Microbiol 9: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R.S. , Meunier, J.‐C. , Takikawa, S. , Faulk, K. , Engle, R.E. , Bukh, J. , et al. (2008) Advantages of a single‐cycle production assay to study cell culture‐adaptive mutations of hepatitis C virus. Proc Natl Acad Sci USA 105: 4370–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman, P.G. , and Delbruck, M. (1975) Brownian motion in biological membranes. Proc Natl Acad Sci USA 72: 3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz, B. , Barretto, N. , Martin, D.N. , Hiraga, N. , Imamura, M. , Hussain, S. , et al. (2012) Identification of the Niemann‐Pick C1‐like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18: 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala‐Valdés, M. , Ursa, A. , Charrin, S. , Rubinstein, E. , Hemler, M.E. , Sánchez‐Madrid, F. , and Yáñez‐Mó, M. (2006) EWI‐2 and EWI‐F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin‐radixin‐moesin proteins. J Biol Chem 281: 19665–19675. [DOI] [PubMed] [Google Scholar]
- Sattentau, Q. (2008) Avoiding the void: cell‐to‐cell spread of human viruses. Nat Rev Microbiol 6: 815–826. [DOI] [PubMed] [Google Scholar]
- Scarselli, E. , Ansuini, H. , Cerino, R. , Roccasecca, R.M. , Acali, S. , Filocamo, G. , et al. (2002) The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21: 5017–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvie, O. , Charrin, S. , Billard, M. , Franetich, J.‐F. , van Clark, K.L., Gemert, G.‐J. , et al. (2006) Cholesterol contributes to the organization of tetraspanin‐enriched microdomains and to CD81‐dependent infection by malaria sporozoites. J Cell Sci 119: 1992–2002. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S. , Kolesnikova, T.V. , and Hemler, M.E. (2001) EWI‐2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 276: 40545–40554. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S. , Kolesnikova, T.V. , and Hemler, M.E. (2003) EWI‐2 regulates alpha3beta1 integrin‐dependent cell functions on laminin‐5. J Cell Biol 163: 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe, J.M. , Stamataki, Z. , Jennings, A. , Hu, K. , Farquhar, M.J. , Harris, H.J. , et al. (2008) Hepatitis C virus cell–cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47: 17–24. [DOI] [PubMed] [Google Scholar]
- Wakita, T. , Pietschmann, T. , Kato, T. , Date, T. , Miyamoto, M. , Zhao, Z. , et al. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveldt, J. , Evans, M.J. , Bitzegeio, J. , Koutsoudakis, G. , Owsianka, A.M. , Angus, A.G.N. , et al. (2009) CD81 is dispensable for hepatitis C virus cell‐to‐cell transmission in hepatoma cells. J Gen Virol 90: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.H. , Kovalenko, O.V. , Kolesnikova, T.V. , Andzelm, M.M. , Rubinstein, E. , Strominger, J.L. , and Hemler, M.E. (2006) Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem 281: 12976–12985. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A. , Lane, W.S. , Charrin, S. , Rubinstein, E. , and Liu, L. (2003) EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res 63: 2665–2674. [PubMed] [Google Scholar]
- Zhang, Y. , Li, X. , Becker, K.A. , and Gulbins, E. (2009) Ceramide‐enriched membrane domains – structure and function. Biochim Biophys Acta 1788: 178–183. [DOI] [PubMed] [Google Scholar]
- Zhong, J. , Gastaminza, P. , Cheng, G. , Kapadia, S. , Kato, T. , Burton, D.R. , et al. (2005) Robust hepatitis C virus infection in vitro . Proc Natl Acad Sci USA 102: 9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Cell surface distribution of CD81 and control proteins. Immunofluorescence images showing the basal membrane of living Huh‐7 clones expressing EWI‐2wint 1, LAL or Qcc. Experiments were performed at 37°C using TIRF microscopy. Cells were stained with Atto647N‐labelled Fab fragments of TS81, SYB‐1 and 11C5 mAbs to observe CD81, CD9 and CD46 respectively.
Fig. S2. EWI‐2wint has no effect on CD46 dynamics. The distribution of all the apparent diffusion coefficients (ADC) was calculated for CD46 in Huh‐7 clones expressing EWI‐2, EWI‐2wint (two clones), LAL or Qcc. Each dot represents one trajectory and 500 trajectories are represented for each cell clone. CD46 molecules were stained usingAtto647N‐labelled Fab fragments of 11C5 mAb.
Fig. S3. EWI‐2/EWI‐2wint expression in CHO cells. CHO cells were stably transfected with hCD81 and either the empty vector (pcDNA3.1), the uncleavable EWI‐2 (EWI‐2, see Experimental procedures) or wild‐type EWI‐2 construct (EWI‐2wint). It has to be noted that the cleavage of EWI‐2 generating EWI‐2wint naturally occurs in CHO cells. Cell surface biotinylation followed by immunoprecipitation assays were performed to control the expression of proteins. FLAG‐tagged EWI‐2 constructs and CD81 were immunoprecipitated with M2 and 5A6 mAbs respectively. Proteins were revealed by Western blotting with HRP‐conjugated streptavidin. Asterisks indicate additional cleavage products of EWI‐2wint, as previously described (Rocha‐Perugini et al., 2008). Triangles indicate an unidentified partner of CD81, which is not related to EWI‐2/EWI‐2wint proteins.


