Abstract
Background and Objectives:
Burn wound infections have emerged as an important cause of morbidity and mortality in patients due to prolonged hospital stay. Pseudomonas aeruginosa, is the second cause of bacterial burn wound infections. Resistance mechanisms among P. aeruginosa are intrinsic or acquired. Intrinsic resistance mechanisms among P. aeruginosa isolates are inducible AmpC cephalosporinase, decrease of specific porin OprD, and overexpression of RND efflux pump. The aim of this study was detection of mutations in nalC gene in carbapenem resistant P. aeruginosa isolated from burn wounds.
Materials and Methods:
In this cross-sectional study, 180 burn-wound specimens were collected. Suspected lactose-negative colonies were identified by conventional biochemical methods. Kirby-Bauer and Etest methods were used for susceptibility testing. PCR and sequencing techniques were used for the detection of nalC mutation.
Results:
Out of 180 specimens received in the laboratory, 54 of isolates were isolated and identified as P. aeroginosa (30%). Of these isolates 20 (37%) were resistant to at least two carbapenems simultaneously. From these carbapenem resistant isolates, 19 (95%), 14 (70%), 14 (70%), 19 (95%) and 16 (80%) were resistant to imipenem, cefepime, piperacillin, ceftizoxime and gentamicin, respectively. Only 1 (2%) isolate was sensitive to all carbapenems and did not has mutation in nalC gene, 20 (37%) isolates were resistant to at least two carbapenems, and had mutations in nalC gene (Gly71▸Glu and Ser209▸Arg).
Conclusion:
As the results showed, mutation in efflux pump was observed in carbapenem resistant isolate and this confirmed that the indiscriminate use of antibiotics for treatment or prophylaxis can increase mutation in efflux pump.
Keywords: Pseudomonas aeruginosa, Carbapenem resistant, Efflux pump, nalc gene
INTRODUCTION
Burn wound infections have emerged as an important cause of morbidity and mortality in patients due to prolonged hospital stay (1). Gram-positive bacteria are some of the first bacteria that colonize burn wounds, followed quickly by Gram-negative. Pseudomonas aeruginosa is the second cause of bacterial burn wound infections (2).
Resistance mechanisms among P. aeruginosa are intrinsic or acquired. Intrinsic resistance mechanisms among P. aeruginosa isolates are inducible AmpC cephalosporinase, decrease of specific porin OprD (3, 4) and overexpression of RND efflux pump. Over-expression of mexAB-OprM has caused the emergence of Multi-Drug Resistance (MDR) P. aeroginosa (5).
RND pumps typically exist as a tripartite system (6), consisting of a RND cytoplasmic membrane transporter, a Membrane Fusion Protein (MFP), and an Outer Membrane Protein (OMF) (7). This complex forms a channel, spanning the entire membrane, allowing for the proton-derived transport of lipophilic and amphiphilic drugs from the cytoplasm of the cell across the cytoplasmic membrane, peptidoglycan, and outer membrane through the periplasmic space (8). The MexAB–OprM system has the broadest substrate range among all of 10 characterized P. aeruginosa efflux pumps (9). Substrates of this pump include ß-lactams, ß-lactamase inhibitors, quinolones, macrolides, tetracyclines, chloramphenicol, novobiocin, sulfonamides, and trimethoprim (9, 10). Several regulatory loci influence the expression of the MexAB-oprM operon (11). The mexR gene is located directly up the stream of mexA and transcribed divergently from MexAB-oprM and encodes a repressor belonging to the MarR family of regulatory proteins. Also, nalC gene (also known as PA3721) (12) encodes a putative repressor of the TetR/AcrR family, whose genes are located up stream of operon-encoded PA3720-PA3719 genes, that is negatively regulated by NalC (13). Loss of NalC resulted in over-expression of PA3720-PA3719, and subsequent experiments demonstrated that PA3719 upregulates MexAB-oprM by interacting with MexR (12, 13).
The purpose of this study was the detection of mutations in nalC gene (second regulatory gene of mex AB-OprM system) in carbapenem resistant P. aeruginosa isolated from burn wounds.
MATERIALS AND METHODS
Bacterial strain and growth conditions. In this cross-sectional study, 180 burn wound specimens were collected from burnt patients in a burn-hospital in Yazd, Iran. These patients stayed over a week in hospital and did not have a burn wound infection at the admission time. Specimens were immediately transferred to microbiology laboratory at School of Medicine, Shahid Sadoughi University of Medical Sciences, inoculated on MacConkey agar media (Merck-Darmstadt, Germany) and Cetrimide agars (Merck-armstadt, Germany), and were incubated for 16–18 h at 35 °C. Suspected lactose-negative colonies were identified by conventional biochemical methods. Sugar utilization in Oxidation-Fermentation (OF) medium (Merck-Darmstadt, Germany), production of oxidase enzyme, growth at 42 °C and pigment production tests were performed for the identification of P. aeruginosa.
Antibacterial agent and susceptibility testing. Susceptibility of P. aeruginosa isolates to Ertapenem (10 μg), Meropenem (10 μg), Imipenem (10 μg), cefepime (10 μg), piperacillin (10 μg), ceftizoxime (10 μg) and gentamicin (10 μg) (Mast, England) were performed by the disk diffusion method (Kirby-Bauer) on Mueller-Hinton agar (Merck-Darmstadt, Germany) according to CLSI protocols (14).
Meropenem Etest strips (biomerieux, France) were used for the performing of the MIC. Briefly, bacterial suspension equivalent to 0.5 McFarland turbidity tube was prepared and inoculated on Mueller-Hinton agar. Etest strips were placed on the medium. After incubation for 16–18 h at 35 °C, Eclipse intersection with strip was considered as the inhibitory concentration. P. aeruginosa ATCC 27853 was used for control of all phenotypic tests.
Bacterial genomic DNA extraction. The salting out method was used for bacterial genomic DNA extraction (15). Briefly, after overnight culturing of bacteria in the Trypticase Soy Broth (TSB; Merck-Darmstadt, Germany), they were washed with phosphate buffer saline in triple. Cells were lysed by NET buffer (NaCl, 50 mM; EDTA, pH 8, 10 mM, Tris-base, pH 7.6, 50 mM) and Sodium dodecyl sulfate (SDS) with final concentration of 1%. Purification and precipitation were performed using saturated salt and cold ethanol, respectively. The quality and quantity of extracted DNA were assessed by agarose gel electrophoresis and spectrophotometer, respectively. The samples were stored at −20 °C for the next steps.
Detection of nalC gene by polymerase chain reaction. PCR technique was performed for detection of nalC gene using specific primers (nalC-L> CCTGGACATGGTGATAGAACG-nalC-R> CGGGTCCTGAACGAACTCT) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The PCR reaction mixture contained 1× master mix (Amlicon, Denmark) (including 10 mM tris-base, 0.2 mM dNTP, 1.5 mM MgCl2, and 1U Taq DNA polymerase), 10 pmol of each primer, and 100 ng template DNA in a total volume of 20 μl. Amplification was performed using thermocycler (Quant biotech, England) with initial denaturation at 94 °C for 5 min, followed by 35 cycles denaturation step at 94 °C for 85 s, annealing at 52 °C for 85 s, and extension at 72 °C for 85s. The final extension was at 72 °C for 5 min. PCR products were analyzed using gel agarose. Electrophoresis alongside with 50 bp DNA ladder. The samples with single amplicon with 724 bp in length were sequenced and analyzed with BLAST and the bioinformatic software necessary for mutation analysis in Sequence Retrieved System (SRS) at ABI (https://www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi) (16).
RESULTS
Out of 180 burn wound specimens, 54 (30%) isolates were identified as P. aeroginosa. These were isolated from 36 men (67.6%) and 18 women (33.3%). Twenty (37%) isolates were resistant to at least two carbapenems (ertapenem, meropenem and imipenem) simultaneously. Among carbapenem resistant isolates, over 70% of them were resistant to ertapenem, meropenem, imipenem, cefepime, piperacillin, ceftizoxime and gentamicin (Table 1).
Table 1.
Antimicrobial susceptibility pattern of carbapenem resistant P. aeruginosa isolates.
| Antibiotic | Piperacilin N (%) | Ertapenem N (%) | Imipenem N (%) | Gentamicin N (%) | Cefepim N (%) | Meropenem N (%) | Ceftizoxime N (%) |
|---|---|---|---|---|---|---|---|
| Sensitive | 6 (30) | 2 (10) | 1 (5) | 3 (15) | 5 (25) | 1 (5) | 1 (5) |
| Intermediate | 0 (0) | 0 (0) | 0 (0) | 1 (5) | 1 (5) | 1 (5) | 0 (0) |
| Resistant | 14 (70) | 18 (90) | 19 (95) | 16 (80) | 14 (70) | 18 (90) | 19 (95) |
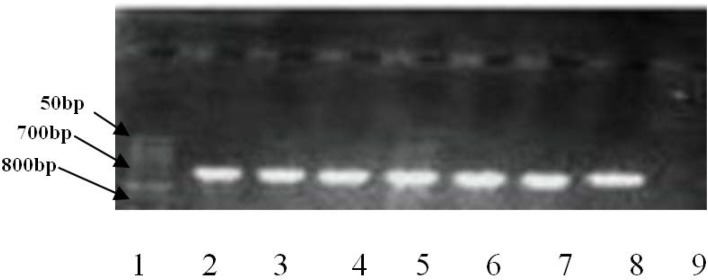
Isolates with MIC>16 μg/ml, according to the manufacturer's instructions, were considered as resistant to meropenem. In this way, out of 54 P. aeroginosa isolates, 35 (64.8%) were resistant to meropenem. Four isolates that were resistant to meropenem by disk diffusion method, had MIC<2 μg/ml, and were, therefore, considered sensitive. Only one sample was sensitive to all carbapenems and did not have mutation in nalC. Twenty (37%) isolates were resistant to at least two carbapenems and had mutation in nalC. Gel electrophoresis of nalC gene is shown in Fig. 1. nalC gene mutations (Gly71▸Glu and Ser209▸Arg) were observed in 22 resistant isolates. nalC mutation was not seen in susceptible isolates.
Fig. 1.
Agarose gel electrophoresis image for amplification analysis of nalC gene. Lane 1: 50 bp DNA ladder; lanes 2–8: Isolates with nalC (724 bp) and lane 9: negative control.
Among the isolates that had mutation in nalC gene, the strains 2ss, 9ss, 23ss were registered at GeneBank under accession numbers KP774795, KT624617 and KT624618, respectively.
DISCUSSION
Burn wound infections as nosocomial infections are an important cause of mortality and disability after burns. P. aeruginosa is an opportunistic pathogen that grows well in moist environments at hospitals. Low sensitivity to antibiotics is one of the characteristics of these bacteria that make them difficult to treat. Carbapenems are used for treatment of infections caused by P. aeruginosa and over-expression of mexAB-OprM has led to the emergence of MDR P. aeroginosa (17). In this study, out of 20 carbapenem-resistant isolates, 18 (90%), 18 (90%), 19 (95%), 14 (70%), 14 (70%), 19 (95%) and 16 (80%) isolates were resistant to ertapenem, meropenem, imipenem, cefepime, piperacillin, ceftizoxime, and gentamicin, respectively. Ahadi et al. (2012) reported that out of 100 clinical isolates, 56, 59, 61, 65, 55, 57, 60, 62, 100 and 48% of them were resistant to ciprofloxacin, gentamicin, tobramycin, amikacin, imipenem, cefepime, ceftazidime, ceftriaxone, cefotaxime, oxacillin and piperacillin, respectively (18). Mir Salehian et al. (2011) reported that among 170 P. aeruginosa strains isolated from burn wounds in Tehran, Iran, the most resistant were observed against aminoglycosides and 52.9% of isolates were resistant to imipenem (2). In another study conducted in Esfahan, out of 98 P. aeruginosa isolates, the most resistance were observed against cefepime (91%), cefotaxime (95%) and ceftizoxime (85.7%) (1). Gill et al. (2011) in Pakistan reported that 90.3% and 85.4% of P. aeruginosa isolates were resistant to imipenem and meropenem, respectively, whereas in this study, 95% and 70% of isolates were resistant to imipenem and meropenem, respectively (20). Considering that in the Burns Hospital of Yazd, imipenem is one of the most frequently prescribed antibiotics for the treatment of hospitalized patients and since imipenem is prescribed as prophylaxis for the prevention of burn wounds infection, therefore, increasing resistance to this antibiotic is inevitable. Yet, it could be indicative of increased resistance to this class of antibiotics because of indiscriminate usage.
In present study, 35 (64%) of isolates had MIC>16 μg/ml for meropenem, whereas in China, more than 70% of P. aeruginosa isolates were susceptible to imipenem and meropenem (19). Gill et al. (2011) showed that 90.3% and 85.4% of P. aeruginosa strains in Pakistan were resistant to imipenem and meropenem, respectively (20). In a study in New York, USA, resistance rate of P. aeruginosa isolates against meropenem was 28% (3). It seems that the resistance pattern to antibiotics based on the method and amount of antibiotic usage is different in the various countries.
In this study, 20 (37%) isolates were resistant to at least two carbapenems and had mutation in nalC. Also, nalC mutations (Gly71▸Glu and Ser209▸Arg) were observed in isolates. In the study by Sadeghifard et al. (2012) in Tehran, Iran, 87.1% of resistant P. aeruginosa isolates had nalC mutation (21). Quale et al. (2006) reported that 33 carbapenem-resistant P. aeruginosa isolates were detected for the presence of nalC mutations in New York, USA. Gly71 ▸Glu, Ala145▸Val and, Ser209▸Arg mutations were observed in their study (3). In our study, substitution mutations (Gly71▸Glu and Ser209▸Arg) were observed in resistant isolates and, nalC mutations were not seen in sensitive isolates. Our substitution mutations that were seen in our study were similar to Quale`s results.
CONCLUSION
The results of this study shows resistance to carbapenem and cephalosporins that are used for the treatment of patients with infections caused by P. aeruginosa are growing in our country. In present study, more than 70% of P. aeruginosa isolates were resistant to ceftizoxime, imipenem, gentamicin, and piperacillin. Detection of these resistant organisms, implementation of strict antimicrobial policies, and infection control programs such as antibiotic stew-ardship may prevent the rapid dissemination of these organisms.
The mutation in efflux pumps causes multi-drug resistant strains. Mutations in efflux pump were observed in resistant isolates and this confirmed that the indiscriminate use of antibiotics for treatment or prophylaxis can increase mutation in efflux pumps. In order to obtain more complete results in this area, it is necessary to conduct more studies about changes in gene expression and its relationship with the nalC mutations.
REFERENCES
- 1.Fazli H, Fatahi Bafghi M, Faghri M. Molecular study of PER and VEB genes is multidrug resistant Pseudomonas aeroginosa isolated from clinical specimens in Isfahan/Iran and their antibiotic resistance patterns. J Kerman Univ Med Sci 2012; 19: 345–353. [Google Scholar]
- 2.Mirsalehian A, Nakhjavani F, Bahador A, Jabalamoli F, Beikvardi R, Goli H. Prevalence of MBL-producing Pseudomonas aeruginosa isolated from burn patients. Tehran Univ Med J 2011; 68: 563–569. [Google Scholar]
- 3.Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2006; 50: 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 4783–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweizer HP. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res 2003; 2: 48–62. [PubMed] [Google Scholar]
- 6.Najar Pirayeh SH, Esmaeili D. Antibiotic efflux pumps. Ann Mil Health Sci Res 2004; 2: 301–306. [Google Scholar]
- 7.Blair J, Piddoc L. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria. Curr Opin Microbiol 2009;12: 512–519. [DOI] [PubMed] [Google Scholar]
- 8.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22: 582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv Drug Deliv Rev 2005; 57: 1486–1513. [DOI] [PubMed] [Google Scholar]
- 10.Sobel ML, Hocquet D, Cao L. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49: 1782–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Yi C, Zhang J, Zhang W, Ge Z, Yang CG, et al. Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR. EMBO Rep 2010; 11: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daigle DM, Cao L, Fraud S, Mark S, Pacey A, Klinoski R, et al. Wilke protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol 2007; 189: 5441–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao L, Srikumar R, Poole K. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol Microbiol 2004; 53: 1423–1436. [DOI] [PubMed] [Google Scholar]
- 14.M07-A8. M07–A8. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard: 8th ed CLSI 2009, Wayne, PA. [Google Scholar]
- 15.Pickaup W, Rhodes G, Saumders J. Moulecolar Microbial Ecology Manual, Wolters Kluwer N.V; 1995; 300–300. [Google Scholar]
- 16.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol 2000; 7: 203–214. [DOI] [PubMed] [Google Scholar]
- 17.Evans K, Poole K. The MexA-B-OprM multidrug efflux system of Pseudomonas aeruginosa is growth phase regulated. FEMS Microbiol Lett 1999; 173: 35–39. [DOI] [PubMed] [Google Scholar]
- 18.Golshani Z, Ahadi AM, Sharifzadeh A. Occurrence of ambler class B metallo-β-lactamase gene in imipenem-resistant Pseudomonas aeruginosa strains isolated from clinical samples. Zahedan J Res Med Sci 2014; 16: 6–9. [Google Scholar]
- 19.Wang H, Chen M, Ni Y, Liu Y, Sun H, Yu Y, et al. Antimicrobial resistance among clinical isolates from the Chinese Meropenem Surveillance Study (CMSS), 2003–2008. Int J Antimicrob Agents 2010; 35: 227–234. [DOI] [PubMed] [Google Scholar]
- 20.Gill MM, Usman J, Kaleem F, Hassan A, Khalid A, Anjum R, et al. Frequency and antibiogram of multi-drug resistant Pseudomonas aeruginosa. J Coll Physicians Surg Pak 2011; 21: 531–534. [PubMed] [Google Scholar]
- 21.Sadeghifard N, Valizadeh A, Zolfaghary MR, Maleki MH, Maleki A, Mohebi R, et al. Relationship between the presence of the nalC mutation and multidrug resistance in Pseudomonas aeruginosa. Int J Microbiol 2012;2012:575193. [DOI] [PMC free article] [PubMed] [Google Scholar]