Abstract
Alzheimer’s disease (AD) is the most common form of dementia globally and is characterized by aberrant accumulations of amyloid-beta (Aβ) and tau proteins. Oligomeric forms of these proteins are believed to be most relevant to disease progression, with oligomeric amyloid-β (oAβ) particularly implicated in AD. oAβ pathology spreads among interconnected brain regions, but how oAβ induces pathology in these previously unaffected neurons requires further study. Here, we use well characterized iPSC-derived human neurons to study the early changes to the proteome and phosphoproteome after 24 h exposure to oAβ 1-42. Using nLC-MS/MS and label-free quantification, we identified several proteins that are differentially regulated in response to acute oAβ challenge. At this early timepoint, oAβ induced the decrease of TDP-43, heterogeneous nuclear ribonucleoproteins (hnRNPs), and coatomer complex I (COPI) proteins. Conversely, increases were observed in 20 S proteasome subunits and vesicle associated proteins VAMP1/2, as well as the differential phosphorylation of tau at serine 208. These changes show that there are widespread alterations to the neuronal proteome within 24 h of oAβ uptake, including proteins previously not shown to be related to neurodegeneration. This study provides new targets for the further study of early mediators of AD pathogenesis.
Subject terms: Cellular neuroscience, Alzheimer's disease, Neurodegeneration, Molecular neuroscience, Neurodegeneration
Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia globally and is expected to grow substantially along with the aging global population. Much of the research in this field has focused on the pathological hallmarks of AD, amyloid-β (Aβ) and tau, but the way in which these mediators initiate neuronal pathogenesis at the molecular level requires further understanding. An important consideration in the study of AD is the different properties of Aβ assemblies: monomers, oligomers and fibrils. In particular, there is growing evidence to suggest that low molecular weight Aβ oligomers (oAβ) and protofibrils seed the aggregation of naïve Aβ, confer neurotoxicity, and also correlate to cognitive decline, as reviewed in1–4. Much of our current understanding of AD comes from the study of long-term exposure to Aβ in animal models, which has provided valuable insight into the effects of chronic exposure to Aβ, but often overlook the initial steps involved in pathogenesis. In order to further our understanding of the acute effects of exposure to oAβ, we challenged human iPSC-derived neurons with oAβ for 24 h and examined the changes to the proteome and phosphoproteome elicited by this acute oAβ treatment. In this way, we aim to elucidate some of the early effects elicited by oAβ upon neurons.
During AD, widespread changes occur in the proteome, including the up- and downregulation of disease-related proteins as well as the post-translational modification (PTM) of proteins. PTMs such as phosphorylation greatly affect the function of proteins and are well known to be important for the pathogenesis of neurodegenerative disorders, where detection of phosphorylated proteins are used to diagnose the severity of pathology5,6. Other types of PTMs include glycosylation, nitration, ubiquitination, truncation, methylation, and acetylation, among others. PTMs have significant influence upon the protein secondary and tertiary structure, which in turn impact its function, and importantly to neurodegeneration, its aggregation potential. During AD, PTMs of the tau protein are thought to lead to alterations in its ability to bind and stabilize microtubules and these PTMs also contribute to its misfolding and aggregation. The homeostasis of tau phosphorylation is delicately regulated by major kinases and phosphatases including the Casein Kinase (CK) and Glycogen Synthase Kinase 3 families (GSK3), as well as the Protein Phosphatase family (PP). In AD, CK1ε and GSK3β are increased, while PP2A and PP5 are decreased, contributing to the hyperphosphorylation of tau7,8. The association between tau and Aβ and their role in the initiation and driving of AD related neurodegeneration is a major topic of debate. Previous studies have demonstrated that Aβ can induce tau PTMs, leading to erroneous distribution of tau and neurotoxicity9,10. Others have shown the ability of oligomeric Aβ to bind tau and promote its fibrillization11–13, indicating a close relationship between Aβ and tau in AD pathogenesis.
While targeted interaction studies like those involving Aβ and tau have been the subject of many studies, unbiased proteomics approaches to understanding changes to the proteome in response to Aβ are lacking, leaving a significant gap in the understanding of how oAβ induces pathology in naïve human neurons. In this study, we use two well characterized human neuronal cellular models derived from human iPSCs: one harbouring wild-type amyloid precursor protein (APP; AF22) and another with the London APP (V717I) mutation (ADP2)14–21. Using LC-MS/MS and label-free quantification, we investigated the early changes to the proteome and phosphoproteome after neurons were challenged with oAβ, simulating a cellular environment comparable to early oAβ pathology in AD22–25. Using this approach, we identified expression alterations in response to oAβ challenge, including the mRNA processing proteins TDP-43 and hnRNP-F/K, vesicle trafficking proteins VAMP1/2 and coatomer subunits delta/gamma-1, among others. Both WT APP and V717I APP neurons displayed a marked decrease in coatomer complex I (COPI) subunit expression, indicating that intracellular retrograde transport pathways are greatly affected by oAβ. Interestingly, we also observed the differential phosphorylation of tau at serine 208 in response to the oAβ challenge. The alterations described in this study provide insight toward the early effects mediated by oAβ, adding new understanding of AD pathogenesis and identifying potential novel targets for early intervention in AD and other amyloidopathies.
Results
Protein identification by nLC-MS/MS
To obtain neuronal phenotypes in our human cellular model, long-term neuroepithelial-like stem cells (ltNES) were differentiated for 60 days prior to addition of 1 µM oligomeric amyloid-β (oAβ). This differentiation process results in cells with phenotypes indicative of functional neurons, including mature electrophysiological activity and the expression of neuronal markers NeuN and β-III tubulin. Differentiated cells were predominantly neurons as determined by their expression of the neuronal nuclear marker NeuN (AF22 = 78.5 ± 5.4%, ADP2 = 84.5 ± 9.0%), consistent with previous studies using these ltNES cells (Supplementary Fig. S1)14–21. After 24 h incubation with the oAβ, cells were collected, and proteins extracted for whole proteome or phosphoproteome analysis by nLC-MS/MS (summarized in Fig. 1). The raw data were analyzed using the Sequest HT algorithm in Proteome Discoverer and exported to Scaffold to perform a comparative differential analysis, employing Student’s t-test using normalized Total TIC. These analyses were performed in 3 datasets: AF22 (Wild-type APP; n = 16 control, n = 14 oAβ treated), AF22 phosphoenriched (PPE) with TiO2 (n = 11 control, n = 8 oAβ treated) and ADP2 (V717I APP mutation; n = 7 control, n = 7 oAβ treated). These analyses resulted in the identification of 2253 proteins (1627 protein groups; AF22), 1724 proteins (1315 protein groups; ADP2) and 577 proteins (498 protein groups, AF22 PPE), respectively.
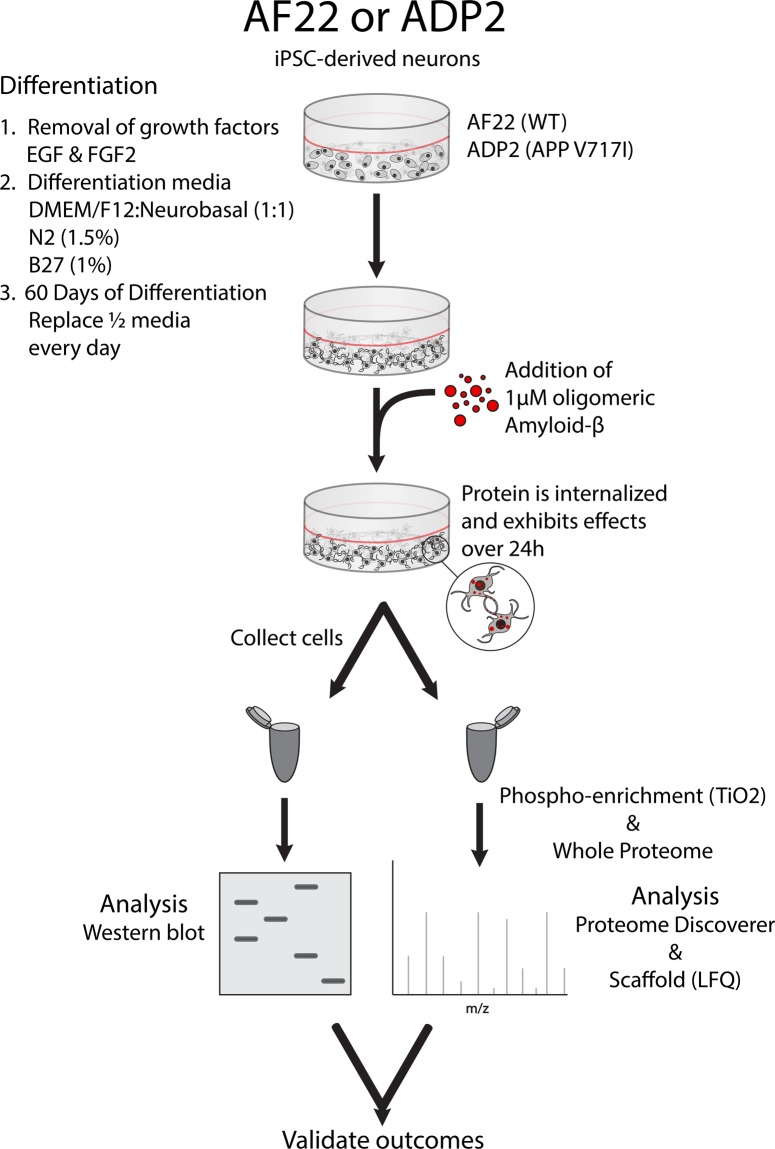
Figure 1.
Depiction of cell treatment models. iPSC-derived neurons are first differentiated for 60 days, then treated with 1 µM oligomeric amyloid-beta for 24 h. Neurons are collected and used for Western blot, or with nLC-MS/MS (whole proteome analysis and phosphoenrichment). Quantification is performed by densitometry (Western blot) and by label-free quantification (LFQ) using Scaffold proteomics software. Fold-change values were compared between both methods to validate the LFQ method.
Protein expression in human iPSC-derived neurons are significantly altered in response to oAβ
In the AF22 proteome, 42 proteins were found to be significantly (p < 0.05) up- or downregulated (Table 1). Several of these were of particular relevance to cellular pathways implicated in AD, including significant upregulations in vesicle-associated protein VAMP2 (fold change 1.5, p = 0.036), the Aβ binding protein SGTA (small glutamine-rich tetratricopeptide repeat-containing protein alpha; fold change 1.9, p = 0.018) and the AD-associated, CNS development and maturation protein DPYL5 (Dihydropyrimidinase-related protein 5, AKA CRMP5, fold change 1.4, p = 0.047). In response to the oAβ challenge, there were also significant downregulations observed in several proteins, including the neurodegenerative disease (ND)-related protein TDP-43 (fold change 0.4, p = 0.038) and COPI retrograde trafficking protein coatomer subunit delta (fold change 0.2, p = 0.026) were observed in response to oAβ. Next, we queried this dataset for several possible PTMs, which identified 10 proteins with significantly altered PTM expression in response to oAβ, including acetylation of 14-3-3ε (fold change 0.3, p = 0.0097), methylation of tubulin beta-3 chain (fold change 0.07, p = 0.036) and phosphorylation of stathmin (fold change 2.1, p = 0.046) (Supplementary Table S2). We have also provided a curated list of proteins of interest with relevance to the studies of NDs, including tau (fold change 1.8, p = 0.25) and protein phosphatase 5 (PP5; fold change 0.2, p = 0.11), α-synuclein (fold change 1.9, p = 0.4), among others (Supplementary Table S3).
Table 1.
Significantly up- and downregulated proteins in the proteome of AF22 cells resulting from 24 h oAβ treatment.
| Identified Proteins | Protein ID | AF22 T-Test (p-value) | AF22 Fold Change | ADP2 T-Test (p-value) | ADP2 Fold Change |
|---|---|---|---|---|---|
| Amyloid beta A4 protein | A4_HUMAN | 0.0035 | 3.3 | 0.067 | 17 |
| Aspartate aminotransferase, cytoplasmic | AATC_HUMAN | 0.015 | 1.8 | 0.58 | 1.5 |
| Cytoplasmic aconitate hydratase | ACOC_HUMAN | 0.049 | 0.3 | 0.19 | 0 |
| Proteasomal ubiquitin receptor ADRM1 | ADRM1_HUMAN | 0.035 | 3.8 | 0.13 | INF |
| Actin-related protein 2/3 complex subunit 3 | ARPC3_HUMAN | 0.032 | 0.3 | 0.18 | 2.8 |
| Cluster of Isoform Beta of Apoptosis regulator BAX | BAX_HUMAN | 0.015 | 0.4 | 0.65 | 1.2 |
| Bleomycin hydrolase | BLMH_HUMAN | 0.04 | 0.6 | ND | ND |
| BRI3-binding protein | BRI3B_HUMAN | 0.011 | 2.6 | ND | ND |
| 60 kDa heat shock protein, mitochondrial | CH60_HUMAN | 0.039 | 1.3 | 0.66 | 1.1 |
| Cluster of Isoform 2 of Clathrin heavy chain 1 | CLH1_HUMAN | 0.041 | 0.7 | 0.52 | 1.2 |
| Cluster of Cofilin-1 | COF1_HUMAN | 0.024 | 0.7 | 0.97 | 1 |
| Coatomer subunit delta | COPD_HUMAN | 0.026 | 0.2 | 0.88 | 1.2 |
| Casein kinase II subunit alpha | CSK21_HUMAN | 0.029 | 0.5 | 0.54 | 1.5 |
| Dihydropyrimidinase-related protein 5 | DPYL5_HUMAN | 0.047 | 1.4 | 0.3 | 1.4 |
| Cluster of Spliceosome RNA helicase DDX39B | DX39B_HUMAN | 0.022 | 0.5 | 0.58 | 0.7 |
| Cluster of Isoform 2 of Elongation factor 1-delta | EF1D_HUMAN | 0.038 | 1.6 | 1 | 1 |
| Cluster of Isoform 2 of Histone deacetylase 2 | HDAC2_HUMAN | 0.027 | 0.4 | 0.21 | 0.05 |
| Heterogeneous nuclear ribonucleoprotein F | HNRPF_HUMAN | 0.0087 | 0.5 | 0.78 | 0.9 |
| LIM and SH3 domain protein 1 | LASP1_HUMAN | 0.038 | 2.8 | 0.17 | 3.6 |
| Leucine-rich PPR motif-containing protein, mitochondrial | LPPRC_HUMAN | 0.017 | 0.07 | 0.43 | 0.4 |
| Microtubule-associated protein RP/EB family member 1 | MARE1_HUMAN | 0.015 | 2 | 0.9 | 1.1 |
| Isoform 2 of RNA-binding protein Musashi homolog 2 | MSI2H_HUMAN | 0.037 | 0.5 | 0.47 | 0.2 |
| Metallothionein-3 | MT3_HUMAN | 0.033 | 9.8 | ND | ND |
| Isoform 2 of NADH dehydrogenase 1 alpha subcomplex subunit 11 | NDUAB_HUMAN | 0.015 | 0.3 | 0.34 | INF |
| Isoform 2 of Nuclear mitotic apparatus protein 1 | NUMA1_HUMAN | 0.03 | 3.8 | ND | ND |
| Isoform 2 of GPI transamidase component PIG-S | PIGS_HUMAN | 0.042 | 0.5 | 0.91 | 0.9 |
| Isoform 2 of Proteasome subunit alpha type-3 | PSA3_HUMAN | 0.038 | 1.8 | 0.13 | 4 |
| Cluster of 26 S proteasome non-ATPase regulatory subunit 4 | PSMD4_HUMAN | 0.046 | 2.6 | 0.21 | 3.3 |
| Multifunctional protein ADE2 | PUR6_HUMAN | 0.04 | 0.5 | 0.99 | 1 |
| Cluster of 60 S ribosomal protein L26 | RL26_HUMAN | 0.018 | 0.3 | 0.22 | 2.4 |
| Small glutamine-rich tetratricopeptide repeat-containing protein alpha | SGTA_HUMAN | 0.018 | 1.9 | 0.31 | 1.5 |
| Synaptogyrin-3 | SNG3_HUMAN | 0.036 | 2.1 | 0.32 | 2.6 |
| Signal recognition particle subunit SRP72 | SRP72_HUMAN | 0.045 | 0 | 0.34 | 0 |
| Translocon-associated protein subunit delta | SSRD_HUMAN | 0.022 | 0.5 | 0.61 | 0.8 |
| Phenylalanine–tRNA ligase alpha subunit | SYFA_HUMAN | 0.037 | 0.5 | 0.8 | 0.8 |
| TAR DNA-binding protein 43 | TADBP_HUMAN | 0.038 | 0.4 | 0.64 | 0.8 |
| Transformer-2 protein homolog beta | TRA2B_HUMAN | 0.047 | 0.4 | 0.27 | 0.5 |
| Cluster of Isoform 3 of Thioredoxin reductase 1, cytoplasmic | TRXR1_HUMAN | 0.041 | 0.4 | 0.11 | 3.8 |
| Isoform 3 of 116 kDa U5 small nuclear ribonucleoprotein component | U5S1_HUMAN | 0.03 | 0.2 | 0.52 | 0.5 |
| Isoform 3 of Vesicle-associated membrane protein 1 | VAMP1_HUMAN | 0.039 | 87 | 0.99 | 1 |
| Cluster of Vesicle-associated membrane protein 2 | VAMP2_HUMAN | 0.036 | 1.5 | 0.29 | 1.5 |
The provided table depicts proteins with significant changes in expression (p < 0.05, T-test with Benjamini-Hochberg correction) in cells challenged with oAβ relative to untreated cells (i.e. AF22 + oAβ relative to AF22 control). AF22: n = 16 (control), n = 14 (oAβ treated). Data represent analysis of whole proteome preparations. ADP2 columns provided for comparison. ADP2: n = 7 (control), n = 7 (oAβ treated). ND = Not detected.
In order to confirm the validity of the LFQ values determined by normalized Total TIC, we analyzed the proteins from AF22 neurons by Western blotting and compared the LFQ outcomes to those determined by densitometry (Table 2). We examined 6 proteins by Western blot (Supplementary Figs. S2 and S3) and found that the fold change values and significance obtained by both methods (LFQ and Western blot) gave similar results, providing confidence in the data obtained by this method.
Table 2.
Verification of the validity of quantifications by Normalized Total TIC (MS/MS LFQ).
| Protein | Protein ID | Fold Change (MS/MS) | p-value (MS/MS) | Fold Change (WB) | p-value (WB) |
|---|---|---|---|---|---|
| Calreticulin | CALR_HUMAN | 1 | 0.89 | 1.21 | 0.67 |
| Ras-related protein Rab-7a | RAB7A_HUMAN | 1 | 0.89 | 0.98 | 0.97 |
| TAR DNA-binding protein 43 | TADBP_HUMAN | 0.4 | 0.038 | 0.49 | 0.04 |
| Mitochondrial import receptor subunit TOM22 homolog | TOM22_HUMAN | 0.6 | 0.12 | 0.48 | 0.15 |
| Vesicle-associated membrane protein 2 | VAMP2_HUMAN | 1.5 | 0.036 | 2.11 | 0.029 |
| Vesicle-associated membrane protein-associated protein B/C | VAPB_HUMAN | 1.5 | 0.27 | 1.25 | 0.25 |
A number of proteins were analyzed by Western blot and quantified for comparison to the fold change values determined by MS/MS LFQ. The fold changes and p-values calculated by both methods gave comparable results, providing confidence in the LFQ quantification method used in this study. The data provided here refer to AF22 cells challenged with oAβ relative to untreated AF22 cells. Fold changes determined by MS/MS are described in Table 1 and Supplementary Table S3. Western blots are provided in Supplementary Fig. S2. For MS/MS data: T-test with Benjamini-Hochberg correction, n = 14-16. For WB data: T-test, n = 6-7 per group.
oAβ treatment elicits significant changes to the neuronal phosphoproteome
Given the importance of phosphorylation in ND-related proteins, we analyzed the phosphoproteome in differentiated human neurons. To achieve this, TiO2 enrichment was performed prior to nLC-MS/MS analysis and the resulting data was analyzed in a similar fashion to that of the proteome. In addition to phosphorylation, the dataset was searched for other PTMs (oxidation, methylation, acetylation), but none of these were found significantly changed in this sample. When searching the dataset for phosphorylation PTMs, we observed a significant alteration in 5 phosphorylated proteins: increases to the α-tubulin acetylating protein isoform 2 of alpha-tubulin N-acetyltransferase 1 (fold change 9.9, p = 0.002), isoform 2 of protocadherin-1 (fold change 11, p = 0.014), isoform 3 of periphilin-1 (fold change 1.7, p = 0.022), eukaryotic translation initiation factor 4B (fold change 1.6, p = 0.031), and a decrease in phosphorylated RNA-binding protein with serine-rich domain 1 (fold change 0.4, p = 0.043) (Table 3). Similar to the experiments with the unenriched proteome, we have also provided a list of proteins of interest to NDs that were identified while searching for phosphorylation PTMs (Supplementary Table S4), or without any search filters (Supplementary Table S5). Of particular interest, was the increase in phosphorylated tau in response to oAβ addition (fold change 1.3, p = 0.10) but this effect did not reach statistical significance. Similarly, the AD-associated tau kinase Casein kinase Iε was also increased (fold change 4.1, p = 0.053).
Table 3.
Changes in the AF22 phosphoproteome resulting from 24 h oAβ treatment.
| Identified Proteins | Protein ID | T-Test (p-value) | Fold Change | PTM |
|---|---|---|---|---|
| Isoform 2 of Alpha-tubulin N-acetyltransferase 1 | ATAT_HUMAN | 0.002 | 9.9 | Phosphorylation |
| Eukaryotic translation initiation factor 4B | IF4B_HUMAN | 0.031 | 1.6 | Phosphorylation |
| Isoform 2 of Protocadherin-1 | PCDH1_HUMAN | 0.014 | 11.0 | Phosphorylation |
| Isoform 3 of Periphilin-1 | PPHLN_HUMAN | 0.022 | 1.7 | Phosphorylation |
| Isoform 3 of RNA-binding protein with serine-rich domain 1 | RNPS1_HUMAN | 0.043 | 0.4 | Phosphorylation |
The provided table depicts proteins with significant changes in expression (p < 0.05, T-test with Benjamini-Hochberg correction) in the phosphoproteome of AF22 cells challenged with oAβ relative to untreated AF22 cells. The proteins detailed in this table were identified after enrichment with TiO2. n = 11 (control), n = 8 (oAβ treated).
Phosphorylation of tau at Ser208 may be an early PTM elicited by oAβ
One of the most important, and therefore most studied, aspects of NDs is the variable phosphorylation of the tau protein, which influences its microtubule binding efficiency, but also misfolding and self-aggregation. We examined the phosphorylation of tau in our PPE samples (Table 4), and identified that serine 208 (defined from the longest tau isoform, 2N4R containing 441 amino acids) was differentially phosphorylated in response to oAβ at this early time point, where 75% of the treated samples displayed phosphorylation at Ser208 (n = 8), while only 27% of the control cells had phosphorylation at this residue (n = 11). Other commonly studied phosphorylation sites such as Thr181 (epitope of antibody AT270) Ser202/Ser205 (epitope of antibody AT8), Thr231 (epitope of antibody AT180), Ser262, Ser396/Ser404 (epitope of antibody PHF-1) did not display appreciable differences in their tau phosphorylation at this early time point. We suggest that phosphorylation of tau at Ser208 could be an early marker of pathology in response to oAβ and warrants further investigation.
Table 4.
Phosphorylation at Serine 208 is differentially expressed after 24 h oAβ challenge and may be a useful residue in the detection of early oAβ-induced effects.
| Tau P10636-8 AA 441 | Control samples with phosphorylation detected at residue: (n = 11) | Percentage of Control with detected residue | oAβ challenged samples with phosphorylation detected at residue: (n = 8) | Percentage of oAβ challenged with detected residue |
|---|---|---|---|---|
| 181 | 1 | 9.09090909 | 1 | 12.5 |
| 197 | 2 | 18.1818182 | 2 | 25 |
| 198 | 2 | 18.1818182 | 2 | 25 |
| 199 | 8 | 72.7272727 | 6 | 75 |
| 202 | 11 | 100 | 8 | 100 |
| 205 | 3 | 27.2727273 | 3 | 37.5 |
| 208 | 3 | 27.2727273 | 6 | 75 |
| 212 | 1 | 9.09090909 | 0 | 0 |
| 217 | 1 | 9.09090909 | 0 | 0 |
| 231 | 2 | 18.1818182 | 0 | 0 |
| 235 | 9 | 81.8181818 | 7 | 87.5 |
| 238 | 2 | 18.1818182 | 0 | 0 |
| 262 | 3 | 27.2727273 | 1 | 12.5 |
| 394 | 1 | 9.09090909 | 0 | 0 |
| 396 | 5 | 45.4545455 | 5 | 62.5 |
| 400 | 9 | 81.8181818 | 8 | 100 |
| 403 | 2 | 18.1818182 | 0 | 0 |
| 404 | 11 | 100 | 8 | 100 |
Canonical phosphorylation sites such as 181, 202, 231, 262 and 404 do not show any appreciable changes at this early stage. The table depicts all detected phosphorylated tau residues for both AF22 cells challenged with oAβ and untreated AF22 cells. Phosphorylations detailed in this table were identified after TiO2 enrichment. Amino acid number corresponds to full-length 2N4R tau (441 residues; UniProt accession P10636-8), n = 11 (control) and n = 8 (oAβ).
oAβ challenge elicits similar responses in V717I London APP mutant neurons as those observed in WT neurons
In addition to changes in the proteome observed in the WT APP neurons (AF22), we also examined the effects of oAβ stimulation on the proteome of iPSC-derived neurons from a donor harbouring the V717I London mutation (ADP2). We observed 11 significantly altered proteins (oAβ challenged ADP2 neurons relative to control ADP2 neurons; Table 5), many of which were related to the changes observed in the AF22 neurons (Table 1). In the ADP2 neurons, we observed the significant increase in proteasome subunit alpha type-4 (PSA4; fold change 4.3, p = 0.0084; Table 5), while its expression partner PSA3 was not significantly upregulated (fold change 4.0, p = 0.13; Table 1). In AF22 neurons, PSA3 that was upregulated (fold change 1.8, p = 0.038; Table 1), while PSA4 was not (fold change 1.0, p = 0.98; Table 5). Proteins belonging to the coatomer protein complex (COPI), which have been identified as risk factors for late-onset AD (LOAD), were differentially regulated in response to oAβ in both AF22 and ADP2 neurons. In ADP2, expression of Coatomer subunit gamma-1 (COPG1) was decreased in response to oAβ (fold change 0.2, p = 0.028; Table 5), while its expression partner Coatomer subunit delta (COPD) was unchanged (fold change 1.2, p = 0.88; Table 1). Conversely, in the AF22 neurons’ response to oAβ, COPG1 was not significantly altered (fold change 0.8, p = 0.66; Table 5), while COPD was significantly decreased (fold change 0.2, p = 0.026; Table 1). Similar to the experiments with the AF22 proteome, we searched the dataset for PTMs, however we only observed 2 proteins with significant changes in the ADP2 dataset: acetylated guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 was found to be increased in the oAβ stimulated group (fold change 4.2, p = 0.019; Supplementary Table S2), while phosphorylated DPYL2, which is associated with NFTs, was observed exclusively after oAβ challenge (fold change INF, p = 0.028; Supplementary Table S2). A curated list of ND relevant proteins that did not reach statistical significance is provided in Supplementary Table S3.
Table 5.
Significantly up- and downregulated proteins in the proteome of ADP2 cells resulting from 24 h oAβ treatment.
| Identified Proteins | Protein ID | AF22 T-Test (p-value) | AF22 Fold Change | ADP2 T-Test (p-value) | ADP2 Fold Change |
|---|---|---|---|---|---|
| Alpha-2-macroglobulin receptor-associated protein | AMRP_HUMAN | 0.33 | 2.1 | 0.021 | INF |
| Coatomer subunit gamma-1 | COPG1_HUMAN | 0.66 | 0.8 | 0.028 | 0.2 |
| Isoform 2 of Serine/threonine-protein kinase DCLK2 | DCLK2_HUMAN | 0.33 | 0.5 | 0.031 | 8 |
| Isoform Cytoplasmic of Fumarate hydratase, mitochondrial | FUMH_HUMAN | 0.14 | 1.5 | 0.012 | 2.6 |
| Cluster of Isoform 2 of Eukaryotic initiation factor 4A-II | IF4A2_HUMAN | 0.93 | 1 | 0.029 | 0.5 |
| Cluster of Isoform Delta 10 of Calcium/calmodulin-dependent protein kinase type II subunit delta | KCC2D_HUMAN | 0.68 | 1.2 | 0.011 | 2.5 |
| Isoform 2 of Nucleosome assembly protein 1-like 1 | NP1L1_HUMAN | 0.55 | 0.9 | 0.041 | 2 |
| Proteasome subunit alpha type-4 | PSA4_HUMAN | 0.98 | 1 | 0.0084 | 4.3 |
| 60 S acidic ribosomal protein P1 | RLA1_HUMAN | 0.85 | 0.9 | 0.032 | 2 |
| Isoform 2 of Neuronal-specific septin-3 | SEPT3_HUMAN | 0.29 | 1.4 | 0.037 | 3.8 |
| Serine/arginine-rich splicing factor 2 | SRSF2_HUMAN | 0.76 | 0.9 | 0.043 | 2.3 |
The provided table depicts proteins with significant changes in expression (p < 0.05, T-test with Benjamini-Hochberg correction) in cells challenged with oAβ relative to untreated cells (i.e. ADP2 + oAβ relative to ADP2 control). ADP2: n = 7 (control), n = 7 (oAβ treated). Data represent analysis of whole proteome preparations. AF22 columns provided for comparison. AF22: n = 16 (control), n = 14 (oAβ treated). ND = Not detected.
Discussion
One of the most studied hallmarks of AD is the progressive propagation of Aβ pathology, thought to be caused by the spreading of oAβ among interconnected brain regions from neuron to neuron5,22, further reviewed in1–4. In this work, we aim to elucidate intracellular protein changes elicited by oAβ upon naïve human neurons, thereby furthering our understanding of the early mediators involved in Aβ-induced pathogenesis. To do this, we used well characterized iPSC-derived ltNES cells, which upon differentiation, are electrophysiologically mature and display mature neuronal markers14–21. Since Aβ oligomers are thought to be the most relevant aggregates in AD pathology, we challenged the differentiated neurons with 1 µM oAβ over 24 h to mimic the early phase of oAβ challenge, similar to previous studies22,23,26–30. Previous studies have shown that concentrations of up to 3 µM oAβ 1-42 exist within neurons of the AD brain31, while the concentrations of oAβ used in this study may reflect those in the immediate vicinity of amyloid plaque cores, the so-called halo region32,33. Using nLC-MS/MS and LFQ, we investigated the changes to the proteome and phosphoproteome elicited by this oAβ challenge. Using this unbiased approach, we were able to identify a number of proteins of interest that support previous findings34,35, but also proteins that extend our current understanding of early amyloid pathology.
Analysis of the AF22 proteome in response to oAβ revealed 42 significantly up- and downregulated proteins. These included a significant reduction to the mRNA processing protein Heterogeneous nuclear ribonucleoprotein F (hnRNP-F). The hnRNP family of proteins are RNA binding proteins that perform many functions, including nucleic acid metabolism, translational regulation and alternative splicing (reviewed in36). In AD, ALS and FTLD, there is dysregulation of hnRNP expression (especially A/B isoforms, but also H/F), which are thought to exacerbate memory impairments37–41. Conversely, we observed increases in the Aβ-binding protein SGTA42 and neuronal growth-related protein DPYL5. The DPYL family of proteins, also known as Collapsin response mediator proteins (CRMPs) are neuronally expressed proteins important to axon and neurite formation and have been linked to neurodegeneration in AD, PD and traumatic brain injury43,44. In a recent study, it was shown that increased DPYL5 exacerbated memory loss in an AD mouse model45. A previous study also identified the expression of DPYL proteins to be significantly altered in response to oAβ 1-42, although this study had the limitations of being performed in neuroblastoma SH-SY5Y cells with an extremely large dose of oAβ (15 µM)35. Nevertheless, in their study Földi et al. found significant changes in proteins related to those observed in this work, including reductions in hnRNP-K, but contrary to our findings, a decrease in DPYL proteins. Previous studies have strongly suggested that Aβ and the ND-related, mRNA processing protein TDP-43 are related in the pathology of neurodegeneration. Animal studies have shown that chronic exposure of Aβ in the brain leads to TDP-43 pathology46 and that TDP-43 can induce the cross-seeding of Aβ into oAβ47. In human AD patients, more than half also display TDP-43 comorbidity48–50. Supporting the link between them, we found a significant reduction in the expression of TDP-43 after challenge with oAβ in the AF22 neurons, but not ADP2. We speculate that the reduction in TDP-43 expression that we observed in response to oAβ after 24 h may reflect the neuron’s attempt to limit the damage caused by TDP-43 to cells during acute oAβ exposure. In animal models of AD, TDP-43 levels are initially indistinguishable from the WT but increase over time during chronic oAβ exposure. Given our observations, we suggest that there is an initial reduction in TDP-43 expression during initial oAβ exposure, which gradually increases over time as the cell is exposed to Aβ chronically51,52. This may also be a contributing factor as to why TDP-43 reduction was not as robust in the ADP2 neurons, owing to their chronic exposure to endogenously generated Aβ 1-42 during differentiation53. Altogether, this model points toward widespread RNA processing dysregulation in the early stages of oAβ challenge54.
Given that hyperphosphorylation is important to the neurodegenerative process in multiple ND’s, we also investigated the phosphoproteome in AF22 neurons after 24 h incubation with oAβ, with a specific interest in the differential phosphorylation of tau. Following phosphoenrichment with TiO2, we found that tau expression was elevated after exposure to oAβ, but that this elevation did not achieve statistical significance (fold change 1.3, p = 0.10). Furthermore, we identified the differential phosphorylation of tau at serine 208. At Ser208, we found that 75% of the oAβ-exposed samples displayed phosphorylation, whereas only 27% of the control samples experienced phosphorylation at the same site. Other canonical tau phosphorylation sites, including early (Thr181, Ser202/Ser205 and Ser262), intermediate (Thr231 and Thr212/Ser214) and late residues (Ser396/Ser404 and Ser422)6,55,56 were not remarkably altered by exposure to oAβ for 24 h. In future studies, it will be imperative to confirm this finding using other methods (e.g. Western blot), however there are currently no commercially available antibodies specific to tau phosphorylation at Ser208 alone. Nevertheless, this finding indicates that phosphorylation of tau at Ser208 may be an early consequence of oAβ exposure. Following independent verification of this finding, this tau phosphoepitope should be further investigated as a potential early marker of tauopathy in Aβ related NDs. In addition to tau itself, we observed an increase (p = 0.053) in CK1ε, a member of the Casein Kinase 1 family (CK1 isoforms include α1, α2, γ1, γ2, γ3, δ, and ε; family also includes Tau-tubulin kinases (TTBK1 and TTBK2)), which are known to be major tau and TDP-43 kinases46,57,58, and phosphorylate tau at Ser20859. Furthermore, CK1ε specifically, has been shown to be significantly elevated in AD and contributes to increased expression of Aβ7,58. CK1ε also functions as a major regulator of circadian rhythms60, leading speculation that these disturbances to CK1ε expression in AD could be related to the sleep disturbances experienced by patients. Previous in vivo experiments have shown that treatment of 5xFAD mice with recombinant nuclease-sensitive element-binding protein 1 (YBOX 1) results in decreased levels of Aβ in the brain, but also inhibits the fibrillization of Aβ 1-42 in vitro61. We show that oAβ exposure caused a decrease in phosphorylated YBOX 1 in the exposed neurons (fold change 0.3, p = 0.054). The effect of phosphorylation on YBOX 1 in relation to AD is unknown, but YBOX 1 phosphorylation is thought to enhance the activation of transcription of NF-κB during colon cancer62,63.
The APP London (V717I) mutation is one of the most common familial APP mutations of AD, with patients experiencing the onset of clinical dementia at an average age of 5964. This mutation results in an increased Aβ42:Aβ40 ratio in the brain and in patient derived iPSCs through increased production of Aβ 1-4253,65–67. oAβ challenge of differentiated ltNES neurons harbouring this mutation (ADP2) resulted in fewer significant changes to the proteome relative to the WT APP (AF22) neurons (11 vs. 42, respectively). It is possible that the additional exposure to 1 µM oAβ did not elicit a robust response, considering the elevated baseline expression of Aβ 1-42 in V717I mutants. Using LFQ, we identified that total Aβ was increased in the oAβ exposed ADP2 cells relative to the control ADP2 cells, however the difference did not achieve statistical significance, presumably relating to the baseline expression of Aβ 1-42 in the ADP2 neurons53. It is worth noting that during differentiation, ADP2 neurons experience a much greater degree of spontaneous cell death compared to AF22, likely a result of the increased expression of endogenous Aβ 1-42. Since whole proteome analysis requires small amounts of protein input, this did not prohibit proteomic analysis, however phosphoenrichment requires the input of large quantities of peptides (≈1000 µg), making analysis of the ADP2 phosphoproteome unattainable. Nevertheless, we identified a number of proteins that were upregulated following oAβ exposure, including the increased expression of the tricarboxylic acid cycle (TCA cycle)-related protein Fumarate hydratase, mitochondrial (FUMH), which was also found to be upregulated in response to oAβ by Földi et al.35. Additionally, in the whole proteome analysis of ADP2, we found that DPYL2 (AKA CRMP2), a member of the aforementioned AD-associated DPYL protein family, was found to be phosphorylated, and this phosphorylated DPYL2 was only observed after the oAβ challenge. DPYL2 phosphorylation is regulated by the neurodegenerative disease related kinase GSK-3β, with the phosphorylated form being an important mediator of axon elongation and repair43,44. Interestingly, phosphorylated DPYL2 was identified as an antigen of the 3F4 monoclonal antibody, which was raised against partially purified PHFs and NFTs, indicating that phosphorylated DPYL2 may be implicated in the formation of PHFs and NFTs in AD68. After oAβ exposure, we observed a statistically significant increase in PSA4 in the ADP2 neurons, while in AF22 neurons, its expression partner PSA3 was significantly increased. The oAβ-induced PSA protein increases observed in this work are further supported by a previous proteomics study that found these 20 s proteasomal proteins were differentially regulated in AD brains69. As the 20 S proteasome is important for the degradation of ubiquitinated and misfolded proteins70, its disruption at early stages of oAβ challenge, like those shown here, should be further investigated for their possible contributions to AD pathogenesis.
In both ADP2 and AF22 neurons, exposure of oAβ caused a significant downregulation of coatomer complex proteins (COPG1 in ADP2 and COPD in AF22). Both COPD and COPG1 are subunits of the larger protein complex COPI. COPI-coated vesicles are responsible for the retrograde transport of proteins from the Golgi to the endoplasmic reticulum (ER, reviewed in71). COPI function is important to AD pathogenesis, as it regulates APP trafficking, maturation and therefore production of Aβ peptides72. Furthermore, SNPs in the COPI subunits gamma-1 and delta have been identified as risk factors for the development of LOAD73. Upon COPI depletion, the LOAD related protein TREM2 fails to mature, impairing its cell-surface expression, instead accumulating in the ER–Golgi intermediate compartment74. Additionally, mice with a missense mutation in COPD develop protein accumulations, ER stress, and neurofibrillary tangles75. COPI downregulation leads to excessive accumulation of APP at the Golgi, thereby hindering its maturation into Aβ and cell surface expression, culminating in less Aβ load, reduced Aβ plaque deposition and improved memory72,73.
As recently shown by Volpato et al., it is important to consider the reproducibility of iPSC-derived cells with respect to expressional variations between laboratories76. In this study, we have utilized thoroughly characterized iPSC-derived ltNES cells, which have been widely used in several different laboratories to derive neurons for in vitro studies14–21. Importantly, these cells have been demonstrated to maintain stable morphology, differentiation potential and gene expression profiles over more than 100 passages14 and display consistent expression profiles across different laboratories15,16,18,19. Using the same differentiation scheme as previously described for these ltNES cells14, we found similar proportions of neurons to glia upon differentiation as those previously described14,16,18,19. While each of these ltNES cells have been shown to retain consistent morphology and gene expression they are each derived from different individual donors, with individual genotype and epigenetic modifications resulting in differences in baseline mRNA and protein levels. This could result both in differences in expression, resilience to oAβ and morphological differences, e.g. levels of RNA expression can affect nuclear size77. In this study we have examined the effects of the oAβ challenge upon each ltNES cell-type separately (i.e. AF22 + oAβ was compared to AF22 control) to avoid the confounding factor of different baselines. Further studies including additional genotypes and other models will be important to confirm the results in this study. In addition, it will be interesting to study the effect of other misfolded protein aggregates, which could activate similar but also additional pathways. Despite these potential future expansions, we believe that the results obtained here give important indications of the early oAβ induced changes.
In this study, we have demonstrated that iPSC-derived ltNES neurons are a useful model for the study of oAβ-induced changes to the proteome and phosphoproteome. Using nLC-MS/MS and subsequent LFQ, we identified a number of proteins that were differentially regulated, some of which have been associated with neurodegenerative disorders including AD, but also several that are previously unknown to be involved in these diseases. In this way, we have shown that TDP-43, hnRNPs, DPYLs and COPI proteins are among the earliest proteins affected by the uptake of oAβ in neurons. Additionally, oAβ induced the phosphorylation of tau at serine 208, which should be further investigated as a potential early marker of tau pathology. Together, these findings indicate that there are widespread alterations to the neuronal proteome within 24 h of oAβ uptake and provides new targets for the further study of early mediators of AD pathogenesis.
Materials and Methods
Cell culture
Well described iPSC-derived neuronal progenitor cells, long term neuroepithelial-like stem cells (ltNES), lineages AF22 (WT) and ADP2 (V717I London APP mutant) were kindly provided by Anna Falk (Karolinska Institute, Sweden)14–21. iPSC-derived cells were expanded in DMEM/F12 with Glutamax (Gibco), supplemented with EGF (10 ng/mL; Peprotech) and FGF2 (10 ng/mL; Peprotech), 1% N2 (Gibco), 0.1% B27 (Gibco), 1% Pen/Strep (Lonza). Once confluent, cells were differentiated for 60 days to obtain neuronal phenotypes14, followed by the addition of 1 µM amyloid beta oligomers (oAβ) for 24 h. Next cells were thoroughly washed, trypsinized and lysed in 4% SDS lysis buffer supplemented with protease inhibitor (HALT protease inhibitor cocktail; Thermo Scientific) and phosphatase inhibitor (PhosSTOP; Roche). Cell lysates were stored at −80 °C until use. Cell differentiation media was as follows: 1:1 mixture of Neurobasal and DMEM/F12 with Glutamax, supplemented with 1.5% N2, 1% B27 (all from Gibco) and 1% Pen/Strep (Lonza), with ½ media changed every day. These cells have been demonstrated to maintain stable morphology, differentiation potential and gene expression profiles over more than 100 passages14, however the cells used in this study did not exceed passage 40.
Oligomerization of Aβ 1-42
Aβ oligomers were generated and confirmed by size exclusion chromatography (SEC) as previously described22,23,27,78,79. Recombinant Aβ 1-42 peptides (Innovagen) were dissolved in 1,1,1,3,3,3-hexafuoro-2-propanol (HFIP, Sigma-Aldrich) and vacuum dried overnight. Aβ 1-42 was first re-suspended in 44 µl DMSO (Sigma-Aldrich) and then diluted to a final concentration of 100 µM in HEPES 20 mM pH 7.4, vortexed, sonicated for 10 min and incubated at 4 °C overnight. After the overnight incubation, the sample was vacuum dried, re-suspended in 500 µl NH4HCO3 50 mM pH 8.5. The oligomeric Aβ 1-42 was separated from the monomeric form with SEC. A Sephadex 75 10/300 GL column coupled to a liquid chromatography system (ÄKTA pure, GE Healthcare) was equilibrated with NH4HCO3 50 mM pH 8.5 and 500 µl of sample was injected into the column. To estimate the molecular weight of the Aβ species, LMW gel filtration calibration kits (GE Healthcare) were used. Oligomeric and monomeric Aβ species were eluted at a flow rate of 0.5 ml/min, collected and lyophilized. Then, Aβ species were re-suspended in PBS and quantified spectrophotometrically at 215 nm by using the Aβ 1-42 extinction coefficient (Aβ 1-42 ɛ214 nm = 75,887 M-1 cm-1) according to Lambert–Beer’s law. Protein aliquots were stored at −80 °C. A representative SEC chromatogram of the Aβ oligomer preparations is provided in Supplementary Fig. S1.
Protein extraction and digestion
Samples were lysed in SDS buffer (4% SDS, 25 mM HEPES pH 7.6, 1 mM DTT in H2O supplemented with protease inhibitor (HALT; Thermo Scientific) and phosphatase inhibitor (PhosStop; Roche)) which sufficiently solubilized all proteins. Solubilized proteins were precipitated with acetone, air dried, and re-suspended in urea solution (8 M urea, 2 M thiourea, 25 mM ammonium bicarbonate (ABC), incubated for 30 min), followed by reduction and alkylation (25 mM DTT (15 min) and 75 mM iodoacetamide (15 min), respectively). To remove residual SDS, solutions were washed five times with urea wash solution (8 M urea, 25 mM ABC) with 3 kDa Amicon spin filters (Merck Millipore), followed by removal of urea by washing the solution five times with 25 mM ABC. At this stage, the protein in solution (25 mM ABC) was digested in trypsin (Sigma-Aldrich; 1:25 w/w trypsin:protein) for 2 h at 37 °C and repeated overnight at 37 °C, then dried in a speed vacuum concentrator. To ensure complete removal of contaminants, peptides were desalted with C18 spin columns according to manufacturer’s instructions (Pierce), dried and stored at −80 °C until use. Peptides were reconstituted in 0.1% formic acid in MilliQ H2O, and approximately 0.25 µg was used in LC-MS/MS analysis.
For analysis of the phosphoproteome, proteins were extracted and digested as described above, but subjected to phosphoenrichment (PPE) with TiO2 spin columns (Pierce) after trypsinization instead of C18 cleanup. PPE was performed according to manufacturer’s instructions.
LC-MS/MS analysis
Peptides were separated by reverse phase chromatography on a 20 mm × 100 μm C18 pre-column followed by a 100 mm × 75 μm C18 column with particle size 5 μm (NanoSeparations) at a flow rate of 300 nL/min. Samples were loaded using an EASY-nLC II (Thermo) by linear gradient of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) (0–100% B in 90 min for proteome analysis, 0–100% B in 120 min for phosphoproteome analysis). Automated online analyses were performed with an LTQ Orbitrap Velos Pro hybrid mass spectrometer (Thermo) with a nano-electrospray source.
Database searches and label-free quantification
Database searches and label-free quantification (LFQ) were performed as described previously80–82. Raw files were searched using Sequest HT in Proteome Discoverer (Thermo; version 1.4.0.288) and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1)). Sequest was searched against a UniProt Human database (available at UniProtKB website: https://www.uniprot.org/taxonomy/9606) assuming the digestion enzyme trypsin. X! Tandem was set up to search a reverse concatenated subset of the Swiss-Prot database. Sequest and X! Tandem were searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 PPM. Carbamidomethyl of cysteine was specified in Sequest and X! Tandem as a fixed modification. Methyl of aspartic acid, glutamic acid, oxidation of methionine, acetyl of lysine and phospho of serine, threonine and tyrosine were specified in Sequest and X! Tandem as variable modifications.
Scaffold (version Scaffold_4.8.7, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Identifications were based on a minimum of 1 unique peptide, 99% protein identification probability and 95% peptide identification probability (using the Scaffold Local FDR algorithm). Protein probabilities were assigned by the Protein Prophet algorithm83, resulting in decoy FDRs: 3.6% AF22 phosphoproteome, 3% AF22 proteome and 1.4% ADP2 proteome. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
LFQ analysis of peptides was performed using the sum of the intensities of the peaks in the MS/MS spectra (Normalized Total TIC; normalized to reduce run-to-run variations), and quantitative differences were statistically analyzed by a T-Test with Benjamini-Hochberg correction. Differences with p-values <0.05 were considered statistically significant.
Western blotting
To verify the validity of the quantification obtained by LC-MS/MS and Scaffold LFQ analysis, we analyzed protein expression in AF22 neurons by Western blotting with several antibodies. Protein concentrations were determined with DC Assay (Bio-Rad), and approximately 15 µg protein was loaded per lane on 4–20% gradient gels (CBS Scientific). After transfer to Nitrocellulose membranes (iBlot; Thermo Scientific), blots were incubated with primary antibodies overnight at 4 °C in TBST, followed by HRP-conjugated secondary antibodies for 90 minutes at ambient temperature in TBST. Membranes were visualized using Clarity ECL on a ChemiDoc MP imaging system (both Bio-Rad), followed by quantification with ImageJ. Membranes were stripped with Restore PLUS Western Blot Stripping Buffer (Thermo Scientific) for 10 minutes at ambient temperature. A list of antibodies used in this study and their dilutions can be found in Supplementary Table S1.
Immunocytochemistry
In order to confirm the that our differentiation of AF22 and ADP2 cells resulted in predominantly neuronal populations, we analyzed the differentiated cells for the expression of neuron-specific markers NeuN and β-III Tubulin (Tuj1). Cells were fixed with 4% PFA for 10-20 minutes and washed thoroughly with PBS prior to permeabilization with 0.1% saponin and 5% FBS (in PBS) for 20 min. Cells were further permeabilized/blocked in a PBS buffer containing 0.02% Triton X-100, 0.1% BSA, and 0.05% Tween-20. Primary antibodies were incubated overnight at 4 °C followed by thorough washing and a 75-minute incubation at ambient temperature with secondary antibody. Cells were mounted using ProLong Gold Antifade Mountant with DAPI (Invitrogen). Antibody dilutions are presented in Supplementary Table S1. To quantify the proportion of neurons, AF22 (150 cells) and ADP2 (130 cells) cells were analyzed for DAPI (all nucleated cells) and NeuN-positivity (neurons). The proportions of NeuN-positive cells were compared between AF22 and ADP2 with an unpaired T-test.
Supplementary information
Acknowledgements
We are grateful to Dr. Anna Falk for kindly supplying the AF22 and ADP2 cell lines and to Dr. Juan F. Reyes for critically reviewing the manuscript. We thank Dr. Emelie Severinsson for generating the amyloid-β oligomers. Mass spectrometry analysis was carried out at the Mass Spectrometry Core Facility, both at the Faculty and imaging at the Microscopy Core Facility, both at the Medicine and Health Sciences, Linköping University. We thank Dr. Maria V. Turkina for assistance in the analysis of mass spectrometry data. This research was made possible by funding from the Swedish Research Council (MH: 523-2013-2735), The Swedish Alzheimer foundation, The Swedish Brain Foundation, the Hans-Gabriel and Alice Trolle-Wachtmeister Foundation for Medical Research, Konung Gustaf V:s och Drottning Victorias Frimurarestiftelse, Swedish Dementia Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access funding provided by Linköping University.
Author contributions
Study concept and design: C.S. and M.H., Experimental work: C.S., Analysis of data: C.S., Interpretation of data: C.S. and M.H., Manuscript preparation: C.S. Both authors critically read and approved the manuscript.
Data availability
All relevant data analysed during this study are included in this published article (and its Supplementary Information files). The raw proteomics data are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63398-6.
References
- 1.Brettschneider J, Del Tredici K, Lee VMY, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 2015;16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira, S. T. & Klein, W. L. The Aβ oligomer hypothesis for synapse failure and memory loss in Alzheimer’s disease. Neurobiology of Learning and Memory10.1016/j.nlm.2011.08.003 (2011). [DOI] [PMC free article] [PubMed]
- 3.Larson, M. E. & Lesné, S. E. Soluble Aβ oligomer production and toxicity. Journal of Neurochemistry10.1111/j.1471-4159.2011.07478.x (2012). [DOI] [PMC free article] [PubMed]
- 4.Sengupta U, Nilson AN, Kayed R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine. 2016;6:42–49. doi: 10.1016/j.ebiom.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 6.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flajolet M, et al. Regulation of Alzheimer’s disease amyloid-beta formation by casein kinase I. Proc. Natl. Acad. Sci. 2007;104:4159–4164. doi: 10.1073/pnas.0611236104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leroy, K., Yilmaz, Z. & Brion, J. P. Increased level of active GSK-3β in Alzheimer’s disease and accumulation in argyrophilic grains and in neurones at different stages of neurofibrillary degeneration. Neuropathol. Appl. Neurobiol. 10.1111/j.1365-2990.2006.00795.x (2007). [DOI] [PubMed]
- 9.Zempel H, Thies E, Mandelkow E, Mandelkow E-M. A Oligomers Cause Localized Ca2+ Elevation, Missorting of Endogenous Tau into Dendrites, Tau Phosphorylation, and Destruction of Microtubules and Spines. J. Neurosci. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zempel H, et al. Amyloid-β oligomers induce synaptic damage via Tau-dependent microtubule severing by TTLL6 and spastin. EMBO J. 2013;32:2920–2937. doi: 10.1038/emboj.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasconcelos B, et al. Heterotypic seeding of Tau fibrillization by pre-aggregated Abeta provides potent seeds for prion-like seeding and propagation of Tau-pathology in vivo. Acta Neuropathol. 2016;131:549–569. doi: 10.1007/s00401-015-1525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busciglio, J., Lorenzo, A., Yeh, J. & Yankner, B. A. β-Amyloid fibrils induce tau phosphorylation and loss of microtubule binding. Neuron10.1016/0896-6273(95)90232-5 (1995). [DOI] [PubMed]
- 13.Do, T. D. et al. Interactions between amyloid-β and tau fragments promote aberrant aggregates: Implications for amyloid toxicity. J. Phys. Chem. B10.1021/jp506258g (2014). [DOI] [PMC free article] [PubMed]
- 14.Falk A, et al. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. Plos One. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, D. et al. A 3D Alzheimer’s disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials 1–9 10.1016/j.biomaterials.2013.11.028 (2013). [DOI] [PubMed]
- 16.Fujimoto Y, et al. Treatment of a mouse model of spinal cord injury by transplantation of human induced pluripotent stem cell-derived long-term self-renewing neuroepithelial-like stem cells. Stem Cells. 2012;30:1163–73. doi: 10.1002/stem.1083. [DOI] [PubMed] [Google Scholar]
- 17.Raciti M, et al. Glucocorticoids alter neuronal differentiation of human neuroepithelial-like cells by inducing long-lasting changes in the reactive oxygen species balance. Neuropharmacology. 2016;107:422–431. doi: 10.1016/j.neuropharm.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Yu NY, et al. Acute doses of caffeine shift nervous system cell expression profiles toward promotion of neuronal projection growth. Sci. Rep. 2017;7:11458. doi: 10.1038/s41598-017-11574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tailor J, et al. Stem cells expanded from the human embryonic hindbrain stably retain regional specification and high neurogenic potency. J. Neurosci. 2013;33:12407–22. doi: 10.1523/JNEUROSCI.0130-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundin A, et al. Human iPS-Derived Astroglia from a Stable Neural Precursor State Show Improved Functionality Compared with Conventional Astrocytic Models. Stem Cell Reports. 2018;10:1030–1045. doi: 10.1016/j.stemcr.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk R, et al. Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods. 2012;58:69–78. doi: 10.1016/j.ymeth.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nath S, et al. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of β-amyloid. J. Neurosci. 2012;32:8767–77. doi: 10.1523/JNEUROSCI.0615-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domert J, et al. Spreading of amyloid-β peptides via neuritic cell-to-cell transfer is dependent on insufficient cellular clearance. Neurobiol. Dis. 2014;65:82–92. doi: 10.1016/j.nbd.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Meyer-Luehmann M, et al. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 25.Eisele YS, et al. Peripherally applied Aβ-containing inoculates induce cerebral β-amyloidosis. Science. 2010;330:980–982. doi: 10.1126/science.1194516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domert J, et al. Aggregated alpha-synuclein transfer efficiently between cultured human neuron- like cells and localize to lysosomes. Plos One. 2016;11:500. doi: 10.1371/journal.pone.0168700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackmann V, et al. Anti-inflammatory (M2) macrophage media reduce transmission of oligomeric amyloid beta in differentiated SH-SY5Y cells. Neurobiol. Aging. 2017;60:173–182. doi: 10.1016/j.neurobiolaging.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Hallbeck, M., Nath, S. & Marcusson, J. Neuron-to-Neuron Transmission of Neurodegenerative Pathology. Neuroscientist10.1177/1073858413494270 (2013). [DOI] [PubMed]
- 29.Agholme L, et al. Proteasome inhibition induces stress kinase dependent transport deficits–implications for Alzheimer’s disease. Mol. Cell. Neurosci. 2014;58:29–39. doi: 10.1016/j.mcn.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Reyes, J. F. et al. Binding of α-synuclein oligomers to Cx32 facilitates protein uptake and transfer in neurons and oligodendrocytes. Acta Neuropathol. 1–25 10.1007/s00401-019-02007-x (2019). [DOI] [PMC free article] [PubMed]
- 31.Hashimoto M, et al. Analysis of microdissected human neurons by a sensitive ELISA reveals a correlation between elevated intracellular concentrations of Abeta42 and Alzheimer’s disease neuropathology. Acta Neuropathol. 2010;119:543–54. doi: 10.1007/s00401-010-0661-6. [DOI] [PubMed] [Google Scholar]
- 32.Koffie, R. M. et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U. S. A. 10.1073/pnas.0811698106 (2009). [DOI] [PMC free article] [PubMed]
- 33.Sadleir KR, et al. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Aβ generation in Alzheimer’s disease. Acta Neuropathol. 2016;132:235–256. doi: 10.1007/s00401-016-1558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savas JN, et al. Amyloid Accumulation Drives Proteome-wide Alterations in Mouse Models of Alzheimer’s Disease-like Pathology. Cell Rep. 2017;21:2614–2627. doi: 10.1016/j.celrep.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Földi I, et al. Proteomic study of the toxic effect of oligomeric Aβ1-42 in situ prepared from ‘iso-Aβ1-42’. J. Neurochem. 2011;117:691–702. doi: 10.1111/j.1471-4159.2011.07238.x. [DOI] [PubMed] [Google Scholar]
- 36.Han, S. P., Tang, Y. H. & Smith, R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 10.1042/bj20100396 (2010). [DOI] [PubMed]
- 37.Berson A, et al. Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 2012;4:730–42. doi: 10.1002/emmm.201100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper-Knock, J. et al. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. (2015). 10.1007/s00401-015-1429-9 [DOI] [PMC free article] [PubMed]
- 39.Appocher, C. et al. Major hnRNP proteins act as general TDP-43 functional modifiers both in Drosophila and human neuronal cells. Nucleic Acids Res. (2017). 10.1093/nar/gkx477 [DOI] [PMC free article] [PubMed]
- 40.Mizukami, K. et al. Immunohistochemical study of the hnRNP A2 and B1 in the hippocampal formations of brains with Alzheimer’s disease. Neurosci. Lett. 10.1016/j.neulet.2005.05.070 (2005). [DOI] [PubMed]
- 41.Prudencio, M. et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 10.1038/nn.4065 (2015). [DOI] [PMC free article] [PubMed]
- 42.Fonte V, et al. Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9439–44. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole, A. R. et al. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J. Biol. Chem. 10.1074/jbc.C400412200 (2004). [DOI] [PMC free article] [PubMed]
- 44.Fang, W. et al. Role of the Akt/GSK-3β/CRMP-2 pathway in axon degeneration of dopaminergic neurons resulting from MPP+ toxicity. Brain Res. 10.1016/j.brainres.2014.08.030 (2015). [DOI] [PubMed]
- 45.Lin, Y. S., Lin, Y. F., Chen, K. C., Yang, Y. K. & Hsiao, Y. H. Collapsin response mediator protein 5 (CRMP5) causes social deficits and accelerates memory loss in an animal model of Alzheimer’s disease. Neuropharmacology10.1016/j.neuropharm.2019.107673 (2019). [DOI] [PubMed]
- 46.Herman AM, Khandelwal PJ, Stanczyk BB, Rebeck GW, Moussa CE-H. β-amyloid triggers ALS-associated TDP-43 pathology in AD models. Brain Res. 2011;1386:191–9. doi: 10.1016/j.brainres.2011.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang Y-SS, et al. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat. Commun. 2014;5:4824. doi: 10.1038/ncomms5824. [DOI] [PubMed] [Google Scholar]
- 48.Robinson JL, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141:2181–2193. doi: 10.1093/brain/awy146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Josephs KA, et al. Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol. 2014;127:441–450. doi: 10.1007/s00401-013-1211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Josephs KA, et al. TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol. 2014;127:811–824. doi: 10.1007/s00401-014-1269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caccamo, A., Magrí, A. & Oddo, S. Age-dependent changes in TDP-43 levels in a mouse model of Alzheimer disease are linked to A oligomers accumulation. Mol. Neurodegener. 10.1186/1750-1326-5-51 (2010). [DOI] [PMC free article] [PubMed]
- 52.Herman, A. M., Khandelwal, P. J., Rebeck, G. W. & Moussa, C. E. H. Wild type TDP-43 induces neuro-inflammation and alters APP metabolism in lentiviral gene transfer models. Exp. Neurol. (2012). [DOI] [PMC free article] [PubMed]
- 53.Muratore, C. R. et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum. Mol. Genet. 18, (2014). [DOI] [PMC free article] [PubMed]
- 54.Raj T, et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 2018;50:1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braak H, Thal DR, Ghebremedhin E, Del Tredici K, Tredici KDel. Stages of the Pathologic Process in Alzheimer Disease: Age Categories From 1 to 100 Years. J. Neuropathol. Exp. Neurol. 2011;70:960–969. doi: 10.1097/NEN.0b013e318232a379. [DOI] [PubMed] [Google Scholar]
- 56.Augustinack JC, Schneider A, Mandelkow E-M, Hyman BT. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2001;103:26–35. doi: 10.1007/s004010100423. [DOI] [PubMed] [Google Scholar]
- 57.Li G, Yin H, Kuret J. Casein kinase 1 delta phosphorylates tau and disrupts its binding to microtubules. J. Biol. Chem. 2004;279:15938–45. doi: 10.1074/jbc.M314116200. [DOI] [PubMed] [Google Scholar]
- 58.Chen C, et al. Up-regulation of casein kinase 1ε is involved in tau pathogenesis in Alzheimer’s disease. Sci. Rep. 2017;7:13478. doi: 10.1038/s41598-017-13791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomizawa, K., Omori, A., Ohtake, A., Sato, K. & Takahashi, M. Tau-tubulin kinase phosphorylates tau at Ser-208 and Ser-210, sites found in paired helical filament-tau. FEBS Lett. 10.1016/S0014-5793(01)02256-6 (2001). [DOI] [PubMed]
- 60.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21359–64. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bobkova, N. V. et al. The Y-box binding protein 1 suppresses Alzheimer’s disease progression in two animal models. PLoS One10.1371/journal.pone.0138867 (2015). [DOI] [PMC free article] [PubMed]
- 62.Martin, M. et al. Novel serine 176 phosphorylation of YBX1 activates NF-κB in colon cancer. J. Biol. Chem. 10.1074/jbc.M116.740258 (2017). [DOI] [PMC free article] [PubMed]
- 63.Prabhu, L. et al. Critical role of phosphorylation of serine 165 of YBX1 on the activation of NF-κB in colon cancer. Oncotarget (2015). [DOI] [PMC free article] [PubMed]
- 64.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 65.Herl L, et al. Mutations in amyloid precursor protein affect its interactions with presenilin/γ-secretase. Mol. Cell. Neurosci. 2009;41:166. doi: 10.1016/j.mcn.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Jonghe C, et al. Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability. Hum. Mol. Genet. 2001;10:1665–71. doi: 10.1093/hmg/10.16.1665. [DOI] [PubMed] [Google Scholar]
- 67.Eckman CB, et al. A new pathogenic mutation in the APP gene (I716V) increases the relative proportion of Abeta 42(43) Hum. Mol. Genet. 1997;6:2087–9. doi: 10.1093/hmg/6.12.2087. [DOI] [PubMed] [Google Scholar]
- 68.Gu, Y., Hamajima, N. & Ihara, Y. Neurofibrillary tangle-associated collapsin response mediator protein-2 (CRMP-2) is highly phosphorylated on Thr-509, Ser-518, and Ser-522. Biochemistry10.1021/bi992323h (2000). [DOI] [PubMed]
- 69.Gillardon F, et al. The 20S proteasome isolated from Alzheimer’s disease brain shows post-translational modifications but unchanged proteolytic activity. J. Neurochem. 2007;101:1483–1490. doi: 10.1111/j.1471-4159.2006.04438.x. [DOI] [PubMed] [Google Scholar]
- 70.Yano M, Koumoto Y, Kanesaki Y, Wu X, Kido H. 20S Proteasome Prevents Aggregation of Heat-Denatured Proteins without PA700 Regulatory Subcomplex Like a Molecular Chaperone. Biomacromolecules. 2004;5:1465–1469. doi: 10.1021/bm049957a. [DOI] [PubMed] [Google Scholar]
- 71.Spang, A. Retrograde traffic from the golgi to the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, (2013). [DOI] [PMC free article] [PubMed]
- 72.Bettayeb K, et al. δ-COP modulates Aβ peptide formation via retrograde trafficking of APP. Proc. Natl. Acad. Sci. USA. 2016;113:5412. doi: 10.1073/pnas.1604156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bettayeb K, et al. Relevance of the COPI complex for Alzheimer’s disease progression in vivo. Proc. Natl. Acad. Sci. USA. 2016;113:5418–23. doi: 10.1073/pnas.1604176113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sirkis DW, Aparicio RE, Schekman R. Neurodegeneration-associated mutant TREM2 proteins abortively cycle between the ER and ER–Golgi intermediate compartment. Mol. Biol. Cell. 2017;28:2723–2733. doi: 10.1091/mbc.e17-06-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu X, et al. Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration. Plos Genet. 2010;6:e1000956. doi: 10.1371/journal.pgen.1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Volpato V, et al. Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-Derived Neurons: A Multi-Site Omics Study. Stem Cell Reports. 2018;11:897–911. doi: 10.1016/j.stemcr.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sato S, Burgess SB, McIlwain DL. Transcription and Motoneuron Size. J. Neurochem. 1994;63:1609–1615. doi: 10.1046/j.1471-4159.1994.63051609.x. [DOI] [PubMed] [Google Scholar]
- 78.Sardar Sinha M, et al. Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018;136:41–56. doi: 10.1007/s00401-018-1868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ansell-Schultz A, Reyes JF, Samuelsson M, Hallbeck M. Reduced retromer function results in the accumulation of amyloid-beta oligomers. Mol. Cell. Neurosci. 2018;93:18–26. doi: 10.1016/j.mcn.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 80.Augier E, et al. A molecular mechanism for choosing alcohol over an alternative reward. Science. 2018;360:1321–1326. doi: 10.1126/science.aao1157. [DOI] [PubMed] [Google Scholar]
- 81.Turkina MV, Ghafouri N, Gerdle B, Ghafouri B. Evaluation of dynamic changes in interstitial fluid proteome following microdialysis probe insertion trauma in trapezius muscle of healthy women. Sci. Rep. 2017;7:43512. doi: 10.1038/srep43512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molinas, A., Turkina, M. V., Magnusson, K.-E., Mirazimi, A. & Vikström, E. Perturbation of Wound Healing, Cytoskeletal Organization and Cellular Protein Networks during Hazara Virus Infection. Front. Cell Dev. Biol. 10.3389/fcell.2017.00098 (2017). [DOI] [PMC free article] [PubMed]
- 83.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data analysed during this study are included in this published article (and its Supplementary Information files). The raw proteomics data are available from the corresponding author on reasonable request.