Graphical abstract
Keywords: Marine food, Musculus senhousei, Pyropheophorbide a, Anti-influenza A virus activity, Enveloped virus
Highlights
-
•
Pyropheophorbide a (PPa) was isolated from the seafood of M. senhousei.
-
•
PPa shows a potent activity against a broad panel of influenza A viral strains.
-
•
The mechanism of PPa is to block the entry of virus in the early stage of infection.
-
•
The target of PPa may be that of lipid bilayer of the enveloped viruses.
Abstract
Marine environments are known to be a new source of structurally diverse bioactive molecules. In this paper, we identified a porphyrin derivative of Pyropheophorbide a (PPa) from the mussel Musculus senhousei (M. senhousei) that showed broad anti-influenza A virus activity in vitro against a panel of influenza A viral strains. The analysis of the mechanism of action indicated that PPa functions in the early stage of virus infection by interacting with the lipid bilayer of the virion, resulting in an alteration of membrane-associated functions, thereby blocking the entry of enveloped viruses into host cells. In addition, the anti-influenza A virus activity of PPa was further assessed in mice infected with the influenza A virus. The survival rate and mean survival time of mice were apparently prolonged compared with the control group which was not treated with the drug. Therefore, PPa and its derivatives may represent lead compounds for controlling influenza A virus infection.
Abbreviations
- PPa
Pyropheophorbide a
- IAV
Influenza A viruse
- NA
neuraminidase
- HA
hemagglutinin
- MDCK
Madin Darby Canine Kidney
- IC50
half maximal inhibitory concentration
- CC50
half maximal cytoxic concentration
- TCID50
50% tissue culture infective dose
- MTT
3-(4,5-di methyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
- CPE
cytopathic effect
- ITC
isothermal titration calorimetry
- TPCK
tosyl-phenylalanine-chloromethyl-ketone
- FITC
fluorescein isothiocyanate
- DAPI
4,6-diamino-2-phenylindole
1. Introduction
Musculus senhousei is a traditional Chinese seafood that is widely distributed all over the Pacific coast including the USA, Australia, Japan, and China, among other countries [1]. This mussel is reported to live in the region ranging from intertidal to shallow subtidal zones at the depth of approximately 30 m, and it is tolerant of low salinity and low oxygen levels during its life span of ca. two years (http://www.exoticsguide.org/musculista_senhousia). The outside shell of M. senhousei is smooth and shiny with a yellow-green color and can grow to a maximum length of 35 mm, while its interior is purplish-gray. Analyses of the habitat and growth conditions indicate that M. senhousei is a passive filter-feeding shellfish. Thus, in addition to a small number of protozoa, the main component of its food is diatoms, which belong to 20 different genera [2].
Marine environments have long been viewed as a major reservoir of bioactive molecules that have the potential to be developed as therapeutic drugs [3]. In our continuous search for anti-influenza A viral compounds from natural sources [4], [5], [6] using the H5N1 pseudo-typed virus screening approach, we identified the traditional Chinese seafood M. senhousei as showing a good inhibitory activity toward influenza A virus (IAV). The preliminary mechanistic study indicated that the antiviral activity of this food resulted from the inhibition of virus entry during early infection. We then investigated the bioactive components of this mussel using a bioassay-guided approach, from which a porphyrin derivative named pyropheophorbide a (PPa) that showed significant anti-IAV activity in vitro was isolated and identified, indicating a potential application of this molecule in the development of new antiviral agents.
IAVs are enveloped viruses, and the viral envelopes are derived from portions of the host cell membranes and function to cover the capsids to protect the packaged viral genome [7]. In addition to the lipid bilayer, the viral envelope also contains viral glycoproteins, such as hemagglutinin (HA), neuraminidase (NA) and the viral M2 protein. These components, including lipid bilayers and associated proteins, play important roles in the process of viral infection [8], [9]. As a result, they are viewed as promising tools for the development of new anti-influenza A drugs [10].
The HA glycoprotein consisting of two subunits, HA1 and HA2, is located on the surface of the envelope and is responsible for binding to receptor sites on the host membrane (HA1) and mediating the fusion between viral and host membranes (HA2). Following fusion, the viral genome is able to enter and subsequently infect the host cells [11].
The lipid bilayer is a major component of the IAV envelope. To date, a number of molecules targeting virion envelope lipids to interfere with the fusion of viral-host cellular membranes have been reported [12], [13]. These molecules convey their antiviral effects through biophysical mechanisms, due to the molecular shapes and amphipathicity, thereby inhibiting the formation of the negative curvature in envelope lipid bilayers [14]. Consequently, these compounds show broad antiviral activity toward many unrelated enveloped viruses.
Similarly, in this study, the identified compound PPa displayed anti-IAV activity in the early stage of virus entry, while further experimental data indicated that PPa did not mainly act on the HA glycoprotein or its HA1 and HA2 subunits. Instead, it showed a strong tendency to interact with envelope lipids, thus suggesting that the fusion of viral and cellular membranes might be interrupted following the interactions between PPa and viral envelope lipids. Herein, we report the anti-IAV activity and the possible mechanisms of action of PPa.
2. Materials and methods
2.1. Chemicals and analytical instruments
NMR spectroscopic data were acquired in CDCl3 solution (Macklin, China) at 400 MHz for 1H and 100 MHz for 13C respectively with a Bruker DRX-400 spectrometer. Chemical shifts were referenced to the corresponding solvent residual signal (7.26/77.23 in CDCl3). Low-resolution ESI-MS were recorded using a Waters 3100 single-quadrupole mass spectrometer. Silica gel (300–400 mesh, Qingdao Marine Chemical Factory, China), Sephadex LH-20 (Amersham Pharmacia Biotech) and reverse-phase silica gel C18 (40–63 μm, Merck) were used for the column chromatography. Positive control of oseltamivir phosphate was purchased from Yichang Dongyangguang Pharmaceutical Co., Ltd.
2.2. Cells and viruses
Madin Darby canine kidney (MDCK) cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Sigma, USA) supplemented with 10% fetal bovine serum (FBS, Sigma, USA) and 1% penicillin/streptomycin. Influenza A subtypes include A/FM-1/1/47 (H1N1) mouse adjustment strain, A/Puerto Rico/8/34 (H1N1) with NA-H274Y mutation, and A/Aichi/2/68 (H3N2) were propagated in 9-day-old chick embryo at 37 °C, and the allantoic fluid containing the above viruses were stored at −80 °C and quantified in a 50% tissue culture infectious dose (TCID50) test until needed.
2.3. Extraction and isolation of pyropheophorbide a
Specimens of Musculus senhousei were collected from the seafood market in Guangzhou, China, in March 2013. A voucher of specimen (No. HGZ) was deposited at the Group of peptides and natural products Research, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou, P. R. China.
The fresh mussels (40 kg) were grounded, and repeatedly extracted for at least three times in 95% EtOH (3 × 20 L) at room temperature. The EtOH extracts were evaporated under reduced pressure to afford a dark brown semi-solid (250 g), which was then suspended in water and partitioned sequentially with petroleum ether (30 g), dichloromethane (18 g), and EtOAc (6.5 g). A portion of the EtOAc fraction (1.06 g) was subjected to silica gel column chromatography and was eluted with a gradient of petroleum ether and EtOAc (2:1 to 1:4) to yield three fractions (Fr. 1–3). The most active fraction was then eluted with a gradient the mixture of CH2Cl2 and MeOH (from 2:1 to 1:2) to afford another three fractions (Fr. 4–6), of which Fr. 5 was further purified by reversed-phase silica gel (RP-18) with an eluent of CH3CN : MeOH: H2O (4:4:2) to afford PPa (25 mg).
2.3.1. Pyropheophorbide a (PPa)
Dark green solids, 1H NMR (CDCl3, 400 Hz) δ: 9.48 (1H, s, H-10), 9.37(1H, s, H-5), 8.54 (1H, s, H-20), 7.99 (1H, m, H-31), 6.28 (1H, dd, J = 1.28, 18.24 Hz, H-32 (trans)), 6.16 (1H, dd, J = 1.32, 15.52 Hz, H-32 (cis)), 5.27 (1H, d, J = 19.88 Hz, H-132), 5.12 (1H, d, J = 19.84 Hz, H-132), 4.48 (1H, m, H-18), 4.30 (1H, m, H-17), 3.67 (1H, q, J = 17.64 Hz, H-81), 3.65 (3H, m, H-121), 3.40 (3H, s, H-21), 3.23 (3H, s, H-71), 2.70 (1H, m, H-171), 2.29 (1H, m, H-172), 1.81 (3H, d, J = 7.28 Hz, H-181), 1.68 (3H, t, J = 7.64 Hz, H-82). 13C NMR (CDCl3, 100 Hz) δ: 196.7 (s, C-131), 177.3 (s, C-173), 171.6 (s, C-19), 160.4 (s, C-16), 155.6 (s, C-6), 151.0 (s, C-14), 149.3 (s, C-9), 145.3 (s, C-8), 141.9 (s, C-1), 138.1 (s, C-11), 136.5 (s, C-4), 136.3 (s, C-7), 136.1 (s, C-3), 131.9 (s, C-2), 130.6 (s, C-12), 129.4 (d, C-31), 128.6 (s, C-13), 122.8 (t, C-32), 106.2 (s, C-15), 104.3 (d, C-10), 97.4 (d, C-5), 93.3 (d, C-20), 51.2 (d, C-17), 50.2 (d, C-18), 48.2(t, C-132), 30.8 (t, C-172), 29.9 (t, C-171), 23.4 (q, C-181), 19.7 (t, C-81), 17.6 (q, C-82), 12.3 (q, C-121), 12.3 (q, C-21), 11.5 (q, C-71). m/z 534.0 [M - H]−. These data are the same as those reported in the literature [15], [16].
2.4. Cytotoxicity assay
The cytotoxicity of PPa on MDCK cells was evaluated by MTT assay as described before [17]. Briefly, MDCK cells were prepared in 96-well plates (1 × 104 cells for each well) for 24 h and exposed to PPa in a 2-fold serial dilution. After incubation for 48 h, 100 μL of MTT (Sigma, USA) solution (0.5 mg/mL DMEM diluent) was added and left at 37 °C for 4 h. Subsequently, the supernatant was removed, and 150 μL of DMSO solution was added to plate to dissolve the formazan product. The absorbance of each well was measured at 570 nm by using a Multiskan FC microplate reader (Thermo Fisher Scientific, Massachusetts, USA).
2.5. Anti-influenza A virus assay
MDCK cells (ATCC) were cultured in 96-wellg plates (2 × 104 cells/well) for 24 h. A series of double-diluted PPa solutions was pre-incubated with 100 TCID50 of the virus at 37 °C for 30 min, and these cells were incubated with a virus-compound mixture for 1 h after two washes with PBS. Then, 1 μg/mL of TPCK-trypsin (trypsin treated with TPCK, Sigma, USA) in serum-free DMEM was added to the cells. Next, cell viability was measured using the MTT method at 48 h after the infection. S-KKWK [17] was used as a positive control, and the experiment was independently repeated at least three times. Virus subtypes such as the A/FM-1/1/47 (H1N1) mouse-adapted strain, A/Puerto Rico/8/34 (H1N1), A/Puerto Rico/8/34 (H1N1) with the NA-H274Y mutation, and A/Aichi/2/68 (H3N2) were selected to evaluate the antiviral effects of PPa.
2.5.1. Anti-SARS-CoV-2 assay
Anti-SARS-CoV-2 assay was processed as reported previously [18]. Briefly, 5 × 104 Vero-E6 cells/well were seeded in 48-well plates at 37 °C overnight. To start the assay, SARS-CoV-2 virus (MOI of 0.05) was pre-incubated with gradiently diluted PPa at 37 °C for 30 min, then the mixture was transferred to the cells and incubated for another 1 h. After incubation, cells were washed with PBS and added with the fresh medium for 24 h. Then cell supernatants were collected and subjected to viral RNA isolation, then qRT-PCR was performed to measure expression of the S gene of SARS-CoV-2 [18].
2.5.2. Inhibition on respiratory syncytial virus (RSV) by immuno-based plaque assay
Generally, serially diluted PPa in 100 μL 2% FBS DMEM were incubated with 200 plaque forming units (PFUs) of RSV (A2 strain) in equal volume of 2% FBS DMEM for 1 h at 37 °C. Then 150 μL mixture were added to each well of 24-well plate seeded with Vero-E6 cell to near confluency. Then the plate was incubated at 37 °C for 1 h before replacing the mixture with 1% methylcellulose DMEM. Five days later, methylcellulose medium was thoroughly removed and cells were fixed with 4% formaldehyde, permeabilized with 0.1% triton and blocked with 5% nonfat-milk. Mouse antisera against RSV Fusion protein diluted at 1:300 w incubated with cells for 1 h at 37 °C and washed three time by PBS-0.1% Tween 20. Cells were then incubated with second antibody-conjugated with horseradish peroxidase (catalogue number: SA00001, Proteintech, China) for 1 h at room temperature and washed three times as usual. Finally, plaques appeared 15 mins later after adding 3, 3′, 5, 5′-tetramethylbenzidine reagent. Plaques in each well were manually counted, no-compound well were made as control in each test.
2.6. Antiviral assay with different drug administration protocols
Four different drug administration protocols were used in this study to assess the possible mechanisms of PPa, as described in a previous study [19]. (1) Pretreatment of cells: After pre-incubating cells with PPa at 37 °C for 30 min, the cells were infected with the influenza virus A/PR/8/34 (H1N1) (100 TCID50). (2) Pretreatment of the virus: Influenza virus A/PR/8/34 (H1N1) (100 TCID50) was added to MDCK cells after a pre-incubation with PPa for 30 min at 37 °C. (3) During infection: The mixture of PPa and virus was added to the cells simultaneously. (4) After infection: PPa was added after the virus adsorbed onto the cells. The cells were washed with PBS and cultured with serum-free serum DMEM (Sigma, USA) containing 1 μg/mL TPCK-trypsin for 48 h. At the end of the incubation, the MTT assay was performed to evaluate cell survival. The cytopathic effect (CPE) on virus-infected cells was observed under the microscope.
2.7. Plaque reduction assay
MDCK cells were cultured in 6-well plates (4 × 105 cells/well) for 24 h. Antiviral effects were evaluated using two drug treatment approaches: “During infection” and “Pretreatment of the virus”, as mentioned above [17]. After treatment, cells were washed with PBS to remove the free virus and then cultured in 3 mL of serum-free MEM (2×) containing 1× TPCK-trypsin (1.6% AGAR for 72 h, as mentioned above). The cells were then fixed with 4% paraformaldehyde for 20 min, and then incubated with a 0.5% (w/v) crystal violet dye solution for 1 h at 37 °C. The effect of PPa on viral plaque formation was determined by counting the number of plaques.
2.8. Quantitative real-time PCR assay
MDCK cells cultured in 6-well plates (4 × 105 cells for each well) were subjected to two modes of drug treatment: “during infection” and “pretreatment virus”. After 24 h of post-infection, total cellular RNA was extracted with Trizol reagent (Sigma, St. Louis, MO, USA) and reverse transcribed into cDNA using the primers as listed below. Real-time quantitative PCR was performed using the two-step PCR amplification standard procedure of ABI7500 system (Applied Biosystems, Massachusetts, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Sigma, USA) was used as an internal control. The relative expression of the HA gene was measured by a classical 2−ΔΔCT method by using 7500 software. Each sample was tested independently more than three times. The primer sequences of target genes are as follows: 5’-TTCCCAAGATCCATCCGGCAA-3’ (HA-Forward), 5’-CCTGCTCGAAGACAGCCACAACG-3’ (HA-Reverse), 5’-AGGGCAATGCC AGCCCCAGCG-3’ (GAPDH-Forward), 5’-AGGCGTCGGAGGGCCCCCTC-3’ (GAPDH-Reverse).
2.9. Immunofluorescence staining and microscopy
MDCK cells were cultured to 80% confluence in 48-well plates and then infected with influenza virus A/PR/8/34 (H1N1) (100 TCID50) using both the “Pretreatment of the virus” and “During infection” protocols. After 24 h, cells were fixed with 4% paraformaldehyde for 20 min and then blocked with 4% bovine serum albumin (BSA) for 4 h at 37 °C. An anti-influenza virus nucleoprotein (NP) antibody (1:250 dilution in 4% BSA, Santa Cruz, Texas, USA) was incubated with the cells overnight at 4 °C, followed by an incubation with a secondary antibody conjugated with fluorescein isothiocyanate (FITC) (Sigma, USA) (diluted 1:250 in 4% BSA) for 4 h at 37 °C. Then, cells were counterstained with 4,6-diamino-2-phenylindole (DAPI) (Sigma, USA) for 10 min, and the plate was monitored under a fluorescence microscope (ZEISS Axio Observer, Germany).
2.10. Time-of-addition assay
To study which phase of the virus life cycle PPA interacted with, the infected MDCK cells were treated with 5 μg/mL of PPa for the indicated time interval (0–2, 2–4, 4–6, 6–8 h), covering the first viral replication cycle. After 24 h, the cells were frozen and thawed three times until the cells were completely disrupted, the supernatant was collected, the cell debris was removed by centrifugation, and the virus titer was determined by a method to measuring TCID50 of virus.
2.11. Polykaryon formation inhibition assay
The polykaryon formation inhibition assay was performed using a protocol described in the literature. Briefly, MDCK cells were transfected with plasmid-encoded HA (from influenza A/Thai/ Kan353/2004 virus) (2 μg DNA/well) using PEI according to the Manufacturer’s instructions. After a 10 h incubation, the transfection medium was replaced with fresh DMEM containing 10% fetal bovine serum. At 48 h after transfection, the cells were washed twice with PBS and treated with tosyl-phenylalanine chloromethyl-ketone (TPCK)-trypsin (5 μg/mL) for 15 min at 37 °C. Then, cells were pretreated with 0.5 mL of PPa or 1% methanol at 37 °C for 15 min followed by an incubation with 0.5 mL of pH 5.0 PBS or 1% methanol at 37 °C for another 15 min. Afterwards, the cells were washed twice with PBS, and then 1 mL of complete medium was added and incubated at 37 °C. After 3 h, the cells were fixed with methanol and incubated with Giemsa stain (Sigma, USA). The formation of syncytia was quantified by counting the number of polynuclei (containing 5 or more nuclei) under the microscope, and the percentage of syncytia that formed was determined and compared to the methanol control.
2.12. Fluorescence spectroscopy analyses
MDCK cells were collected, washed twice with PBS, and resuspended in 10 mL of PBS. Afterwards, 20 mL of chloroform–methanol (1:2, v/v) were added. After shaking for 90 min, the supernatant was removed and 20 mL of chloroform:water (1:1, v/v) were added. Then the mixture was shaken for another 30 min. Then, the chloroform phase was separated using a separating funnel. The solvent was removed with a rotary evaporator to obtain lipids.
A lipid suspension was obtained ultrasonically by adding a certain amount of PBS to 100 μg/mL of lipids and then 1 mL aliquots were transferred to several Eppendorf tubes to measure the binding of PPa to a lipid bilayer. PPa was next added to the lipid suspension at a final concentration of 5 μg/mL. After extensive mixing, the mixture was incubated at 37 °C for 45 min, the supernatant was removed, and the lipids were washed twice with PBS. Afterwards, 1 mL of methanol was added to the lipids and shaken for 20 min to dissolve the PPa absorbed on the lipids. The fluorescence intensity was then measured using a fluorescence spectrophotometer (FluoroMax-4, HORIBA, France) at an excitation wavelength of 365 nm and a scanning range of 500 to 800 nm.
The effects of the PPa interaction with lipids were further investigated by measuring the fluorescence intensity. Briefly, lipids were suspended in PBS at a concentration of 1 mg/mL and incubated with 1 μg/mL of PPa in the dark for 2 h. The fluorescence emission spectrum was detected using a fluorescence spectrophotometer and compared with the sample lacking lipids.
2.13. Isothermal titration calorimetry (ITC) assay
Lipids extracted from MDCK cells were dissolved in 5% DMSO at a concentration of 1 mg/mL, and then degassing for 10 min. The change in caloric value when PPa (75 μg/mL) interacted with lipids was recorded using isothermal titration calorimetry (MicroCal PEAQ -ITC, Malvern, UK). The ITC reaction conditions were set as follows: Temperature: 25 °C; Number of injections: 19; Volume: 2 μL; Spacing: 100 s; Reference power: 10 μcal/s; Stirring speed: 750 rpm. The experimental data were analyzed by MicroCal peak - ITC analysis software which can provided a thermal parameter calculation model. The average MW of lipids was set to be 3,000 [20].
2.14. Anti-influenza virus test in vivo
2.14.1. Animals
Specific-pathogen-free (SPF) male KM mice aged approximately four weeks with an average weight of 19–22 g were purchased from the Laboratory Animal Center of Southern Medical University (SMU) (Guangzhou, China). All mice were fed standard laboratory chow and provided ad libitum access to water during the animal experiments. The experiments were monitored in accordance with the Standard Operating Procedures of the facility and the Animal Welfare Act.
In the anti-influenza A virus activity test of PPa in vivo, the mice were randomly divided into five groups, including the uninfected and water-treated group (Blank), the infected and water-treated group (Viruses), the group that was infected and treated with 10 mg/mL oseltamivir (Oseltamivir), and the groups that were infected and treated with PPa at concentrations of 10 mg/Kg (PPa-H) and 2.5 mg/Kg (PPa-L). Oseltamivir was dissolved in water, while PPa was initially dissolved in polyethylene glycol 400 and then diluted four-fold with water. The infected mice were anaesthetized with ethyl ether and then intranasally challenged two times with influenza A/PR/8/34 (H1N1) virus at the titer of 2 × 105 TCID50 in a volume of 30 μL. Subsequently, the mice were orally administered different agents once a day for five days. On the sixth day, the mice were provided access to the food and water ad libitum. Body weights, mortality and the general behavior of the mice were recorded daily for 15 consecutive days.
2.14.2. Lung index
Using the same protocol as the in vivo anti-influenza virus test described above, the intragastric administration was stopped in three mice from each group on the fourth day after infection with the virus and the animals were sacrificed. Their lungs were harvested, washed with normal saline, dried with gauze and weighed. The lung index was calculated using the following equation:
2.14.3. Histopathologic observation
After evaluating the lung index, all of the lung tissues obtained from mice were immersed in 4% paraformaldehyde solution (PFA, LEAGENE) as soon as possible. Then the lung tissues were embedded in paraffin, cut into thin sections (about 4 μm thick), and stained using the Hematoxylin and Eosin staining method before histopathologic analysis under the microscope.
2.15. Statistical analysis
Statistics were performed using GraphPad Prism 5 software (San Diego, CA). Data expressed as the means ± standard deviation (SD) were repeated at least three times. ImageJ was used to quantized the Fluorescent images. Data were determined by one-way ANOVA using SPSS 22.0 software. The mortality rates were analyzed by Log-rank (Mantel-Cox) test (P < 0.01) using GraphPad Prism 5 software. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001.
3. Results
3.1. Pyropheophorbide a was isolated and identified from M. senhousei
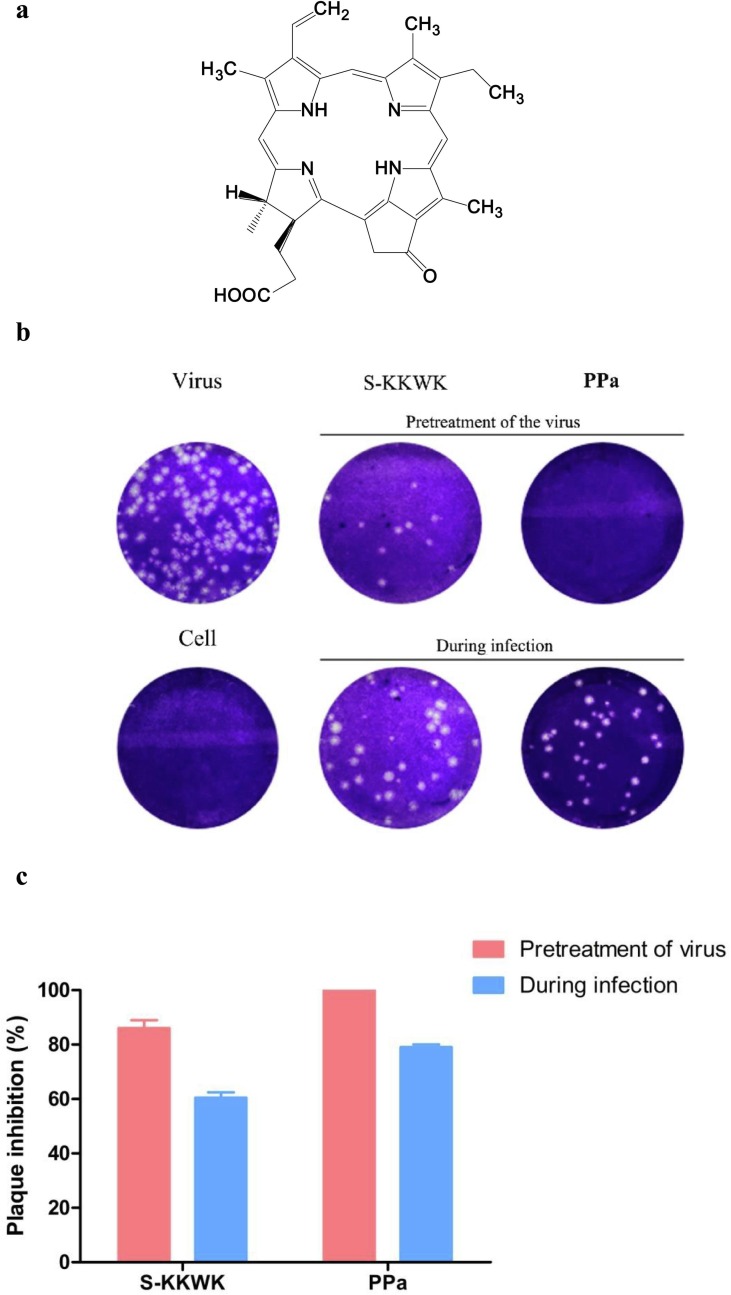
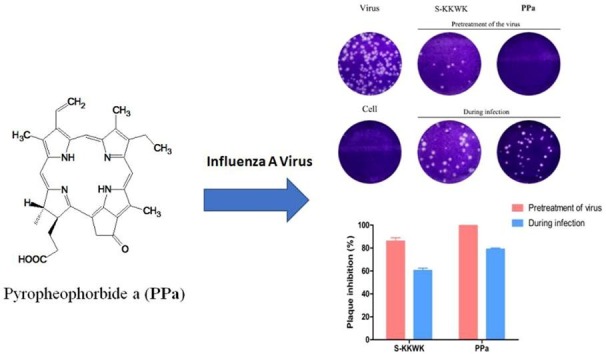
The mussel M. senhousei was collected in the area of the South China Sea, Guangdong Province, China. The fresh mussels were grounded and extracted with ethanol at least three times. The crude extracts were then isolated and purified using a bioassay-guided approach with influenza A/Puerto Rico/8/34 (H1N1) virus as the test strain. Consequently, a compound with a green color was isolated and subsequently identified using NMR and MS spectroscopic interpretation, and proven to be a known compound, 3-[(3S,4S)-14-ethyl-4,8,13,18-tetramethyl-20-oxo-9- vinyl-3-phorbinyl] propanoic acid, also named pyropheophorbide a (PPa) [15], as shown in Fig. 1 a.
Fig. 1.
a The structure of PPa. 1b Assay examining the ability of PPa to reduce the number of plaques of the influenza A/Puerto Rico/8/34 (H1N1) virus. The assay was performed using the “Pretreatment of the virus” and “During infection” approaches. In the first approach, 5 μg/mL PPa was pre-incubated with 100 TCID50 of the virus for 30 min at 37 °C before being added into the cells, while in the latter approach, the same titer of virus mixed with PPa (5 μg/mL) was simultaneously added to cells. After an incubation at 37 °C in a 5% CO2 atmosphere for 48 h, the cells were fixed and stained. 1c The number of plaques from each group were counted, and the plaque inhibition rate of PPa against the influenza A/Puerto Rico/8/34 (H1N1) virus was calculated.
3.2. PPa actively inhibited the infectivity of a broad range of influenza A viruses
We then tested the antiviral activity of PPa toward a panel of influenza A viruses, including influenza A/Puerto Rico/8/34 (H1N1), A/FM/1/47 (H1N1) mouse-adapted viral strain, A/Puerto Rico/8/34 (H1N1) with the NA-H274Y mutation, and A/Aichi/2/68 (H3N2) viral strains. In addition, the compound S-KKWK that was previously reported as an entry inhibitor of IAVs was used as a positive control [17], and chlorophyll was used for comparison due to it is structurally similar to PPa. The antiviral activity was evaluated by pretreating host MDCK cells with the virus [20], and the survival of the host cells was observed under microscope by evaluating the cytopathic effects (CPE) and then quantified with the MTT assay. As shown in Table 1 , PPa showed broad activity against all tested strains, with IC50 values ranging from 0.17 to 0.88 μg/mL. Notably, PPa also displayed a similar activity against A/Puerto Rico/8/34 (H1N1) with the NA-H274Y mutation, an influenza A viral strain that is resistant to the clinically used drug oseltamivir, indicating a different action mechanism from oseltamivir.
Table 1.
Inhibitory activity of PPa against influenza A viruses and toxicity in MDCK cells.
| Name | IC50 ± SD (μg/mL)a |
CC50 ± SD (μg/mL)b | MWg | |||
|---|---|---|---|---|---|---|
| PR8c | FM-1d | 4274e | H3N2f | |||
| S-KKWK | 1.88 ± 0.93 | 1.99 ± 0.23 | NTh | NT | NT | 1014.84 |
| PPa | 0.17 ± 0.15 | 0.88 ± 0.30 | 0.56 ± 0.53 | 0.66 ± 0.59 | 74.84 ± 1.93 | 534.00 |
| Chlorophyll | 24.91 ± 1.67 | >50 | >50 | >50 | 56.05 ± 2.62 | NT |
The anti-influenza A virus (IAV) activity was assessed after the virus was pretreated with PPa.
The toxicity in MDCK cells was evaluated using the MTT assay.
A/Puerto Rico/8/34.
A/FM/1/47 mouse-adapted viral strain.
A/Puerto Rico/8/34 with the NA-H274Y mutation.
A/Aichi/2/68.
MW: molecular weight
NT: not tested.
In addition, the MTT assay was performed to evaluate the cytotoxicity of the tested molecules and to determine whether the anti-IAV activity resulted from the cytotoxicity of PPa. As shown in Table 1, the CC50 value of PPa was 74.84 ± 1.93 μg/mL, a value that is much higher than its viral inhibitory activity.
The anti-IAV activity of PPa was next confirmed using the plaque reduction assay. As indicated in Fig. 1b and c, under the testing condition of “Pretreatment”, 5 µg/mL of PPa suppressed the formation of all plaques compared with an ca. 80% inhibition rate under the condition of simultaneous treatment (“During infection”), which was further assessed using indirect immunofluorescence staining and qRT-PCR, as shown in Fig. 2 .
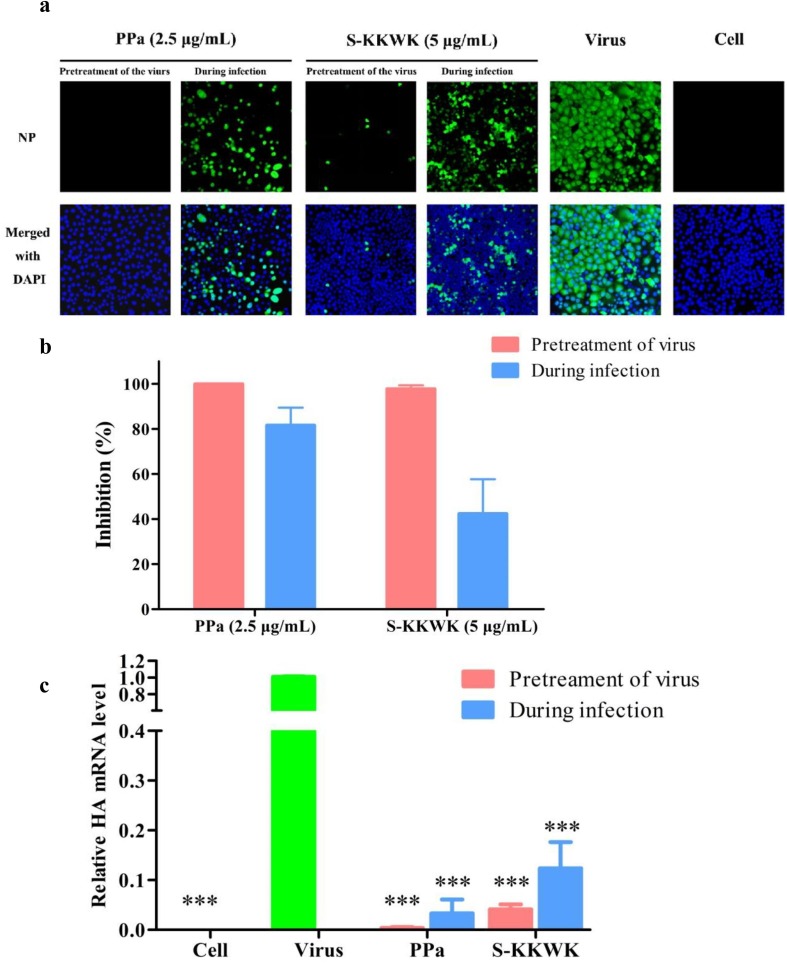
Fig. 2.
a The effect of PPa on nucleoprotein (NP) expression in cells was detected using indirect immunofluorescence staining. MDCK cells were treated with 100 TCID50 of influenza A/PR/8/34(H1N1) virus and 2.5 μg/mL PPa using the “Pretreatment of the virus” and “During infection” approaches. At 24 h post-infection, NP in the cytoplasm (green fluorescence) of MDCK cells was immunostained and detected under a fluorescence microscope. DAPI was used to indicate the position of the nucleus (blue fluorescence), and 5 μg/mL S-KKWK was administered using the same methods as a positive control. 2b The green fluorescence of NP expression in each group was quantified using ImageJ software, and the rate at which PPa inhibited NP expression from the influenza A/Puerto Rico/8/34 (H1N1) virus was calculated. 2c The antiviral effects of PPa on influenza A/PR/8/34 (H1N1) were evaluated by measuring the mRNA level of the HA gene after treatment with PPa using the “Pretreatment of virus” or “During infection” drug administration approaches. One-way analysis of variance (one-way ANOVA) was employed to assess the statistical significance of differences in the data between the virus-infected groups: * p < 0.05, ** p < 0.01, and *** p < 0.001. 2d. The antiviral effects of PPa against RSV (left) and SARS-CoV-2 (right). The green and red bars indicate the inhibition of PPa against RSV or SARS-CoV-2, while the black bars refer to the cytotoxicity of PPa on Vero-E6 cells at the indicated concentrations. The anti-RSV and anti- SARS-CoV-2 assays were as shown in the experimental section. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Indirect immunofluorescence staining was used to detect the effects of PPa on the expression of the viral nucleoprotein (NP) in MDCK cells. After cells were infected with the virus using the “Pretreatment” and “During infection treatment” methods, the expression of NP from the A/Puerto Rico/8/34 (H1N1) influenza virus was observed under a fluorescence microscope at 24 h post-infection (Fig. 2a) and then quantified using ImageJ software [21] (Fig. 2b). The green fluorescence associated with NP expression was significantly decreased compared with the “virus” group without drug treatment. Notably, the “Pretreatment” group showed greater activity of the drug than the simultaneous treatment group (“During infection”).
A similar conclusion was obtained by measuring the mRNA level of the HA gene from influenza A/PR/8/34 (H1N1) after treatment with PPa using two drug administration approaches (Fig. 2c). The level of HA gene was dramatically decreased when treated with 5 µg/mL PPa compared with the group without drug treatment (P < 0.001), and the HA level observed in cells subjected to the “Pretreatment of the virus” was lower than in cells subjected to the “During infection” treatment.
Furthermore, the antiviral activity of PPa was also assessed by testing viruses that induce respiratory disease, including respiratory syncytial virus (RSV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As indicted in Fig. 2d, at 25 µM, PPa inhibited the formation of RSV plaques up to 75% detected by immuno-based plaque assay, and inhibited almost 100% of the expression of S gene of SARS-CoV-2 detected by qRT-PCR (Fig. 2d). Therefore, in addition to influenza A viruses, PPa was also active against other viruses that cause respiratory diseases.
3.3. The anti-IAV activity of PPa results from the blockade of the early infection of influenza A virus
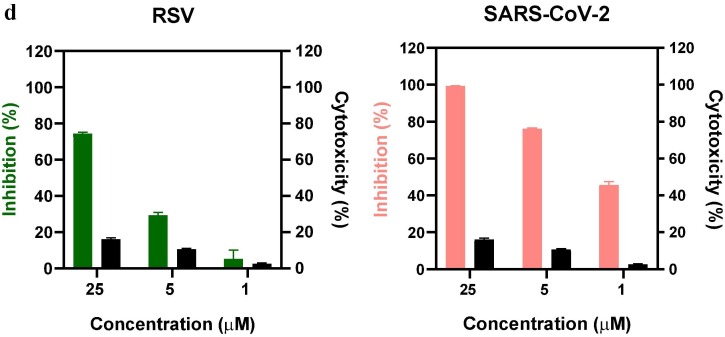
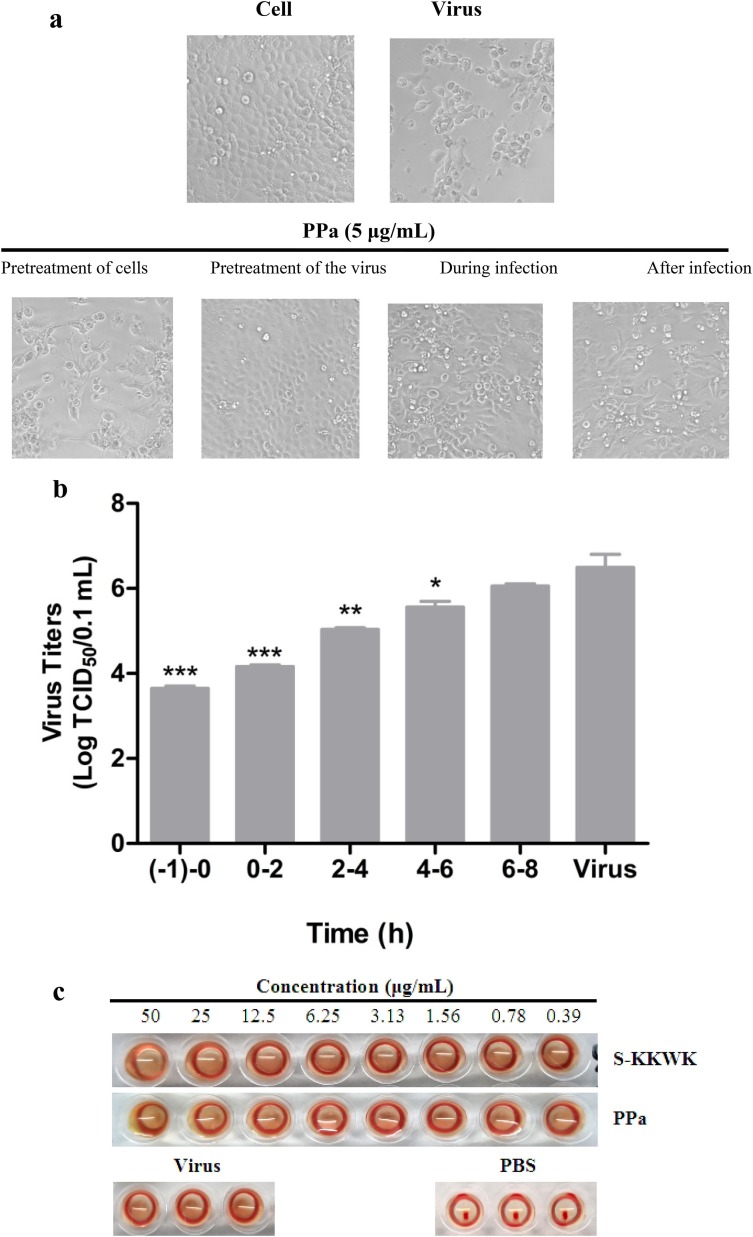
Next, we investigated the inhibitory effects of PPa on the influenza virus life cycle. Four different drug administration approaches, including Pretreatment of cells, Pretreatment of the virus, During infection, and After infection, were employed as described above [19] to test the inhibitory effects of the drug. The CPEs were observed under a microscope and used to evaluate the antiviral effects of PPa (Fig. 3 a), which were further quantified using the MTT assay. As shown in Table 2 , the “Pretreatment of the virus” was the most active drug administration approach and resulted in an IC50 value of 0.17 ± 0.15 µg/mL compared with 4.47 ± 0.23 µg/mL for the “During infection” treatment. On the other hand, the “Pretreatment of cells” and “After infection” approaches showed no activity in the tested range, clearly indicating that PPa may act on the early stage of IAV infection by blocking the entry of viruses into host cells.
Fig. 3.
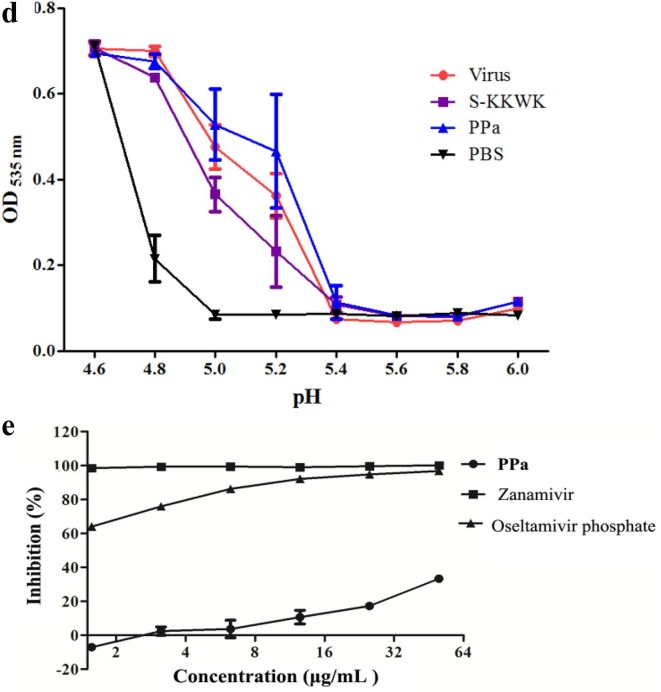
a Cytopathic effects (CPE) on MDCK cells infected with influenza A/PR/8/34 (H1N1) (100 TCID50). Four different drug administration protocols were employed to treat the virus or cells at the concentration of 5 μg/mL of PPa. Phase contrast images were obtained using a microscope at 48 h post-infection. 3b Time-of-addition assay. MDCK cells infected with A/PR/8/34(H1N1) (100 TCID50) were treated with 5 μg/mL PPa at the indicated time intervals. The virus titer was determined by measuring the TCID50 of virus in the supernatant at 48 h post-infection; *p < 0.05, **p < 0.01, and ***p < 0.001. 3c Hemagglutinin inhibition assay designed to measure the inhibitory effect of PPa on the binding of the virus to target cells. An equal volume of influenza A/Puerto Rico/8/34(H1N1) virus (4HAU) was transferred to a microwell plate containing two-fold serially diluted PPa in PBS. Then, erythrocytes (1% v/v in saline) were added to each well. After an incubation at room temperature for 1 h, the hemagglutination reaction was analyzed. PBS without virus served as a positive control, while virus served as the negative control. 3d Hemolysis inhibition assay. A suspension of freshly prepared chicken erythrocytes (2%) was added to the mixture of PPa (15 μg/mL) with the influenza virus A/PR/8/34 (H1N1) in allantoic fluid. The mixture was then acidified to a pH ranging from 4.6 to 6.0 (sodium acetate, 0.5 M), and incubated at 37 °C for 30 min. At the end of the incubation, the mixture was centrifuged and the absorbance of the supernatants containing released hemoglobin was measured at OD535using a Multiskan FC microplate reader. 3e Neuraminidase (NA) inhibition assay. A reaction mixture consisting of PPa and influenza A/Puerto Rico/8/34(H1N1) virus in MES buffer was incubated for 45 min. Afterwards, 4-MU-NANA was added to each well, the mixture was incubated for another 1 h, and the reaction was terminated with NaOH (83% ethanol). Finally, an excitation wavelength of 340 nm and an emission wavelength of 440 nm were used to measure resulting fluorescence of the mixture. Oseltamivir phosphate and zanamivir were employed as positive controls.
Table 2.
Assessment of the anti-influenza virus activity of PPa using four different drug treatment modes.
| Name | IC50 ± SD (μg/mL) a |
|||
|---|---|---|---|---|
| Pretreatment of cells | Pretreatment of the virus | During infection | After infection | |
| S-KKWK | >10 | 1.88 ± 0.93 | 5.84 ± 1.89 | >10 |
| PPa | >10 | 0.17 ± 0.15 | 4.47 ± 0.23 | >10 |
The A/PR/8/34 (H1N1) influenza virus was used in this experiment.
The inhibitory effect of PPa on virus entry was again assessed using a time of addition experiment. As shown in Fig. 3b, 5 μg/mL PPa was added to MDCK cells infected with the A/PR/8/34 (H1N1) virus (100 TCID50) at the indicated time intervals, and the titers of the virus in the supernatant were then monitored at 48 h post-infection. The highest percentage of inhibition was observed at the interval of (-1)-0h of treatment of the virus (equal to “Pretreatment of the virus”), consistent with the observations obtained from different drug administration experiments.
3.4. PPa does not act on the surface glycoproteins HA and NA
Due to the important role of HA in the entry of IAV into host cells, we studied whether HA was the possible target of PPa. We used the hemagglutinin inhibition assay to measure the inhibitory effect of PPa on viral adsorption into target cells and the hemolysis inhibition assay to determine whether PPa inhibited the hemolytic effect of PPa on chicken erythrocytes [19]. The negative results obtained in both experiments shown in Fig. 3c and d indicated that PPa was unable to block the agglutination of erythrocytes and failed to reduce the hemolytic effect of the virus on erythrocytes under both acidic and neutral pH conditions. Therefore, hemagglutinin (HA) is not likely the major target of PPa.
In addition to HA, we also investigated if the surface glycoprotein neuraminidase (NA) was the possible target of PPa. As shown in Fig. 3e, PPa did not noticeably inhibit NA inhibition at the concentrations tested. In the NA inhibition assay performed with a concentration of PPa as high as 32 µg/mL, the percent inhibition of the activity of NA toward the substrate 4-MU-NANA was still less than 25%.
3.5. PPa interacts with the viral envelope lipid bilayer to block the entry of IAV
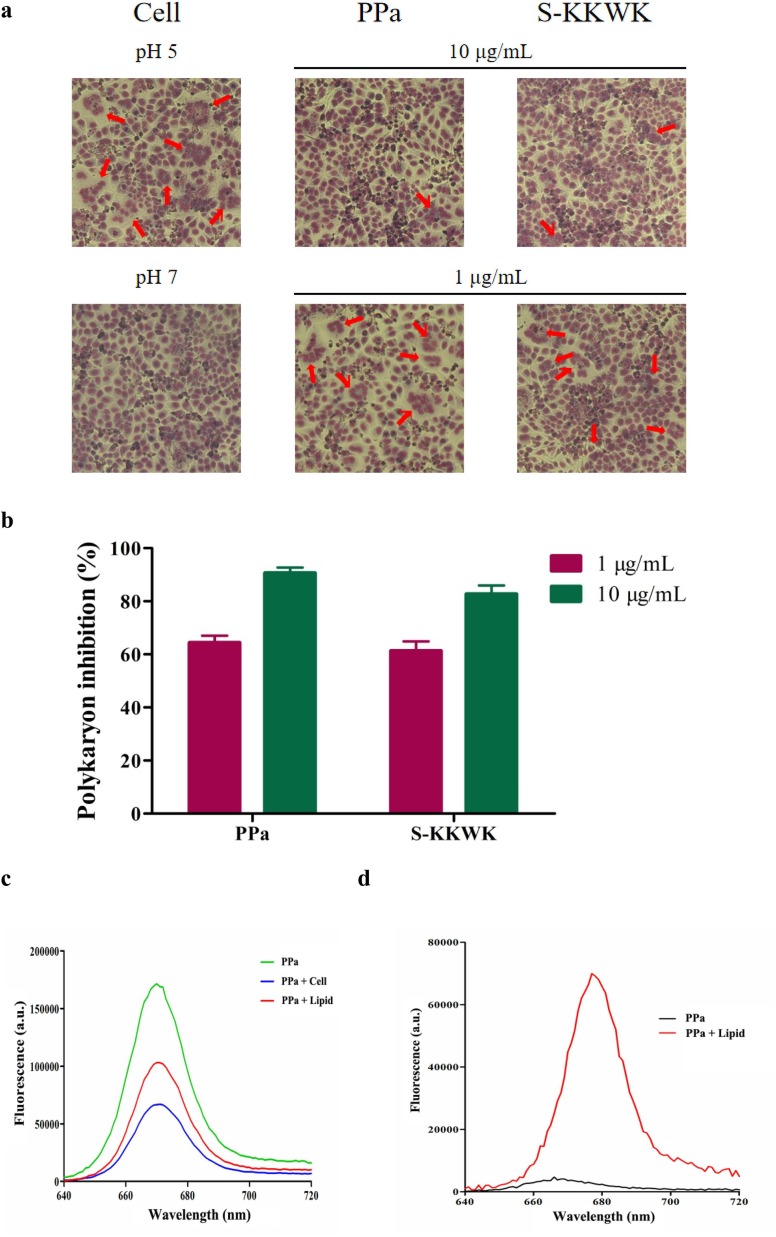
Considering the early inhibitory effect of PPa on IAV and the important role of the viral envelope lipid bilayer in mediating the entry of viruses [22], we speculated whether the viral envelope lipids were the possible target of PPa. Initially, we investigated the ability of PPa to inhibit fusion by blocking the interaction between HA and lipids using the polykaryon formation inhibition assay [23], as shown in Fig. 4 a and b. At a higher concentration of 10 µg/mL, PPa inhibited the formation of polykaryons by up to 95% compared with the samples lacking the drug treatment, indicating the presence of interactions among PPa, HA and cell membrane lipids that inhibited the formation of a polykaryon to some extent.
Fig. 4.
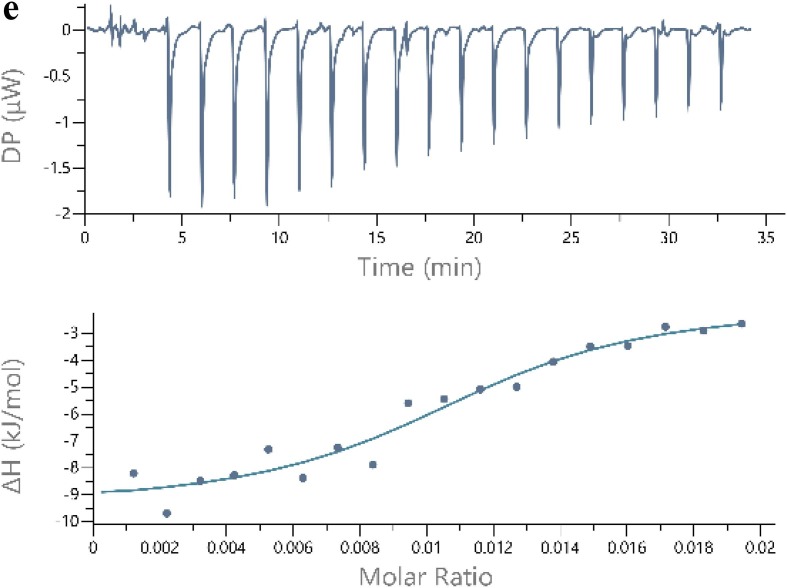
a Polykaryon formation inhibition assay. MDCK cells expressing HA from influenza A/Thailand/Kan353/2004 were treated with PPa and then acidified to pH 5.0 for 15 min at 37 °C. Afterwards, the cells were incubated at for 3 h. At the end of the incubation, the cells were fixed and stained with Giemsa solution. Syncytium formation was visualized and counted using a microscope. Polykaryons are indicated by red arrowheads. 4b The results were quantified by counting the number of polykaryons. Syncytium formation was quantified by counting the number of polykaryons in whole fields of the plate. Each of the polykaryons contains at less three nuclei. The polykaryon inhibition was compared to the pH 5.0 control.4c Lipids were extracted from MDCK cells, mixed with PBS to form a suspension, and then PPa was added to a final concentration of 5 μg/mL. The mixture was incubated at 37 °C for 45 min, the supernatant was removed, and the lipids were washed with PBS twice. The bound PPa was then extracted with 1 mL of methanol for 1 h, and the fluorescence intensity was measured at the excitation wavelength of 365 nm. 4d Lipids were suspended in PBS at a concentration of 1 mg/mL, and then PPa was added at a final concentration of 1 μg/mL. After an incubation for 2 h, the fluorescence intensity was measured at the excitation wavelength of 365 nm. 4e The thermodynamic curve of the binding of 75 μg/mL PPa to 1 mg/mL of lipids was obtained by performing an ITC assay. The thermal changes occurring in response to the interactions between PPa and cellular lipids were detected. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. PPa may interact with the envelope lipid bilayer to inhibit the entry of viruses
Since the viral envelope lipid bilayer was derived from the host cell membrane, we next investigated the interactions between PPa and lipids that were extracted from MDCK cells. First, we tested whether PPa bound to the surface of the lipid bilayer. In this experiment, 5 μg/mL PPa was incubated with lipid extracts or with MDCK cells at 37 °C for 45 min. After extensive rinses with PBS to remove the unbound PPa, the PPa absorbed on lipid or cells was then extracted with methanol, and the fluorescence intensity was subsequently measured at the excitation wavelength of 365 nm. As shown in Fig. 4c, an intense fluorescence emission of PPa at a wavelength of ca. 666 nm was observed from lipid extracts (60% remaining) and cell extracts (38% remaining), indicating a strong binding affinity of PPa for the lipid bilayer.
Next, fluorescence spectroscopy was employed to measure the interactions between PPa and lipids. First, 1 µg/mL of PPa in PBS or mixed with 1 mg/mL of lipids in PBS was excited at a wavelength of 365 nm, and then the emission fluorescence was measured at wavelengths ranging from 640 to 720 nm, as indicated in Fig. 4d. The data revealed a ca. 15-fold increase in the fluorescence intensity when the assay was performed in the presence of lipids. Furthermore, an apparent red shift of 11 nm from 666 nm to 677 nm in the maximum emission wavelength was observed when lipids were added to PPa.
Inspired by these data, we then semi-quantitatively measured the lipid-PPa interactions using isothermal titration calorimetry (ITC) to obtain the thermodynamic curve of the binding of PPa to lipids [24]. The lipids extracted from MDCK cells were added at a concentration 1 mg/mL, while the concentration of PPa was 75 μg/mL. The thermal changes due to the interactions between PPa and cellular lipids were detected and presented in the thermogram shown in Fig. 4e. The ITC peaks were negative, indicating that the interactions between PPa and lipids were exothermic. Based on the measured ΔH and TΔS values (Table 3 ), the binding affinity Kd (dissociation constant) between PPa and lipids was calculated to be 8.71 µM.
Table 3.
Thermodynamic parameters of the interaction between PPa and lipids from MDCK cells.
Kd: dissociation constant.
Ka: binding constant.
△H: enthalpy.
△G: Gibbs free energy.
△S: entropy.
3.7. The anti-IAV activity decreases upon the addition of a lipid bilayer
Because PPa interacts with the lipid bilayer, we next examined the effect of lipids on the anti-IAV activity of PPa in vitro. As indicated in Table 4 , the antiviral activity of PPa was apparently reduced after the addition of lipids, further supporting the presence of interactions between PPa and lipids.
Table 4.
Anti-Influenza virus inhibition rate after PPa interacted with lipids from MDCK cellsa.
| Lipidb (C/C) |
Inhibition Rate ± SD (%) |
|
|---|---|---|
| PPa (1 μg/mL) | PPa (2 μg/mL) | |
| 0 | 100.76 ± 0.66 | 101.23 ± 1.89 |
| 10× | 97.03 ± 1.34 | 98.52 ± 0.94 |
| 50× | 54.69 ± 2.02 | 72.14 ± 1.33 |
| 100× | 0.10 ± 1.05 | 58.96 ± 2.08 |
Influenza virus A/PR/8/34 (H1N1) was tested using the “Pretreatment” administration protocol.
Lipid was extracted from MDCK cells.
3.8. PPa increases the survival rate and prolongs the mean survival time of IAV-infected mice
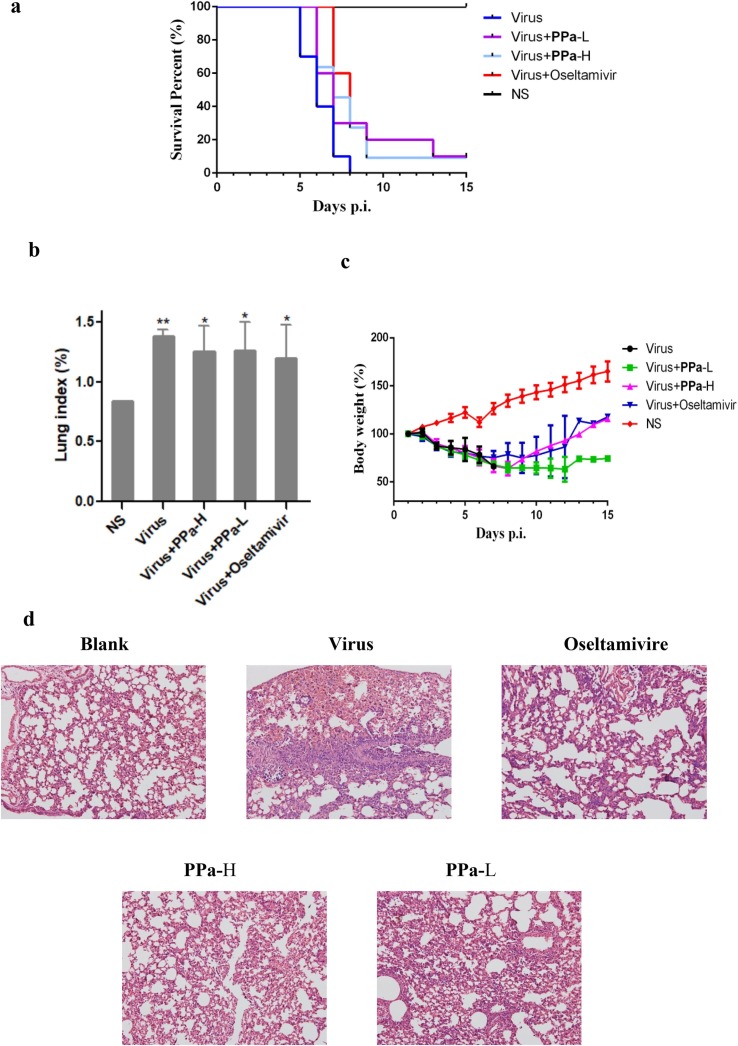
The anti-IAV activity of PPa was further assessed by analyzing IAV-infected mice. In the experiment, 4-week-old male Kunming mice (the average body weight was ca. 19–22 g) were divided into five groups of 10 mice each, as indicated in Fig. 5 a. The mice were intranasally challenged twice with 2 × 105 TCID50 of influenza virus A/PR/8/34 (H1N1) in a volume of 30 µL of normal saline (NS). After the viral infection, the mice were orally administered one dose of PPa or oseltamivir. The compounds were administered once a day until the fifth day. The mean number of days the animals survived and survival rate were measured for 15 days to evaluate the efficacy of PPa.
Fig. 5.
The anti-IAV activity of PPa in mice. 5a Analysis of the percentages of surviving influenza virus-infected and PPa- or oseltamivir-treated mice compared with the infected mice. The mice were intranasally challenged twice with 106 TCID50 of influenza virus A/PR/8/34 (H1N1) in a volume of 30 µL of normal saline (NS). Immediately after the viral infection, the mice were orally administered PPa or oseltamivir. The compounds were administered once a day until the fifth day. The mortality rates were analyzed with Pearson’s Chi-square test (P < 0.05). 5b Lung index of infected mice treated with PPa. The lung index of the mice was determined after the viral infection and PPa treatment for 3 days. Data were analyzed with one-way ANOVA, *p < 0.05, **p < 0.01, and ***p < 0.001. 5c The body weight of the infected mice was recorded. 6d Histopathological changes (200 × ) of the lung tissues obtained from mice.
As shown in Fig. 5a, compared with the virus control group, the survival rate and mean survival time of infected mice treated with both high and low dosages of PPa were noticeably increased. In parallel, a significant increase in the lung index was observed for all virus-challenged mice compared with the blank control group. In addition, the average lung indices for compound-treated groups were lower than the virus control group; nevertheless, no significant difference between PPa-H group and PPa-L group was observed in this experiment (Fig. 5b).
Furthermore, the body weight of all virus-infected mice decreased until the eighth to ninth days compared with the slow increase in body weight observed in the blank control group. The body weights of compound-treated groups slowly increased, while all the mice in virus control group had died (Fig. 5c). In addition, the histopathological examination of the lungs of mice (Fig. 5d) showed that as compared with the blank control group, a large amount of alveolar collapse or deformation around bronchioles occurred, and many inflammatory cell infiltrations were observed in the virus-infected mice. In contrast, the symptoms were dramatically alleviated in the compound-treated group (Fig. 5d), further supporting the protective effect of PPa.
4. Discussion
Influenza A virus is an infectious pathogen that induces respiratory tract infections in humans and affects 5–15% of the global population with a mild to severe illness each year [25]. Currently, only two types of anti-IAV drugs are available, but the emergence of viral strains resistant to these compounds has been reported [26], in addition to some side effects. Therefore, it is imperative to develop new and effective anti-IAV drugs that employ a different mechanism by targeting other viral proteins or cellular factors involved in the influenza virus life cycle. In this study, we identified a porphyrin derivative, pyropheophorbide a (PPa), with broad anti-IAV activity in vitro. PPa was isolated from the marine mussel M. senhousei, and it has also been isolated from abalone [27]. Structurally, PPa is a derivative of chlorophyll. Therefore, a reasonable deduction that the original sources of PPa might be phytoplankton, which were subsequently transformed by mussels to produce PPa [28].
As shown in Fig. 1a, the structure of PPa consists of a porphyrin ring and a carboxyl group, which shows a typical amphipathic structure. This characteristic facilitates PPa to interact with hydrophobic lipids of enveloped virions or cell membrane, by which to block the entry of virus [29]. When the carboxyl group connected with a lipid chain as that in chlorophyll, which decreased the amphipathicity of PPa, and as a result, decreased the antiviral activity of PPa. However, more and extensive structure-activity relationship studies should be conducted to support this observation.
In the present study, the anti-IAV activity of PPa was assessed using various experiments, including CPE, RT-PCR, a plaque reduction assay, and indirect immunofluorescence staining. These data, together with the results from experiments using different drug administrations and time-of-addition assay, indicated that the anti-IAV effects of PPa resulted from the inhibition of early stage of virus infection, namely, a possible blockade of the entry of the virus into host cells. Furthermore, we also test other viral strains that cause respiratory disease such as RSV and SARS-CoV-2. The results showed that PPa was active against these viruses and therefore proved to be a broad antiviral compound.
As reported in previous studies, the process of IAV entry involves multiple steps, including virus binding to host cells, entering host cell compartments through endocytosis (internalization), virus trafficking to the perinuclear region, and fusion of viral and host cell membranes [30]. Due to the critical roles of these steps in the process of virus entry, each step may represent a potential target for anti-IAV drug development. To date, a number of influenza A virus entry inhibitors with different mechanisms have been identified, such as the cell attachment blocker [31], internalization inhibitor [32], [33], and fusion inhibitor [34].
In contrast to those reported viral entry inhibitors, our data showed that although PPa and its derivatives functioned as virus entry inhibitors and exhibited strong binding affinities for hemagglutinin (HA) and HA2 subunit in an experiment using surface plasmon resonance (SPR) (data not shown), the negative results of the hemagglutinin inhibition assay and hemolysis inhibition assay suggested that its major target may not be HA, as a consequence, PPa and its derivatives were unable to block the attachment of IAV to host cells and fusion of the viral envelope with the host cell membrane, indicating that the interactions with HA may not contribute significantly to the antiviral activity compared with the interactions with other possible antiviral targets.
On the other hand, the results of experiments using different drug administration protocols, fusion inhibition assay, fluorescence spectroscopy and ITC analyses indicated that PPa is a membrane-binding compound that interacts with the viral membrane or biophysically interferes with the virus-cell membrane fusion process. Consequently, PPa may block virus entry into cells by physically damaging the viral membrane integrity, thereby inhibiting the entry of an enveloped virus in the early stage of infection, consistent with the mechanism hypothesized by Bouslama and colleagues [35]. Furthermore, similar mechanisms in which a compound physically interacts with virion envelope lipids to inhibit fusion of the influenza virus with the cell membrane have also been reported in other articles, such as the studies by Colpitts [29], Speerstra [36], St Vincent [37], Kim [32], and He [38].
In conclusion, in this paper, we identified a porphyrin derivative of PPa from the marine mussel M. senhousei that shows broad anti-IAV activity in vitro and in vivo, as well as other viral strains that cause respiratory diseases including RSV and SARS-CoV-2. PPa interacted with the viral lipid bilayer to alter membrane-associated functions, thereby inhibiting the entry of enveloped viruses. However, additional, more extensive experiments are still needed to further assess the antiviral efficacy, the detailed mechanism by which PPa inhibits virus entry into the host cells, and particularly, the in vivo anti-IAV effects of PPa.
Nevertheless, as we have known, porphyrins are a group of conjugated planar molecules that possess broad applications in the field of biomedical science, due to their unique structural features and electrochemical performance [39]. Our findings in this study expand the application of porphyrins, and significantly, by targeting an essential component in the virus life cycle and using PPa or its derivatives as a leading compound, we believe that a more potent antiviral agent may be generated.
CRediT authorship contribution statement
Daiwei Chen: Investigation, Formal analysis. Shengsheng Lu: Investigation, Formal analysis. Guang Yang: Data curation. Xiaoyan Pan: Data curation. Sheng Fan: Validation. Xi Xie: Data curation. Qi Chen: Data curation. Fangfang Li: Validation. Zhonghuang Li: Methodology. Shaohua Wu: Writing - review & editing. Jian He: Conceptualization, Funding acquisition, Supervision, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (81773556), Southern Medical University (B1040903), China, and Science and Technology Department of Guangdong Province of China (2015A020211010).
References
- 1.R.C. Willan, Successful Establishment of the Asian Mussel Musculista senhousia (Benson in Cantor, 1842) in New Zerland. Rec. Auckland Inst. Museum, 22 (1985) 85–96.
- 2.Hawkins A.J.S., Pascoe P.L., Parry H., Brinsley M., Cacciatore F., Black K.D. Comparative feeding on chlorophyll-rich versus remaining organic matter in bivalve shellfish. J. Shellfish Res. 2013;32(3):883–897. [Google Scholar]
- 3.Suleria H.A., Masci P., Gobe G., Osborne S. Current and potential uses of bioactive molecules from marine processing waste. J. Sci. Food Agr. 2016;96(4):1064–1067. doi: 10.1002/jsfa.7444. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S.P., Huang R., Li F.F., Wei H.X., Fang X.W., Xie X.S. Antiviral anthraquinones and azaphilones produced by an endophytic fungus Nigrospora sp. from Aconitum carmichaeli. Fitoterapia. 2016;112:85–89. doi: 10.1016/j.fitote.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Ma K.X., Shen X.T., Huang R., Wang T., Xie X.S., Liu S.W. Bioactive metabolites produced by the endophytic fungus Phomopsis sp. YM355364. Nat. Pro. Com. 2014;9(5):669–670. [PubMed] [Google Scholar]
- 6.Li F.F., Chen D.W., Lu S.S., Yang G., Zhang X.L., Chen Z. Anti-influenza a viral butenolide from Streptomyces sp. Smu03 inhabiting the intestine of Elephas maximus. Viruses. 2018;10(7) doi: 10.3390/v10070356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L., Li Y.H., Li S.H., Li H.D., Qiu Z.X., Lee C.C. Inhibition of influenza A virus (H1N1) fusion by benzenesulfonamide derivatives targeting viral hemagglutinin. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0029120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnab B., Aleksandar A., Minxiu W., Bing L., Mills D.M., Ames J.A. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J. Virol. 2014;88(3):1447–1460. doi: 10.1128/JVI.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas W. John Wiley & Sons, Ltd; 2001. Viral Capsids and Envelopes: Structure and Function, eLS. [Google Scholar]
- 10.Loredana V., Cross K.J., Jens K., Straus S.K., Thomas D.J., Wharton S.A. Plasticity of influenza haemagglutinin fusion peptides and their interaction with lipid bilayers. Biophys. J. 2005;88(1):25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q., Korte T., Rachakonda P.S., Knapp E.W., Herrmann A. Energetics of the loop-to-helix transition leading to the coiled-coil structure of influenza virus hemagglutinin HA2 subunits. Proteins. 2009;74(2):291–303. doi: 10.1002/prot.22157. [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Li M.M., Shen X.T., Liu S.W. Influenza A virus entry inhibitors targeting the hemagglutinin. Viruses. 2013;5(1):352–373. doi: 10.3390/v5010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobusawa E., Aoyama T., Kato H., Suzuki Y., Tateno Y., Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182(2):475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 14.Reed M.L., Bridges O.A., Seiler P., Kim J.K., Yen H.L., Salomon R. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J. Virol. 2010;84(3):1527–1535. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z.M., Wang L., Zhu W., Gao Y.H., Wu H.M., Wang M. Preparation of a chlorophyll derivative and investigation of its photodynamic activities against cholangiocarcinoma. Biomed. Pharmacother. 2017;92:285–292. doi: 10.1016/j.biopha.2017.05.052. [DOI] [PubMed] [Google Scholar]
- 16.Kentaro M., Kei O., Shunichi F. Efficient two-electron reduction of dioxygen to hydrogen peroxide with one-electron reductants with a small overpotential catalyzed by a cobalt chlorin complex. J. Am. Chem. Soc. 2013;135(7):2800–2808. doi: 10.1021/ja312199h. [DOI] [PubMed] [Google Scholar]
- 17.Lin D.G., Luo Y.Z., Yang G., Li F.F., Xie X., Chen D.W. Potent influenza A virus entry inhibitors targeting a conserved region of hemagglutinin. Biochem. Pharmacol. 2017;144:35–51. doi: 10.1016/j.bcp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin D.G., Li F.F., Wu Q.Y., Xie X., Wu W., Wu J. A “building block” approach to the new influenza A virus entry inhibitors with reduced cellular toxicities. Sci. Rep. 2016;6:22790. doi: 10.1038/srep22790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G., Wang J.Y., Lu S.S., Chen Z., Fan S., Chen D.W. Short lipopeptides specifically inhibit the growth of Propionibacterium acnes with a dual antibacterial and anti-inflammatory action. Br. J. Pharmacol. 2019;176(13):2321–2335. doi: 10.1111/bph.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zu M., Yang F., Zhou W., Liu A., Du G., Zheng L. In vitro anti-influenza virus and anti-inflammatory activities of theaflavin derivatives. Antiviral. Res. 2012;94(3):217–224. doi: 10.1016/j.antiviral.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Wu W., Lin D., Shen X., Fang Y., Li K., Xun T. New influenza A virus entry inhibitors derived from the viral fusion peptides. PLoS ONE. 2015;10(9) doi: 10.1371/journal.pone.0138426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai K.K., Cheung N.N., Yang F., Dai J., Liu L., Chen Z. Identification of novel fusion inhibitors of influenza A virus by chemical genetics. J. Virol. 2015;90(5):2690–2701. doi: 10.1128/JVI.02326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myszka D.G. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Method. Enzymol. 2000;323 doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 25.W.D. Li, M.D. Joshi, S. Smita, K.H. Ramsey, A.K. Murthy, Peptide Vaccine: Progress and Challenges. Vaccines, 2 (3) (2014) 515–536. [DOI] [PMC free article] [PubMed]
- 26.Hurt A.C., Tawee C., Cox N.J., Rod D., Fry A.M., Gubareva L.V. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect. Dis. 2012;12(3):240–248. doi: 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
- 27.Allen B.J., Williams S.L. Native eelgrass Zostera marina controls growth and reproduction of an invasive mussel through food limitation. Marine Ecol. Progress. 2003;254:57–67. [Google Scholar]
- 28.Wang J., Liu Q., Zhang Y., Shi H., Liu H., Guo W. Folic acid-conjugated Pyropheophorbide a as the photosensitizer tested for In Vivo targeted photodynamic therapy. J. Pharm. Sci. 2017;106(6):1482–1489. doi: 10.1016/j.xphs.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Colpitts C.C., Ustinov A.V., Epand R.F., Epand R.M., Korshun V.A., Schang L.M. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J. Virol. 2013;87(7):3640–3654. doi: 10.1128/JVI.02882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edinger T.O., Pohl M.O., Stertz S. Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 2014;95(Pt 2):263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 31.Londrigan S.L., Turville S.G., Tate M.D., Deng Y.M., Brooks A.G., Reading P.C. N-linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J. Virol. 2011;85(6):2990–3000. doi: 10.1128/JVI.01705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M., Kim S.Y., Lee H.W., Shin J.S., Kim P., Jung Y.S. Inhibition of influenza virus internalization by (-)-epigallocatechin-3-gallate. Antivir. Res. 2013;100(2):460–472. doi: 10.1016/j.antiviral.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 33.E. de. Vries, D.M. Tscherne, M.J. Wienholts, V. Cobos-Jiménez, F. Scholte, A. García-Sastre, et al., Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. Plos Pathog., 7 (3) (2011) e1001329. [DOI] [PMC free article] [PubMed]
- 34.Corti D., Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 35.Bouslama L., Hayashi K., Lee J., Ghorbel A., Hayashi T. Potent virucidal effect of pheophorbide a and pyropheophorbide a on enveloped viruses. J. Nat. Med. 2011;65(1):229–233. doi: 10.1007/s11418-010-0468-8. [DOI] [PubMed] [Google Scholar]
- 36.Speerstra S., Chistov A.A., Proskurin G.V., Aralov A.V., Ulashchik E.A., Streshnev P.P. Antivirals acting on viral envelopes via biophysical mechanisms of action. Antivir. Res. 2018;149:164–173. doi: 10.1016/j.antiviral.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 37.M.R.St. Vincent, C.C. Colpitts, A.V. Ustinov, M. Muqadas, M.A. Joyce, N.L. Barsby, et al., Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. P Natl. Acad. Sci. USA, 107 (40) (2010) 17339–17344. [DOI] [PMC free article] [PubMed]
- 38.He J., Sun E., Bujny M.V., Kim D., Davidson M.W., Zhuang X. Dual function of CD81 in influenza virus uncoating and budding. Plos Pathog. 2013;9(10) doi: 10.1371/journal.ppat.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballut S., Naud-Martin D., Loock B., Maillard P. A Strategy for the targeting of photosensitizers. Synthesis, characterization, and photobiological property of porphyrins bearing glycodendrimeric moieties. J. Org. Chem. 2011;76(7):2010–2028. doi: 10.1021/jo102185d. [DOI] [PubMed] [Google Scholar]