Abstract
The human body contains many microorganisms, including a large number of bacteria, viruses, fungi, and protozoa, which are referred to as the microbiota. Compared with the number of cells comprising the human body, that of the microbiota has been found to be much larger. The microbiome is defined as microorganisms and their genomes have been shown to contain about 100 times more genes than the human genome. The microbiota affects many vital functions in the human body. It contributes to regulation of the immune system, digestion of food, production of vitamins such as B12 and K, metabolization of xenobiotic materials, and many other tasks. Many factors affect the microbiota biodiversity, such as diet, medicines including antibiotics, relationships with the environment, pregnancy, and age. Studies have shown that the lack of microbiota diversity leads to many diseases like autoimmune diseases such as diabetes type I, rheumatism, muscular dystrophy, problems in blood coagulation due to lack of vitamin K, and disturbances in the transfer of nerve cells due to lack of vitamin B12, in addition to its involvement in a number of conditions such as cancer, memory disorders, depression, stress, autism, and Alzheimer’s disease. The aim of this review is to summarize the latest studies discussing the relationship between the microbiota and the human body in health and diseases.
Keywords: gut microbiota, dysbiosis, immune system, infectious diseases, probiotic, metabolic disorder
INTRODUCTION
All plants and animals, including humans contain many microorganisms, which invoke many relationships with the host, such as symbiosis, commensalism, and parasitism [1, 2]. The cells of the human body, 90% of which are alien to it [3], are referred to as the microbiota or microbiome. The microbiota is an assemblage of microorganisms that form an ecological community in a specific area. The first scientists to mention this term were Lederberg and McCray when they referred to the role of microorganisms in human health and diseases [1]. The term microbiome, on the other hand, focuses on the genomes of all microorganisms in a specific environment nook [4]. Prebiotics are selectively fermented components that lead to specific changes in the composition and/or activity of the intestinal microbiota, thereby providing benefits to host health [3]. The word probiotic came from the Latin word pro (meaning for) and Greek word biōtikós (meaning of life) Probiotics are live, beneficial, nonpathogenic bacteria such as Lactobacillus and Bifidobacterium and yeasts like Saccharomyces that provide health benefits to the host when administered in sufficient amounts [5]. On the other hand, prebiotics are selectively fermented components that lead to specific changes in the composition and/or activity of the intestinal microbiota, thereby providing benefits to host health [6].
There are more than 100 trillion microorganisms in the human gut alone, and they have 150-times more genes than the entire human genome [7]. The development of molecular methods that rely on 16S rRNA, 18S rRNA, and other marker genes has helped in determining of microbes found in a specific area. These methods have opened the doors to studying and clarifying the roles of microorganisms in the human body [8].
The publication of the human genome sequence in 2003 is considered a remarkable biological achievement. However, this achievement is considered incomplete because of the impact of a large number of microbes on the human body and its genes, and this impact is still not understood. So, the “Human Microbiome Project” was established to study the microbiome in the skin, vagina, mouth, and gut by random shotgun sequencing procedures that targeted large-insert clone sequencing and by using high-density microarrays. These methods gave great insight into the role of the microbiota in health and diseases [9].
Recently, many studies have demonstrated the important role of the human gut microbiota in boosting the ability to extract energy from food, in increasing the harvest of nutrients [10], in changing the appetite signal [11], in producing vitamins [12], and in the ability to metabolize many materials including xenobiotics [13] because it contains varied, unique, and specific enzymes and has miscellaneous biochemical pathways [7]. The gut microbiota is involved in many basic biological processes, including regulation of epithelial development, modulation of the metabolic phenotype, and stimulation of innate immunity [3]. In addition, the microbiota protects the body from external pathogens through competitive colonization or production of antimicrobial agents like bacteriocins that kill pathogens [14].
The host’s genes and lifestyle, type of food, and consumed drugs and antibiotics have an impact on the microbiota, which affects the health of the host by modifying physiological systems like immune system development, secretions of the endocrine, metabolism regulation, or even genes within the host’s genome [4]. Studies have proven the existence of four dominant phyla in the gut microbiota, with Firmicutes and Bacteroidetes accounting for 90% of the total population and Actinobacteria and Proteobacteria accounting for less than 1–5% [4, 15]. Alteration of this balance is called dysbiosis. Gut microbiota dysbiosis leads to many diseases [16], like auto-immunity diseases such as asthma and arthritis [4], chronic diseases such as inflammatory bowel disease (IBD), and metabolic and cardiovascular diseases like obesity, diabetes, atherosclerosis [7], and liver diseases [8]. The consequences of microbiota dysbiosis can extend to as far as cancer and psychological diseases like depression, anxiety, autism, and Alzheimer’s disease [17].
In this review, we will discuss the role of the human gut microbiota in health and disease and the consequences of gut microbiota dysbiosis in human growth, the immune system, exposure to a xenobiotic, metabolic disorders, and psychiatric diseases.
THE MICROBIOTA AND HUMAN GROWTH
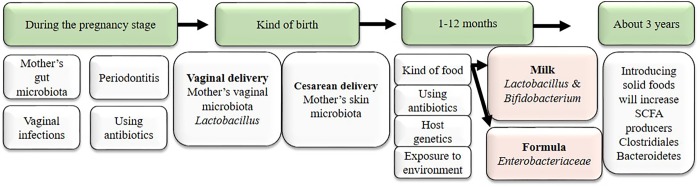
The belief that the fetal gastrointestinal environment is sterile comes from the hypothesis that the placenta barrier protects the fetus from any microbes that would threaten its life. The defense for this hypothesis is based on the fact that the existence of any microbes in the uterus was considered to be a potential risk for the fetus and to be associated with premature birth and fetal abnormalities [18]. On the other hand, some studies have mentioned the existence of commensal microbiota in healthy pregnancy in both placental tissues and amniotic fluid. Further, they found them to be similar to mouth clusters. These studies proved the existence of Fusobacteria, Tenericutes, Firmicutes, Bacteroidetes, and Proteobacteria in the placenta [19] and the existence of Streptococcus, Enterococcus, and Staphylococcus in umbilical cord blood [19]. Therefore, the idea that microbiota colonize the human body immediately after birth is not considered correct anymore [18]. The microbiota acquired in early life affects the development of the immune system and its responsibility for health or development of diseases in later life [18]. Many studies have shown that microbiota dysbiosis in infants leads to many diseases, such as lung diseases, asthma [20], food allergy [18], diabetes, obesity, atopic diseases, Crohn’s disease, and autoimmune diseases [7]. For example, lack of exposure to microorganisms in early life leads to allergies. In fact, one previous study showed the difference between children who grow up in natural environments and those who grow up in urban areas. Children from natural environments have fewer allergy symptoms than children who grow up in urban areas due to the lower microbiota diversity in urban areas [4]. Many factors affect the formation of the infant microbiota, such as pregnancy, kind of birth, and feeding mode. Kind of birth plays an important role in the composition of the microbiota that settles in the infant. In vaginal delivery, vaginal microbes like Lactobacillus spp, Bifidobacterium, and Prevotella colonize the child [21], whereas newborns delivered by Cesarean section are colonized by microbiota of the mother’s skin, Corynebacterium, Staphylococcus, and Propionibacterium spp. [21]. In addition, feeding mode has an important role in forming a robust gut microbiota in infants. Breast milk contains more than 700 kinds of bacteria [22] and oligosaccharides [23] that reinforce some specific bacteria like bifidobacteria [22], immunoglobulins like IgG and IgA, and cytokines like TGF-β and interleukin 10 (IL-10), and the combined effects of these factors affect the selection of the bacteria that will colonize the gastrointestinal tract in infants that depend on breast milk [24].
Weaning and introduction of solid food increase gut microbiota diversity and enhance species that produce butyrate, like Clostridium species [25], Prevotella, and Ruminococcus [26]. By three years of age, the gut microbiota composition has become similar to that of an adult (Fig. 1) [24].
Fig. 1.
Factors influencing the pediatric microbiota up to 3 years of age.
During human growth the immune system may need training by good microbiota to do its job in regular manner, and a lack of training may lead to dysregulation and weak tolerance towards noncommunicable diseases [4].
ROLE OF THE MICROBIOTA IN IMMUNITY
The microbiota plays a key role in the training and induction of the immune system. When the system works optimally, it stimulates an immune response to pathogens and tolerates non-harmful antigens [3]. The immune system consists of a complex network (innate and adaptive) which has the ability to adapt and respond to many challenges and works to preserve tissue and restore it in the context of microbial encounters [3]. The immune system mechanism that is used to maintain the relationship with the microbiota is similar to that used to restrict microorganism-causing pathogens [27] for many diseases that affect humans, such as allergies, autoimmune diseases, and infections. These diseases arise from failure of a proper immune response against autoantigens, microbes, or environment-derived antigens [3]. Host strategies are adapted to maintain a balance with microorganisms in order to reduce communication between microorganisms and the surfaces of epithelial cells, thus reducing inflammation and preventing the spread of bacteria throughout the body. So, there is an abundance of immune cells in areas of direct contact with microorganisms, such as the skin and small intestine [3].
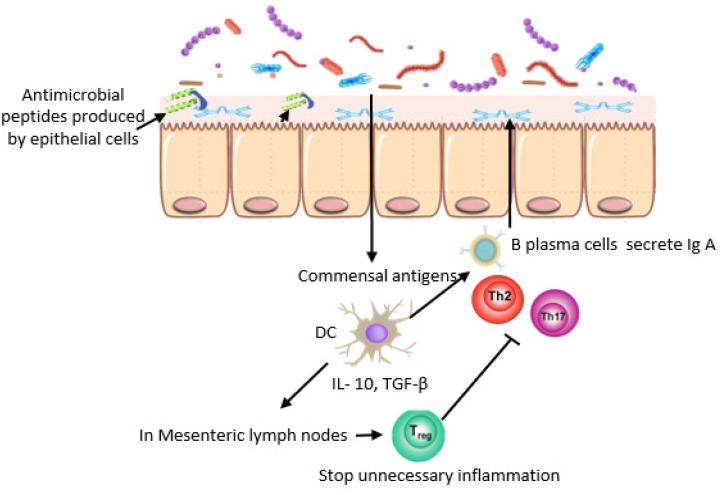
In the human intestine, there is massive synergy between mucus, antimicrobial peptides, immune cells, and IgA, which are referred to as the “mucosal firewall”. This synergy plays an important role in preventing pathogens from crossing the lamina and stimulating inflammation [28], thus maintaining intestinal homeostasis [29]. Goblet cells secrete mucin glycoproteins that form a mucus layer with a thickness of 150 μm to prevent direct contact between the microbiota and host tissue. While epithelial cells produce antimicrobial peptides (α-defensins, cathelicidins, and C-type lectins) that kill bacteria by attacking their cell walls or by disrupting their inner membranes [30], the expression of some antimicrobial peptides, like RegIIIγ, which is the best antimicrobial peptide expressed immediately after birth or after microbes are given to experimental animals, requires bacterial signals [31]. As for the others, most α-defensins do not require signals like this [29]. The accumulation of antimicrobials in the mucus creates a physical barrier between the microbiota and epithelial cells, which is referred to as the “demilitarized zone” [27], while dendritic cells (DCs) that are located under the Peyer’s patches and in the lamina propria play an important role in the immune response. DCs reduce unnecessary inflammation by promoting Treg that is transported to the lamina propria and secretes Il-10, which suppresses unnecessary inflammation by inducing B cells to differentiate into IgA+ plasma cells in the mesenteric lymph node. IgA+ plasma cells are transported to the lamina propria and secrete IgA, which is transported across the epithelial cell via transcytosis and prevents bacterial penetration (Fig. 2) [3, 30]. In inflammation, DCs produce pro-inflammatory cytokines such as Il-6, Il-12, and Il-23 that lead to the reduction of Il-10 and increase inflammatory cytokines like TNF α, INF γ, and Il-17 [32].
Fig. 2.
Immune system mechanisms that maintain intestinal homeostasis.
Lifestyle, diet, aging, and intake of antibiotics change the gut microbiota, which alters intestinal homeostasis and leads to many diseases, such as rheumatism, diabetes type II, obesity, and autoimmune diseases [32].
THE MICROBIOTA AND XENOBIOTIC MATERIALS
The increased use of industrial chemicals in various industries and agricultural applications (insecticides, herbicides, and fertilizers) [33], in addition to the increased intake of pharmaceuticals that contain large numbers of xenobiotic substances, have coincided with concern among health researchers about how xenobiotic substances are metabolized and what their outcomes are in the human body [34].
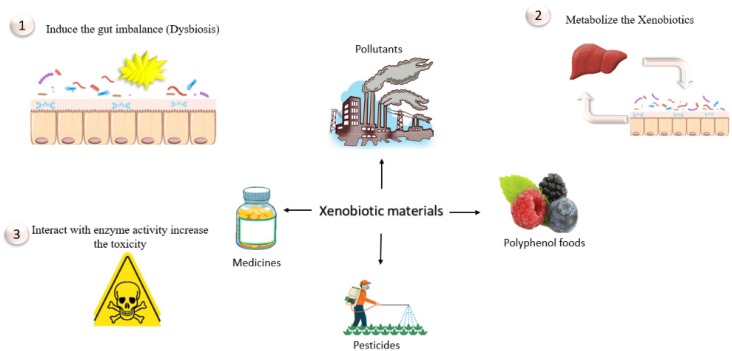
It is known that hepatic enzymes can take apart foreign substances [13]. These substances are modified either by mitigating their toxicity or by facilitating their ejection from the body [34]. On another hand, research suggests that modification of some substances increases their toxicity [35]. These substances are modified either by oxidation (cytochrome P450 monooxygenases), reduction (cytochrome P450 reductases), or hydrolysis (esterases and epoxide hydrolases) [36]. It has long been known that the intestinal microbiota has the ability to break up 40 substances [37]. Recent studies suggest that the intestinal microbiota plays an important role in metabolizing xenobiotic materials because of its ability to produce enzymes that have the ability to disassemble or neutralize many substances [38]. Therefore, the lack of bacteria capable of removing exposed xenobiotic materials may be attributed to many diseases that affected human health [13]. The role of the gut microbiota was clearly observed in low-molecular-weight substances (less than 325 kDa). These substances poorly react with bile. Thus, the gut microbiota metabolizes these substances to nonpolar materials, which are substances modified either by oxidation (cytochrome P450 monooxygenases) or reduction (through reabsorption) and return to the liver by a process called enterohepatic circulation (Fig. 3) [13].
Fig. 3.
Interaction between xenobiotic materials and the gut microbiota.
Polycyclic aromatic hydrocarbons (PAHs) are produced from incomplete combustible organic substances. The largest sources of them are inhalation of air from vehicle exhausts and tobacco smoke, in addition to some foods, such as grilled and smoked meats. The gut microbiota has the ability to metabolize PAHs and even raise their toxicity. The toxicity of these substances varies depending on their final structure. Whereas some of them possess estrogenic properties, others have carcinogenic properties. Exposure to PAHs increases the risk of lung and bladder cancer [39]. The gut microbiota also plays an important role in increasing the toxicity of melamine, metals, and organic pollutants [10]. Furthermore, the consumption of artificial sweeteners (AS) has the ability to change the gut microbial diversity, which can lead to development of metabolic disorders. Sucralose is one of the most common AS used, and it increases anaerobic bacteria species while encouraging decrease of beneficial ones like bifidobacteria and lactobacilli in both experimental animals and humans [40].
THE MICROBIOTA AND ANTIBIOTICS
Antibiotics play a very important role in the microbiota dysbiosis. Antibiotics not only affect pathogenic bacteria but also have an impact on beneficial bacteria, which leads to biological imbalances that lead to a number of diseases, like obesity, diabetes type II, asthma, and Crohn’s disease [41]. For example, use of broad-spectrum antibiotics is the main cause of diarrhea caused by opportunistic infection with Clostridium difficile [4]. In addition, the intestinal environment provides a suitable environment for the horizontal transfer of multiple resistance genes, which increases the risk of indiscriminate use of antibiotics [4]. Moreover, antibiotic consumption, especially in early life, has negative consequences because this phase is important to the formation of a healthy microbial community. Studies on infants have shown that the use of antibiotics or even exposure to them during the intrauterine stage is linked to an increase in the Proteobacteria phylum, as it contains resistance genes for many antibiotics, and a decrease in bifidobacteria, which are considered beneficial bacteria [42]. Likewise, a decrease in Actinobacteria and increase in Bacteroidetes and Proteobacteria, in addition to an increase in resistance genes, were noted in a Finnish study of 1,000 children who used macrolides in their first year of life. This dysbiosis positively correlated with development of asthma or increased body mass in these children [43]. In the long-term, the study aimed to examine the effect of antibiotics on the gut microbiota by using clindamycin in healthy volunteers. A change in the gut microbiota was observed that persisted over two years after the course, in addition to loss of many species and increased expression of resistance genes for clindamycin in the gut bacteria [44].
On the other hand, microorganisms are short-lived, with many mutations affecting their genomes, making them more resilient in adaptation to parasites. Thus, they are faster to respond to parasites than their host, either through interference competition by producing toxins, or antibiotics; by parasitizing parasites, which is referred to as “hyperparasitism”; or through resource competition by competing for nutrients in order to kill parasites and prevent them from staying. Microbiota may indirectly induce the host to have a stronger immune response to destroy a parasite or even enhance the host’s endurance which reduces the host’s response toward the parasites and thus protects tissue from damage [45].
THE HUMAN MICROBIOTA IN INFECTIOUS DISEASES
Numerous studies have demonstrated the relationship between microbial dysbiosis and infectious diseases [46]. It is difficult for pathogens to penetrate the gut ecosystem, and this is referred to as “colonization resistance”. Thus, the absence of robust flora increases a pathogen’s chance to colonize and cause disease [47]. Intake of antibiotics is the main cause of dysbiosis, as mentioned above, and the most common example of this is false colitis caused by an increased rate of C. difficile in the intestine after use of antibiotics (ampicillin, clindamycin, and cephalosporin) [48]. Moreover, a number of studies have shown that the reduction of microbial diversity in the intestine caused by antibiotics leads to an increased ability of pathogens to seize the opportunity for colonization [46,47,48].
In a study aimed at investigating the role of the gut microbiota in suppressing the colonization of pathogens like Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa when introduced with food, van der Waaij et al. noted that when an antibiotic was being used, colonization resistance decreased, leading to colonization by these species and causing diseases [46]. Likewise, in a study by Hapfelmeier and Hardt in which streptomycin was administered to observe its role in Salmonella typhimurium infection, it was noted that streptomycin leads to a higher rate of acute infection than usual [49]. In the next two paragraphs, we will discuss the most important bacterial infections caused by gastrointestinal microbiota dysbiosis.
C. difficile infection
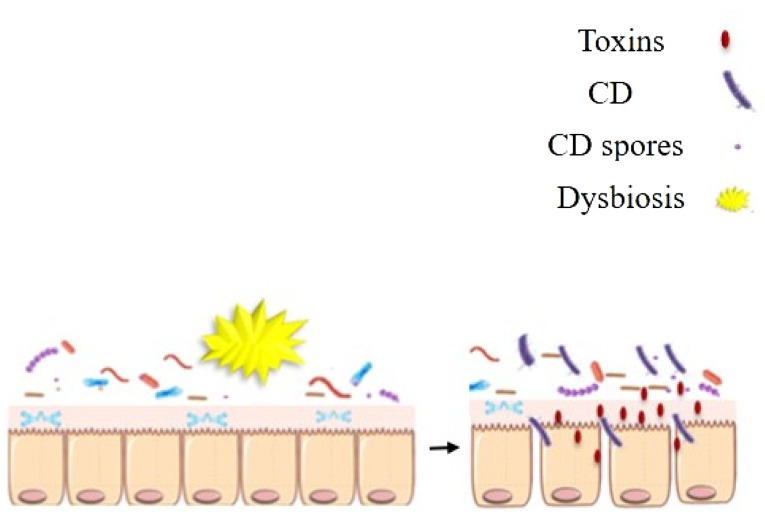
C. difficile (CD) is an anaerobic, gram-positive, spore-forming bacterium that colonizes the intestine. However, dysbiosis due to use of broad-spectrum antibiotics leads to nosocomial diarrhea caused by CD infection [48]. “Furthermore, antibiotics as well as several other factors, like age over 56, immunodeficiency, hospital residence, and some gastrointestinal issues, are associated with the risk of CD infection” [50]. Broad-spectrum antibiotics drain the variety of intestinal microbiota that inhibit the growth of CD. In addition, the gut environment under the influence of antibiotics acts to displace competitors, alter metabolites in the intestines, and decrease short-chain fatty acids such as butyrate, which is an energy source for epithelial cells [51]. CD takes advantage of the surrounding conditions to grow, reproduce, and secrete toxins [52]. Its pathogenicity starts by penetrating the mucus layer to reach the epithelial cells (Fig. 4). In that way, CD colonizes the gastrointestinal tract. Then it produces toxins like toxin A and toxin B that change the cytoskeleton of epithelial cells, as well as inhibit cell division and membrane transport that leads to induction of inflammation. The most common treatments for CD are vancomycin and metronidazole, although a number of patients develop recurrent illnesses [52]. The microbial diversity has a key role in preventing CD infection through fecal microbiota transplantation (FMT) from healthy donors to patients, and restoration of the intestinal microbial diversity has been shown to be effective in up to 95% of patients [53].
Fig. 4.
Gut dysbiosis allows C. difficile (CD) to grow and secrete toxins that destroy epithelial cells.
Helicobacter pylori infection
As mentioned above, gastrointestinal homeostasis is very important for elimination of pathogens, regulation of immune response, and even halting cancer development [54]. H. pylori is one of the very few microorganisms that have been proven to cause gastrointestinal cancer by inducing chronic inflammation [55, 56]. H. pylori is a microaerophilic, spiral, gram-negative bacterium first isolated by Warren and Marshall in 1982 [57] from a patient who was suffering from gastric ulcers [56]. Research has shown that H. pylori is almost everywhere and is found in about 50–90% of the population in developing countries, while in developed countries, it is found in less than 50% of the population due to the level of health awareness in these countries [55]. Although H. pylori is present in half of the human population, its means of transmission have not been definitively clarified to date. It is likely that transition from one person to another is greater among individuals in the same family in one of the assumed methods of transmission (gastro-oral transmission, oral-oral transmission, transmission by water and food, and fecal-oral transmission). Environmental factors, genetic predisposition of the host, as well as virulence factors like urease production, flagella, and adhesion factors contribute to colonization and induction of disease (Fig. 5) [55]. It was observed that inhibition of virulence factors prevents H. pylori from colonizing host cells. Thus, targeting virulence factors in the light of the spread of antibiotics resistance is a feasible of treating H. pylori [58, 59].
Fig. 5.
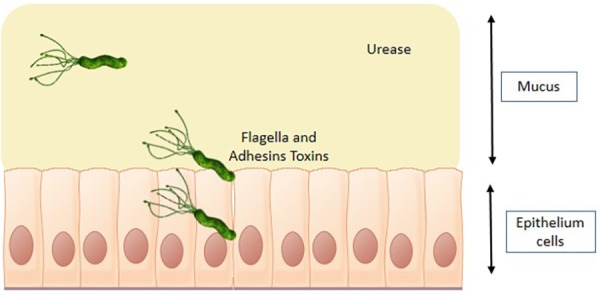
H. pylori virulence factors induce disease. Urease secretion reduces gastric acidity, and flagella and toxins help penetrate stomach epithelial cells and cause infection.
On the other hand, it was noted that H. pylori infection leads to a change in the stomach microbiota. In a study that included healthy volunteers and H. pylori patients, it was found that H. pylori DNA accounted for more than 90% of all sequence reads and that Proteobacteria, Firmicutes, and Actinobacteria accounted for very few reads [60], whereas the healthy stomach shows good biodiversity with respect to Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria and the Streptococcus genus regardless of acidic medium [61].
THE MICROBIOTA AND METABOLIC DISORDERS
The unique dynamic ecosystem of the gastrointestinal microbiota is regarded as a metabolically active “organ”. Thus, interchanges between microbiota and the immune system regulate gastrointestinal homeostasis [54]. However, loss of gastrointestinal homeostasis induces many diseases, such as obesity and type II diabetes.
The microbiota and obesity
Several factors are involved in the development of obesity, including drug interventions, genetic predisposition, hormonal disorders, lifestyle, as well as the role of the microbial gut [15]. Studies have shown the role of the gut microbiota in gaining weight and insulin resistance. It has been noted that germ-free animals do not gain weight despite high-carbohydrate and high-fat diets, while a weight gain of 60% was observed when microbiota were transplanted from other mice that showed an association between increased weight and the proportion of Firmicutes/Bacteroidetes. This means that weight gain does not depend on the amount of food eaten but instead depends on the quality of its metabolism [10]. It was also observed that lipopolysaccharide (LPS) inside the gut lumen of healthy weight people did not cross epithelial cells, whereas it did in obese people and stimulated inflammation [62]. This was attributed to consumption of a high-fat diet (HFD) that reduces proteins like claudin and zonula occludens-1 (ZO-1), which are responsible for the interconnection of epithelial cells [15]. In addition, an HFD plays an important role in the distribution of the gut microbiota. It was demonstrated that feeding normal-weight mice an HFD for 8 weeks led to an increase in Firmicutes and a reduction in Bacteroidetes [63]. Similarly, it was noted that genetically obese mice had more Firmicutes and fewer Bacteroidetes [11]. The production of more energy from food is linked to the presence of more Firmicutes and fewer Bacteroidetes, and this suggests the hypothesis that the microbiota of obese people are more efficient at obtaining energy from food than those of thin people [64].
On the other hand, we can consider the Bacteroidetes and bifidobacteria to be protectors against the development of obesity [65]. We have to be more wary of the development of obesity, especially in early life, because disruption of the gut microbiota may cause many diseases, such as obesity, types I and II diabetes, and cardiovascular diseases, at the adult stage [66].
The microbiota and diabetes
Studies have demonstrated that genetic factors, dietary pattern, and the gut microbiota all affect both type I and type II diabetes [67]. Type I diabetes (TID) is an autoimmune disease in which T cells destroy the pancreas’s beta cells, which produce insulin [68]. Type II diabetes (TIID) is known to reduce insulin sensitivity or insulin deficiency. Studies on interventions for the gut microbiota in subjects with metabolic diseases have increased in recent years, especially with respect to complications with TID and TIID. The gut microbiota and their metabolites play an important role in the development of these diseases [69]. A lot of studies have connected gut microbiota dysbiosis and diabetes and found a disruption in the rate of Firmicutes to Bacteroidetes in diabetic patients [68, 69]. There is a significant decrease in Faecalibacterium prausnitzii, which belongs to the Firmicutes phylum, in TIID patients [69]. On the other hand, a diabetic’s diet plays a role in encouraging symptoms of the disease, as mentioned above: an HFD increases inflammatory cytokines which disrupt insulin-producing beta cells in TID [67] and raise insulin resistance in TIID [68, 70].
An increase of Bacteroidetes (gram-negative bacteria) results in the production of propionate, succinate, and acetate from lactate that decomposes the myosin layer, leading to alteration of epithelial cell permeability and allowing pathogens to enter [68]. It also causes a reduction in bifidobacteria which produce butyrate from lactate, which is considered an anti-inflammatory substance, as well an enhancer of tight junctions between epithelial cells and an increaser of the efficiency of their mission [71, 72]. The other beneficial short-chain fatty acids resulting from bacterial metabolism link to GPR41 and GPR43 thus reduce the inflammation in immune cells, while the linking to L cells in the intestine increases peptide YY and the glucagon-like peptide GLP-1, which improve insulin sensitivity (Fig. 6) [68]. Moreover, microbiota can metabolize deconjugated bile acids to secondary bile, which binds to the G protein-coupled receptor TGR5 and leads to an increase in GLP-1 and insulin sensitivity [68]. In addition, in the case of TID, it was suggested that some bacteria species stimulate the promotion of self-antigens, leading to strengthening of the attack on beta cells in the pancreas [68], thus confirming the role of dysbiosis in the gut microbiota in the development of TID and TIID. Therefore, improving our understanding of the effects of the microbial gut in the development of both types of diabetes may provide the potential for preventive interventions [68].
Fig. 6.
Relationship between the intestinal microbiota and metabolic disorders.
THE MICROBIOTA AND LIVER DISEASES
The liver-gut axis drew the attention of researchers due to its effect on all the organs of the body. Furthermore, dysbiosis of the gut microbiota has an important influence on liver diseases in particular compared with those of other organs as previously mentioned. The gut-liver relationship interferes with all the substances produced as a result of metabolism, cytokines, and immune cells, in addition to the effects of endocrine products that reach the liver and intestines by blood circulation [73].
Microbiota effects on chronic hepatitis B and C
Hepatitis B and C are the most common medical problems leading to development of liver cancer in 4–5% of cases [8]. In some cases, hepatitis B virus (HBV) does not result in the development of chronic hepatitis due to the strength of the immune system, as well as factors unknown to date [74, 75]. In an experiment to investigate the role of the gut microbiota in the elimination of HBV, it was noted that adult mice with an intact microbiota had the ability to dispose of HBV and that when a microbial imbalance was caused by antibiotics, the mice could not get rid of HBV [76]. Another study showed that implantation of feces from healthy donors into hepatitis B patients stimulated the removal of HBeAg, which proved the importance of the gut microbiota in stimulating an immune response capable of protecting the liver from this virus [77].
As was noted, patients with hepatitis B and C were characterized by a lowering of bacterial diversity, especially a reduction in the genus Bifidobacterium [8]. Likewise, according to an Egyptian study, there was a high percentage of Bacteroidetes and a low percentage of Bifidobacterium in hepatitis C virus (HCV) patients [78]. Similarly, in HBV patients, there were increases in Escherichia, Shigella, and Enterococcus and decreases in Bifidobacterium, Faecalibacterium, Ruminococcus, and Ruminiclostridium [79].
The resulting microbiota imbalance leads to impaired intestinal permeability and the introduction of pathogens in the bloodstream, which increases the activation of Toll-like receptors (TLRs) and NOD-like receptors (NLRs), increases the inflammatory cytokines that reach the liver by bloodstream, and causes development of cirrhosis in both B and C cases (Fig. 7) [8].
Fig. 7.
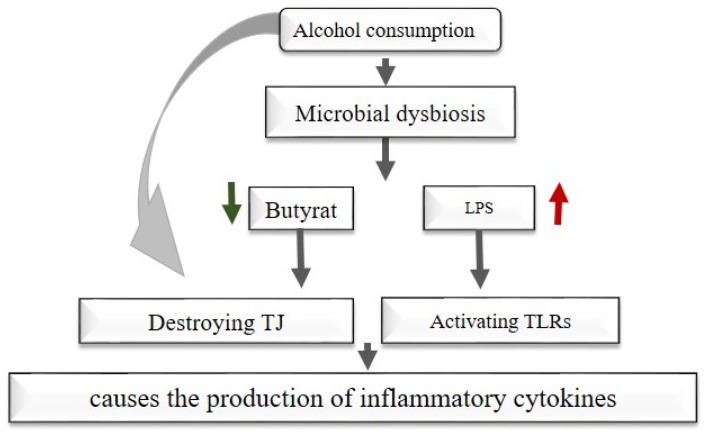
Effects of microbiota dysbiosis and alcohol consumption on the liver.
Microbiota effects on other liver diseases
The gut microbiota plays a role in liver diseases like alcoholic liver diseases and non-alcoholic fatty liver disease (NAFLD). Alcohol consumption induces microbial dysbiosis that reduces species that produce beneficial short-chain fatty acids (SCFAs), especially butyrate, which is considered an energy source for epithelial cells in the intestine, as an increase in Proteobacteria leads to an increase in LPS [80]. Likewise, alcohol consumption or alcohol resulting from gut microbiota metabolism increases the permeability of the intestine by destroying (TJs), leading to endotoxin penetration (Fig. 7) [81]. Alcohol consumption also increases the exposure of liver cells to oxidative stress due to the increase in endogenous ethanol, subsequently activating TLRs in the liver by pathogen-associated molecular patterns (PAMPs) and leading to the production of inflammatory cytokines. In addition, the gut microbiota dysbiosis in NAFLD patients stimulates insulin resistance and fat generation in the liver (de novo lipogenesis), which is the main reason for this disease [8].
Bile acids are synthesized in the liver and excreted into the intestines to digest and breakdown fats so that they can be absorbed by the small intestine, where the gut microbiota convert them into their secondary form. Bile acids activate the expression of bile acid receptors such as TGR5 and farnesoid X receptor (FXR) in the liver and intestine that play an important role in regulating the levels of triglyceride and bile acids in the liver. Besides, the FXR activation reduces the triglyceride levels and reduces insulin resistance by increasing glycogenesis [8].
Gut microbiota dysbiosis reduces the conversion of bile acids into their secondary form due to the reduction of the responsible species. This, in turn, reduces the expression of FXR, which leads to liver diseases [8]. This drew attention to the manufacture of drugs like obeticholic acid that activate FXR and reduce unnecessary inflammation in the liver [82].
THE MICROBIOTA AND PSYCHIATRIC DISORDERS
Scientific progress has rehashed old concepts about the brain and mental health and specifically the concept that mental health is not only related to the brain but is also related to the gut microbiota [73]. In recent years, potential effects of microbiota products on the brain have been demonstrated either directly by producing regulatory hormones or neurotransmitters or indirectly by affecting the gastrointestinal tract, autonomic nervous system, or intestinal nervous system or by stimulating the immune system [17].
Some bacteria, such as Lactobacillus and Bifidobacterium, produce gamma-aminobutyric acid (GABA), which is one of the most important transmitters in the central nervous system [83, 84]. On the other hand, Bacillus and Escherichia produce noradrenaline and dopamine, which play significant roles in the central and peripheral nervous systems [85]. Production of serotonin was also observed by Candida, Streptococcus, Escherichia, and Enterococcus. This hormone is responsible for regulating mood, appetite, and sleep, in addition to its role in stimulating memory and learning [17].
Several studies have shown that microbial effects on the brain are mediated by the vagus nerve. In a study on Lactobacillus rhamnosus aimed at determining its effects on an animal model’s emotions, it was found that feeding on L. rhamnosus leads to a reduction in corticosterone, which is responsible for anxiety and stress, and the effect path was via the vagus nerve [86].
Studies in major depressive disorder (MDD) patients have shown that gut microbiota dysbiosis is connected to psychiatric diseases via an increase in Bacteroidetes, Proteobacteria, and Actinobacteria or a decrease in Firmicutes [87]. Furthermore, researchers noted an increasing ratio of Bacteroidetes in an autistic group, while Firmicutes was dominant in a healthy group [88]. This drew attention to the use of probiotic bacteria like Bacteroides fragilis to alleviate autism symptoms in an autistic mouse model [89]. In another study, microbiota transfer therapy (MTT) was used to treat autism-like behaviors. Kang et al. found improvement in behavioral symptoms in autistic children when the children received transplanted commensal microbes from healthy donors [90].
The subtleties knowledge of our microbiota has led us to reconsider the factors that control human behavior. That our behavior is not only shaped by our genes and their interactions with the environment it is also shaped by our microbiota, their genes, and interactions with their environment that is, with us [17].
CONCLUSION
Studies have shown that the human microbiota is simultaneously both an extremely complex and orderly community. Human microbiota imbalance is referred to as dysbiosis. Many factors lead to dysbiosis, but the way it affects human health remains unclear.
Microorganisms start to colonize the human body immediately after birth, though new studies have demonstrated their existence in the intrauterine environment. A good microbiota composition plays an important role in educating the immune system to fight against pathogens and tolerate nonpathogenic microorganisms. On the other hand, a bad composition leads to many diseases, such as autoimmune diseases, metabolic disorders, and increased exposure to infectious diseases, and dysbiosis in adults is correlated with neuropsychiatric disorders.
Humans have sought to recover a good gut microbiota through live microorganisms in the form of probiotics or even through MTT, which has a long history in ancient Chinese medicine [91]. The history of probiotics extends back to a long time ago. In 1899, Tissier from the Pasteur Institute was the first to mention the role of a specific microorganism in relieving diarrhea. In 1917, Alfred Nissle isolated a subspecies of E. coli and used it to treat shigellosis [92]. Subsequently, studies have accumulated prove the importance of probiotics in restoring beneficial gut microbiota compositions.
In the near future, extensive and important research will make it possible to use the gut microbiota as a biomarker for many diseases, including cancer, and by using probiotics, we will be able to treat dysbiosis and prevent diseases from developing.
REFERENCES
- 1.Marchesi JR, Ravel J. 2015. The vocabulary of microbiome research: a proposal. Microbiome 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung TLF, Poulin R. 2008. Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Vie Milieu 58: 107–115. [Google Scholar]
- 3.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157: 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flandroy L, Poutahidis T, Berg G, Clarke G, Dao MC, Decaestecker E, Furman E, Haahtela T, Massart S, Plovier H, Sanz Y, Rook G. 2018. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci Total Environ 627: 1018–1038. [DOI] [PubMed] [Google Scholar]
- 5.Islam SU. 2016. Clinical uses of probiotics. Medicine (Baltimore) 95: e2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, Berenjian A, Ghasemi Y. 2019. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B, Yao M, Lv L, Ling Z, Li L. 2017. The human microbiota in health and disease. Engineering (Beijing) 3: 71–82. [Google Scholar]
- 8.Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, Gmizic I, Stevanovic O, Djordjevic V, Lekic N, Russo E, Amedei A. 2019. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: a review of the literature. Int J Mol Sci 20: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M, NIH HMP Working Group.2009. The NIH human microbiome project. Genome Res 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raoult D. 2008. Obesity pandemics and the modification of digestive bacterial flora. Eur J Clin Microbiol Infect Dis 27: 631–634. [DOI] [PubMed] [Google Scholar]
- 11.Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev 90: 859–904. [DOI] [PubMed] [Google Scholar]
- 12.LeBlanc JG, Laiño JE, del Valle MJ, Vannini V, van Sinderen D, Taranto MP, de Valdez GF, de Giori GS, Sesma F. 2011. B-group vitamin production by lactic acid bacteria—current knowledge and potential applications. J Appl Microbiol 111: 1297–1309. [DOI] [PubMed] [Google Scholar]
- 13.Clarke G, Sandhu KV, Griffin BT, Dinan TG, Cryan JF, Hyland NP. 2019. Gut reactions: breaking down xenobiotic–microbiome interactions. Pharmacol Rev 71: 198–224. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. 2019. Gut microbiota as a source of novel antimicrobials. Gut Microbes 10: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manchanayake LN. 2019. The impact of gut microbiota on host obesity. J Gastrointest Dig Syst 9: 591. [Google Scholar]
- 16.Trishna S, Vijeev V, Himanshi T. 2018. Gut microbiota in health and disease—an overview. Gastro Med Res 2 (2): GMR.000531. [Google Scholar]
- 17.Treisman GJ. 2017. The role of the brain-gut-microbiome in mental health and mental disorders. In The Microbiota in Gastrointestinal Pathophysiology, Floch MH, Ringel Y, Walker WA (eds), Academic Press, Cambridge, pp. 389–397. [Google Scholar]
- 18.Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC. 2015. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26: 26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med 6: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lal CV, Travers C, Aghai ZH, Eipers P, Jilling T, Halloran B, Carlo WA, Keeley J, Rezonzew G, Kumar R, Morrow C, Bhandari V, Ambalavanan N. 2016. The airway microbiome at birth. Sci Rep 6: 31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. 2012. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96: 544–551. [DOI] [PubMed] [Google Scholar]
- 23.Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. 2011. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 6: e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka M, Nakayama J. 2017. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int 66: 515–522. [DOI] [PubMed] [Google Scholar]
- 25.Vallès Y, Artacho A, Pascual-García A, Ferrús ML, Gosalbes MJ, Abellán JJ, Francino MP. 2014. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet 10: e1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Doré J, Other Members of the INFABIO Team2010. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 51: 77–84. [DOI] [PubMed] [Google Scholar]
- 27.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson AJ, Slack E, Geuking MB, McCoy KD. 2009. The mucosal firewalls against commensal intestinal microbes. Semin Immunopathol 31: 145–149. [DOI] [PubMed] [Google Scholar]
- 29.Corfield AP. 2018. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms 6: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper LV, Macpherson AJ. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10: 159–169. [DOI] [PubMed] [Google Scholar]
- 31.Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rea D, Coppola G, Palma G, Barbieri A, Luciano A, Del Prete P, Rossetti S, Berretta M, Facchini G, Perdonà S, Turco MC, Arra C. 2018. Microbiota effects on cancer: from risks to therapies. Oncotarget 9: 17915–17927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jourova L, Anzenbacher P, Anzenbacherova E. 2016. Human gut microbiota plays a role in the metabolism of drugs. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 160: 317–326. [DOI] [PubMed] [Google Scholar]
- 34.Das A, Srinivasan M, Ghosh TS, Mande SS. 2016. Xenobiotic metabolism and gut microbiomes. PLoS One 11: e0163099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grilo NM, Charneira C, Pereira SA, Monteiro EC, Marques MM, Antunes AM. 2014. Bioactivation to an aldehyde metabolite—possible role in the onset of toxicity induced by the anti-HIV drug abacavir. Toxicol Lett 224: 416–423. [DOI] [PubMed] [Google Scholar]
- 36.Jakoby WB, Ziegler DM. 1990. The enzymes of detoxication. J Biol Chem 265: 20715–20718. [PubMed] [Google Scholar]
- 37.Jeong HG, Kang MJ, Kim HG, Oh DG, Kim JS, Lee SK, Jeong TC. 2013. Role of intestinal microflora in xenobiotic-induced toxicity. Mol Nutr Food Res 57: 84–99. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Jia W. 2013. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin Pharmacol Ther 94: 574–581. [DOI] [PubMed] [Google Scholar]
- 39.Bosetti C, Boffetta P, La Vecchia C. 2007. Occupational exposures to polycyclic aromatic hydrocarbons, and respiratory and urinary tract cancers: a quantitative review to 2005. Ann Oncol 18: 431–446. [DOI] [PubMed] [Google Scholar]
- 40.Abou-Donia MB, El-Masry EM, Abdel-Rahman AA, McLendon RE, Schiffman SS. 2008. Splenda alters gut microflora and increases intestinal p-glycoprotein and cytochrome p-450 in male rats. J Toxicol Environ Health A 71: 1415–1429. [DOI] [PubMed] [Google Scholar]
- 41.Arboleya S, Sánchez B, Solís G, Fernández N, Suárez M, Hernández-Barranco AM, Milani C, Margolles A, de Los Reyes-Gavilán CG, Ventura M, Gueimonde M. 2016. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int J Mol Sci 17: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francino MP. 2016. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6: 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7: 10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jernberg C, Löfmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1: 56–66. [DOI] [PubMed] [Google Scholar]
- 45.Ford SA, King KC. 2016. Harnessing the power of defensive microbes: evolutionary implications in nature and disease control. PLoS Pathog 12: e1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Waaij D, Berghuis-de Vries JM, Lekkerkerk Lekkerkerk-v Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 69: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stecher B, Hardt WD. 2008. The role of microbiota in infectious disease. Trends Microbiol 16: 107–114. [DOI] [PubMed] [Google Scholar]
- 48.Bartlett JG, Gerding DN. 2008. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis 46 Suppl 1: S12–S18. [DOI] [PubMed] [Google Scholar]
- 49.Hapfelmeier S, Hardt WD. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol 13: 497–503. [DOI] [PubMed] [Google Scholar]
- 50.Keller JM, Surawicz CM. 2014. Clostridium difficile infection in the elderly. Clin Geriatr Med 30: 79–93. [DOI] [PubMed] [Google Scholar]
- 51.Ling Z, Liu X, Jia X, Cheng Y, Luo Y, Yuan L, Wang Y, Zhao C, Guo S, Li L, Xu X, Xiang C. 2014. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep 4: 7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Cobas AE, Moya A, Gosalbes MJ, Latorre A. 2015. Colonization resistance of the gut microbiota against Clostridium difficile. Antibiotics (Basel) 4: 337–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18: 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao JJ, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, Vieth M, Ma JL, Bajbouj M, Suchanek S, Liu WD, Ulm K, Quante M, Li ZX, Zhou T, Schmid R, Classen M, Li WQ, You WC, Pan KF. 2018. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol 8: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin Microbiol Rev 10: 720–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischbach W, Malfertheiner P. 2018. Helicobacter pylori infection. Dtsch Arztebl Int 115: 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren JR, Marshall B. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1: 1273–1275. [PubMed] [Google Scholar]
- 58.Kao CY, Sheu BS, Wu JJ. 2016. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed J 39: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang WL, Yeh YC, Sheu BS. 2018. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci 25: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiotani A, Matsumoto H, Fukushima S, Katsumata R, Kawano M, Saito M. 2018. H. pylori and human gut microbiota. J Bacteriol Mycol 5: 1086. [Google Scholar]
- 61.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3: e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boutagy NE, McMillan RP, Frisard MI, Hulver MW. 2016. Metabolic endotoxemia with obesity: is it real and is it relevant? Biochimie 124: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73: 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Z, Zhang Y, Li Z, Yu Y, Kang W, Han Y, Geng X, Ge S, Sun Y. 2016. Effect of Helicobacter pylori infection on chronic periodontitis by the change of microecology and inflammation. Oncotarget 7: 66700–66712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- 66.Manco M, Putignani L, Bottazzo GF. 2010. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev 31: 817–844. [DOI] [PubMed] [Google Scholar]
- 67.Gülden E, Wong FS, Wen L. 2015. The gut microbiota and type 1 diabetes. Clin Immunol 159: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munro N. 2016. Gut microbiota: its role in diabetes and obesity. Diabetes & Primary Care 18: 1–6. [Google Scholar]
- 69.Greiner T, Bäckhed F. 2011. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab 22: 117–123. [DOI] [PubMed] [Google Scholar]
- 70.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56: 1761–1772. [DOI] [PubMed] [Google Scholar]
- 71.Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, Wang RX, Onyiah JC, Kominsky DJ, Colgan SP. 2017. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of claudin-2. J Immunol 199: 2976–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hague A, Butt AJ, Paraskeva C. 1996. The role of butyrate in human colonic epithelial cells: an energy source or inducer of differentiation and apoptosis? Proc Nutr Soc 55: 937–943. [DOI] [PubMed] [Google Scholar]
- 73.Zuo T, Ng SC. 2018. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol 9: 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morikawa K, Shimazaki T, Takeda R, Izumi T, Umumura M, Sakamoto N, Hepatitis B. 2016. Hepatitis B: progress in understanding chronicity, the innate immune response, and cccDNA protection. Ann Transl Med 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chisari FV. 2000. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am J Pathol 156: 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou HH, Chien WH, Wu LL, Cheng CH, Chung CH, Horng JH, Ni YH, Tseng HT, Wu D, Lu X, Wang HY, Chen PJ, Chen DS. 2015. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc Natl Acad Sci USA 112: 2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren YD, Ye ZS, Yang LZ, Jin LX, Wei WJ, Deng YY, Chen XX, Xiao CX, Yu XF, Xu HZ, Xu LZ, Tang YN, Zhou F, Wang XL, Chen MY, Chen LG, Hong MZ, Ren JL, Pan JS. 2017. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology 65: 1765–1768. [DOI] [PubMed] [Google Scholar]
- 78.Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. 2016. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Q, Li F, Zhuang Y, Xu J, Wang J, Mao X, Zhang Y, Liu X. 2019. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cassard AM, Ciocan D. 2018. Microbiota, a key player in alcoholic liver disease. Clin Mol Hepatol 24: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szabo G. 2015. Gut-liver axis in alcoholic liver disease. Gastroenterology 148: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abenavoli L, Falalyeyeva T, Boccuto L, Tsyryuk O, Kobyliak N. 2018. Obeticholic acid: a new era in the treatment of nonalcoholic fatty liver disease. Pharmaceuticals (Basel) 11: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. 2019. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 4: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pokusaeva K, Johnson C, Luk B, Uribe G, Fu Y, Oezguen N, Matsunami RK, Lugo M, Major A, Mori-Akiyama Y, Hollister EB, Dann SM, Shi XZ, Engler DA, Savidge T, Versalovic J. 2017. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil 29: e12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neuman H, Debelius JW, Knight R, Koren O. 2015. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 39: 509–521. [DOI] [PubMed] [Google Scholar]
- 86.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 108: 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B. 2015. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48: 186–194. [DOI] [PubMed] [Google Scholar]
- 88.Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linløkken A, Wilson R, Rudi K. 2014. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil 26: 1155–1162. [DOI] [PubMed] [Google Scholar]
- 89.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang DW, Adams JB, Coleman DM, Pollard EL, Maldonado J, McDonough-Means S, Caporaso JG, Krajmalnik-Brown R. 2019. Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci Rep 9: 5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi HH, Cho YS. 2016. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc 49: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sonnenborn U. 2016. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett 363: fnw212. [DOI] [PubMed] [Google Scholar]