Abstract
The facultative anaerobic bacterium Lactobacillus casei IGM394 is used as a host for drug delivery systems, and it exhibits the same growth rate under aerobic and anaerobic conditions. L. casei strains carry several genes that facilitate oxygen and reactive oxygen species (ROS) tolerance in their genomes, but their complete functions have not been uncovered. To clarify the oxygen and ROS tolerance mechanisms of L. casei IGM394, we constructed 23 deficient mutants targeting genes that confer oxidative stress resistance. Significantly decreased growth and high H2O2 accumulation were observed in the NADH peroxidase gene-mutated strain (Δnpr) compared with the findings in the wild type. The H2O2 degradation capacity of Δnpr revealed that NADH peroxidase is a major H2O2-degrading enzyme in L. casei IGM394. Interestingly, ΔohrR, a mutant deficient in the organic hydroperoxide (OhrA) repressor, exhibited higher H2O2 resistance than the wild-type strain. Increased Npr expression and H2O2 degradation ability were observed in ΔohrR, further supporting the importance of OhrA to ROS tolerance mechanisms. The other mutants did not exhibit altered growth rates, although some mutants had higher growth in the presence of oxygen. From these results, it is presumed that L. casei IGM394 has multiple oxygen tolerance mechanisms and that the loss of a single gene does not alter the growth rate because of the presence of complementary mechanisms. Contrarily, the H2O2 tolerance mechanism is solely dependent on NADH peroxidase in L. casei IGM394.
Keywords: NADH peroxidase, oxidative stress, deficient mutants, Lactobacillus casei, H2O2
INTRODUCTION
Lactic acid bacteria are facultative anaerobic bacteria that do not require oxygen for growth, and they do not have a respiratory chain and catalase; thus, they rely on anaerobic fermentation to produce energy. There is a wide range of variations in tolerance to oxygen and reactive oxygen species (ROS) stress among lactic acid bacteria, even in the same species. This means that stress tolerance in bacteria depends on the genes present in their genomes. ROS are produced via the conversion of oxygen to the superoxide anion radical, which is further converted to hydrogen peroxide (H2O2), and Fe2+ in cells induces production of the more toxic hydroxyl radical via the Fenton reaction. ROS damage intracellular proteins and DNA and cause cell death [1]. Many studies have examined the tolerance mechanisms of lactic acid bacteria to oxygen and ROS. Enzymes such as NADH oxidase and pyruvate oxidase, which degrade molecular oxygen [2,3,4,5,6,7], superoxide dismutase (SOD), which targets superoxide as a substrate [8, 9], and NADH peroxidase, which degrades H2O2 [10], are involved in the tolerance mechanisms. Further, lactic acid bacteria in the Lactobacillus casei group possess multiple types of peroxidase, including NADH peroxidase, glutathione peroxidase, thiol peroxidase and iron-dependent peroxidase. Therefore, several antioxidant enzymes are involved in oxidative stress tolerance in lactic acid bacteria. In addition to enzymes that confer direct resistance to oxygen and ROS, some enzymes contribute to oxidative tolerance. Thioredoxin reductase (TrxB2), which maintains the intracellular redox state balance, has been reported to be involved in oxygen tolerance in Lactococcus lactis, Lactobacillus plantarum WCFS1, and Lactobacillus casei Shirota [11,12,13], and similar findings have been reported for Escherichia coli [14]. Streptococcus mutans carries an iron-binding protein (Dpr) to avoid the Fenton reaction, in addition to antioxidant enzymes such as NADH oxidase [15, 16]. Furthermore, L. casei Shirota expresses the iron-binding protein HprA1, which is involved in H2O2 resistance via a different mechanism than Dpr [17]. HprA1 is involved in H2O2 resistance, but it does not exhibit H2O2-decomposing activity. It has also been reported that the disruption of the NADH peroxidase gene (npx) of L. casei Shirota results in a decreased growth rate under shaking and the loss of H2O2-decomposing activity. On the contrary, in recent studies, some lactic acid bacteria in which the electron transfer system is activated by the addition of heme alone or together with menaquinone (vitamin K2) and oxygen is consumed as the final electron acceptor have been reported [18,19,20]. Compared with anaerobic fermentation, in which ATP is obtained only from glycolysis, use of the electron transfer system increases the amount of ATP and improves growth rates. Additionally, organic hydroperoxide resistance protein transcriptional regulator (OhrR), which was initially found in the gram-negative bacterium Xanthomonas campestris, is involved in resistance to organic peroxide and H2O2 [21]. There are similar reports in the gram-positive bacterium Bacillus subtilis [22]. However, OhrR has not been reported in lactic acid bacteria. According to the information on lactobacilli published in KEGG, L. casei and L. plantarum, which are generally considered oxygen-resistant, carry ohrR, but oxygen-sensitive species such as L. acidophilus and L. delbrueckii subsp. bulgaricus do not possess the gene. Thus, the antioxidant factors possessed by lactic acid bacteria vary depending on the genus and species, and the response to oxygen stress differs accordingly.
Comparative genomic analysis of L. casei and L. paracasei revealed that several genes involved in oxidative stress tolerance are shared between the species [23,24,25,26,27,28,29,30,31]. However, the functions of these genes remain to be clarified.
L. casei IGM394 has high immunostimulatory capacity, and it is used as a host for drug delivery systems [32, 33]. The bacterium also exhibits an extremely good growth rate under aerobic conditions. Similar to other L. casei group bacteria, this strain has multiple oxidative stress tolerance genes, and thus, it is predicted that it has complex mechanisms of oxygen stress tolerance. However, the details of these mechanisms are unclear. It is important to clarify the functions of genes involved in tolerance to oxidative stress in conducting applied research with this strain as the host.
In this study, we constructed 23 deficient mutants (deficient in a single gene, 14 strains; deficient in multiple genes, 9 strains) targeting antioxidant genes reported in other bacteria via a double-crossover method. The oxidative stress tolerance mechanisms of these strains were evaluated by examining oxygen resistance in shaking culture as well as based on the consumption and resistance to H2O2 generated in metabolic processes. As a result, although no differences were observed in the growth of most of the deficient mutants, the Δnpr strain had a decreased growth rate. We found that NADH peroxidase is an essential enzyme for H2O2 degradation in L. casei IGM394.
METHODS
Strains, plasmids, media, and growth conditions
The strains and plasmids used in this study are listed in Table 1. L. casei IGM394 was used as the wild type. The L. casei IGM394 was a derivative of L. casei ATCC 393, and the L. casei ATCC 393 was distributed by a European collaborator. The L. casei IGM394 exhibits high transformation efficiency. Escherichia coli DH5α (Toyobo, Osaka, Japan) was used as the competent cells for DNA transformation. The plasmid pBTE was used as a cloning vector for deficient mutants. Lactic acid bacteria were grown at 37°C in MRS medium (Becton, Dickinson and Company, Sparks, MD, USA) and LAPTg medium (2% glucose, 1% yeast extract, 1% Bacto Proteose Peptone No. 3, 0.1% Bacto Tryptone, 0.1% Tween 80, and 0.01% MgSO4·7H2O). E. coli was grown at 37°C in LB Miller medium (Becton, Dickinson and Company). Erythromycin was added at a final concentration of 5 µg/mL for lactic acid bacteria. Ampicillin was added at a final concentration of 100 µg/mL for E. coli. The optical density of the culture was measured at 600 nm (OD600) using a UV-1200 UV-VIS spectrophotometer (Shimadzu, Kyoto, Japan). The medium was dispensed into test tubes with loose aluminum caps (static condition) or silicon caps that allowed free air exchange (shaking condition). The cells were cultured with shaking at 180 rpm for the shaking condition. Growth analysis of wild-type and mutant strains was performed in three independent experiments under the static or shaking condition. The data are shown as the mean ± SE of three independent experiments.
Table 1. Bacterial strains and plasmids used in this study.
| Strains or plasmid | Phenotype of genotype | Source or reference | ||
|---|---|---|---|---|
| Strains | ||||
| L. casei | ||||
| IGM394 | Wild-type | our collection | ||
| Δnox | deficient of nox gene | This study | ||
| Δnox5 | deficient of nox5 gene | This study | ||
| ΔpoxF | deficient of poxF gene | This study | ||
| ΔcidC | deficient of cidC gene | This study | ||
| ΔahpC | deficient of ahpC gene | This study | ||
| ΔohrR | deficient of ohrR gene | This study | ||
| Δsod | deficient of sod gene | This study | ||
| Δsuf | deficient of suf gene | This study | ||
| Δflp | deficient of flp gene | This study | ||
| ΔdpsB | deficient of dpsB gene | This study | ||
| ΔcydAB | deficient of cydAB gene | This study | ||
| ΔgshR1 | deficient of gshR1 gene | This study | ||
| Δipr | deficient of ipr gene | This study | ||
| Δnpr | deficient of npr gene | This study | ||
| Δnox::Δnpr | deficient of nox and npr gene | This study | ||
| Δnox5::Δnpr | deficient of nox5 and npr gene | This study | ||
| Δsod::Δnpr | deficient of sod and npr gene | This study | ||
| ΔgshR1::Δnpr | deficient of gshR1 and npr gene | This study | ||
| ΔgshR2::Δnpr | deficient of gshR2 and npr gene | This study | ||
| Δipr::Δnpr | deficient of ipr and npr gene | This study | ||
| ΔgshR1::ΔgshR2::Δnpr | deficient of gshR1, gshR2 and npr gene | This study | ||
| Δsod::ΔgshR1::ΔgshR2::Δnpr | deficient of sod, gshR1, gshR2 and npr gene | This study | ||
| Δipr::ΔgshR1::ΔgshR2::Δnpr | deficient of ipr, gshR1, gshR2 and npr gene | This study | ||
| E. coli | ||||
| DH5a | Commercial strain purchased from Toyobo | |||
| Plasmids | ||||
| pBTE | E. coli-gram positive bacteria shuttle vector carrying pBT2 ori region, pAMb1 erythromycin resistance gene, multi cloning sites and temperature sensitivity | our collection | ||
| pBTE::Δnox | pBTE carrying deficient fragment of nox | This study | ||
| pBTE::Δnox5 | pBTE carrying deficient fragment of nox5 | This study | ||
| pBTE::ΔpoxF | pBTE carrying deficient fragment of poxF | This study | ||
| pBTE::ΔcidC | pBTE carrying deficient fragment of cidC | This study | ||
| pBTE::ΔahpC | pBTE carrying deficient fragment of ahpC | This study | ||
| pBTE::ΔohrR | pBTE carrying deficient fragment of ohrR | This study | ||
| pBTE::Δsod | pBTE carrying deficient fragment of sod | This study | ||
| pBTE::Δsuf | pBTE carrying deficient fragment of suf | This study | ||
| pBTE::Δflp | pBTE carrying deficient fragment of flp | This study | ||
| pBTE::ΔdpsB | pBTE carrying deficient fragment of dpsB | This study | ||
| pBTE::ΔcydAB | pBTE carrying deficient fragment of cydAB | This study | ||
| pBTE::ΔgshR1 | pBTE carrying deficient fragment of gshR1 | This study | ||
| pBTE::ΔgshR2 | pBTE carrying deficient fragment of gshR2 | This study | ||
| pBTE::Δipr | pBTE carrying deficient fragment of ipr | This study | ||
| pBTE::Δnpr | pBTE carrying deficient fragment of npr | This study | ||
Construction of deficient mutants
pBTE is a derivative of the shuttle and thermosensitive plasmid vector pBT2. The origin of replication for lactic acid bacteria cannot function at 42°C. Recombinant plasmids for deficient mutants were constructed as follows. The upstream and downstream fragments of the target gene were amplified by PCR using L. casei IGM394 genomic DNA as a template, PrimeSTAR Max DNA polymerase (Takara, Shiga, Japan), and the primer pairs listed in Table 2. The fragments were digested at both ends using appropriate restriction enzymes (Table 2). The fragments were cloned into pBTE, which had previously been digested using the same restriction enzymes. Recombinant plasmids were purified using NucleoSpin® Plasmid (Macherey-Nagel, Bethlehem, PA, USA) and transferred into L. casei IGM394 via electroporation. Cells were grown in 10 mL of MRS broth to the stationary phase and harvested via centrifugation, and they were then suspended in 10 mL of MRS broth containing 8% (w/v) glycine and incubated at 37°C for 90 min. The cells were subsequently washed twice with an equal volume of sterile water, followed by washing with an equal volume of 50 mM EDTA solution and washing twice with an equal volume of 0.3 M sucrose solution. They were then suspended in 1 mL of 0.3 M sucrose solution. Electroporation was done with a Gene Pulser (BTX, San Diego, CA, USA) using 100 µL of competent cells and 10 µL of plasmid DNA solution in a 2-mm electroporation cuvette at a capacitance, resistance, and voltage of 25 µF, 48 Ω, and 1.5 kV, respectively. Cells were transferred to 1 mL of MRS broth and then incubated at 37°C for 2 hr. After incubation, cells were plated onto MRS agar containing 5 µg/mL erythromycin and incubated at 37°C for 3 or 4 days under anaerobic conditions using AnaeroPouch®-Anaero (MGC, Tokyo, Japan). Erythromycin-resistant colonies were selected, and plasmid introduction was confirmed by PCR with appropriate primers (Table 2). To induce plasmid integration, transformants were incubated at 42°C in MRS broth containing 5 µg/mL erythromycin. After several cycles of subculture, cells were plated onto MRS agar containing 5 µg/mL erythromycin and incubated at 37°C for 3 or 4 days under anaerobic conditions using Anaero Pouch®-Anaero. A colony was selected at random, and plasmid integration was confirmed by PCR with appropriate primers (Table 2). The integrants were incubated at 37°C in MRS broth. After several cycles of subculture, cells were plated onto MRS agar and incubated at 37°C for 3 or 4 days under anaerobic conditions using AnaeroPouch®-Anaero. Colonies were selected at random, and gene disruption was confirmed by PCR with appropriate primers (Table 2).
Table 2. Primers sequence used in this study.
| Target gene | Primer name | Primer sequence (5’ to 3’) | Restriction enzyme site | |
|---|---|---|---|---|
| Construction of deficient mutants | ||||
| nox | nox_A-forward | CAACCTGCAGTTTTTGCTGTTGATTAATATGTTTGAAAAT | Pst I | |
| nox_A-reverse | TGTGAAGGAGTGTTTAACTATCCATTCGAATTGCAAACAA | |||
| nox_B-forward | TTGTTTGCAATTCGAATGGATAGTTAAACACTCCTTCACA | |||
| nox_B-reverse | TTC GAAGCTTGTTATCCGCAACGTGCCGT | Hind III | ||
| nox5 | nox5_A-forward | CCATGGATCCCCCGTGAAGCGTAGTTGTTG | BamH I | |
| nox5_A-reverse | ACGAATTCATAATTTTCCCCCAGCATCTGCCTTCCTTTCA | |||
| nox5_B-forward | TGAAAGGAAGGCAGATGCTGGGGGAAAATTATGAATTCGT | |||
| nox5_B-reverse | GCC AAAGCTTCTTGATCGGCTCGTCTGATC | Hind III | ||
| poxF | poxF_A-forward | GTTGGGATCCAGCCAATGGCGACTTCTGGA | BamH I | |
| poxF_A-reverse | ACTTTTTGGGAGGGATTCTTCTAGTGATTAAAAAAGAGAT | |||
| poxF_B-forward | ATCTCTTTTTTAATCACTAGAAGAATCCCTCCCAAAAAGT | |||
| poxF_B-reverse | TGCACTGCAGGGCTTGGCAGTGCCGAA | Pst I | ||
| cidC | cidC_A-forward | GCTAGGATCCCAGCGTGACGGCTTTTTATA | BamH I | |
| cidC_A-reverse | TGATACAAGCTAATCGAAAAATCAAAATCTCCTTTATCGC | |||
| cidC_B-forward | GCGATAAAGGAGATTTTGATTTTTCGATTAGCTTGTATCA | |||
| cidC_B-reverse | GACAAAGCTTGGGACACAATATGCTGAGGC | Hind III | ||
| ahpC | ahpC_A-forward | GCTAGGATCCGCTACATTCTCGATATCGGT | BamH I | |
| ahpC_A-reverse | CTAAAAATGGAGGTAATATCAACGGTGCTTTGATCAGCTA | |||
| ahpC_B-forward | TAGCTGATCAAAGCACCGTTGATATTACCTCCATTTTTAG | |||
| ahpC_B-reverse | GATTAAGCTTATGTCCGTGATCGTCCGTTTATTAAGAATC | Hind III | ||
| ohrR | ohrR_A-forward | CAAGCTGCAGGAGCGATGATGCCTAG | Pst I | |
| ohrR_A-Reverse | CCTTATTTTTGGGCGGCTGCTTCCTCCTAA | |||
| ohrR_B-forward | TTAGGAGGAAGCAGCCGCCCAAAAATAAGG | |||
| ohrR_B-Reverse | GCACAAGCTTAAGGTTGATGGATCAGGATG | Hind III | ||
| sod | sod_A-forward | CTGACTGCAGCGCTGAAATTGCGCAAAATC | Pst I | |
| sod_A-reverse | AGTGACCAAACATCGAGGAATCAACCTTTC | |||
| sod_B-forward | GAAAGGTTGATTCCTCGATGTTTGGTCACT | |||
| sod_B-reverse | GGTTAAGCTTGGTAAATTACCATCCCCAAAAGGTG | Hind III | ||
| suf | suf_A-forward | AATAGGATCCTTGTGGTCCAAGCATCAGGAC | BamH I | |
| suf_A-reverse | TTGGAGGCAAGTGTCCTGGTTGACTTAAAA | |||
| suf_B-forward | TTTTAAGTCAACCAGGACACTTGCCTCCAA | |||
| suf_B-reverse | GCATCTGCAGAATCATGCTAAAATGTTGGC | Pst I | ||
| flp | flp_A-forward | GCTAGGATCCCAAGCACAGACCCATTTTG | BamH I | |
| flp_A-reverse | CATCACTCGGCAACCGTTGCCACCTCCTAA | |||
| flp_B-forward | TTAGGAGGTGGCAACGGTTGCCGAGTGATG | |||
| flp_B-reverse | TCGGCTGCAGGCCCGATGACCTCAAC | Pst I | ||
| dpsB | dpsB_A-forward | GGTCGGATCCAATGCCTTACGGTTACGGCA | BamH I | |
| dpsB_A-reverse | CGCTCTGTCAACAAGCTTGAGTTCTTCTCCTCTTAACCGC | |||
| dpsB_B-forward | GCGGTTAAGAGGAGAAGAACTCAAGCTTGTTGACAGAGCG | |||
| dpsB_B-reverse | GCGGCTGCAGGAGAAGCAATTGTTGAAAGTCAGT | Pst I | ||
| cydAB | cydAB_A-forward | CCTCGGATCCCCTACTCACTATCATTC | BamH I | |
| cydAB_A-reverse | TCTTTTCGCCGCCGCTTTGATCACCTCTAT | |||
| cydAB_B-forward | ATAGAGGTGATCAAAGCGGCGGCGAAAAGA | |||
| cydAB_B-reverse | TAGACTGCAGCCTGAACACGTGCATACTG | Pst I | ||
| gshR1 | gshR-1_A-forward | CAATGTCGACGCTTCGGTATCCCGG | Sal I | |
| gshR-1_A-reverse | ACAAGGAGGATCAATGGTTGTAGCGGCAAT | |||
| gshR-1_B-forward | ATTGCCGCTACAACCATTGATCCTCCTTGT | |||
| gshR-1_B-reverse | GTTGAAGCTTTGATCGGCGCGCCG | Hind III | ||
| gshR2 | gshR-2_A-forward | AACAGGATCCCAATTAAGGTCATATCATCCC | BamH I | |
| gshR-2_A-reverse | CACAGAGACGAAGGAAGGCGTAATTGATAC | |||
| gshR-2_B-forward | GTATCAATTACGCCTTCCTTCGTCTCTGTG | |||
| gshR-2_B-reverse | GCGGGTCGACGCATCACGGCCTAAG | Sal I | ||
| ipr | ipr_A-forward | CCGACTGCAGCGTATTAATCACGTTGC | Pst I | |
| ipr_A-reverse | TTTGGAGGCGTAACGATAAAGTTGCTTGAT | |||
| ipr_B-forward | ATCAAGCAACTTTATCGTTACGCCTCCAAA | |||
| ipr_B-reverse | GGCCAAGCTTATGAGGTACAGCTTCGC | Hind III | ||
| npr | npr_A-forward | CAATGGATCCATCGGAGCATATCCCTTCAG | BamH I | |
| npr_A-reverse | AATTAGGAGGAATTTACTTTTAAAAAGACA | |||
| npr_B-forward | TGTCTTTTTAAAAGTAAATTCCTCCTAATT | |||
| npr_B-reverse | GATACTGCAGATTTGGCCGGGACAAGTG | Pst I | ||
| Confirmation of deficient mutants | ||||
| nox | 279962_F | GTAGCATCGGCAATTGTCATGTAGTGTCAC | ||
| 282934_R | CTGTTTTGAGTCATACCGTGCAACCCG | |||
| nox5 | 177892_F | CTGCGGTTCGATGGTGCTAAGGTCACCTTC | ||
| 180877_R | GTTTTGACGCATTCATCGAATCGAGTCGCG | |||
| poxF | 2268013_F | GTCTGACTAATATGCAGTGGCGCAAAGTGAG | ||
| 2270930_R | CGAGGCAGCCAAAGCTTTCGTTAAGAAGCAC | |||
| cidC | 498129_F | CGTTGCTTCGATCATGGTCTGGCAGAATTC | ||
| 501457_R | GGCCAGTGGCATTCCTGATTACACCGAG | |||
| ahpC | 2611951_F | GAATAACCATAGAAAGAAGGGAGGCAGTTG | ||
| 2614120_R | AATTATTACCAGCCGGACCCGAGCACAAAG | |||
| ohrR | 1040892_F | CAATTTTAGATCCGGATACCATGGCGATTTC | ||
| 1043371_R | CTCCATTGCACACAAATTGCACACAAATTC | |||
| sod | 2004800_F | CAATCGCATGCTCGGAAATGAGTTTCAAAC | ||
| 2007409_R | GGAAATAGGTATGCGATATTCATTTACGAC | |||
| flp | 69484_F | CTTATGGAGGAGGTTTCGATCCTATAGAAC | ||
| 71774_R | GCAGTATACCAACGTTCCAACCGCTATC | |||
| dpsB | 68755_F | GAAAAGGTGATGTTTGTCGGTGACGGGATC | ||
| 70973_R | GTATTTAAAAAACATCACTCGGCAACCTCACCAAG | |||
| cydAB | 14573_F | GAAGCTTAGAGTGACGGCTAATGAAC | ||
| 18661_R | CCGCAAAATGGACGGGTATTATCCATC | |||
| gshR1 | 2311599_F | CAATGGGTTGCGGTTCGCATTCCTGAC | ||
| 2314729_R | CTGTCGGAACGTTACTCGTCATGCTTG | |||
| gshR2 | 2748254_F | CAGTGACCAAAGATTTTGACCATCATAAAC | ||
| 2751189_R | GTTGATCCAACGAGCGGCGTCATC | |||
| ipr | 706255_F | GGGTAATAAACCAGCAATGACCACAAGACG | ||
| 708795_R | CTAGAATTCAATCGAAATAATATTCGGATTGTCGG | |||
| npr | 464266_F | CCAATTTTTTCTGCAAAGTCCTTTTGAGAG | ||
| 467233_R | CGTTTTACAAGCATGGGAAAATACGGC | |||
| qRT-PCR | ||||
| NADH peroxidase | ACGGCAATCCACAAGTTTGC | |||
| TTGTTGTTGAACGGCGAGTG | ||||
| Elongation factor Tu | AACCGCGAACAAGTTGAACG | |||
| ACGGCCACCTTCTTCTTTTG | ||||
| Glyceraldehyde 3-phosphate dehydrogenase | AACACGATTCCTCACAGCAC | |||
| ACAACAGAAACACGCTGTGC | ||||
Quantification of H2O2
A mixture of the chromogenic reagent DA64 (100 µM in PIPES buffer [0.1 M, pH 6.8, 0.5% Triton-X 100]) and horseradish peroxidase (100 units/mL) was used to measure H2O2 concentrations. Cultures of each strain were harvested via centrifugation (10,000 × g, 3 min). Each supernatant (20 µL) were added to the mixture, which was incubated at 37°C for 5 min. After incubation, OD727 was measured, and H2O2 content was quantified using the standard curve.
H2O2 consumption
Cells precultured at 37°C were inoculated into 10 mL of MRS medium at OD600 = 0.05. The cells were used after static culture at 37°C for 5 hr. They were then washed twice with PIPES buffer (pH 6.8) and resuspended in 10 mL of H2O2 adjusted to 50 µM, 100 µM, 300 µM with PIPES buffer. After incubation at 37°C for 1 hr under a static condition, the cells were harvested via centrifugation (10,000 × g, 3 min). Supernatants were used to measure H2O2 concentrations.
Repressive effect of H2O2 on bacterial growth
Cells precultured overnight at 37°C were inoculated into fresh MRS medium at OD660 = 0.05. These bacterial cultures were aliquoted (180 µL/well) into a 96-well plate, and 20 µL of H2O2 solution was added to a final concentration of 0.5, 1.0, or 2.0 mM; the plate was then incubated at 37°C for 24 hr. The OD600 was measured using a multiplate reader, and the value at each concentration was calculated on the basis of the value at 0 mM for each strain. Significance was indicated by p<0.05 (for each wild-type concentration).
RNA isolation
H2O2 treatment was performed as follows. Samples precultured at 37°C were inoculated into 20 mL of MRS medium at OD600 = 0.05. The cultures were grown for 5 hr at 37°C under static conditions and divided into two 10 mL aliquots. The cells were harvested via centrifugation (10,000 × g, 3 min) and washed twice with PIPES buffer (pH 6.8). One aliquot was resuspended in 10 mL of PIPES buffer and incubated at 37°C for 1 hr. The other aliquot was resuspended in H2O2 adjusted to 0.5 mM with PIPES buffer and incubated at 37°C for 1 hr. The 10 mL cultures were added to 20 mL of RNAprotect Bacteria Reagent (Qiagen). The mixtures were kept at room temperature for 5 min. The cells were then harvested via centrifugation for 10 min at 5000 × g, suspended in 500 µL of TE buffer (50 mM Tris–HCl, pH 8.0) containing 5 mg/mL lysozyme and 20 µL/mL mutanolysin, and incubated at 37°C for 30 min. Total RNA was purified using a Direct-zol™ RNA MiniPrep kit (Zymo Research) according to the manufacturer’s protocol. DNA was digested using DNase I in the purification step. RNA was isolated from three independent cultures.
Quantitative real-time PCR assays
cDNA was synthesized using a PrimeScript RT reagent kit (Takara) according to the manufacturer’s protocol. In total, 0.1 mg of total RNA was used as a template. Quantitative real-time PCR assays were performed using a CFX96 Real-Time PCR Detection System (Bio-Rad) with THUNDERBIRD SYBR qPCR Mix (Toyobo). The primers were designed to amplify products of approximately 80 bp in length (Table 2). The reaction mixture contained 25 µL of THUNDERBIRD SYBR qPCR Mix, 1 µL of 15 µM forward primer, 1 µL of 15 µM reverse primer, 1 µL of 50× ROX reference dye, 20 µL of dH2O, and 2.5 µL of diluted cDNA templates. All reactions were run in duplicate for each of the three independent RNA samples. The gene expression values were normalized using the elongation factor Tu and glyceraldehyde 3-phosphate dehydrogenase as an internal standard. Standard curves for both the internal standard and target genes were generated by amplifying 10-fold serial dilutions of cDNA. The gene expression data from quantitative real-time PCR were analyzed using Student’s t-test.
RESULTS
Construction of mutants deficient in genes involved in oxygen and ROS tolerance in L. casei IGM394
To elucidate the mechanisms of oxygen and ROS tolerance in L. casei IGM394, we constructed gene-deficient mutants targeting enzymes or factors that are expected to be involved in oxygen tolerance (Table 3). The target gene was completely deleted via the double-crossover method using a thermosensitive suicide vector. Fourteen out of 16 targeted genes were successfully disrupted in mutants. However, disruptants were not obtained for trxB2 and the chaperone protein gene groEL.
Table 3. Targeting enzymes or factors that are expected to be involved in oxidative stress tolerance.
| Gene name | |
|---|---|
| NADH peroxidase | npr |
| Organic hydroperoxide resistance protein transcriptional regulator | ohrR |
| NADH oxidase (H2O - forming) | nox |
| NADH oxidase (H2O2 - forming) | nox5 |
| Pyruvate oxidase | poxF |
| Pyruvate oxidase | cidC |
| Alkyl hydroperoxide reductase subunit C | ahpC |
| Superoxide dismutase | sod |
| Fe-S cluster assembly protein | suf |
| Probable transcriptional regulator | flp |
| DNA binding protein | dpsB |
| Cytochrome bd ubiquinol subunit I, II | cydAB |
| Glutathione reductase | gshR1 |
| Iron-dependent peroxidase | ipr |
| Thioredoxin reductase | trxB2 |
| Chaperon protein | groEL |
Growth of deficient mutants under static and shaking conditions
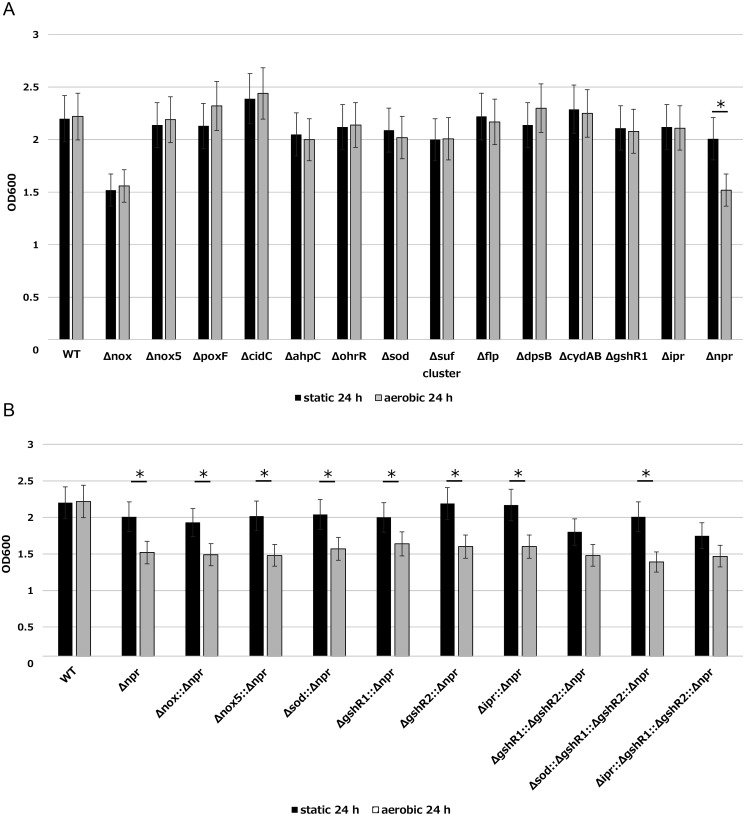
We evaluated the growth rates of 23 deficient mutants under static and shaking conditions (Fig. 1A, 1B). The growth rate of Δnox, which is an NADH oxidase (nox, H2O-forming) gene-deficient mutant, was decreased under both culture conditions. Compared with the findings for the wild type, the OD decreased from 2.2 to 1.5 under the static condition and from 2.2 to 1.6 under the shaking condition after 24 hr of culture. In the Δnpr mutant, the growth rate was decreased only under shaking culture. Under the static condition, the OD of Δnpr was 2.0, which was similar to that of the wild type (2.2). However, under the shaking condition, the OD of Δnpr was 1.5, whereas that for the wild type was 2.2. Conversely, the ODs of four strains, namely the NADH oxidase (nox5, H2O2-forming) gene-deficient mutant Δnox5, pyruvate oxidase gene-deficient mutants ΔpoxF and ΔcidC, and DNA-binding protein gene-deficient mutant ΔdpsB, were slightly increased (approximately 0.1–0.2) under the shaking condition. The other eight deficient mutants did not exhibit different growth rates versus the wild type under either of the culture conditions. Decreased growth under shaking was observed only for Δnpr. Therefore, we constructed mutants deficient in multiple genes using Δnpr as a host and evaluated the effect on viability. We constructed six double-deficient mutants, one triple-deficient mutant, and two quadruple-deficient mutants. The target genes of the six double-deficient mutants were nox, nox5, sod, glutathione reductase (gshR1 or gshR2), and iron-dependent peroxidase (ipr). The triple-deficient mutant featured mutations of gshR1 and gshR2. Finally, the quadruple-deficient mutants featured mutations of sod or ipr using the triple-deficient mutant ΔgshR1::ΔgshR2::Δnpr as a host. However, these deficient mutants exhibited the same growth rate as Δnpr (Fig. 1B). These results indicated that the Npr gene is important for the growth of L. casei IGM394 under the shaking condition. Measuring the amount of H2O2 accumulated in LAPTg medium after 24 hr, H2O2 was detected only in Δnpr and nine multiple-deficient mutants (data not shown). In addition, the evaluation by bacterial turbidity included dead cells, and there was a possibility that the results might be inaccurate. Therefore, we measured colony-forming units, and the results were in line with the measured OD values. Thus, only the results for turbidity are presented.
Fig. 1.
(A) Growth rates of wild type and deficient mutants under static and shaking conditions. Strains precultured overnight at 37°C were inoculated into fresh LAPTg medium at a final OD600 of 0.05. After 24 hr, we measured the OD600 using a spectrophotometer. The black bar shows the static condition, and the gray bar shows the shaking condition. (LAPTg medium has no ability to consume H2O2.) The data are shown as the mean ± SE of three independent experiments. asterisk (*) Student’s t-test; p<0.05. (B) Growth rates of Δnpr and multiple deficient mutants under static and shaking conditions. Strains precultured overnight at 37°C were inoculated into fresh LAPTg medium at a final OD600 of 0.05. After 24 hours, we measured the OD600 using a spectrophotometer. The black bar shows the static condition, and the gray bar shows the shaking condition. (LAPTg medium has no ability to consume H2O2.) The data are shown as the mean ± SE of three independent experiments. asterisk (*) Student’s t-test; p<0.05.
Growth of the wild-type and Δnpr strains under the shaking condition and H2O2 concentrations in the culture
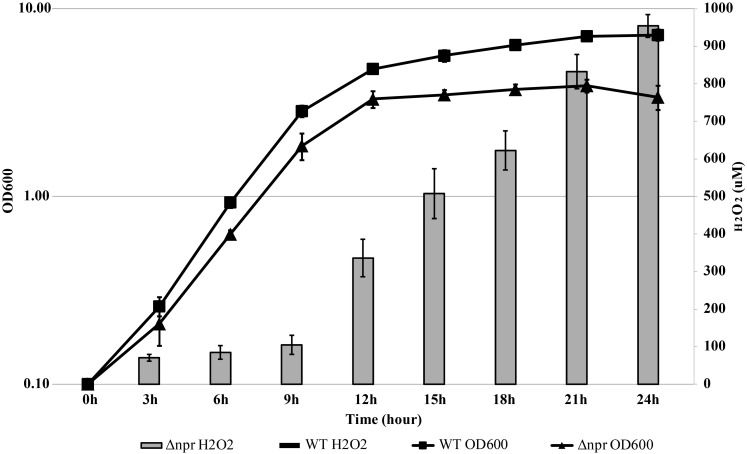
H2O2 concentrations in the MRS culture medium of wild-type and Δnpr cultures were measured over time under the shaking condition (Fig. 2). Although H2O2 was not detected in the wild-type culture, following overnight culture of Δnpr, approximately 1,000 µM H2O2 had accumulated. In addition, no accumulation of H2O2 was observed in either strain under the static condition (data not shown). These results suggest that L. casei IGM394 converts oxygen in its growth process under shaking and that the generated H2O2 is degraded by NADH peroxidase.
Fig. 2.
Growth and accumulated H2O2 concentration of wild type and Δnpr under the shaking condition.
Strains precultured overnight at 37°C were inoculated into fresh MRS medium at a final OD600 of 0.05. We measured the OD600 using a spectrophotometer every 3 hr. Subsequently, 1 mL of the culture was collected and centrifuged (10,000 g, 3 min), and 20 µL of the supernatant was used for measuring the H2O2 concentration. After measuring the wavelength at 727 nm, chromogenic reagent DA64 was used to quantify H2O2 based on the standard curve. The black square represents the OD value of wild type, and the black triangle represents that of Δnpr. The gray bar shows the concentration of H2O2 in the Δnpr culture. The data are shown as the mean ± SE of three independent experiments.
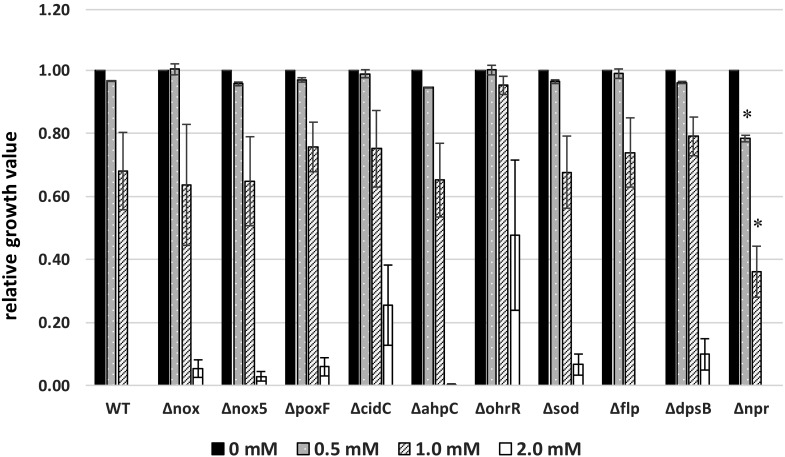
Growth suppression by H2O2
The effect of H2O2 on the growth of each mutant was evaluated under various H2O2 concentrations (0–2.0 mM) in MRS medium (Fig. 3). The suppressive effect of H2O2 on growth was concentration dependent. In particular, 0.5 mM H2O2 had little effect on growth, whereas 1.0 mM H2O2 reduced proliferation. Bacterial growth was completely inhibited by 2.0 mM H2O2. The influence of 0.5 and 1.0 mM H2O2 on the growth rates of nine mutants was similar to that observed in the wild-type strain. However, the growth rate of Δnpr was reduced by 0.5 mM H2O2, and the rate was significantly lower than that of the wild type in the presence of 1.0 mM H2O2. The growth of ΔohrR was not suppressed by 1.0 mM H2O2. In addition, ΔohrR also proliferated in the presence of 2.0 mM H2O2, and H2O2 resistance was improved by the deletion of ohrR. From these findings, it was presumed that ohrR is one of the tolerance mechanisms in L. casei IGM394.
Fig. 3.
Effects of adding H2O2 on the growth of the wild type and ten mutants.
Strains precultured at 37°C were inoculated into MRS medium at a final OD600 of 0.05, and these culture media were aliquoted (180 µL/well) to a 96-well plate. Twenty microliters of H2O2 solution was added to a final concentration of 0.5, 1.0, or 2.0 mM, and the plate was incubated at 37°C. After 24 hr, the OD600 was measured using a multiplate reader. Relative growth values were calculated using the following equation. The data are shown as the mean ± SE of three independent experiments.
Relative growth value = OD600 value at each H2O2 concentration/OD600 value at 0 mM H2O2.
*Student’s t-test; p<0.05 (for each wild type concentration).
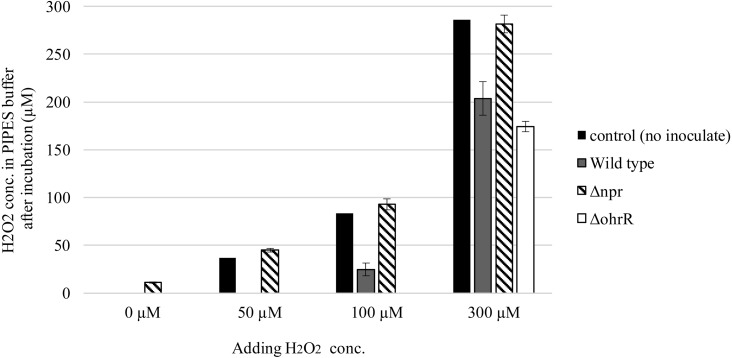
H2O2 consumption in PIPES buffer
H2O2 accumulation in the medium was estimated for 14 deficient mutants under the shaking condition, and H2O2 was only detected in the Δnpr culture. This illustrated that only Δnpr could not consume H2O2. H2O2 accumulation in the medium under the shaking condition was confirmed for Δnpr, and Δnpr could not consume H2O2 generated during the growth process (Fig. 4). Therefore, the ability to consume added H2O2 in PIPES buffer was measured. In this experiment, the wild-type, Δnpr, and ΔohrR strains were used after 5 hr of logarithmic growth. After the cells were exposed to 0, 50, 100, or 300 µM H2O2 for 1 hr under the static condition, H2O2 concentrations in PIPES buffer were measured. On average, the H2O2 concentration was decreased by 84% compared with the initial concentration in the presence of wild-type cells. The wild-type strain decreased the supplemented H2O2 concentration from 50 to 0 µM, from 100 to 25 µM, and from 300 to 204 µM. The Δnpr strain could not consume H2O2 efficiently. The H2O2 concentration for the Δnpr strain after 1 hr of incubation was similar to or slightly higher than the control level. In ΔohrR culture buffer, H2O2 could not be detected after adding 50 or 100 µM H2O2. The ΔohrR strain completely consumed 100 µM H2O2 and decreased the H2O2 concentration from 300 to 174 µM. This indicated that the H2O2 consumption ability of the ΔohrR strain was greater than that of the wild-type strain.
Fig. 4.
H2O2 concentration of wild type and Δnpr in PIPES buffer.
Strains precultured overnight at 37°C were inoculated into fresh MRS medium at a final OD600 of 0.05. The cells were used after static culture at 37°C for 5 hr. They were washed twice with PIPES buffer (pH 6.8) and resuspended in 10 mL H2O2 adjusted to 0 to 300 µM with PIPES buffer. After incubation at 37°C for 2 hr, the cells were harvested by centrifugation (10,000 g, 3 min). Twenty microliters of the supernatant was used for measuring the H2O2 concentration. After measuring the wavelength at 727 nm, the chromogenic reagent DA64 was used to quantify H2O2 based on the standard curve. The data are shown as the mean ± SE of three independent experiments.
Gene expression analysis of NADH peroxidase via quantitative real-time PCR
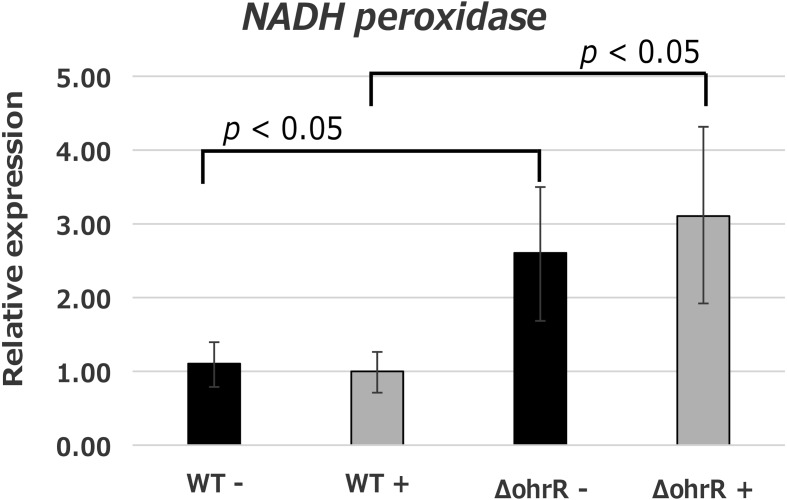
The aforementioned results revealed that H2O2 consumption was mainly performed by Npr and that deletion of ohrR eliminated the growth-suppressing effects of H2O2 in L. casei IGM394. It was presumed that OhrR was involved in H2O2 consumption; therefore, we examined the expression level of npr in the wild-type and ΔohrR strains via quantitative real-time PCR. In addition, we observed that the expression level changed depending on the presence or absence of H2O2. In this experiment, the treatment conditions were exposure to H2O2 adjusted to 0.5 mM with PIPES buffer for 1 hr at 37°C. In the wild-type strain, the expression level of npr was constant regardless of the presence of H2O2. However, the expression level of npr in the ΔohrR strain was 2.5-fold higher in the absence of H2O2 and 3-fold higher than that of the wild-type strain in the presence of H2O2.
DISCUSSION
Aerobic organisms have various tolerance mechanisms against oxygen and ROS. Lactic acid bacteria, which are facultative anaerobes, do not require oxygen to grow, but they can grow in the presence of oxygen. Several factors have been reported to be involved in oxidative stress tolerance, but the mechanisms differ by species and strain. The similar growth of L. casei IGM394 (wild type) under static and shaking conditions observed in this study indicated that this strain has multiple mechanisms to respond to oxidative stress.
SOD, which converts highly toxic superoxide substrates into H2O2, is important in the mechanism of oxidative tolerance. Serata et al. reported that sod of L. casei Shirota was transcribed but that its protein was inactive, and they reported that superoxide was eliminated via the intracellular accumulation of Mn2+ [34]. The possibility that L. casei IGM394 has the same Mn2+ accumulation mechanism as L. casei Shirota could explain why sod disruption did not affect the growth rate of the former bacterium (Fig. 1A).
Higuchi et al. reported that AhpC degrades H2O2 into water and that H2O2 is produced by Nox as a byproduct of oxygen consumption in S. mutans [3]. L. casei IGM394 expresses Nox5, which produces H2O2 in a manner similar to that observed in S. mutans. However, H2O2 was not detected in the culture medium of ΔahpC under shaking. It is predicted that L. casei IGM394 carries a number of enzymes for degrading H2O2, such as NADH peroxidase, and that these enzymes complement the function of AhpC to degrade H2O2 under shaking.
There are reports that Fnr-like protein (Flp) is a potential sensor protein and regulator, although the genes it regulates in Lc. lactis and L. casei remain to be clarified [35, 36]. When Flp is oxidized, an intramolecular disulfide bond is formed, thereby conferring the ability to bind to the promoter region. Although double deletion of flpA and flpB leads to hypersensitivity to H2O2 in Lc. lactis ssp. cremoris MG1363 [37], Δflp of L. casei IGM394 exhibited the same growth rate as the wild type under static and shaking conditions (Fig. 1A) or in the presence of 2 mM H2O2. It is unclear whether the different responses of the two strains are due to different functions of Flp. The DNA-binding protein Dps is an H2O2 resistance factor in E. coli that has been identified as a nonspecific DNA-binding protein accumulated in stationary cells [38]. It has been reported that Dps forms a ferritin-like complex, binds to DNA to form an extremely stable complex, and protects DNA against H2O2 [39, 40]. The suf cluster may participate in Fe-S cluster assembly or repair. Under oxidative stress, OxyR (regulator protein) activates the expression of the suf cluster in E. coli [41]. Cytochrome bd oxidase (CydAB) is the terminal electron acceptor that finally reduces oxygen to water. In all lactic acid bacteria, cydAB is clustered [42, 43]. GshR is one of the enzymes constituting the glutathione-ascorbic acid cycle, a metabolic pathway that detoxifies H2O2 generated in the process of metabolism. Yamamoto et al. reported that GshR may be important in protecting S. mutans against oxidative stress [44, 45]. Despite reports of their involvement in oxidative stress resistance, deletion mutants of these genes (ΔdpsB, Δsuf, ΔcydAB, ΔgshR) had similar growth rates as the wild-type strain under static and shaking conditions. It is presumed that the mechanisms of oxygen consumption or tolerance were complemented by other mechanisms in L. casei IGM394.It should be noted, however, that we predicted the presence of genes that are essential for growth under oxidative stress conditions but are not complemented by other mechanisms. In addition, we tried to disrupt trxB2, which plays a significant role in cellular redox processes, including protein repair and defense against oxidative stress, but these efforts were unsuccessful. Serata et al. succeeded in constructing a trxB2-deficient mutant in L. casei Shirota, and the ability of this strain to grow under aerobic conditions was significantly reduced. This suggested that TrxB2 is an important enzyme for oxygen tolerance in L. casei Shirota [13]. Our different findings may be due to the fact that we did not perform experimentation under an anaerobic condition.
Our finding that only Δnox exhibited decreased growth versus the wild type indicated that Nox may have different functions than other oxygen-consuming enzymes. One reason for this is that Nox converts oxygen to water without producing H2O2. Other oxygen-consuming enzymes, such as NADH oxidase (Nox5, H2O2-forming) and pyruvate oxidase (PoxF, CidC), convert oxygen to H2O2. It is considered that the oxygen consumption function depends on the H2O2-generating enzyme following the deletion of Nox, and that the influence of H2O2 or ROS produced by the Fenton reaction explains the decreased growth rate of Δnox. One other reason is that Nox works to maintain the redox potential in cells. Futhermore, the increased growth rates observed for Δnox5, ΔpoxF, and ΔcidC suggested that the decreased production of H2O2 by these proteins eased the stress on cells.
Δnpr, in which the NADH peroxidase gene was disrupted, only displayed decreased growth under the shaking condition relative to the wild type. As NADH peroxidase is an H2O2-degrading enzyme, it was considered that Δnpr could not degrade the H2O2 generated as a byproduct of oxygen consumption under the shaking condition. A high concentration of H2O2 was detected in the Δnpr culture under the shaking condition. The H2O2 concentration increased with the incubation time and reached about 500 µM after 15 hr and about 950 µM after 24 hr. However, in cultures of the wild-type and all other deficient mutant strains, H2O2 was not detected under either condition (Fig. 2). This revealed that the loss of H2O2 degradation could not be compensated for by other genes in L. casei IGM394. Other mutants featuring deficiencies of multiple genes displayed no changes in phenotype under shaking. From this finding, it was suggested that the oxidative stress tolerance mechanisms of L. casei IGM394 are multiple and diverse, and thus, no effect on growth was observed because missing functions could be complemented by other genes. However, the data indicated that H2O2 consumption is critical for the oxidative stress tolerance mechanism in this strain because decreased growth under the shaking condition was only observed for Δnpr. As lactic acid bacteria cannot synthesize heme, there is no catalase-based H2O2 degradation system. Previous studies reported that L. plantarum carries manganese catalase, which uses manganese as a cofactor [46], and that L. sakei synthesizes heme catalase when heme is added to the medium [47]. Genomic data revealed that L. casei IGM394 possesses four peroxidases: NADH peroxidase, iron-dependent peroxidase, glutathione peroxidase, and thiol peroxidase; however, the bacterium does not carry manganese catalase. Our findings revealed that peroxidases other than NADH peroxidase cannot efficiently degrade H2O2.
Interestingly, ΔohrR, which is a deficient mutant of the transcriptional regulator gene (ohrR), showed strong resistance to H2O2 (Fig. 3). The ΔohrR could grow under 0.5 mM and 1.0 mM H2O2 supplemented conditions as well as 0 mM supplemented conditions. Furthermore, ΔohrR could grow under even 2.0 mM supplemented conditions, that is, conditions in which wild type could not grow. OhrR is a transcriptional repressor of organic hydroperoxide resistance protein (OhrA). As the ohrR disruption resulted in the constitutive expression of the OhrA protein, it induced strong resistance to H2O2 in ΔohrR. The OhrR gene was first identified in Xanthomonas campestris [20] and subsequently reported in many gram-negative bacteria. In gram-positive bacteria, the Ohr family was reported to be involved in resistance to organic peroxide and H2O2 in Bacillus subtilis [21], and OhrA overexpression induced H2O2 tolerance. In B. subtilis, OhrR repressed ohrA expression by binding to the inverted repeat (IR) sequence (TACAATT-N-AATTGTA) presented upstream of ohrA. However, there is no detailed report on the Ohr family in lactic acid bacteria, and a similar IR sequence upstream of ohrA was not detected in L. casei IGM394. Our results suggested that deletion of ohrR induced greater H2O2 resistance, and these effects appear to be related to the constitutive expression of ohrA. Meanwhile, deletion of ohrR induced higher expression of NADH peroxidase (Fig. 5). In L. casei IGM394, ohrA expression might be regulated by the recognition of different IR sequences or a different mechanism from that observed for ohrR in Bacillus. The association between the constitutive expression of ohrA and the higher expression of NADH peroxidase is unclear at present. However, it was suggested that the OhrR protein participates in the mechanisms combating oxygen and ROS in lactic acid bacteria. Analyses of ohrA expression control and function will be required in the future.
Fig. 5.
NADH peroxidase expression level with and without H2O2 in the wild type and ΔohrR.
Target gene: NADH peroxidase gene (npr).
Housekeeping gene: elongation factor Tu gene and glyceraldehyde 3-phosphate dehydrogenase gene.
The relative expression levels were calculated using 2 housekeeping genes. Black bars represent untreated; gray bars represent H2O2 treated. The data are shown as the mean ± SE of three independent experiments.
The findings of decreased growth under the shaking condition and the loss of H2O2 consumption following disruption of npr were similar to those reported for L. casei Shirota [16]. Although npx expression was increased by approximately 10-fold in L. casei Shirota in response to H2O2 exposure according to quantitative real-time PCR, the expression level of npr in L. casei IGM394 was constant under the shaking condition. Serata et al. reported that L. casei Shirota exhibited the ability to consume H2O2 only after 1 hr of pretreatment with 0.5 mM H2O2 added to the culture medium. However, L. casei IGM394 could consume H2O2 without this pretreatment. This difference in the regulation of H2O2 consumption remains to be clarified.
According to our study, L. casei IGM394 has multiple oxygen consumption mechanisms, and disruption of a single gene is not sufficient to eliminate the ability to consume oxygen or alter growth. It is presumed that NADH oxidase efficiently converts oxygen to water in the wild-type strain, and thus, multiple H2O2 consumption mechanisms may not be necessary. NADH peroxidase plays a key role in H2O2 consumption, and other genes could not compensate for its function. Thus, it was concluded that the NADH peroxidase has a critical role in the oxidative stress response mechanisms in L. casei IGM394.
REFERENCES
- 1.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77: 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol 139: 2343–2351. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, Takahashi N, Kamio Y. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol 181: 5940–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorquet F, Goffin P, Muscariello L, Baudry JB, Ladero V, Sacco M, Kleerebezem M, Hols P. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J Bacteriol 186: 3749–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedewitz B, Schleifer KH, Götz F. 1984. Physiological role of pyruvate oxidase in the aerobic metabolism of Lactobacillus plantarum. J Bacteriol 160: 462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki Y, Horiuchi H, Kawashima H, Mukai T, Yamamoto Y. 2014. NADH Oxidase of Streptococcus thermophilus 1131 is required for the effective yogurt fermentation with Lactobacillus delbrueckii subsp. bulgaricus 2038. Biosci Microbiota Food Health 33: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patton TG, Rice KC, Foster MK, Bayles KW. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol Microbiol 56: 1664–1674. [DOI] [PubMed] [Google Scholar]
- 8.Chang SK, Hassan HM. 1997. Characterization of superoxide dismutase in Streptococcus thermophilus. Appl Environ Microbiol 63: 3732–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders JW, Leenhouts KJ, Haandrikman AJ, Venema G, Kok J. 1995. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol 177: 5254–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi A, Rochat T, Gratadoux JJ, Le Loir Y, Oliveira SC, Langella P, Azevedo V. 2003. Oxidative stress in Lactococcus lactis. Genet Mol Res 2: 348–359. [PubMed] [Google Scholar]
- 11.Vido K, Diemer H, Van Dorsselaer A, Leize E, Juillard V, Gruss A, Gaudu P. 2005. Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J Bacteriol 187: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrano LM, Molenaar D, Wels M, Teusink B, Bron PA, de Vos WM, Smid EJ. 2007. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb Cell Fact 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serata M, Iino T, Yasuda E, Sako T. 2012. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology 158: 953–962. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren A. 1985. Thioredoxin. Annu Rev Biochem 54: 237–271. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol 182: 3740–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. 2000. Identification of a new gene responsible for the oxygen tolerance in aerobic life of Streptococcus mutans. Biosci Biotechnol Biochem 64: 1106–1109. [DOI] [PubMed] [Google Scholar]
- 17.Serata M, Kiwaki M, Iino T. 2016. Functional analysis of a novel hydrogen peroxide resistance gene in Lactobacillus casei strain Shirota. Microbiology 162: 1885–1894. [DOI] [PubMed] [Google Scholar]
- 18.Brooijmans RJ, de Vos WM, Hugenholtz J. 2009. Lactobacillus plantarum WCFS1 electron transport chains. Appl Environ Microbiol 75: 3580–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sijpesteijn AK. 1970. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie van Leeuwenhoek 36: 335–348. [DOI] [PubMed] [Google Scholar]
- 20.Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C. 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol 182: 3863–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol 180: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol 183: 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA 103: 15611–15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 1: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazé A, Boël G, Zúñiga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casarégola S, Auffray Y, Pérez-Martínez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol 192: 2647–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Yu D, Sun Z, Wu R, Chen X, Chen W, Meng H, Hu S, Zhang H. 2010. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J Bacteriol 192: 5268–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ai L, Chen C, Zhou F, Wang L, Zhang H, Chen W, Guo B. 2011. Complete genome sequence of the probiotic strain Lactobacillus casei BD-II. J Bacteriol 193: 3160–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Ai L, Zhou F, Wang L, Zhang H, Chen W, Guo B. 2011. Complete genome sequence of the probiotic bacterium Lactobacillus casei LC2W. J Bacteriol 193: 3419–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochwind K, Weinmaier T, Schmid M, van Hemert S, Hartmann A, Rattei T, Rothballer M. 2012. Draft genome sequence of Lactobacillus casei W56. J Bacteriol 194: 6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koryszewska-Baginska A, Aleksandrzak-Piekarczyk T, Bardowski J. 2013. Complete genome sequence of the probiotic strain Lactobacillus casei (formerly Lactobacillus paracasei) LOCK919. Genome Announc 1: e00758–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Zhu H, He F, Luo Y, Kang Z, Lu C, Feng L, Lu X, Xue Y, Wang H. 2014. Whole genome sequence of the probiotic strain Lactobacillus paracasei N1115, isolated from traditional Chinese fermented milk. Genome Announc 2: e00059–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajikawa A, Igimi S. 2011. Development of recombinant vaccines in lactobacilli for elimination of Salmonella. Biosci Microflora 30: 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu A, Igimi S, Kawana K. 2018. Optimization of human papillomavirus (HPV) type 16 E7-expressing lactobacillus-based vaccine for induction of mucosal E7-specific IFNγ-producing cells. Vaccine 36: 3423–3426. [DOI] [PubMed] [Google Scholar]
- 34.Serata M, Yasuda E, Sako T. 2018. Effect of superoxide dismutase and manganese on superoxide tolerance in Lactobacillus casei strain Shirota and analysis of multiple manganese transporters. Biosci Microbiota Food Health 37: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott C, Guest JR, Green J. 2000. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol Microbiol 35: 1383–1393. [DOI] [PubMed] [Google Scholar]
- 36.Gostick DO, Green J, Irvine AS, Gasson MJ, Guest JR. 1998. A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology 144: 705–717. [DOI] [PubMed] [Google Scholar]
- 37.Scott C, Rawsthorne H, Upadhyay M, Shearman CA, Gasson MJ, Guest JR, Green J. 2000. Zinc uptake, oxidative stress and the FNR-like proteins of Lactococcus lactis. FEMS Microbiol Lett 192: 85–89. [DOI] [PubMed] [Google Scholar]
- 38.Almirón M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6 12B: 2646–2654. [DOI] [PubMed] [Google Scholar]
- 39.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol 5: 294–303. [DOI] [PubMed] [Google Scholar]
- 40.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. 1999. DNA protection by stress-induced biocrystallization. Nature 400: 83–85. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giuffrè A, Borisov VB, Arese M, Sarti P, Forte E. 2014. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta 1837: 1178–1187. [DOI] [PubMed] [Google Scholar]
- 43.Cesslein B, Derrē-Bobillot A, Fernandez A, Lamberet G, Lechardeur D, Yamamoto Y, Pedersen MB, Garrigues C, Gaudu P. 2010. Respiration, a strategy to avoid oxidative stress in Lactococcus lactis, is regulated by the heme status. Lactic Acid Bacteria 21: 10–15. [Google Scholar]
- 44.Sherrill C, Fahey RC. 1998. Import and metabolism of glutathione by Streptococcus mutans. J Bacteriol 180: 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto Y, Kamio Y, Higuchi M. 1999. Cloning, nucleotide sequence, and disruption of Streptococcus mutans glutathione reductase gene (gor). Biosci Biotechnol Biochem 63: 1056–1062. [DOI] [PubMed] [Google Scholar]
- 46.Barynin VV, Whittaker MM, Antonyuk SV, Lamzin VS, Harrison PM, Artymiuk PJ, Whittaker JW. 2001. Crystal structure of manganese catalase from Lactobacillus plantarum. Structure 9: 725–738. [DOI] [PubMed] [Google Scholar]
- 47.Hertel C, Schmidt G, Fischer M, Oellers K, Hammes WP. 1998. Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sakei LTH677. Appl Environ Microbiol 64: 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]