SUMMARY
Interhomolog crossovers (COs) are a prerequisite for achieving accurate chromosome segregation during meiosis [1, 2]. COs are not randomly positioned, occurring at distinct genomic intervals during meiosis in all species examined [3, 4, 5, 6, 7, 8, 9, 10]. The role of CO position as a major determinant of accurate chromosome segregation has not been previously directly analyzed in a metazoan. Here we use spo-11 mutants, which lack endogenous DNA double-strand breaks (DSBs), to induce a single DSB by Mos1 transposon excision at defined chromosomal locations in the C. elegans germline, and show that the position of the resulting CO directly affects the formation of distinct chromosome subdomains during meiotic chromosome remodeling. CO formation in the typically CO-deprived center region of autosomes leads to premature loss of sister chromatid cohesion and chromosome missegregation, while COs at an off-centered position, as in wild type, can result in normal remodeling and accurate segregation. Ionizing radiation (IR)-induced DSBs lead to the same outcomes and modeling of IR dose-response reveals that the CO-unfavorable center region encompasses up to 6% of the total chromosome length. DSBs proximal to telomeres rarely form COs, likely due to formation of unstable recombination intermediates that cannot be sustained as chiasmata until late prophase. Our work supports a model in which regulation of CO position early in meiotic prophase, is required for proper designation of chromosome subdomains and normal chromosome remodeling in late meiotic prophase I, resulting in accurate chromosome segregation and providing a mechanism to prevent aneuploid gamete formation.
Keywords: Crossover, DNA double-strand break, DNA repair, germline, meiosis, C. elegans
Graphical Abstract
eTOC Blurb
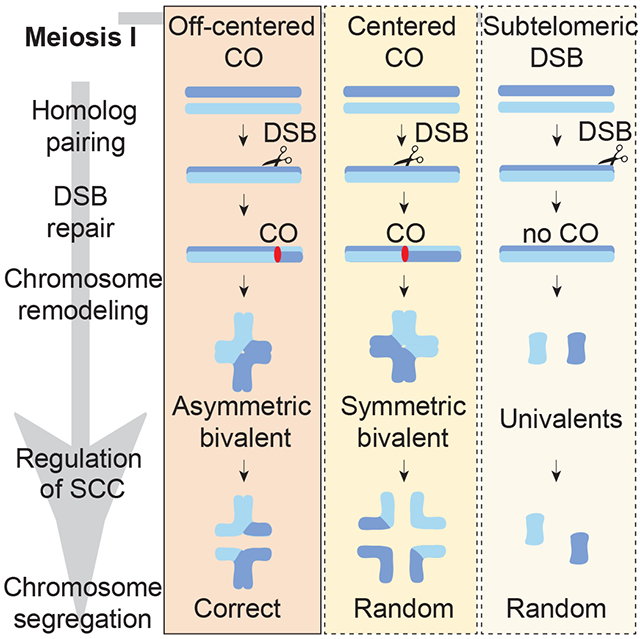
Altendorfer et al. show that placing a crossover (CO) at a typically CO-deprived chromosome region results in a chromosome configuration susceptible to nondisjunction. Moreover, CO suppression at chromosome ends is a conserved feature of meiosis. This study proposes that CO placement is highly regulated to ensure productive chromosome remodeling.
RESULTS AND DISCUSSION
Crossover (CO) recombination provides genetic diversity and also results in the formation of physical linkages (chiasmata) between homologous chromosomes that ensure accurate chromosome alignment and segregation during meiosis [1, 2]. As a consequence, CO formation is a highly regulated process in which only a very small number of DNA double-strand breaks (DSBs) are converted into mature COs [3, 4, 5]. The distribution of recombination events is not random as shown in human, mouse, fly, plant and yeast meiosis, where COs are suppressed near centromeric and telomeric regions, most likely to reduce the risk of aneuploidy [5, 6, 7, 8, 9, 10]. However, how CO position can affect accurate chromosome segregation is not well understood and has not been directly examined in a metazoan. In this study, we assess this in the nematode C. elegans and show that the position of the CO directly affects the formation of distinct chromosome subdomains during chromosome remodeling in late meiotic prophase, which in turn is critical for faithful chromosome segregation.
CO distribution is tightly regulated such that a CO in one location inhibits the occurrence of adjacent COs, a phenomenon referred to as CO interference [11, 12]. C. elegans, which has holocentric chromosomes, exhibits complete CO interference, which gives rise to a single off-centered CO per homologous chromosome pair [12]. This off-centered CO has been proposed to lay the foundation for the formation of bivalents at diakinesis with a cross-shaped configuration consisting of two chromosomal axes, corresponding to the short and long arms, that intersect perpendicularly at the chiasma [13, 14] (Figure S1A). These subdomains are occupied by various proteins that are essential for accurate chromosome segregation at meiosis I, including LAB-1, the functional analog of Shugoshin, which localizes to the long arms of the bivalents to inhibit premature sister chromatid separation through phosphorylation of cohesin by Aurora B kinase (AIR-2), which in turn is restricted to the short arms of the bivalents [15, 16, 17]. To assess the impact of CO position on chromosome remodeling and segregation we generated a single DSB by heat shock-induced Mos1 transposon excision [18, 19, 20] in a spo-11 mutant background, which lacks the meiosis-specific endonuclease required for DSB formation, but is still capable of chromosome pairing and synaptonemal complex (SC) formation [21], at the following defined genomic positions: (1) the physical center of the chromosomes, where CO formation levels are low in wild type, (2) the terminal thirds of the chromosomes, herein referred to as “arms”, which are the off-centered regions preferred for CO formation in wild type, and (3) the subtelomeric region, where CO formation is not detected in human oocytes [22, 23] (Figure 1A and Figure 1B).
Figure 1. The position of a single DSB/CO affects late prophase I chromosome remodeling.
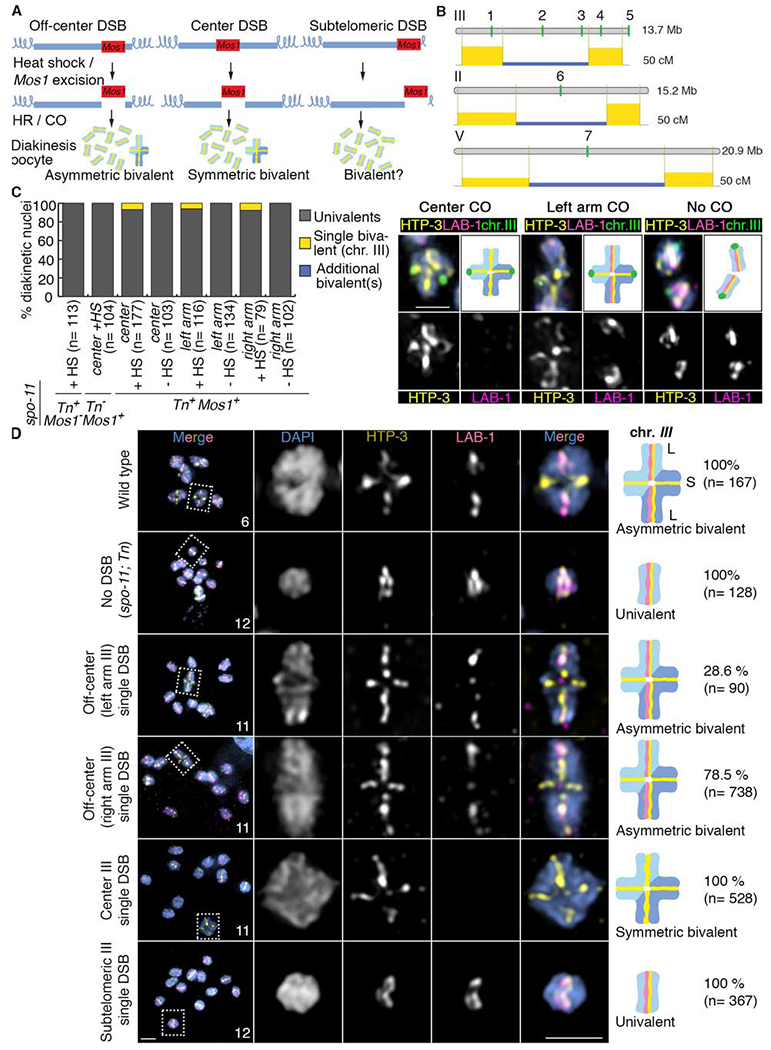
(A) Schematic drawing of the single inducible DSB assay. Blue lines represent the chromosomes and the red boxes correspond to the Mos1 transposon inserted at different chromosomal locations. After heat shock-induced DSB formation by transposon excision and subsequent repair via homologous recombination (HR), diakinesis nuclei were analyzed for the presence of either a single asymmetric or symmetric bivalent (all other chromosomes were univalents given that this analysis was performed in a spo-11 mutant background that lacks endogenous DSBs). (B) Schematic drawing of the locations of the Mos1 transposon insertions on chromosomes III, II and V. 1 indicates the location of ttTi1730, 2 the location of ttTi23808, 3 the location of ttTi2384, 4 the location of ttTi13055 and 5 the location of ttTi17604 on chromosome III. 6 indicates the location of ttTi5605 on chromosome II and 7 the location of ttTi35006 on chromosome V. Numbers 1 and 4 indicates DSBs occurring in an off-center position, 2, 6 and 7 indicates DSBs occurring in a center position, 5 indicates a DSB occurring in a subtelomeric position and 3 indicates a DSB occurring at the border of the center region. The physical (Mb) and genetic (cM) maps are shown one above of the other for every chromosome. Genetic maps are based on Rockman and Kruglyak [42]. The yellow rectangles represent the recombination frequencies on the chromosome arms compared to frequencies on the center region in blue. (C) Quantification of bivalent formation for chromosome III by immunoFISH analysis of late diakinesis oocytes following Mos1 transposon excision (+ or − heat shock; HS) from the indicated locations on chromosome III. All lines carry the spo-11 mutation and either presence or absence of a Mos1 transposon insertion (Mos1+ or Mos1−, respectively) on chromosome III and presence or absence of the transposase (Tn+ or Tn− respectively). Representative immunoFISH images with LAB-1 (magenta), HTP-3 (yellow) and a FISH probe for chromosome III (green). (D) Quantification of asymmetric or symmetric bivalent formation in late diakinesis oocytes based on coimmunostaining for the HORMA domain-containing protein HTP-3 (yellow) and LAB-1 (magenta). Immunofluorescence images from wild type, spo-11;Tn (transposase+) mutants and spo-11 mutants in which a single Mos1-induced DSB was generated in one of the following locations on chromosome III: off-center (at either the right or left arms), the center, and the subtelomeric region. Both chromosomal axes are highlighted by HTP-3 while LAB-1 is restricted to the long arm of the bivalents in wild type. Total numbers of DAPI-stained bodies (bivalents/univalents) observed in the oocytes at diakinesis are shown in the first column. Dashed boxes indicate the bivalents/univalents shown at higher magnification. Illustrations on the right depict the chromosome configuration observed at this stage and the localization of HTP-3 (yellow) and LAB-1 (magenta). Long (L) and short (S) arms of the asymmetric bivalent are indicated on wild type. n= number of nuclei scored. Bar, 2 μm. See also Figures S1, S2 and Data S1 and S2.
First, we determined that DSBs are successfully formed following heat shock-induced Mos1 excision at all three positions given that increased levels of RAD-51 foci were observed following rad-54 depletion by RNAi, which blocks removal of RAD-51 from DSB repair sites [4, 24] (Figure S1B). Moreover, quantification of these RAD-51 foci revealed that levels of DSBs are indistinguishable whether DSBs were induced at the center, an arm or a subtelomeric region. Next, fluorescence in situ hybridization (FISH) analysis showed that chiasma formation occurs only following heat shock-induced Mos1 excision and specifically on the chromosomes harboring the transposon insertion. We observed a single bivalent in 6.7% of diakinesis nuclei (n=177) when a DSB is formed at the center of chromosome III and in 5.2 to 7.6% of nuclei when a single off-centered break is formed at either the left (n=116) or right (n=79) arms of chromosome III, compared to 0% bivalents without heat-shock (n=103 for center; n=134 for left arm; n=102 for right arm) (Figure 1C, STAR Methods: Specificity and efficiency of bivalent formation following Mos1-induced DSB formation, and Data S1B).
To test whether CO location influences the successful formation of chromosome subdomains that are required for accurate chromosome segregation, we focused on proteins that specifically localize either to the long or short arms of the bivalents at diakinesis. Strikingly, the formation of a single CO at the center of chromosome III gave rise to a configuration we refer to herein as a “symmetric bivalent” where both chromosome axes were of similar lengths and which failed to undergo accurate chromosome remodeling as evidenced by the complete absence of LAB-1 signal in 100% of single bivalents detected at diakinesis (n=28/28 bivalents from 528 diakinesis nuclei scored) (Figure 1D and Data S1B). In contrast, when inducing an off-centered DSB on either the left or right arm of chromosome III, we observed a single asymmetric bivalent with LAB-1 signal only at the long arm in 28.6% and 78.5%, respectively, of all single bivalents detected (n= 2/7 bivalents from 90 diakinesis nuclei and n= 51/65 bivalents from 738 diakinesis nuclei scored respectively) (Figure 1D, Figures S2A and S2B and Data S1B). The remaining bivalents produced by off-centered DSB induction were symmetric. In addition, there were two categories of asymmetric bivalents: those with strong LAB-1 signal along the long arm (14.3% and 21.5%, left and right arm induced DSB, respectively) and those with weak or discontinuous LAB-1 signal on the long arm (14.3% and 57%, left and right arm induced DSB, respectively) (Figure S2B). These results indicate that while a CO at the physical center of a chromosome fails to undergo normal chromosome remodeling in late prophase, an off-centered CO induced by Mos1 excision can result in the formation of short and long arm domains as observed in wild-type worms. Consistent with this, phosphorylation of histone H3 (pH3), a chromosomal substrate of AIR-2 normally restricted to the short arms by LAB-1-mediated recruitment of the PP1 phosphatase to the long arms [15, 16], was observed on both arms in 100% of the bivalents upon formation of a centered CO (n= 8/8 bivalents from 138 diakinesis nuclei). Whereas an increase in the number of bivalents exhibiting pH3 properly restricted to the short arms was observed following a single off-centered CO on either the left or right arms of chromosome III (7.14%, n= 1/14 bivalents from 209 diakinesis nuclei and 16.6%, n= 2/12 bivalents from 292 diakinesis nuclei, respectively) (Figure S2C and S3D, Data S1B). However, the levels of asymmetric bivalents with pH3 restricted to the short arm were lower compared to those with LAB-1 signal (strong and weak combined) restricted to the long arm albeit similar to the levels of asymmetric bivalents only with strong LAB-1 signal. This suggests that LAB-1 may not always be fully functional (i.e. cases with weak LAB-1 signal). Alternatively, additional factors may be required to promote robust asymmetric bivalent formation following a single off-center DSB. Analysis of a line where two DSBs are produced on chromosome III by excision of Mos1 at the center and right arm, showed a trend towards increase both in the total number of bivalents detected and of asymmetric bivalents with strong LAB-1 signal compared to a single DSB at the center or the right arm of that chromosome (Figure S3A–C). This suggests that chromosomes must undergo multiple DSBs (a minimum “threshold” level of DSBs may need to be produced) for efficient conversion of a DSB into a CO (also see STAR Methods: Assessment of chromosome remodeling and segregation upon Mos1-induced DSB formation). In line with previous observations that mislocalization of both LAB-1 and AIR-2/pH3 at diakinesis result in premature sister chromatid separation [15, 17], we observed evidence of increased premature sister separation when a single DSB is induced at the center of chromosome III compared to the right arm of chromosome III both by loss of sister chromatid cohesion by SMC-1 (Structural Maintenance of Chromosomes) staining in diakinesis stage oocytes (6% compared to 0%, n=200, respectively; Figure 2A) and FISH analysis of metaphase to anaphase I transition (72.7% compared to 50%, respectively; Figure 2B). Finally, consistent with suppression of COs close to telomeres reported in yeast and female mammalian and plant meiosis [7, 10, 22], bivalents were not detected when a single DSB was induced at a subtelomeric region on chromosome III (100% univalents; n=367 diakinesis nuclei) (Figure 1D and Data S1B), suggesting that CO suppression close to telomeres is a conserved feature.
Figure 2. A single centered crossover results in premature sister separation.
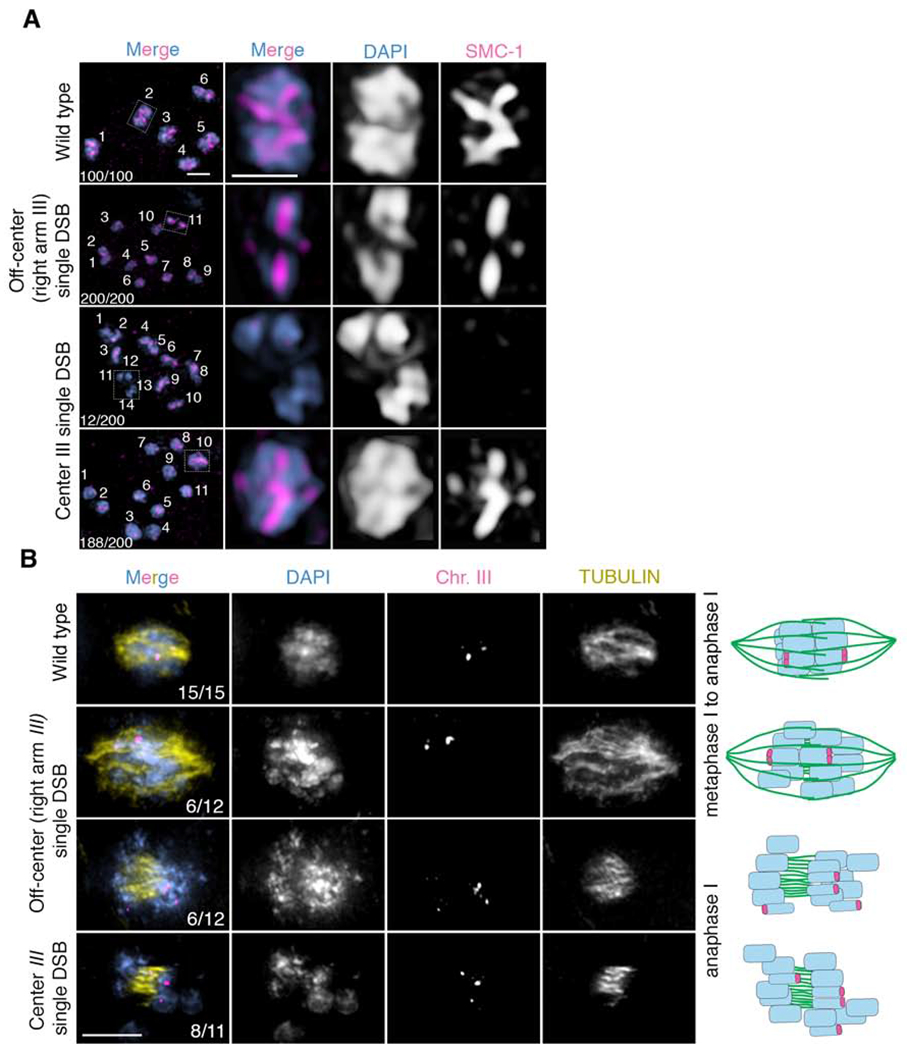
(A) Immunolocalization of SMC-1 (red) and DAPI (blue) in diakinesis stage oocytes in wild type, spo-11 mutants with a single Mos1 insertion at an off-center position (right arm) on chromosome III, and spo-11 mutants with a single Mos1 insertion at the center of chromosome III after heat shock, all of which underwent emb-30 depletion by RNAi to induce arrest of oocytes before the meiotic division to facilitate this analysis. 12 out of 200 oocytes in spo-11 mutants with a single Mos1 insertion at the center of chromosome III showed premature separation of sister chromatids after heat shock as evidenced by the presence of 14 DAPI-stained bodies (10 univalents + 4 separated sister chromatids) and absence of SMC-1 staining in 4 DAPI-stained bodies (presumably the 4 prematurely separated sister chromatids). (B) Immunofluorescence-FISH images of metaphase to anaphase I nuclei. On the first row are representative images of wild-type nuclei at the metaphase to anaphase I transition with a chromosome III FISH probe (magenta) and tubulin (yellow). The two FISH signals (foci) show separating homologs but joined sister chromatids (n=15/15 nuclei examined). The second and third rows are representative images of metaphase to anaphase I and anaphase I nuclei, respectively, from worms harboring a single Mos1 insertion at an off-center position (right arm) on chromosome III. Both nuclei with separating homologs but joined sister chromatids (n=6/12), as seen in wild type, and nuclei with premature sister separation (n=6/12), as depicted by more than two FISH signals, were detected. The last row depicts anaphase I for worms with a single Mos1 insertion at the center of chromosome III. Sister chromatid separation is evidenced by the presence of more than two FISH signals (n=8/11). Numbers in the first column represent number observed/total number scored. Bar, 2 μm.
Given that a single DSB at the center of chromosome III fails to produce long and short arm subdomains, we used two different approaches to examine whether additional DSBs along the chromosome could rescue this defect by providing an alternative position for CO recombination. First, given that IR-induced DSBs are distributed along chromosomes without bias towards specific regions (Figures S3D, S4C and [25, 26]), we exposed worms undergoing a single Mos1-induced DSB at the center of chromosome III to low dose IR (10 Gy) resulting in approximately 4 DSBs per homologous chromosome pair and close to 90% rescue of CO formation [26]. Second, since Mos1-induced DSBs are also subject to interference and can compete with endogenous SPO-11-induced DSBs for CO formation [20], we combined a single Mos1-induced DSB at the center of chromosome III with endogenous SPO-11-induced DSBs to analyze the outcome on chromosome remodeling. As expected, in both cases we observed 100% rescue of the remodeling defects, as demonstrated by the restricted localization of LAB-1 to the long arm of the bivalents (n= 114 and 100 diakinesis nuclei, respectively) (Figures S3E and S4A). These findings suggest that DSBs occurring outside of the center region of the chromosomes are preferred as a substrate for repair resulting in CO formation. Importantly, the remodeling defects observed upon CO formation at the chromosome center are not specific to chromosome III and instead are a general feature of autosomes, as both lack of LAB-1 signal along the long arm and presence of pH3 signal on both short and long arms were recapitulated when a single DSB was induced following Mos1 excision at the center of either chromosome II or V (Figures S4A, S4B and Data S1B).
To determine whether the defects observed at diakinesis result from problems during earlier stages of chromosome remodeling, we monitored the presence of LAB-1 and the CO precursor marker CNTD1/COSA-1 in late pachytene nuclei. In wild type, prior to the initiation of chromosome remodeling, LAB-1 is localized throughout the full length of chromosomes in late pachytene nuclei [15]. Similar to wild type, LAB-1 signal was detected along chromosome III in 98% (n=124, +HS) of late pachytene stage nuclei in spo-11 mutants (Figure 3A and Data S2A and S2B). However, this decreased to 73% (n= 277) and 60% (n=203) when a single Mos1-induced DSB was introduced either at the right arm or the center, respectively, of chromosome III (Figure 3A and Data S2A and S2B). The loss of LAB-1 signal specifically on the chromosome in which a single DSB is induced suggests that the defect in LAB-1 localization observed in late diakinesis originates earlier in prophase and that chromosome subdomains may be shaped prior to late pachytene. This is further supported by analysis of CO precursor formation, which revealed similar levels of late pachytene nuclei with a single COSA-1 focus regardless of whether the Mos1-induced DSB was at the center or on the right arm of chromosome III (22%, n=299, and 27%, n=116, respectively) (Figure 3B and Data S3 and S4). This suggests that events taking place prior to CO precursor formation, either at DSB formation or at an early step of repair, are impinging on the ability to regulate the chromosomal association of LAB-1, with direct impact on the chromosome remodeling taking place later in prophase. Following a single DSB at the subtelomeric region of chromosome III, only 8.6% of late pachytene nuclei (n=261) showed a single subtelomeric COSA-1 focus on chromosome III (Figure 3B and Data S3 and S4). This result indicates that breaks induced at subtelomeric loci undergo CO precursor formation, but due to the lack of bivalents detected later at diakinesis, this repair fails to result in functional chiasmata. Finally, both LAB-1 and COSA-1 localization in late pachytene nuclei are fully rescued by exogenous DSB formation (10 Gy) (Figure 3C and Data 5A). Importantly, these remodeling defects are not a peculiarity of transposon excision since they are fully recapitulated by utilizing IR as an alternative source for DSB formation. Specifically, an exposure to 2.5 Gy, which results in ~1 DSB per homologous chromosome pair [26], resulted in 3.4% of oocytes at diakinesis (n=207) with the same remodeling defects observed by Mos1-induced DSB formation at the center of an autosome (Figure 4A and B, Figure S4D–F, Data S5B and S5D). Further increases in the numbers of IR-induced DSBs dramatically restored asymmetric bivalent formation (Figure 4B and C, and Data S5C), suggesting that some fraction occurs at off-center positions and are sufficient to outcompete the designation of any DSBs occurring at the center region of the chromosomes as COs that would result in abnormal chromosome remodeling. Therefore, the IR-induced DSB analysis further supports our hypothesis that the position of the CO is crucial for the proper implementation of chromosome remodeling independent of the source of DSB formation.
Figure 3. Chromosome remodeling defects are already observed at late pachytene stage and can be rescued by additional exogenous DSBs.
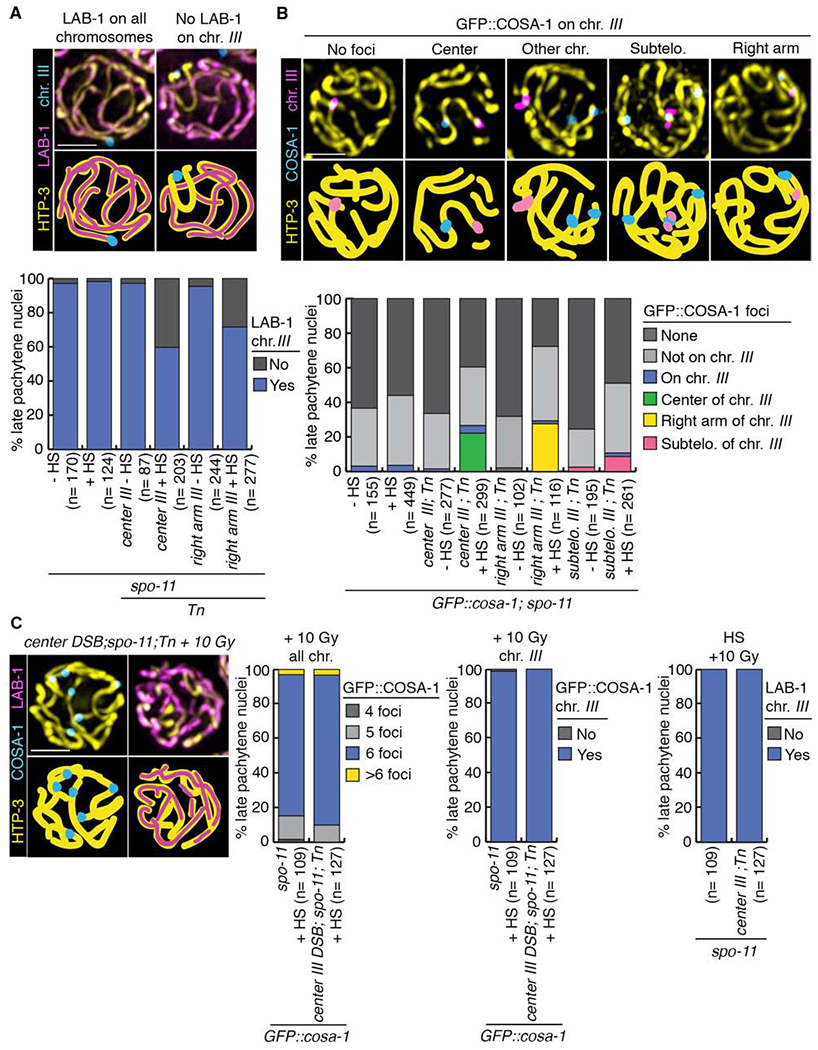
(A) Immunolocalization of LAB-1 in late pachytene on chromosome III following heat shock-induced DSB formation. HTP-3 (yellow), LAB-1 (magenta) and a FISH probe for chromosome III (blue) are shown. Illustrations for each observed localization pattern are shown in the lower panel. Histogram shows the quantification of various LAB-1 localization patterns in late pachytene nuclei subjected to heat shock-induced DSBs at different locations on chromosome III. (B) Crossover precursor marker COSA-1 localizes at high frequency to Mos1-induced CO sites. Late pachytene nuclei of whole mounts hybridized with a FISH probe recognizing chromosome III (magenta) showing the localization of chromosome axis marker HTP-3 (yellow) and GFP::COSA-1 (blue). Illustrations depict the different localizations observed for COSA-1. Histogram shows the quantification of COSA-1 foci in late pachytene nuclei from lines with DSBs induced at the indicated positions on chromosome III (identified by FISH). Similar to other studies, GFP::COSA-1 foci were also detected on chromosomes in spo-11 mutants, potentially reflecting spontaneous DNA lesions [43, 44]. n, number of late pachytene nuclei scored; HS, heat shock. (C) Exogenous DSB formation by γ-IR rescues CO formation and LAB-1 localization defects in late pachytene nuclei subjected to a single Mos1-induced DSB at the center of chromosome III. HTP-3 (yellow), COSA-1 (blue) and LAB-1 (magenta). Histograms show quantifications of COSA-1 foci and LAB-1 localization pattern on chromosome III (identified by FISH). Bars, 2 μm. See also Figure S3 and Data S3, S4 and S5.
Figure 4. Remodeling defects are independent of the source of DSB formation.
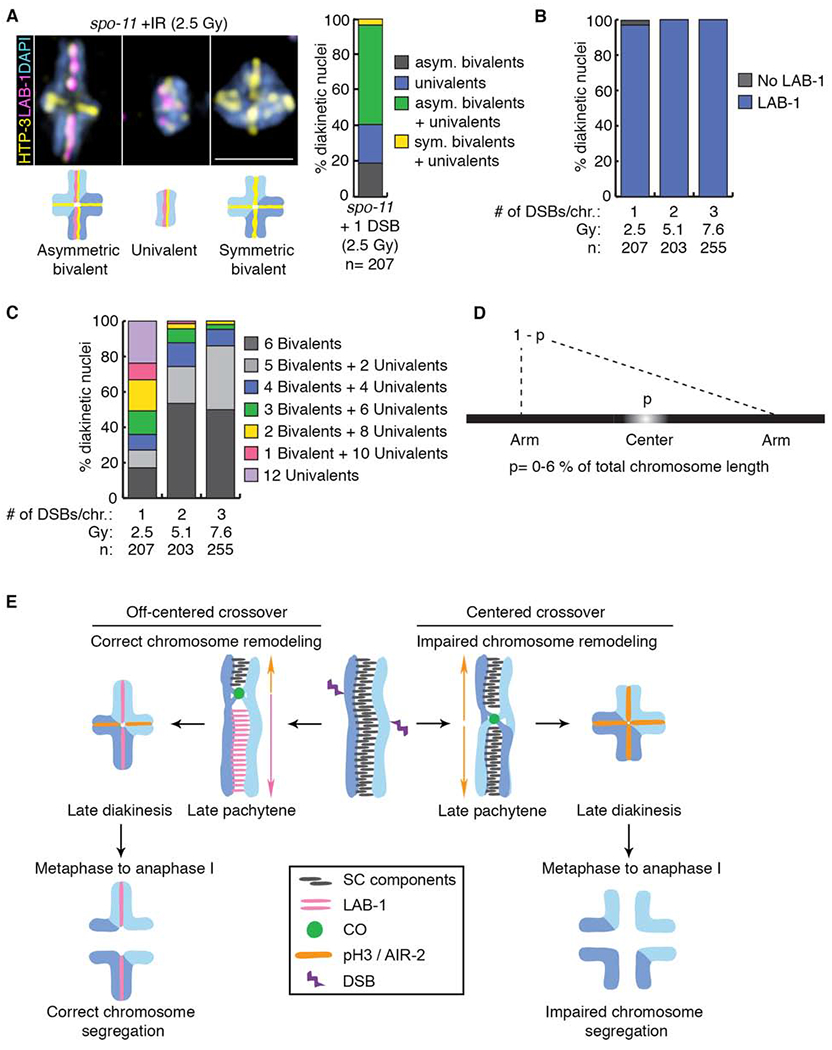
(A) Representative DAPI-stained bodies (blue) showing the localization of HTP-3 (yellow) and LAB-1 (magenta) observed in diakinesis nuclei of spo-11 mutants subjected to γ-IR doses producing 1, 2 or 3 DSBs per chromosome pair. Illustrations are shown below the immunofluorescence images. Bar, 2 μm. Histogram depicting the categories of bivalent/univalent configurations observed at diakinesis after inducing 1 DSB per homolog pair by γ-IR (2.5 Gy), n indicates the number of diakinesis nuclei examined. (B) Quantification of LAB-1 localization in diakinesis nuclei following exposure to indicated γ-IR doses. (C) Histogram showing quantification of the observed number of bivalents and/or univalents at different γ-IR doses. (D) Mathematical modeling used to determine the length of the center region leading to chromosome remodeling defects upon CO formation. (E) Model for how CO position influences late prophase chromosome remodeling and subsequent chromosome segregation. DSBs occur throughout the length of the chromosomes (up to 10 per homolog pair in C. elegans [37]), but only 1 introduced at an off-centered position is preferentially used for CO formation leading to correct chromosome remodeling resulting in asymmetric bivalents and accurate chromosome segregation. In contrast, COs occurring at the center of the chromosomes result in impaired chromosome remodeling resulting in symmetric bivalents. This chromosome configuration lacks LAB-1 localization and this in turn fails to restrict AIR-2/pH3 to a single chromosome axis resulting in premature loss of sister chromatid cohesion and errors in chromosome segregation. See also Figure S4 and Data S5.
We next applied mathematical modeling to our data from both Mos1- and IR-induced DSBs to predict the size of the center region that fails to promote remodeling when subject to CO formation for chromosomes III and V, respectively, one of the smallest (13.7 Mb) and one of the largest (20.9 Mb) chromosomes in the C. elegans genome. Our analysis assumes that both chromosomes are unique and independent of each other given the complex nature of gene expression and differences in histone modifications between chromosomes. This is exemplified by chromosome III, which harbors more genes that are highly expressed in the germline and therefore more active histone marks compared to chromosome V [27, 28]. Based on this assumption and the different lengths of these two chromosomes, we extrapolate that the center region encompasses only up to 6% of the total chromosome lengths for both chromosomes III and V (Figure 4D, STAR Methods: Assessment of chromosome remodeling and segregation upon Mos1-induced DSB formation, and Data S5E). This analysis predicts that the center region of the chromosomes that should be avoided to obtain productive chromosome remodeling is very small, which could explain why chromosome remodeling defects are rarely observed in wild- type worms. Only in situations where DSBs are limiting and/or recombination is shifted towards the center of the chromosomes would defects in the formation of proper chromosome subdomains be observed. Given that chromosomes in C. elegans are holocentric, we also note that the mechanism operating to suppress COs near the center of the chromosomes in this nematode is not due to the centromere effect observed in monocentric organisms [29, 30, 31].
COs are indispensable for achieving accurate chromosome alignment and segregation during meiosis. In this study, we provide evidence that the position of the CO is a major determinant of proper chromosome remodeling. We propose a mechanism that disfavors CO formation at regions that lead to symmetric bivalents due to their inability to form distinct chromosome domains necessary to retain proteins that are indispensable for accurate chromosome segregation (Figure 4E). The retention of proteins at distinct chromosomal subdomains at this stage of prophase is not unique to C. elegans, having also been observed for both SC components and cohesin proteins in yeast and mammals [32, 33, 34, 35]. While the mechanism that favors or disfavors CO formation at certain chromosome regions remains unclear we show that altering the position of the single CO to a typically recombination “cold” region can lead to chromosome configurations susceptible to nondisjunction. Given that altered CO patterning has been detected in aneuploid oocytes during human female meiosis [22, 36, 37, 38, 39, 40, 41], our study provides direct evidence that CO placement in metazoans needs to be a highly regulated non-random process.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Monica P. Colaiacovo (mcolaiacovo@genetics.med.harvard.edu). Worm strains (spo-11 ;transposon and spo-11;transposase) are available for sharing.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Worm strains
All strains are from the N2 Bristol background and were maintained at 20°C under standard conditions [45]. Mos1 insertion lines were obtained from the “Biology of Caenorhabditis elegans” Facility at CNRS, France [46] and screened positive for Mos1 transposon insertion and subsequent excision. Briefly, 150 worms of each line were screened for the Mos1 transposon insertion by PCR. Lines positive for the insertion were outcrossed at least 5 times against wild- type N2 and verified for a single Mos1 insertion site as in Boulin and Bessereau [47] with the following modification: fragments obtained from nested PCR were isolated by gel extraction and subcloned using TOPO® TA Cloning® kit (Thermo Fisher Scientific) prior to sequencing. Not all lines that scored positive for the Mos1 insertion underwent successful Mos1 transposon excision. We speculate that potential DNA sequence and/or chromatin environment might contribute to successful transposon mobilization. Strains used in this study are indicated in Data S1.
Specificity and efficiency of bivalent formation following Mos1-induced DSB formation
Our experiments require the use of spo-11 homozygous worms which produce >99.4% dead embryos and result in high levels of larval lethality among the very few surviving progeny [21]. However, the low frequency of single bivalents observed at diakinesis upon Mos1-induced DSB formation requires the analysis of a high number of nuclei (n values). To overcome these challenges, we used lines homozygous for the Mos1 transposon insertion, which allowed us to obtain a reasonable number of worms for analysis. We carefully monitored for any leakiness of the heat shock promoter-controlled transposase for transposition events and transposon duplications to any other chromosome by FISH analysis of diakinesis oocytes (see Figure S1C). We only detected bivalent formation for the chromosome carrying the Mos1 transposon. This indicates that the transposon, at least in the germline, is not subject to spontaneous excision and transposition without heat shock.
Although Robert et al. [19] used Mos1 transposon homozygous worms to study nonhomologous end joining (NHEJ) in the C. elegans germline, homologous recombination (HR) events could not be detected in their assay given that the progeny would be phenotypically indistinguishable from the parental worms (repair from the homolog would also result in “Unc” F1 progeny). In our assay, we can detect HR events by the formation of a single bivalent, indicating that a Mos1-induced DSB can be channeled into HR-directed repair. We cannot rule out the possibility that some of the Mos1-induced DSBs are processed through alternative pathways like NHEJ leading to a reduction in HR events. However, although we used worms homozygous for the Mos1 transposon, the numbers of bivalents we observed at diakinesis are similar to those reported by Rosu et al. where they used Mos1 heterozygous worms [20]. Furthermore, Rosu et al. [20] inferred from the analysis of spo-11 homozygous (Mos1 heterozygous) worms that most likely only one chromatid had been subjected to Mos1-induced DSB formation giving rise to a CO outcome practically 100% of the time compared to a 50% CO outcome when both chromatids were cut. However, in our hands, the Mos1 transposon heterozygous line for a DSB at the center of chromosome III did not show an increase in the frequency of bivalent formation (4.4%, n=8/182) compared to the Mos1 homozygous at the same position (4.9%, n=36/729). This result argues that there is no difference in using homozygous or heterozygous lines for the Mos1 transposon at least in the spo-11 mutant background and that DSBs are being preferentially repaired as COs. In contrast, studies in yeast meiosis using HO-induced DSBs in spo11Δ mutants have shown that COs are detected following DSB formation on both sister chromatids, but at lower levels compared to HR events derived from a DSB at only one sister chromatid [48]. However, spo11Δ mutants undergoing HO-induced DSBs failed to form extensive SCs (SC-like structures were observed <1%) while synapsis is SPO-11-independent in C. elegans [21], which could account for the higher CO frequency observed with Mos1-induced breaks in worms.
In summary, given that the efficiency of cytologically-detected bivalent formation at the unc-5 locus on chromosome IV in a spo-11 background is 9% based on Rosu et al., we conclude that even when assessing bivalent formation conservatively after Mos1-induced DSB formation at either the center or on the arms of chromosome III, COs are induced efficiently in the Mos1 transposon homozygous lines used in our study.
METHOD DETAILS
Experimental conditions for heat shock
Heat shock experiments were performed as described previously [20], except that all strains used for this study were homozygous for both the Mos1 transposon and the Mos1 transposase. Between 100 to 200 homozygous progeny from heterozygous parents for each genotype were grown until 24 hours post-L4 at 15°C. Plates were heat shocked for 1 hour and 15 min at 34°C. Worms were then incubated at 20°C for either 40 hours for analysis of bivalent formation at diakinesis or for 20-24 hours for analysis of nuclei at late pachytene. The time points for analysis are based on previously published meiotic progression studies [20, 49]. See Data S1A.
γ-irradiation experiments
20 hours post-L4 spo-11 homozygous worms grown at 20°C were exposed to a Cs137 source using the following doses based on Yokoo et al. [26]: 256 Rad corresponding to ~1 DSB/homologous chromosome pair, 512 Rad corresponding to ~2 DSBs/homologous chromosome pair, and 1000 Rad corresponding to ~4 DSBs/homologous chromosome pair. After irradiation, worms were incubated at 20°C for an additional 36 hours. For combined heat shock and irradiation experiments, 24 hours post-L4 worms grown at 15°C were heat shocked for 1 hour and 15 min at 34°C and then allowed to recover for 3 hours at 20°C followed by a Cs137 exposure at 1000 rad. After irradiation, worms were grown for an additional 36-38 hours at 20°C for analysis of bivalent formation at diakinesis or 20 hours for analysis of late pachytene stage nuclei. See Data S5A–E.
Immunofluorescence and imaging
Immunostaining was performed as described previously [50], with modifications. Gonads were fixed with 2% formaldehyde for 5 minutes prior to freeze cracking. Primary and secondary antibody incubations were performed overnight at 4°C. The following antibodies were used: rabbit anti-LAB-1 (1:2000; [15, 16]), guinea pig anti-HTP-3 (1:500; [51]), rabbit anti-phospho-Histone H3 (Ser10) (1:500; Millipore, cat. #06-570), rat anti-SMC-1 (1:200; [52]), mouse anti- GFP (1:300; Roche, cat. #11814460001), donkey anti-rabbit Cy5 (1:100; Jackson Immunoresearch, cat. #711-175-152), donkey anti-guinea pig Alexa488 (1:500; Jackson Immunoresearch, cat. #706-545-148), donkey anti-rabbit Cy3 (1:200; Jackson Immunoresearch, cat. #711-165-152), goat anti-mouse Alexa488 (1:500; Jackson Immunoresearch, cat. #115-546-146), and donkey anti-guinea pig Cy3 (1:200; Jackson Immunoresearch, cat. #706-165-148). DAPI (Thermo Fisher Scientific, cat. #D1306) was used to counterstain DNA and slides were mounted with VectaShield (Vector Laboratories, cat. #H-1000). SMC-1 immunostaining was performed as mentioned above except primary antibody was incubated overnight at room temperature and secondary antibody was incubated for 2 hours at room temperature.
Gonads were imaged using an IX-70 microscope (Olympus) with a cooled CCD camera (model CH350; Roper Scientific) controlled by the DeltaVision system (Applied Precision). Images were collected at 0.2 μm intervals and deconvolved using the SoftWorx 3.0 program (Applied Precision) and processed with Fiji ImageJ [53, 54]. See Data S1B, S2A, S3, and S5.
ImmunoFISH
FISH probes were generated as in Smolikov et al. [55] and labeled with either Fluorescein-12-dCTP (Perkin Elmer, cat. #NEL-424) or Cyanine 3-CTP (Perkin Elmer, cat. #NEL580001EA). The following pooled cosmids were used: T03F6, F11F1, T28A8 (for chromosome III, right arm) and T17A3, F40G9, W05G11 (for chromosome III, left arm). The 5S rDNA probe was generated by PCR with the following primers: 5’-TACTTGGATCGGAGACGGCC-3’ and 5’-CTAACTGGACTCAACGTTGC-3’.
Immunostaining was performed as described above with the following modifications: Slides were charged with Poly-L-Lysine (0.01%, Sigma cat. #P8920). Gonads were dissected in egg buffer containing 0.1% Tween and fixed with 2% formaldehyde for 5 minutes. After freeze cracking, slides were incubated in ice-cold methanol for 1 minute. Both primary and secondary antibody incubations were performed overnight at 4°C. Samples were post-fixed with PBS containing 3.7% Formaldehyde for 15 min and washed 1 time with PBST and 2 times with 2xSSC. Slides were incubated in 2xSSC containing 50% Formamide for 2 hours prior to adding the FISH probes at 37°C. Slides were sealed with CytoBond (SciGene 2020-00-1) and DNA was denatured for 90 sec at 93°C. Hybridization was carried out overnight at 37°C in a water bath. After hybridization, slides were washed 2 times with 2xSSC containing 50% Formamide for 30 min at 37°C, 1 time with 2xSSCT containing 25% Formamide at room temperature for 15 min, and 2 times with 2xSCCT for 10 min. DNA was counterstained with DAPI (Thermo Fisher Scientific D1306), destained with 2xSSCT for 1 hour, and mounted with VectaShield (Vector Laboratories H-1000). For metaphase analysis, gonad dissections were carried out directly on Poly-L-Lysine (0.01%, Sigma, cat. #P8920) charged slides by cutting the worm at the vulva. Slides were incubated for 30 min to 1 hour in ice-cold methanol. Primary monoclonal mouse anti-tubulin antibody (Sigma, cat. #T0926) was used at a 1:500 dilution and incubated overnight at room temperature. The following modifications were made for FISH: slides were incubated overnight at 37°C in SSCT containing 50% Formamide. DNA was denatured for 3 min at 93°C and probe was hybridized at 37°C for at least 48 hours. See Data S1B, Data S2, Data S3 and Data S4.
RNA interference by feeding
RNAi was carried out as previously described [56] with rad-54, emb-30 and pL4440 (empty vector) clones from the Ahringer Lab RNAi library [57]. To obtain a synchronized worm population, 10 plates with >1000 gravid balanced adults each were subjected to mild bleach (0.25M NaOH and 1% bleach non-germicidal) and incubated overnight in M9 with rocking. L1 larvae were seeded onto 20 fresh RNAi plates, grown until spo-11 homozygous mutants reached the L4 stage, then transferred to fresh RNAi plates and incubated 20*24h (for rad-54 RNAi) or 36 hours (for emb-30 RNAi) at 20°C. Heat shock and immunofluorescence analysis was carried out as described above. Worms were analyzed 24 (rad-54) or 36 (emb-30) hours post-heat shock.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Prism 8.0 (Graphpad) and R. The statistical analysis details (statistical test used, n value, mean, SD, SEM and confidence intervals) for each experiment can be found in the figure legends, supplemental figures and Data S1–S5. No data were excluded from analysis and the experiments were not randomized. Statistical significance was determined using p-values [0.033 (*), 0.002 (**), and <0.001 (***)].
Metaphase and anaphase I analysis
Metaphase and anaphase I stages were identified by immunofluorescence of alpha-tubulin. Analysis was performed only for nuclei in which the chromosomes started to separate and which exhibited low levels of FISH probe background signal.
Bivalent count at diakinesis
Bivalent formation was assessed by analyzing unprojected z-stacks encompassing whole nuclei. Only bivalents where the axis marker HTP-3 was observed localized along both chromosome axes (cruciform pattern) and the DAPI signal was observed on the same focal plane were scored. This excludes scoring of closely aligned univalents, but will also exclude bivalents oriented sideways on the slides.
GFP::COSA-1 foci count and LAB-1 andpH3 localization analysis
Quantification of GFP::COSA-1 foci and LAB-1 and pH3 localization studies were performed by analyzing unprojected z stacks encompassing whole nuclei at late pachytene.
Time course analysis for RAD-51 foci
Quantitative analysis of RAD-51 foci for mid-pachytene nuclei was performed as in [50]. The experiment was performed in duplicate and nuclei were scored from 4 gonads for each strain and condition. Statistical comparisons were performed using the two-tailed Mann-Whitney test, 95% C.I.
Analysis of premature sister separation at diakinesis
emb-30, which encodes for a component of the anaphase promoting complex and is required for metaphase to anaphase transition during meiosis I [58], was depleted by RNAi as described above. Whole-mounted gonads were co-stained with DAPI and anti-SMC-1 as described above. 30, 54, and 67 gonad arms were analyzed for wild type, spo-11 mutants with single Mos1 insertion at an off-center position (right arm) on chromosome III, and spo-11 mutants with single Mos1 insertion at the center of chromosome III after heat shock, respectively, from four biological repeats.
Computational straightening of chromosomes
Immunofluorescence using the axis marker HTP-3 was performed as described above. Chromosomes axes were traced in 3D and straightened employing Priism [4, 25].
Mathematical modeling
The region at the center of each chromosome occupies a proportion p of the whole chromosome. A break in this region produces a symmetric bivalent if there are no breaks elsewhere on the chromosome. One or more breaks elsewhere on the chromosome produce an asymmetric bivalent, while a complete lack of DSB formation results in a univalent. We analyzed the outcomes from two different types of experiments. In experiment one we scored the number of chromosomes observed in each configuration (symmetric bivalent, asymmetric bivalent or univalent) for every nucleus, without identifying specific chromosomes. In experiment two, FISH probes were utilized allowing us to determine the specific configurations observed for chromosomes III and V. The irradiation dosage in Rads is D and we assumed that this dose produces B breaks on each chromosome, where B is Poisson distributed with rate and λ is a nuisance parameter indicating the number of Rads needed to produce one break per chromosome on average. There are 3 possible outcomes for each chromosome:
There are no breaks on the chromosome, and we see a univalent. This occurs with probability .
There are B breaks on the chromosome where B>0 and they are all in the center. We observe a symmetric bivalent. This occurs with probability
There is at least 1 break outside the center of the chromosome, and we see an asymmetric bivalent. Since there are only three outcomes, this has probability ps = 1-pa-pu
Given these probabilities, the distribution of observation is multinomial with log-likelihood given by ℓ(p, λ, D, S, A, U) = S log(ps) + A log(pa) + U log(pu) where S, A and U are the number of symmetric, asymmetric and univalent chromosomes respectively. We used this log-likelihood to make inferences about p in two different ways. First, we obtained a maximum likelihood estimate (MLE) by maximizing the likelihood over p and λ, constraining p ∈ (0,1) and λ>0. Approximate Wald confidence intervals obtained through the difference in log- likelihood have poor coverage. For experiment 2, we obtained 95% confidence intervals (CI) by computing the binomial confidence interval for pa using the Wilson method, fixing λ at the MLE and inverting the formula for pa to obtain values for p at the end of the confidence interval. We also performed a Bayesian analysis. We assumed a uniform U(0,1) prior for p and a n independent Gaussian prior for λ with mean 250 and standard deviation 50. Using the likelihood above, we sampled from the posterior distribution using the Metropolis-Hastings algorithm with a multivariate Gaussian proposal density with standard deviations 0.01 and 2 and covariance 0. We ran the MCMC for one million iterations, starting at (p, λ) = (0.01,250), and report the posterior mean and the 95% equal-tailed credible interval (CrI). We ran this analysis on experiments one and two separately (Data S5C).
Assessment of chromosome remodeling and segregation upon Mos1-induced DSB formation
LAB-1 signal is not observed on chromosome III in 27% (n=277) of late pachytene nuclei following an off-centered Mos1-induced DSB. Consequently, lack of LAB-1 signal was also observed on a fraction of single bivalents at diakinesis. This outcome could be due to a surveillance mechanism proposed to play a role in sensing DSB or CO levels in the C. elegans germline which is turned off only when sufficient levels are detected on all chromosomes [59]. Alternatively, SPO-11-induced DSBs may be required in addition to exogenous DSBs to promote proper repair via HR [60]. Yet another explanation might be that the kinetics of DSB end resection/repair may be altered in the presence of only a single DSB compared to wild-type levels of DSBs, as observed in yeast [61]. We excluded the possibility that the lack of LAB-1 at diakinesis is a result of Mos1 transposon-induced DSBs since we observe the same outcome when low levels of DSBs are formed by γ-irradiation (Figure 4), which are distributed randomly along chromosomes (Figure S3D). Importantly, the exposure of spo-11 worms to γ-irradiation has been shown to increase embryonic viability [21], arguing that these breaks can compensate for the lack of SPO-11-induced DSBs.
Strikingly, analysis of the CO precursor marker COSA-1 revealed a dramatic increase in the formation of GFP::COSA-1 foci after heat shock-induced Mos1 excision at either the center or the arm of chromosome III. These results further validate that Mos1-induced DSBs are proficient in inducing HR. The slight differences in the levels of foci formed between DSBs at the center and arm locations could originate either from the local chromatin environment favoring the accessibility of the transposon for excision or silencing, but could also derive from antibody performance variability during FISH experiments. However, the difference in the number of COSA-1 foci observed in nuclei at late pachytene compared to the number of bivalents observed in oocytes at diakinesis raises the question of whether some of these CO precursor sites are eventually converted to noncrossover (NCO) events and/or if a fraction of COSA-1 foci could also mark intersister repair intermediates. It is also possible that some of the late pachytene nuclei are eliminated by heat shock-induced apoptosis [62] and that some COSA-1 foci show ectopic localization (background) since we observe a general increase in COSA-1 foci formation after heat shock potentially due to increased expression of the GFP::COSA-1 transgene at higher temperature [63].
The existing published evidence supporting the necessity of correct LAB-1 and AIR- 2/pH3 localization for accurate maintenance of cohesion between homologs and sister chromatids [15, 17] prompted us to further corroborate our findings by directly showing a loss of sister chromatid cohesion after inducing crossover formation at the center of chromosome III. The low frequency of bivalent formation at diakinesis coupled with the low levels of viable homozygous progeny yielded by the transgenic line was a bottleneck for live imaging analysis through metaphase and anaphase I. Therefore, we analyzed immunostained fixed gonads in combination with FISH, which allowed us to identify chromosome III, with the caveat of a low yield of observed late metaphase and anaphase I. We screened through more than 900 gonads per genotype and only analyzed nuclei in which chromosomes were observed starting to separate after metaphase I alignment. We monitored the FISH probe signal to assess premature sister chromatid separation (evidenced by two to four foci instead of a single focus) and observed a higher frequency of loss of sister chromatid cohesion for a center CO event compared to an arm CO event, which emphasizes again that a center CO will result more likely in chromosome missegregation.
Based on our data, we used mathematical modeling to calculate the length of the region at the center of chromosomes III (one of the smallest chromosomes in the C. elegans genome) and V (one of the largest) that is avoided for CO formation. Our estimates for this region for both chromosomes are up to 6% of the total chromosome length. This result is supported by our findings in two ways. First, the Mos1 insertion site for chromosome II (ttTi5605) is located approximately 5% away from the physical center and its excision only resulted in single symmetric bivalents without detectable LAB-1. Given that the length of the center region underlies a Gaussian distribution, we cannot exclude the possibility that screening a very high number of diakinesis nuclei could potentially unveil a single bivalent with LAB-1 localization similar to bivalents in wild type. Second, one of our Mos1 insertions for chromosome III (ttTi2384) is, based on the genetic map, located within the CO-deprived center region but at very close proximity to the CO-enriched arm region (Figures 1b and S5a; border of center/off-center). In this scenario, excision of the Mos1 transposon leads to normal LAB-1 localization 44% of the time. Taken together, these results support the modeling predictions that the region at the center of the chromosomes that can lead to the highest frequency of abnormal chromosome remodeling following DSB induction and CO formation encompasses 5-6% of the total chromosome length.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets/code.
Supplementary Material
Data S1: Strains, bivalent frequency and LAB-1 and pH3 localization. Related to STAR Methods and Figures 1, S2A, S2B, S4A, and S4B. (A) List of strains used in this study. (B) Raw data for bivalent frequency and LAB-1 and pH3 localization analysis.
Data S2: Statistical analysis on LAB-1 localization. Related to STAR Methods and Figures 1, S2A, S2B, S4A, and S4B. (A) Mean, SD and N for LAB-1 localization at late pachytene stage. (B) 2 way ANOVA with multiple comparison on LAB-1 localization at late pachytene stage.
Data S3: Statistics of GFP::COSA-1 foci. Related to STAR Methods and Figure 3A and 3B. Mean, SD and N values for GFP::COSA-1 foci count at late pachytene stage.
Data S4: Statistical analysis on GFP::COSA-1 foci. Related to STAR Methods and Figure 3A and 3B. 2 way ANOVA with multiple comparison on GFP::COSA-1 foci count at late pachytene stage.
Data S5: Statistics and data of mathematical modeling. Related to STAR Methods and Figures 3C, 4 and S4D–F. (A) Mean and SD on GFP::COSA-1 foci at late pachytene stage from center CO formation rescued by IR. (B) Mean and SD on LAB-1 localization at bivalents. (C) Mean and SD on bivalent formation obtained after γ-IR exposure of spo-11 mutants. (D) Mean and SD on pH3 localization and on corresponding bivalent formation obtained after γ-IR exposure of spo-11 mutants. (E) Data of mathematical modeling on asymmetric and symmetric bivalent formation obtained after γ-IR exposure of spo-11 mutants.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-LAB-1 | [15] | N/A |
| guinea pig anti-HTP-3 | [51] | N/A |
| rabbit anti-phospho-Histone H3 (Ser10) | Millipore | Cat# 06-570; RRID: AB_310177 |
| rat anti-SMC-1 | [52] | N/A |
| mouse anti-GFP | Roche | Cat# 11814460001; RRID: N/A |
| donkey anti-rabbit Cy5 | Jackson Immunoresearch | Cat# 711-175-152; RRID: AB_2340607 |
| donkey anti-guinea pig Alexa488 | Jackson Immunoresearch | Cat# 706-545-148; RRID: AB_2340472 |
| donkey anti-rabbit Cy3 | Jackson Immunoresearch | Cat# 711-165-152; RRID: AB_2307443 |
| goat anti-mouse Alexa488 | Jackson Immunoresearch | Cat# 115-546-146; RRID: AB_2338868 |
| donkey-anti-guinea pig Cy3 | Jackson Immunoresearch | Cat# 706-165-148; RRID: AB_2340460 |
| mouse anti-tubulin | Sigma Aldrich | Cat# T0926; RRID: N/A |
| rabbit anti-RAD-51 | Novus Biological (SDI) | Cat# 29480002; RRID: AB_2616441 |
| Bacterial and Virus Strains | ||
| Ahringer RNAi library clone: pL4440-W06D4.6 | Source Bioscience | N/A |
| Ahringer RNAi library clone: pL4440-F54C8.3 | Source Bioscience | N/A |
| Ahringer RNAi library clone: pL4440 | Source Bioscience | N/A |
| cosmid for chromosome III FISH probe: T03F6 (III, right) | Sanger Center | WB ID: T03F6 |
| cosmid for chromosome III FISH probe: cosmid F11F1 (III, right) | Sanger Center | WB ID: F11F1 |
| cosmid for chromosome III FISH probe: cosmid T28A8 (III, right) | Sanger Center | WB ID: T28A8 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fluorescein-12-dCTP | Perkin Elmer | Cat# NEL-424 |
| Cyanine 3-CTP | Perkin Elmer | Cat# NEL580001EA |
| Poly-L-Lysine (0.01%) | Sigma | Cat# P8920 |
| CytoBond | SciGene | Cat# 2020-00-1 |
| DAPI | Thermo Fisher Scientific | Cat# D1306 |
| VectaShield | Vector Laboratories | Cat# H-1000 |
| TOPO® TA Cloning® kit | Thermo Fisher Scientific | Cat#451641 |
| Experimental Models: Organisms/Strains | ||
| N2 (wild type) | https://cgc.umn.edu/strain/N2 | WB ID: WBStrain00000001 |
| CV620: F14E5.8(ttTi5605); spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (II;IV;V) | this paper | N/A |
| AV630: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] (II) | [26] | WB strain: AV630 WB ID: WBStrain00000296 |
| CV451: meIs8[unc-119(+) Ppie-1::gfp::cosa-1]; T07E3.4(ttTi23808); spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (II;III;IV;V) | this paper | N/A |
| CV613: meIs8[unc-119(+) Ppie-1::gfp::cosa-1]; ttTi13055; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (II;III;IV;V) | this paper | N/A |
| CV614: meIs8[unc-119(+) Ppie-1::gfp::cosa-1]; ttTi17604; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (II;III;IV;V | this paper | N/A |
| CV686: meIs8[unc-119(+) Ppie-1::gfp::cosa-1]; spo-11(ok79) IV/]/nT1 [unc-?(n754) let-?] (II,IV) | this paper | N/A |
| CV616: ttTi1730; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this paper | N/A |
| CV405: T07E3.7(ttTi23808); spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this paper | N/A |
| CV760: T07E3.7(ttTi23808); krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this paper | N/A |
| CV771: tTi2384; ; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this paper | N/A |
| CV561: ttTi13055; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this paper | N/A |
| CV683: ttTi17604; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;IV;V) | this study | N/A |
| CV838: T07E3.7(ttTi23808); ttTi13055; spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (III;III;IV;V) | this paper | N/A |
| AV106: spo-11(ok79) IV/nT1 [unc-?(n754) let-?] (IV) | [21] | WB strain: AV106; WB ID: WBVar00091464 |
| CV394: spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (IV;V) | this paper | N/A |
| EN909: krls14[Prom(hsp)::transposase; lin-15B; cc::GFP] | [20] | N/A |
| CV485: spo-11(ok79) IV/nT1 [unc-?(n754) let-?]; C35A5.14(ttTi35006); krls14[Prom(hsp)::transposase; lin-15B; cc::GFP]/nT1 [unc-?(n754) let-?] (IV;V;V) | this paper | N/A |
| Oligonucleotides | ||
| FISH probe for chromosome V: 5S_F: 5’-TACTTGGATCGGAGACGGCC-3’ | [21] | N/A |
| FISH probe for chromosome V: 5S_R: 5’-CTAACTGGACTCAACGTTGC-3’ | [21] | N/A |
| genotyping primer: ttTi23808_F: 5’-CTGCGCGAAGATTATCCCGACGA-3’ | this paper | N/A |
| genotyping primer: ttTi23808_R: 5’-CGCGAATCGTACTCCATCAATGC-3’ | this paper | N/A |
| genotyping primer: ttTi35006_F: 5’-CATCATGCTCCTAACACGTGTGG-3’ | this paper | N/A |
| genotyping primer: ttTi35006_R: 5’-TCTGAAAGCTCGGATATGAGCTAAC-3’ | this paper | N/A |
| genotyping primer: ttTi2384_F: 5’-GACGTAGAAATCAACTGGAC-3’ | this paper | N/A |
| genotyping primer: ttTi2384_R: 5’-AAGACTCCCATTATTTGGTC-3’ | this paper | N/A |
| genotyping primer: ttTi1730_F: 5’-TTCCCTAGTCCCATGTTTTGGTT-3’ | this paper | N/A |
| genotyping primer: ttTi1730_R: 5’-CCACTTCCCAAACCACCAAAAAT-3’ | this paper | N/A |
| genotyping primer: ttTi13055_F: 5’-CTTAAAGGCGATTTGATGCACCA-3’ | this paper | N/A |
| genotyping primer: ttTi13055_R: 5’-GGGCCTAATTATACCTAAGCCCC-3’ | this paper | N/A |
| genotyping primer: ttTi5605_F: 5’-CGGCCGAAATTTTACTTGATCGA-3’ | this paper | N/A |
| genotyping primer: ttTi5605_R: 5’-ACACCCGGGTTTGTCTAGATATG-3’ | this paper | N/A |
| genotyping primer: ttTi17604_F: 5’-CTCTCTCTGAAATGAAAACAAG-3’ | this paper | N/A |
| genotyping primer: ttTi17604_R: 5’-CTTGGACCAGTTACAACATC-3’ | this paper | N/A |
| Software and Algorithms | ||
| SoftWoRx | GE Healthcare Life Sciences | N/A |
| Priism | [4, 25, 64] | https://msg.ucsf.edu |
| XQuartz | [4, 25, 64] | https://www.xquartz.org |
| ImageJ Fiji | [53, 54, 65] | https://imagej.net/Fiji |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| R Studio | [66] |
https://www.r-project.org https://rstudio.com |
Highlights.
CO position affects the formation of chromosome subdomains.
COs at the center of autosomes lead to premature loss of sister chromatid cohesion.
The CO-unfavorable center region encompasses up to 6% of the total chromosome length.
CO formation close to telomeres is highly suppressed.
ACKNOWLEDGEMENTS
We thank members of the Colaiacovo laboratory for discussions. We thank Barbara J. Meyer for the SMC-1 antibody, Monique Zetka for the HTP-3 antibody, Anne Villeneuve and the Biology of Caenorhabditis elegans Facility-CNRS UMS 3421 for strains, and Verena Jantsch for protocols. This work was supported by a CONACYT-Mexico (No. 275396) postdoctoral fellowship to L.I.L.-L. and a National Institutes of Health grant R01GM072551 to M.P.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hunter N (2015). Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol 7, a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray S, and Cohen PE (2016). Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet 50, 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeney S, Lange J, and Mohibullah N (2014). Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet 48, 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mets DG, and Meyer BJ (2009). Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, and Fung JC (2008). Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauppi L, Jeffreys AJ, and Keeney S (2004). Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet 5, 413–424. [DOI] [PubMed] [Google Scholar]
- 7.Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, and Mézard C (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7, e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Massy B (2013). Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet 47, 563–599. [DOI] [PubMed] [Google Scholar]
- 9.Hillers KJ, and Villeneuve AM (2003). Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol 13, 1641–1647. [DOI] [PubMed] [Google Scholar]
- 10.Lamb NE, Sherman SL, and Hassold TJ (2005). Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet Genome Res 111, 250–255. [DOI] [PubMed] [Google Scholar]
- 11.Muller HJ (1916). The mechanism of crossing-over. Am. Nat 50, 193–221. [Google Scholar]
- 12.Sturtevant AH (1915). The behavior of the chromosomes as studied through linkage. Z.Ver-erbungslehre 13, 234–287. [Google Scholar]
- 13.Nabeshima K, Villeneuve AM, and Colaiácovo MP (2005). Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol 168, 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, and Villeneuve AM (2008). Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev 22, 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Carvalho CE, Zaaijer S, Smolikov S, Gu Y, Schumacher JM, and Colaiacovo MP (2008). LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev 22, 2869–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzur YB, Egydio de Carvalho C, Nadarajan S, Van Bostelen I, Gu Y, Chu DS, Cheeseman IM, and Colaiácovo MP (2012). LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 10, e1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers E, Bishop JD, Waddle JA, Schumacher JM, and Lin R (2002). The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol 157, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robert V, and Bessereau J-L (2007). Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26, 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert VJ, Davis MW, Jorgensen EM, and Bessereau J-L (2008). Gene conversion and end-joining-repair double-strand breaks in the Caenorhabditis elegans germline. Genetics 180, 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosu S, Libuda DE, and Villeneuve AM (2011). Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334, 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, and Villeneuve AM (1998). Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398. [DOI] [PubMed] [Google Scholar]
- 22.Hassold T (2000). Counting cross-overs: characterizing meiotic recombination in mammals. Hum. Mol. Genet 9, 2409–2419. [DOI] [PubMed] [Google Scholar]
- 23.Tease C, Hartshorne GM, and Hultén MA (2002). Patterns of meiotic recombination in human fetal oocytes. Am. J. Hum. Genet 70, 1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solinger JA, Kiianitsa K, and Heyer W-D (2002). Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 25.Saito TT, Mohideen F, Meyer K, Harper JW, and Colaiácovo MP (2012). SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLoS Genet. 8, e1002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, and Villeneuve AM (2012). COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJM, Davis EB, Scherer S, Ward S, et al. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- 28.Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung M-S, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. (2011). Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam I, and Keeney S (2015). Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol 7, a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambie EJ, and Shirleen Roeder G (1988). A yeast acts in (Cis) to inhibit meiotic gene conversion of adjacent sequences. Cell 52, 863–873. [DOI] [PubMed] [Google Scholar]
- 31.Talbert PB, and Henikoff S (2010). Centromeres convert but don’t cross. PLoS Biol. 8, e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan P, Copsey A, Newnham L, Kolar E, Lichten M, and Hoffmann E (2009). Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis. Genes Dev 23, 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obeso D, Pezza RJ, and Dawson D (2014). Couples, pairs, and clusters: mechanisms and implications of centromere associations in meiosis. Chromosoma 123, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, and Hunter N (2012). Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 8, e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisig CG, Guiraldelli MF, Kouznetsova A, Scherthan H, Höög C, Dawson DS, and Pezza RJ (2012). Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet. 8, e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamb NE, Yu K, Shaffer J, Feingold E, and Sherman SL (2005). Association between maternal age and meiotic recombination for Trisomy 21. Am. J. Hum. Genet 76, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagaoka SI, Hassold TJ, and Hunt PA (2012). Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet 13, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver TR, Tinker SW, Allen EG, Hollis N, Locke AE, Bean LJH, Chowdhury R, Begum F, Marazita M, Cheung V, et al. (2012). Altered patterns of multiple recombinant events are associated with nondisjunction of chromosome 21. Hum. Genet 131, 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver TR, Middlebrooks CD, Tinker SW, Allen EG, Bean LJH, Begum F, Feingold E, Chowdhury R, Cheung V, and Sherman SL (2014). An examination of the relationship between hotspots and recombination associated with chromosome 21 nondisjunction. PLoS ONE 9, e99560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lui DY, and Colaiácovo MP (2013). Meiotic development in Caenorhabditis elegans In Germ Cell Development in C. elegans, Schedl T, ed. (New York, NY: Springer New York; ), pp. 133–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middlebrooks CD, Mukhopadhyay N, Tinker SW, Allen EG, Bean LJH, Begum F, Chowdhury R, Cheung V, Doheny K, Adams M, et al. (2014). Evidence for dysregulation of genome-wide recombination in oocytes with nondisjoined chromosomes 21. Hum. Mol. Genet 23, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockman MV, and Kruglyak L (2009). Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5, e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadarajan S, Lambert TJ, Altendorfer E, Gao J, Blower MD, Waters JC, and Colaiácovo MP (2017). Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife 6, e23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattabiraman D, Roelens B, Woglar A, and Villeneuve AM (2017). Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13, e1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallin E, Gallagher J, Granger L, Martin E, Belougne J, Maurizio J, Duverger Y, Scaglione S, Borrel C, Cortier E, et al. (2012). A genome-wide collection of Mos1transposon insertion mutants for the C. elegans research community. PLoS ONE 7, e30482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulin T, and Bessereau J-L (2007). Mos1-mediated insertional mutagenesis in Caenorhabditis elegans. Nat. Protoc 2, 1276–1287. [DOI] [PubMed] [Google Scholar]
- 48.Malkova A, Klein F, Leung W-Y, and Haber JE (2000). HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc. Natl. Acad. Sci. USA. 97, 14500–14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaramillo-Lambert A, Ellefson M, Villeneuve AM, and Engebrecht J (2007). Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol 308, 206–221. [DOI] [PubMed] [Google Scholar]
- 50.Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, and Villeneuve AM (2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474. [DOI] [PubMed] [Google Scholar]
- 51.Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, and Zetka M (2008). HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14, 263–274. [DOI] [PubMed] [Google Scholar]
- 52.Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, and Meyer BJ (2003). Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423, 1002–1009. [DOI] [PubMed] [Google Scholar]
- 53.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smolikov S, Eizinger A, Schild-Prufert K, Hurlburt A, McDonald K, Engebrecht J, Villeneuve AM, and Colaiácovo MP (2007). SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176, 2015–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Govindan JA, Cheng H, Harris JE, and Greenstein D (2006). Gαo/i and Gαs signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol 16, 1257–1268. [DOI] [PubMed] [Google Scholar]
- 57.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 58.Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, Rose AM, and Greenstein D (2000). EMB-30: An APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. MBoC 11, 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machovina TS, Mainpal R, Daryabeigi A, McGovern O, Paouneskou D, Labella S, Zetka M, Jantsch V, and Yanowitz JL (2016). A surveillance system ensures crossover formation in C. elegans. Curr. Biol 26, 2873–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medhi D, Goldman AS, and Lichten M (2016). Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5, e19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neale MJ, Ramachandran M, Trelles-Sticken E, Scherthan H, and Goldman ASH (2002). Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Molecular Cell 9, 835–846. [DOI] [PubMed] [Google Scholar]
- 62.Salinas LS, Maldonado E, and Navarro RE (2006). Stress-induced germ cell apoptosis by a p53 independent pathway in Caenorhabditis elegans. Cell Death Differ. 13, 2129–2139. [DOI] [PubMed] [Google Scholar]
- 63.Merritt C (2010). Transgenic solutions for the germline. WormBook, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H, Clyborne WK, and JSIP. (1992). PRIISM: an integrated system for display and analysis of 3-D microscope images. Proc. SPIE 1660, Biomedical Image Processing and Three-Dimensional Microscopy. 10.1117/12.59604 [DOI]
- 65.Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Core Team (2011). A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Strains, bivalent frequency and LAB-1 and pH3 localization. Related to STAR Methods and Figures 1, S2A, S2B, S4A, and S4B. (A) List of strains used in this study. (B) Raw data for bivalent frequency and LAB-1 and pH3 localization analysis.
Data S2: Statistical analysis on LAB-1 localization. Related to STAR Methods and Figures 1, S2A, S2B, S4A, and S4B. (A) Mean, SD and N for LAB-1 localization at late pachytene stage. (B) 2 way ANOVA with multiple comparison on LAB-1 localization at late pachytene stage.
Data S3: Statistics of GFP::COSA-1 foci. Related to STAR Methods and Figure 3A and 3B. Mean, SD and N values for GFP::COSA-1 foci count at late pachytene stage.
Data S4: Statistical analysis on GFP::COSA-1 foci. Related to STAR Methods and Figure 3A and 3B. 2 way ANOVA with multiple comparison on GFP::COSA-1 foci count at late pachytene stage.
Data S5: Statistics and data of mathematical modeling. Related to STAR Methods and Figures 3C, 4 and S4D–F. (A) Mean and SD on GFP::COSA-1 foci at late pachytene stage from center CO formation rescued by IR. (B) Mean and SD on LAB-1 localization at bivalents. (C) Mean and SD on bivalent formation obtained after γ-IR exposure of spo-11 mutants. (D) Mean and SD on pH3 localization and on corresponding bivalent formation obtained after γ-IR exposure of spo-11 mutants. (E) Data of mathematical modeling on asymmetric and symmetric bivalent formation obtained after γ-IR exposure of spo-11 mutants.
Data Availability Statement
This study did not generate/analyze datasets/code.


