Abstract
We directly compared two different approaches used for Circulating Tumor Cell (CTC) isolation, a size-dependent microfluidic system versus an EpCAM-dependent positive selection for downstream molecular characterization of CTC both at the gene expression and DNA methylation level in Head and Neck Squamous Cell Carcinoma (HNSCC). A size-dependent microfluidic device (Parsortix, ANGLE) and an EpCAM-dependent positive immune-magnetic isolation procedure were applied in parallel, using 10 mL PB from 50 HNSCC patients and 18 healthy donors. Total RNA was isolated from enriched CTCs and RT-qPCR was used to study the expression levels of CK-19, PD-L1, EGFR, TWIST1, CDH2 and B2M (reference gene). Real time methylation specific PCR (MSP) was used to study the methylation status of RASSF1A and MLL3 genes. In identical blood draws, the label-free size-dependent CTC-isolation system was superior in terms of sensitivity when compared to the EpCAM-dependent CTC enrichment, since a significantly higher percentage of identical PB samples was found positive at the gene expression and DNA methylation level, while the specificity was not affected. Our results indicate that future studies focused on the evaluation of clinical utility of CTC molecular characterization in HNSCC should be based on size-dependent enrichment approaches.
Subject terms: Biomarkers, Oral cancer
Introduction
Liquid biopsy provides a valuable source of biomarkers on prognosis and response to treatment of cancer patients1 and has recently shown a significant potential even for early cancer diagnosis and screening2. Isolation of circulating tumor cells (CTCs) from peripheral blood (PB) and their further downstream molecular characterization at the DNA, RNA and protein level is very important for reliable liquid biopsy analysis3. However, the identification and molecular characterization of CTCs is very challenging since these cells are extremely rare, and the amount of available sample for analysis in most cases is very limited1,3.
A variety of molecular assays have been developed for CTCs detection and molecular characterization. Molecular assays are based on total RNA isolation from CTCs and subsequent mRNA quantification of specific genes, and gDNA isolation for mutation analysis and DNA methylation studies4. CTC molecular characterization at the gene expression level has the potential to elucidate the critical signaling pathways involved in metastasis biology and even improve patient management. We have shown many years ago that the detection of CK-19 expression in CTCs has prognostic significance in both early and metastatic breast cancer5–7. Beyond gene expression, DNA methylation analysis in CTCs has a high potential to provide novel epigenetic biomarkers for diagnosis, prognosis, risk assessment, and disease monitoring in many types of cancer4. Based on this, we selected two tumor-suppressor genes namely, KMT2C/MLL3 and RASSF1A and evaluated their methylation status in CTCs of HNSCC patients. Recently, it was shown that down-regulation of lysine-specific methyltransferase 2 C (KMT2C/MLL3), a putative tumor suppressor leads to epigenetic and expression changes of DNA repair genes8. We also selected RASSF1A, since it has been shown that a high frequency of RASSF1A methylation is associated with more aggressive tumour phenotype among different cancer types9, while a recent meta-analysis suggested that there was a significant association between aberrant RASSF1A methylation and Head and Neck Squamous Cell carcinoma (HNSCC)10.
CTC-enrichment procedures are necessary prior to their molecular characterization, mainly for the reduction of background due to the presence of peripheral blood mononuclear cells (PBMCs). However, CTCs are highly heterogeneous and can have a different profile in different types of cancer. CTC heterogeneity and their phenotypic variation complicate their enrichment and subsequent phenotypic and molecular characterization1,3. Moreover, the effect of pre-analytical conditions and the establishment of quality control procedures in each analytical step is very important and very critical for CTC molecular characterization both at the gene expression and DNA methylation level11.
Over the past few years, a plethora of CTC isolation technologies based on their different biological and physical characteristics have been developed, including immune-magnetic, microfluidic, size dependent12 and function-based methods13. In the CellSearch® system (Menarini Silicon Biosystems), the only Food and Drug Administration (FDA) approved approach for CTC detection and enumeration so far14, CTCs enrichment is based on a positive selection targeting EpCAM-positive cells. However, it is obvious that in all EpCAM-based CTC-capture systems, EpCAM-negative CTC subpopulations are non-detectable. This can have serious implications in cases where CTCs are characterized by a phenotypic plasticity that mainly reflects an epithelial to mesenchymal transition state (EMT)15,16. EMT results in down-regulation of epithelial markers like EpCAM and simultaneous up-regulation of mesenchymal markers17,18. The lack of expression of epithelial markers on CTCs due to EMT has resulted in the development of novel procedures for CTCs isolation that are based on label-free microfluidic devices, size-based filtration, or combination of microchips and positive selection through specific antibodies1,3,13,19. Antibody combinations on immunomagnetic beads when compared to anti-EpCAM antibodies enable capturing of a larger number of CTCs20.
Head and Neck Squamous Cell carcinoma (HNSCC) is a devastating disease and novel treatments are urgently needed. Molecular characterization of CTCs can be a powerful prognostic tool while serial assessments of CTCs at different time points during treatment may guide treatment decisions. EMT is a common phenomenon in HNSCC progression, thus EpCAM-based approaches can be suboptimal for CTC isolation. We have recently shown that in HNSCC the detection of CTCs overexpressing PD-L1 provides important prognostic information21 and could be used for the selection of personalized medicine and treatment monitoring22–24.
The aim of the present study was to select the optimal enrichment system for CTC downstream molecular characterization in HNSCC. For this reason we directly compared the performance of a label-independent size-based microfluidic device versus an EpCAM-based CTC enrichment system using identical blood draws, and downstream molecular characterization both at the gene expression and DNA methylation level.
Results
RNA-based analysis
Comparison between size-dependent and EpCAM-dependent CTC-enrichment at the gene expression level
We first evaluated in a quantitative way the performance of these two different CTC enrichment approaches by downstream RNA-based CTC analysis run in parallel, using exactly the same procedure and the same RT-qPCR assays in material extracted from 50 HNSCC patients and 18 HD. Our results clearly reveal a significant difference in the expression levels of B2M (reference gene) in CTCs isolated through Parsortix versus EpCAM-dependent CTC enrichment. In all molecular assays, reference genes are selected on the basis that they are expressed at the same level both in the CTCs and in the PBMC fraction, as we have previously described 11,20. More specifically, as shown in Fig. 1A, Cq values for B2M were significantly lower in the EpCAM-based CTC-enrichment, indicating a higher number of B2M transcripts that definitely are due to a higher number of non-specific contaminant cells which are in their absolute majority PBMCs co-isolated with CTCs. B2M expression did not differ between HD and HNSCC within each CTC enrichment system as expected, since this reference gene is expressed at similar levels both in CTCs and PBMC, and the number of CTCs is very low (Fig. 1A). Our results also clearly reveal a significant difference in the expression levels of CD45 (specific leukocyte marker) in CTC-fractions isolated through Parsortix versus EpCAM-dependent CTC enrichment. More specifically, as shown in Fig. 1B, Cq values for CD45 were significantly lower in the EpCAM-based CTC-enrichment, indicating a higher number of CD45 transcripts that definitely are due to a higher number of contaminant leucocytes co-isolated with the CTC fractions. CD45 expression did not differ between HD and HNSCC within each CTC enrichment system as expected (Fig. 1B).
Figure 1.
RT-qPCR results using size-dependent enrichment (Parsortix) and EpCAM-dependent CTC enrichment for: (A) Β2Μ (reference gene); (B) CD-45 (specific leukocyte marker) (C) Relative fold change values (2-ΔΔCq) for reference gene (Β2Μ) and target gene (PD-L1) for healthy individuals and HNSCC patients’ CTC samples; (D) Relative fold change values (2-ΔΔCq) for reference gene (Β2Μ) and target gene (TWIST1); (E) Relative fold change values (2-ΔΔCq) for reference gene (Β2Μ) and target gene (CDH2) Red horizontal line: cutoff value. Different colors represent individual patients.
Size-dependent CTC-enrichment
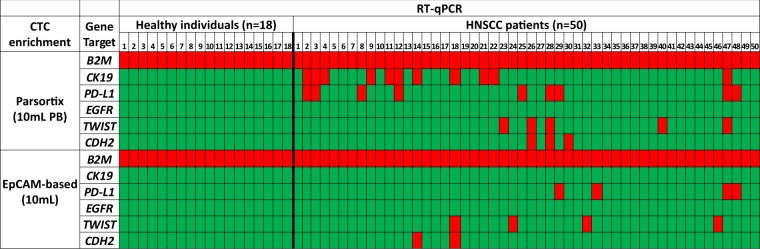
All samples isolated with the size-dependent microfluidic device (Parsortix) were of excellent RNA quality as this was verified by our RT-qPCR assay for B2M. Relative fold change values (2–ΔΔCt) for PD-L1 normalized according to B2M for individual samples are shown in Fig. 1B. Relative PD-L1 expression in the Parsortix was 0.48 (range: 0–2.39) in HD, 0.71 (range: 0–2.48) in PD-L1 negative HNSCC patients and 5.94 (range: 3.07–11.63) in PD-L1 positive HNSCC patients. Relative fold change values (2–ΔΔCt) for TWIST1, and CDH2 normalized according to B2M for individual samples are shown in Fig. 1C,D. Mean fold change of TWIST1 expression in the Parsortix was 0.75 (range: 0–5.35) in HD, 0.10 (range: 0–2.04) in TWIST1 negative HNSCC patients and 227.19 (range: 18.90–982.29) in TWIST1 positive HNSCC patients. Mean fold change of CDH2 expression in the Parsortix was 0.45 (range: 0–1.95) in HD, 0.13 (range: 0–1.35) in CDH2 negative HNSCC patients and 22.26 (range: 4.08–58.49) in CDH2 positive HNSCC patients. Our results for all genes tested are also shown in a heatmap (Fig. 2); PD-L1 was overexpressed in 9/50 (18.0%) and CK-19 in 11/50 (22.0%) cases. In 4/11 (36.4%) of CK-19 positive samples PD-L1 was also overexpressed. EGFR expression was not detected in any sample 0/50 (0.0%). Concerning EMT markers, TWIST1 was overexpressed in 5/50 (10%), and CDH2 in 3/50 (6%). None of 18 samples of the control group (HD) was found positive for mRNA expression for all genes tested.
Figure 2.
Direct comparison of gene expression in HNSCC and HD in CTC enriched using size-dependent enrichment (Parsortix) vs EpCAM-dependent CTC enrichment system using identical blood draws. (Red: gene expression, green: no expression).
EpCAM-dependent CTC enrichment
Using EpCAM-dependent CTC enrichment, all samples were also of excellent RNA quality as this was certified by RT-qPCR for Β2Μ. Relative fold change values (2–ΔΔCt) for TWIST1 normalized according to B2M are shown in Fig. 1C. Mean fold change of TWIST1 expression in the EpCAM-based enrichment was 0.92 (range: 0–3.16) in HD, 0.12 (range: 0–1.48) in TWIST1 negative HNSCC patients and 10.63 (range: 3.2–31.12) in TWIST1 positive HNSCC patients. Relative fold change values (2–ΔΔCt) for CDH2 normalized according to B2M are shown in Fig. 1D. Mean fold change of CDH2 expression in the EpCAM-dependent enrichment was 0.28 (range: 0–1.42) in HD, 0.07 (range: 0–1.41) in CDH2 negative HNSCC patients and 2.29 (range: 2.25–2.33) in CDH2 positive HNSCC patients. Mean fold change of PD-L1 expression in the EpCAM-dependent enrichment was 0.84 (range: 0–1.54) in HD, 0.55 (range: 0–1.66) in PD-L1 negative HNSCC patients and 3.24 (range: 1.72–4.99) in PD-L1 positive HNSCC patients. Our results are shown for all genes tested in a heatmap (Fig. 2); CK-19 and EGFR expression were not detected in any sample tested 0/50 (0%), while TWIST1 was overexpressed in 4/50 (8%), and CDH2 in 2/50 (4%) samples, PD-L1 overexpression was detected in 4/50 (8%) cases. None of 18 samples of the control group (HD) was found positive for mRNA expression for all genes tested.
In HNSCC CK-19 positive CTCs were only detected when enriched using the label-independent Parsortix approach but not in the EpCAM-dependent immunomagnetic CTC enrichment (Fig. 2). PD-L1 overexpression was detected in 9/50 (18.0%) cases after using the Parsortix device but only in 4/50 (8.0%) cases after using EpCAM-dependent CTC-enrichment. Three (#29, #47 and #48) out of four samples found positive for PD-L1 overexpression after using EpCAM-based CTC enrichment were also positive after size-dependent enrichment. CTCs are heterogeneous, so it is possible that in these patients there were populations of PD-L1 positive CTCs that were both EpCAM+ and EpCAM-. EGFR mRNA was not detected in any sample tested, using both approaches.
DNA methylation analysis
Size-dependent CTC-enrichment
Ten peripheral blood samples from healthy donors were processed following exactly the same procedure as the HNSCC samples in order to verify the diagnostic specificity of the DNA methylation analysis assays. None of the HD samples was found positive for RASSF1A and MLL3 methylation (0/10, 0%). DNA methylation analysis was performed in 43 samples isolated using the Parsortix microfluidic device; RASSF1A methylation in 3/43 (7%) and MLL3 methylation in 7/43 (16.3%) samples.
EpCAM-dependent CTC enrichment
Ten peripheral blood samples from healthy donors were processed following exactly the same way as the HNSCC samples in order to verify the diagnostic specificity of the DNA methylation analysis assays. None of the HD samples was found to be positive for RASSF1A and MLL3 methylation (0/10, 0%). DNA methylation analysis was performed in 31 samples isolated using the EpCAM-dependent immunomagnetic enrichment; RASSF1A in 0/31 (0%) and MLL3 promoter methylation in 4/31 (12.9%).
Direct comparison between size-dependent and EpCAM-dependent CTC-enrichment for DNA methylation analyses
For 29 of these patients we had available material for a direct comparison between size-based and EpCAM-dependent CTC-enrichment at the DNA methylation level. The assessment of correlation for DNA methylation profile of each gene of interest between CTCs isolated using the label-independent versus the EpCAM-dependent CTC-enrichment method was performed using the χ2 test for these 29 matched samples. We did not observe any statistically significant correlation for the above samples in none of the tumor-suppressor genes tested (Fig. 3).
Figure 3.
Direct comparison of DNA methylation analysis markers in HNSCC (n = 29) and HD (n = 10) in CTC enriched using a size-dependent enrichment (Parsortix) vs EpCAM-dependent CTC enrichment system, using identical blood draws. (Red: positive for DNA promoter methylation, green: negative for DNA promoter methylation).
Discussion
CTC enrichment technologies based on EpCAM present the risk of missing EpCAM-negative CTCs. Recent technological advances have now enabled CTC-enrichment based on different biological and physical properties of CTCs. Label-independent enrichment microfluidic devices for CTC enrichment have the potential to isolate successfully CTCs in an EMT state, since EMT leads to down-regulation of EpCAM.
In order to evaluate the most appropriate system for downstream molecular characterization at the gene expression and DNA methylation analysis level in CTCs from HNSCC patients, we performed for the first time a direct comparison study, using identical blood draws, between a label-independent size-based microfluidic device and an EpCAM-based CTC enrichment system. More specifically, we studied the expression levels of CK-19, PD-L1, EGFR, TWIST1, CDH2 and B2M (reference gene) and the methylation status of RASSF1A and MLL3 genes, that are all indicating the presence of CTCs. Our data clearly indicate that the CTCs population enriched using the label-independent size-based microfluidic device is of higher purity than that of CTCs isolated using EpCAM-based CTC enrichment; Β2Μ levels revealed a much lower PBMC contamination in the size-dependent approach compared to EpCAM-dependent CTC enrichment. Our results were in concordance with two recent studies; Obermayer et al. reported that the Parsortix system was more appropriate to remove leukocytes and allow for the subsequent molecular analysis in a high purity of the enriched cells25. The Parsortix system was also evaluated as having the highest recovery rate and the lowest leukocyte contamination, compared to two different CTC isolation methods26.
Our results indicate that distinct populations of CTCs are isolated when these two different enrichment approaches are used. It is clear though that when the size dependent isolation was used a lot more positive events for the presence of CTCs are detected, eg the same sample is characterized as CTC negative using EpCAM isolation. We report a higher percentage of PD-L1 positive samples following CTC isolation with the Parsortix system in comparison to the EpCAM-dependent approach. Three out of four samples found positive for PD-L1 overexpression after using EpCAM-dependent CTC enrichment were also positive after using the Parsortix device. The clinical significance of this finding will be soon evaluated in an independent study, involving a large number of HNSCC patients under specific treatments. According to a recent meta-analysis, a high PD-L1 expression in the tumour cells did not correlate with poor prognosis of patients suffering for oral squamous cells carcinoma27. Most studies published on PD-L1 expression are performed in the primary tumour and are based on immunohistochemistry, and have shown a significant variation in results, limiting the use of PD-L1 expression by immunohistochemistry as a prognostic biomarker in clinical practice27. Our results on PD-L1 mRNA overexpression in CTCs after Parsortix enrichment indicate towards a clinical evaluation of this finding in a large number of clinical samples.
EGFR mRNA was not detected in any sample tested, using both approaches. We did not observe any correlation in the expression of EMT markers (TWIST1 and CDH2) between the Parsortix and EpCAM-dependent CTC enrichment. According to our results, the number of patients positive for CDH2 expression is very low, therefore, to draw any conclusion studying this gene is inappropriate, so we simply report our finding.
In HNSCC several preliminary studies have detected CTCs by using various enrichment-isolation methods that yielded varying results with respect to the number of CTCs and the frequency of patients with positive CTCs28,29. In our recent study21 when we evaluated the expression of PD-L1 in CTCs of HNSSC patients using EpCAM isolation, we did not find any CK-19 mRNA expression in CTCs (data not shown). Similar to our results, Bozec et al. have shown that according to the results obtained by the CellSearch system, CTCs are in a relatively low proportion in patients with locally advanced HNSCC29. Thus, HNSCC is not considered as an exclusively EpCAM positive type of cancer.
In CTC samples isolated using the Parsortix microfluidic device a significantly higher percentage of samples positive for MLL3 and RASSF1A promoter methylation was detected when compared to paired samples isolated using the EpCAM-positive immunomagnetic approach. RASSF1A and MLL3 were found to be methylated in CTCs at various percentages, confirming the presence of heterogeneity, even in CTCs isolated from the same patient.
There is an urgent need for the evaluation of technologies investigating the expression, mutation and DNA methylation status of CTCs and it is important to select the best system in every type of cancer. Lampignano et al. developed a workflow using the Parsortix system for single CTC analysis, permitting for the first time assessment of the heterogeneity of PIK3CA mutational status within patient-matched EpCAMhigh and EpCAMl°w/negative CTCs30. Furthermore, Gorges et al. established reliable workflows in order to study multi-marker profiles of single CTCs by qPCR approaches. These workflows were combined with Parsortix system enabling the recovery of higher quality RNA31. El-Heliebi et al. compared three different CTC isolation systems for gene expression and DNA mutation analysis in CTCs of prostate cancer patients and showed also differences32. The incorporation of microfluidics into CTC isolation is now emerging for clinical applications33. Many microfluidic technologies have reported high sensitivity and specificity for capturing CTCs, however, the question still remains as to the superiority in comparison to immunoaffinity based approaches, specifically to identify different CTC populations34.
In conclusion, distinct populations of CTCs are isolated with these two different enrichment technologies. It is clear though that when the size dependent isolation was used a lot more positive events for the presence of CTCs are detected. The clinical significance of this finding will be soon evaluated in an independent prospective study, involving a large number of HNSCC patients under specific treatments.
Materials and Methods
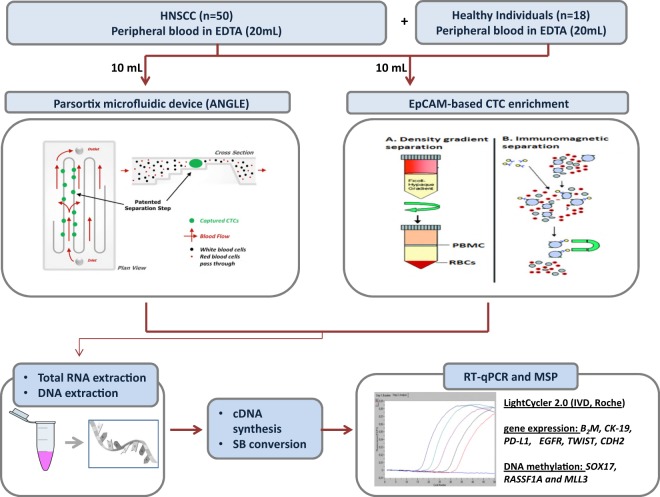
The experimental flowchart of the study is outlined in Fig. 4.
Figure 4.
Experimental flowchart of the study.
Peripheral blood samples collection
Fifty patients with squamous cell carcinoma of the oral cavity, oropharynx, hypopharynx or larynx and eighteen healthy donors were included in this study. Two of these patients, heavy smokers, were positive for HPV. In oral cavity and hypopharynx no HPV testing is indicated, while in some oropharynx cases HPV testing is not feasible because we have only cytology. Blood specimens were obtained before initiation of treatment. The first 5 ml of blood were discarded to avoid skin epithelial cells contamination. Peripheral blood was then collected into two 10 mL K2EDTA tubes (BD Vacutainer, Plymouth, UK) and mixed immediately after blood draw by inverting gently 10 times, maintained at room temperature (RT) and processed within 3 h, to CTC isolation. A size-dependent microfluidic device (Parsortix, ANGLE plc, UK) and an EpCAM-dependent positive immunomagnetic enrichment procedure were applied simultaneously, using 10 mL PB in each case. All patients gave their informed consent for participating in this study. IRB approval for the collection of samples was obtained from Attikon University Hospital, National and Kapodistrian University of Athens, Greece, (EΒΔ472/30–10–14). Attikon University Hospital approved the research and all methods were performed in accordance with the relevant guidelines and regulations.
CTCs enrichment using a size-based microfluidic device
The Parsortix CE-marked system (ANGLE plc, UK), based on micro-fluidics35 was used to capture and then harvest CTCs from 10 mL whole blood collected in K2EDTA tubes. Separation of blood components took place in a microscope slide sized disposable cassette, which contains a series of steps leaving a 6.5 μm measuring gap between the top cover and the final step36–38. Following enrichment, CTCs were harvested in a total volume of 200 μL of PBS deposited into 1.5 mL Eppendorf tubes by applying a reverse flow to the cassette using a specific software protocol. Spiking experiments were performed using 100 cells spiked in 10 mL peripheral blood of a healthy donor using the SCC-47 NHSCC cell line. According to our results, b2M (reference gene) and CK-19 (epithelial marker) transcripts were quantified in these cells before and after spiking. B2M and CK-19 trancripts were detected in these spiked samples after enrichment using the Parsortix.
CTCs enrichment using EpCAM-dependent immunomagnetic approach
Magnetic beads, coated with the monoclonal antibody BerEP4 against the human epithelial antigen, EpCAM, were used for CTCs enrichment (Dynabeads® Epithelial Enrich, Life Technologies, USA) from 10 mL whole blood collected in K2EDTA tubes as previously described21,39.
RNA extraction and cDNA synthesis
Total RNA from CTCs was isolated using TRIZOL-LS (ThermoFisher Scientific, USA), followed by cDNA synthesis as previously described21.
RT-qPCR
We used our previously developed and analytically validated RT-qPCR assays for PD-L1, keratin 19 (CK-19), epidermal growth factor receptor (EGFR) and beta-2-microglobulin (Β2Μ) (used as a reference gene)7,40. A multiplex RT-qPCR was performed for the quantitative determination of EMT markers, cadherin 2 (CDH2), and TWIST family transcription factor 1 (TWIST1) as previously reported41. All RT-qPCR reactions were performed in the LightCycler® 2.0 (IVD instrument, Roche Diagnostics, Germany) following the MIQE guidelines42. The amplification reaction mix for B2M contained 1 μL of the PCR Synthesis Buffer (5Χ), 1.2 μL of MgCl2 (25 mM), 0.15 μL dNTPs (10 mM), 0.3 μL BSA (10 μg/μL), 0.1 μL Hot Start DNA polymerase (HotStart, 5 U/μL, Promega, USA), 0.25 μL of forward and reverse primer (10μΜ), 0.83 μL of hydrolysis probe (3 μM). 1 μL of cDNA was added in the PCR mix and dH2O was added to a final volume of 9 μL. Protocol conditions: 1 cycle at 95 °C for 2 min, followed by 45 cycles of: 95 °C for 10 s, annealing at 58 °C for 20 s and extension at 72 °C for 20 s, and a final cooling cycle at 40 °C for 30 s. In each RT-qPCR run we used the same cDNA as a positive control in order to evaluate the accuracy and reproducibility of the results. For this purpose we aliquoted cDNA from MCF-7 cells and then stored these aliquots at −80 °C. The expression levels of PD-L1, CDH2, and TWIST1 were normalized using the 2−ΔΔCt approach in respect to the expression of Β2Μ43.
gDNA isolation from CTCs
gDNA was extracted from CTCs using the TRIZOL-LS reagent (ThermoFisher Scientific, USA) as previously described. Isolated gDNA44 was dissolved in 30 μL of 8 mmol/L NaOH.
Sodium Bisulfite (SB) treatment
gDNA samples were treated with SB, to convert all non-methylated cytosines to uracil, while methylated cytosines were not converted, using the EZ DNA Methylation Gold Kit (ZYMO Research, USA). SB-treated DNA was stored at −70 °C until use. In each SB reaction, dH2O and 100% methylated DNA were included as negative and positive control respectively.
Real-time MSP
We used our previously designed and analytically validated real time MSP assays for each gene of interest. All experiments for RASSF1A45 methylation analyses were performed in the LightCycler 2.0 (IVD instrument, Roche, Germany), whereas MLL3 methylation analysis was performed in the 96-well plates of LightCycler® 480 system (IVD, Roche Molecular Diagnostics, Switzerland) in a total volume of 10 μL. We report a sample as methylation positive, when we detect an MSP amplification signal (Cq<40.00) and as methylation negative only in the complete absence of amplification signal.
Quality control
Quality control checks were performed in all steps prior to sample analysis. In each step of the analytical procedure we included appropriate positive and negative controls in order to ensure the quality and reproducibility of results as previously described11. Before proceeding to the SB-treatment and real-time MSP, we assessed the gDNA integrity of all samples by amplifying the PIK3CA exon 20. Only samples that were positive for PIK3CA exon 20 amplification were further processed to SB treatment. After SB-treatment, converted DNA was also checked by a real-time PCR assay for β-actin (ACTB) and samples that were not amplified were excluded from the study11,46. Human placental genomic DNA (gDNA; Sigma-Aldrich, USA) was used as a real-time MSP negative control after SB-treatment, while Universal Methylated Human DNA Standard (ZYMO Research, USA) was used as fully methylated (100%) positive control.
Statistical analysis
We performed statistical evaluation of data using SPSS (SPSS Statistics 25.0). We used the chi-square test of independence, and the Mann Whitney test (SPSS, version 25.0) to make comparisons between groups. A level of P < 0.05 is considered statistically significant.
Conclusions
Our data indicate that molecular characterization of CTCs based on a label-free size-dependent isolation CTC system, gives superior results compared to the EpCAM-dependent approach, in HNSCC patients. The clinical significance of CTC detection using this approach remains to be elucidated in prospectively collected well-defined patient cohorts. Our results indicate that future studies focused on the evaluation of clinical utility of CTC molecular characterization in HNSCC should be based on size-dependent enrichment approaches.
Acknowledgements
We would like to thank all patients participating in this study. We would also like to thank M. O Brien and A.Newland (Angle) for providing the Parsortix instrument for evaluation in our lab.
Author contributions
Conceptualization, E.L.; methodology, S.M. and A.S.; validation, M.Z.; formal analysis, M.Z.; investigation, M.Z. and A.S.; resources, G.K. and A.P.; writing—original draft preparation, M.Z.; writing—review and editing, M.Z., S.M., A.S; visualization, E.L., A.K., A.P.; supervision, E.L.; project administration, A.S.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alix-Panabieres C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 2.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lianidou, E. & Hoon, D. Circulating Tumor Cells and circulating Tumor DNA. in Tietz Textbook of Clinical Chemistry and Molecular Diagnostics (eds. Nader, R., Horrath, A. R. & Wittwer, C.) 1111–44 (Elsevier Ltd, 2017).
- 4.Lianidou ES. Gene expression profiling and DNA methylation analyses of CTCs. Mol. Oncol. 2016;10:431–42. doi: 10.1016/j.molonc.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xenidis N, et al. Peripheral blood circulating cytokeratin-19 mRNA-positive cells after the completion of adjuvant chemotherapy in patients with operable breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2003;14:849–55. doi: 10.1093/annonc/mdg259. [DOI] [PubMed] [Google Scholar]
- 6.Stathopoulou, A. et al. Real-Time Quantification of CK-19 mRNA-Positive Cells in Peripheral Blood of Breast Cancer Patients Using the Lightcycler System Real-Time Quantification of CK-19 mRNA-Positive Cells in Peripheral Blood of Breast Cancer Patients Using the Lightcycler Syst. 9, 5145–5151 (2003). [PubMed]
- 7.Stathopoulou A, et al. A highly specific real-time RT-PCR method for the quantitative determination of CK-19 mRNA positive cells in peripheral blood of patients with operable breast cancer. Int. J. cancer. 2006;119:1654–9. doi: 10.1002/ijc.22017. [DOI] [PubMed] [Google Scholar]
- 8.Rampias, T. et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. e46821 10.15252/embr.201846821 (2019). [DOI] [PMC free article] [PubMed]
- 9.Grawenda AM, O’Neill E. Clinical utility of RASSF1A methylation in human malignancies. Br. J. Cancer. 2015;113:372–81. doi: 10.1038/bjc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng R-W, et al. Aberrant Methylation of RASSF1A Closely Associated with HNSCC, a Meta-Analysis. Sci. Rep. 2016;6:20756. doi: 10.1038/srep20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zavridou, M. et al. Evaluation of Preanalytical Conditions and Implementation of Quality Control Steps for Reliable Gene Expression and DNA Methylation Analyses in Liquid Biopsies. Clin. Chem. clinchem.2018.292318 10.1373/clinchem.2018.292318 (2018). [DOI] [PubMed]
- 12.Ribeiro-Samy S, et al. Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci. Rep. 2019;9:8032. doi: 10.1038/s41598-019-44401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alix-Panabières C, Pantel K. Technologies for detection of circulating tumor cells: facts and vision. Lab Chip. 2014;14:57–62. doi: 10.1039/C3LC50644D. [DOI] [PubMed] [Google Scholar]
- 14.Andree KC, van Dalum G, Terstappen LWMM. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016;10:395–407. doi: 10.1016/j.molonc.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alix-Panabières C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J. Mol. Med. 2017;95:133–142. doi: 10.1007/s00109-016-1500-6. [DOI] [PubMed] [Google Scholar]
- 16.Pastushenko I, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 17.Hyun, K.-A. et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget7, (2016). [DOI] [PMC free article] [PubMed]
- 18.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 19.Gorges TM, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila A, et al. EGFR-Based Immunoisolation as a Recovery Target for Low-EpCAM CTC Subpopulation. PLoS One. 2016;11:e0163705. doi: 10.1371/journal.pone.0163705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strati A, et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 2017;28:1923–1933. doi: 10.1093/annonc/mdx206. [DOI] [PubMed] [Google Scholar]
- 22.Schehr JL, et al. High specificity in circulating tumor cell identification is required for accurate evaluation of programmed death-ligand 1. PLoS One. 2016 doi: 10.1371/journal.pone.0159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satelli, A. et al. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. 10.1038/srep28910 (2016). [DOI] [PMC free article] [PubMed]
- 24.Mazel, M. et al. Frequent expression of PD-L1 on circulating breast cancer cells. 9, 1–7 (2015). [DOI] [PMC free article] [PubMed]
- 25.Obermayr E, et al. Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget. 2018;9:812–823. doi: 10.18632/oncotarget.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maertens Y, et al. Comparison of isolation platforms for detection of circulating renal cell carcinoma cells. Oncotarget. 2017;8:87710–87717. doi: 10.18632/oncotarget.21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troiano G, et al. High PD-L1 expression in the tumour cells did not correlate with poor prognosis of patients suffering for oral squamous cells carcinoma: A meta-analysis of the literature. Cell Prolif. 2019;52:e12537. doi: 10.1111/cpr.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grisanti S, et al. Circulating Tumor Cells in Patients with Recurrent or Metastatic Head and Neck Carcinoma: Prognostic and Predictive Significance. PLoS One. 2014;9:e103918. doi: 10.1371/journal.pone.0103918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bozec A, et al. Significance of circulating tumor cell detection using the CellSearch system in patients with locally advanced head and neck squamous cell carcinoma. Eur. Arch. Oto-Rhino-Laryngology. 2013;270:2745–2749. doi: 10.1007/s00405-013-2399-y. [DOI] [PubMed] [Google Scholar]
- 30.Lampignano R, et al. A Novel Workflow to Enrich and Isolate Patient-Matched EpCAMhigh and EpCAMlow/negative CTCs Enables the Comparative Characterization of the PIK3CA Status in Metastatic Breast Cancer. Int. J. Mol. Sci. 2017;18:1885. doi: 10.3390/ijms18091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorges TM, et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016;62:1504–1515. doi: 10.1373/clinchem.2016.260299. [DOI] [PubMed] [Google Scholar]
- 32.El-Heliebi A, et al. In Situ Detection and Quantification of AR-V7, AR-FL, PSA, and KRAS Point Mutations in Circulating Tumor Cells. Clin. Chem. 2018;64:536–546. doi: 10.1373/clinchem.2017.281295. [DOI] [PubMed] [Google Scholar]
- 33.Kozminsky M, Wang Y, Nagrath S. The incorporation of microfluidics into circulating tumor cell isolation for clinical applications. Curr. Opin. Chem. Eng. 2016;11:59–66. doi: 10.1016/j.coche.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murlidhar V, Rivera-Báez L, Nagrath S. Affinity Versus Label-Free Isolation of Circulating Tumor Cells: Who Wins? Small. 2016;12:4450–63. doi: 10.1002/smll.201601394. [DOI] [PubMed] [Google Scholar]
- 35.Hvichia GE, et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. cancer. 2016;138:2894–904. doi: 10.1002/ijc.30007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, L. et al. Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PLoS One10, (2015). [DOI] [PMC free article] [PubMed]
- 37.Chudziak J, et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst. 2016;141:669–678. doi: 10.1039/C5AN02156A. [DOI] [PubMed] [Google Scholar]
- 38.Porras TB, Kaur P, Ring A, Schechter N, Lang JE. Challenges in using liquid biopsies for gene expression profiling. Oncotarget. 2018;9:7036–7053. doi: 10.18632/oncotarget.24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markou A, Strati A, Malamos N, Georgoulias V, Lianidou ES. Molecular Characterization of Circulating Tumor Cells in Breast Cancer by a Liquid Bead Array Hybridization Assay. Clin. Chem. 2011;57:421–430. doi: 10.1373/clinchem.2010.154328. [DOI] [PubMed] [Google Scholar]
- 40.Strati A, et al. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 2011;11:422. doi: 10.1186/1471-2407-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markou A, et al. Multiplex Gene Expression Profiling of In Vivo Isolated Circulating Tumor Cells in High-Risk Prostate Cancer Patients. Clin. Chem. 2018;64:297–306. doi: 10.1373/clinchem.2017.275503. [DOI] [PubMed] [Google Scholar]
- 42.Bustin SA, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 43.Kenneth J. Livak* and Thomas D. Schmittgen†. METHODS 25, 402–408 (2001) doi:10.1006/meth.2001.1262, available online at http://www.idealibrary.com on Analysis of Relative Gene Expression Data Using Real- Time Quantitative PCR and the 2 2 DD C T Method. [DOI] [PubMed]
- 44.Chimonidou M, et al. DNA Methylation of Tumor Suppressor and Metastasis Suppressor Genes in Circulating Tumor Cells. Clin. Chem. 2011;57:1169–1177. doi: 10.1373/clinchem.2011.165902. [DOI] [PubMed] [Google Scholar]
- 45.Giannopoulou L, Chebouti I, Pavlakis K, Kasimir-Bauer S, Lianidou ES. RASSF1A promoter methylation in high-grade serous ovarian cancer: A direct comparison study in primary tumors, adjacent morphologically tumor cell-free tissues and paired circulating tumor DNA. Oncotarget. 2017;8:21429–21443. doi: 10.18632/oncotarget.15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastoraki S, et al. ESR1 Methylation: A Liquid Biopsy–Based Epigenetic Assay for the Follow-up of Patients with Metastatic Breast Cancer Receiving Endocrine Treatment. Clin. Cancer Res. 2018;24:1500–1510. doi: 10.1158/1078-0432.CCR-17-1181. [DOI] [PubMed] [Google Scholar]