Abstract
The blood-tumor barrier limits the delivery of therapeutic drugs to brain tumor tissues. Selectively opening the blood-tumor barrier is considered crucial for effective chemotherapy of glioma. RNA-binding proteins have emerged as crucial regulators in various biologic processes. This study found that RNA-binding Fox-1 homolog 1 (RBFOX1) was downregulated in glioma vascular endothelial cells derived from glioma tissues, and in glioma endothelial cells obtained by co-culturing endothelial cells with glioma cells. Overexpression of RBFOX1 impaired the integrity of the blood-tumor barrier and increased its permeability. Additionally, RBFOX1 overexpression decreased the expression of tight junction proteins ZO-1, occludin, and claudin-5. Subsequent analysis of the mechanism indicated that the overexpression of RBFOX1 increased musculoaponeurotic fibrosarcoma protein basic leucine zipper [bZIP] transcription factor F (MAFF) expression by downregulating LINC00673, which stabilized MAFF messenger RNA (mRNA) through Staufen1-mediated mRNA decay. Moreover, MAFF could bind to the promoter region and inhibit the promoter activities of ZO-1, occludin, and claudin-5, which reduced its expression. The combination of RBFOX1 upregulation and LINC00673 downregulation promoted doxorubicin delivery across the blood-tumor barrier, resulting in apoptosis of glioma cells. In conclusion, this study indicated that overexpression of RBFOX1 increased blood-tumor barrier permeability through the LINC00673/MAFF pathway, which might provide a new useful target for future enhancement of blood-tumor barrier permeability.
Keywords: glioma, blood-tumor barrier, RBFOX1, LINC00673, MAFF, STAU1, stability
Graphical Abstract

In this article, the authors systematically elucidate the molecular network between RNA-binding proteins and lncRNAs and its functional role in regulating the permeability of the blood-tumor barrier at the molecular level. Additionally, this article provides a new theoretical basis and strategies for the clinical treatment of glioma.
Introduction
Glioma is a primary intracranial tumor with rapid progression and invasiveness. Despite continuous improvement of treatments, the median survival time of patients with high-grade gliomas is still less than 15 months.1 Chemotherapy is an essential part of glioma therapy. The presence of the blood-tumor barrier (BTB) severely limits the delivery of chemotherapeutic drugs to tumor tissues and affects the efficacy of chemotherapy.2 Specialized brain microvascular endothelial cells (ECs) in glioma tissues are the structural basis for BTB permeability. Therefore, selectively opening of the BTB is considered one of the effective means to increase the efficacy of chemotherapy for glioma.
Although there is a large repertoire of RNA-binding proteins (RBPs) present in tumor cells, little is known about the functional role of RBPs in endothelial cell biology. The RBP Quaking (QKI) has been shown to regulate the development of embryonic blood vessels.3 Additionally, RBP HuR has been shown to inhibit Sirtuin 1 mRNA stabilization to repress the endothelial inflammatory response.4 RNA-binding Fox (RBFOX) protein is involved in many RNA metabolism processes.5 RBFOX1, also known as A2BP1 or FOX1, with a high affinity with the (U) GCAUG domain, plays a crucial role in regulating RNA metabolism during mammalian development.6 In addition, RBFOX1 regulates mRNA stability, translation, and microRNA (miRNA) processing.5 RBFOX1 has been shown to profoundly inhibit glioma tumorigenesis.7 However, the expression level of RBFOX1 in glioma endothelial cells (GECs) and its potential role in the regulation of BTB permeability remain unclear.
Long non-coding RNAs (lncRNAs) are ncRNAs with transcripts of more than 200 nt. Studies have reported that lncRNA can regulate the biological processes of cells at the level of epigenetics, transcription and post-transcription.8 Long intergenic non-protein coding RNA673 (LINC00673), located in human chromosome 17q25.1, is upregulated in lung adenocarcinoma.9 Moreover, LINC00673 is highly expressed in pancreatic ductal adenocarcinoma and enhances the risk of disease.10 Also, recent data indicate that LINC00673 is upregulated in gastric cancer and acts as an oncogenic function in the progression of gastric cancer.11 However, little is known about its expression in GECs. Bioinformatics software (RBPDB) predicts that LINC00673 harbors a binding site of RBFOX1. Accordingly, the potential interaction of the two above factors and their roles in the regulation of BTB permeability have attracted our interest.
Small musculoaponeurotic fibrosarcoma protein (sMaf) is a basic leucine zipper (bZIP) transcription factor that plays a crucial role in gene regulation, differentiation, tumorigenesis, and in the development of many organisms.12 Musculoaponeurotic fibrosarcoma proteins (MAFs) are divided into large MAFs (MAF, MAFA, MAFB, NRL) containing activation domains, and small MAFs (MAFF, MAFG, MAFK) lacking active activation domains.13 Small MAFs exert transcriptional activation through forming heterodimers by binding to other factors of the bZIP family; inversely, by forming homodimers themselves, they play a transcriptional inhibitory role to further affect gene expression.14 Recent studies have shown that MAFF transcripts are highly expressed in the myometrium and are involved in the regulation of uterine genes at the beginning of labor.15 Moreover, MAFF is downregulated in type I diabetes.16 However, the expression and function of MAFF in GECs remain unclear. Analyses using bioinformatics software (RepeatMasker and IntaRNA) suggest that both LINC00673 and MAFF 3′ UTRs contain the Alu elements. Also, the binding site between LINC00673 and MAFF detected by bioinformatics information provides further impetus to study the mechanism of the interaction between LINC00673 and MAFF.
Tight junction-related proteins (TJPs) are essential molecular basis for BTB function, including the primary component proteins ZO-1, occludin, and claudin-5.17 The adhesion protein ZO-1 forms the basis of support structure for TJPs; the transmembrane protein occludin directly participates in the formation of TJPs on microvascular endothelial cells; and claudin-5 is abundant in brain endothelial cells and exerts an important role in regulating their permeability.18 ZO-1, occludin, and claudin-5 are crucial proteins that explicitly define the paracellular pathways affecting BTB permeability, while their expressions are downregulated when BTB permeability increases.19 In silico analysis using JASPAR reveals that there are binding sites between MAFF and the respective promoters of ZO-1, occludin, and claudin-5, which indicates that MAFF may regulate the expression of these three genes at the transcriptional level.
In this study, we first investigated the expression of RBFOX1, LINC00673, and MAFF in GECs and their effects on BTB permeability. Furthermore, we determined their interaction in the regulation of BTB permeability. Therefore, this study aims to explore the mechanism on selectively opening the BTB as well as to provide new theoretical and experimental evidence for improving the efficacy of treatment against glioma.
Results
Expression of RBFOX1 in Glioma Vascular Endothelial Cells
According to The Cancer Genome Atlas (TCGA) analysis, RBFOX1 had low expression levels in glioma tissue, and the patients who had relatively lower levels of RBFOX1 had poor overall survival (Figure 1A). Laser-capture microdissection (LCM) is capable of isolating cells of interest from archived paraffin-embedded tissue specimens, and it permits microscopic localization of cells at the site of a pathological process. Thus, LCM could permit cells to be isolated from that site and sites not involved in the pathological process for further comparative studies. To study the BTB at the molecular level, RBFOX1 mRNA expression was detected at first in brain microvessels captured from a panel of surgical human brain specimens of brain tissues (normal brain tissues [NBTs]), low-grade glioma (LGG, World Health Organization [WHO] grade I–II), and high-grade glioma (HGG, WHO grade III–IV) by using LCM. As shown in Figure 1B, we captured microvessels from NBT, LGG, and HGG using LCM. Subsequently, after isolating the RNAs from cells, we conformed qRT-PCR assay to detected RBFOX1 mRNA expression. The results showed that the RBFOX1 mRNA expression was downregulated in the captured brain microvessels from LGG and HGG compared with NBT. Additionally, RBFOX1 expression was significantly downregulated in the captured brain microvessels from HGG compared with those from LGG (Figure 1C). Thus, RBFOX1 was also represented in glioma microvessels, and the expression of RBFOX1 was negatively correlated with the pathological grades of glioma. Furthermore, after in vitro BTB models were established, western blot and qRT-PCR analyses showed that the expression of RBFOX1 in GECs was significantly downregulated compared to astrocyte-exposed endothelial cells (AECs) (Figures 1D and 1E). Thus, we hypothesize that RBFOX1 might regulate the function of GECs as tumor suppressor genes.
Figure 1.
Upregulation of RBFOX1 Increased BTB Permeability in GECs and Inhibited the Expression Levels of ZO-1, Occludin, and Claudin-5
(A) The expression levels of RBFOX1 in glioma and patient survival were analyzed through TCGA database. (B) Staining vessel from glioma or normal brain tissues with UEA-I. LCM capture of UEA-I-stained vessel from human brain sections is shown. Scale bar represents 40 μm. (C) qRT-PCR analysis of RBFOX1 expression in brain microvessels captured by laser capture microdissection (LCM) from a panel of surgical human brain specimens of normal brain tissues (NBTs), low-grade glioma (LGG, WHO I–II), and high-grade glioma (HGG, WHO III–IV). Data represent means ± SD (n = 8, each). ∗p < 0.05, ∗∗p < 0.01 versus the NBTs group; ##p < 0.01 versus the LGG group. (D) The expression levels of RBFOX1 protein in AECs and GECs were detected by Western bolt assay. (E) The expression levels of RBFOX1 mRNA in AECs and GECs were detected by qRT-PCR assay. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the AECs group. (F) Effects of RBFOX1 upregulation or knockdown on TEER values. (G) Effects of RBFOX1 upregulation or knockdown on HRP flux. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the vector group; #p < 0.05 versus the shNC group. (H) Effect of RBFOX1 upregulation or knockdown on ZO-1, occludin, and claudin-5 protein expression levels as determined by western blot. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the vector group; #p < 0.05 versus the shNC group. (I) Effects of RBFOX1 upregulation or knockdown on ZO-1, occludin, and claudin-5 distribution and expression levels as determined by immunofluorescence staining (n = 3, each). ZO-1, occludin, and claudin-5 (red) were labeled with secondary antibodies anti-ZO-1, anti-occludin, and anti-claudin-5, respectively, and nuclei (blue) were labeled with DAPI. Scale bar represents 30 μm.
Overexpression of RBFOX1 Increased the Permeability of the BTB and Decreased the Expression of the Tight Junction-Related Protein in GECs
To explore the functional role of endothelial RBFOX1 in BTB permeability, we established GEC lines with stable overexpression or silencing of RBFOX1 successfully. Subsequently, the transendothelial electric resistance (TEER) value and horseradish peroxidase (HRP) flux were used to test the integrity and permeability of the BTB, respectively.20 Figures 1F and 1G show that the TEER value was significantly decreased whereas HRP flux was significantly increased in the RBFOX1 overexpression (OE) group compared with vector group. However, the opposite results were observed in the short hairpin (sh)RBFOX1 group. Those results indicated that overexpression of RBFOX1 in GECs impaired BTB integrity and increased BTB permeability. ZO-1, occludin, and claudin-5 are members of tight junction-related proteins. Then, we focused on these three proteins to explore the mechanism of action of RBFOX1 on BTB permeability. The western blot analysis showed that the expression of ZO-1, occludin, and claudin-5 was significantly decreased in the RBFOX1 OE group compared with vector group in GECs. However, the expression of ZO-1, occludin, and claudin-5 increased in the shRBFOX1 group compared with the shNC group (Figure 1H) in GECs. Subsequently, immunofluorescence staining further confirmed that RBFOX1 overexpression suppressed ZO-1, occludin, and claudin-5 expression, with discontinuous distribution on the boundaries of GECs (Figure 1I). The above results indicate that RBFOX1 overexpression increased BTB permeability.
LINC00673 Was Upregulated in GECs, and LINC00673 Knockdown Increased the Permeability of BTB and Reduced the Expression of the Tight Junction-Related Protein in GECs
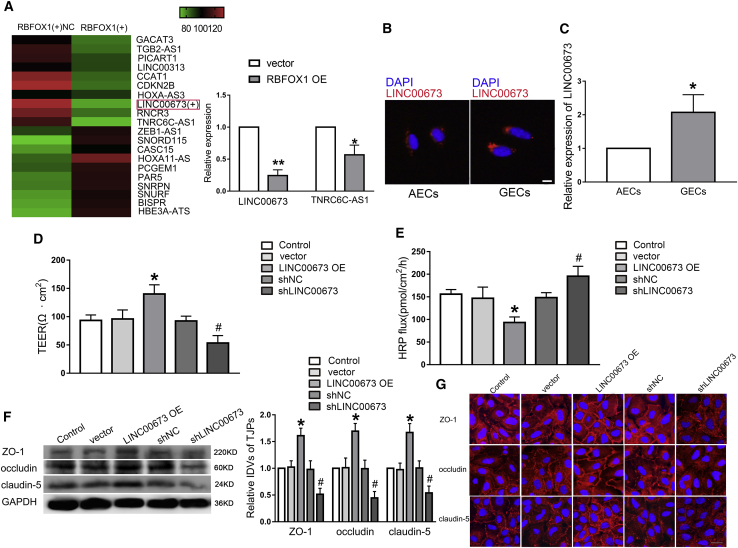
Accumulating studies have demonstrated that RBPs may exert their biological functions through mediating lncRNAs.21,22 lncRNA array analysis was performed in GECs with overexpression of RBFOX1. The results showed that LINC00673 and TNRC6C-AS1 were two of the most downregulated lncRNAs in the RBFOX1 OE group. Meanwhile, qRT-PCR analysis showed that expression of LINC00673 and TNRC6C-AS1 was downregulated, and the LINC00673 expression level was lower than that of TNRC6C-AS1 in GECs with upregulation of RBFOX1 (Figure 2A). Accordingly, we selected LINC00673 to perform the subsequent analyses. In addition, the expression and location of LINC00673 in AECs and GECs were investigated by qRT-PCR and fluorescence in situ hybridization (FISH). LINC00673 was significantly upregulated in GECs compared with AECs (Figure 2C). In addition, the quantitative expression of TNRC6C-AS1 is shown in Figure S1D. Also, LINC00673 was located primarily in the cytoplasm of cells (Figure 2B). Hence, we hypothesized that LINC00673 might be involved in the regulation of BTB function.
Figure 2.
LINC00673 Was Highly Expressed in GECs, and Knockdown of LINC00673 Increased BTB Permeability and Inhibited the Expression Levels of ZO-1, Occludin, and Claudin-5
(A) lncRNA microarray analysis of total RNAs isolated from GECs treated with overexpression of RBFOX1. Red indicates high relative expression, and green indicates low relative expression. qRT-PCR was used to detected the selected molecules. Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the vector group. (B) FISH was conformed to investigate expression and location of LINC00673 in AECs and GECs (red, LINC00673; blue, DAPI nuclear staining). Scale bar represents 20 μm. (C) The expression levels of LINC00673 in AECs and GECs were detected by a qRT-PCR assay. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the AECs group. (D and E) Effects of LINC00673 upregulation or knockdown on TEER values (D) and HRP flux (E). (F) Effects of LINC00673 upregulation or knockdown on ZO-1, occludin, and claudin-5 expression levels determined by western blot. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the vector group; #p < 0.05 versus the shNC group. (G) Effects of LINC00673 upregulation or knockdown on ZO-1, occludin, and claudin-5 distribution and expression levels determined by immunofluorescence staining (n = 3, each). ZO-1, occludin, and claudin-5 (red) were labeled with secondary antibodies anti-ZO-1, anti-occludin, and anti-claudin-5, respectively, and nuclei (blue) were labeled with DAPI. Scale bar represents 30 μm.
The significant upregulation of LINC00673 in GECs prompted us to explore the essential function of LINC00673 in GECs by investigating the effects of LINC00673 knockdown or overexpression on BTB integrity and permeability. Accordingly, stably LINC00673-overexpressed or LINC00673-silenced GEC lines were established successfully. The efficiency of LINC00673 is shown in Figure S1B. As shown in Figures 2D and 2E, the TEER value was increased whereas HRP flux was decreased in the LINC00673 OE group, compared with vector group. Additionally, the TEER value was decreased whereas HRP flux was increased in the shLINC00673 group, compared with the short hairpin negative control (shNC) group. These results suggest that LINC00673 knockdown could impair BTB integrity and increase BTB permeability. To further explore the mechanisms in regulating BTB permeability, the expression levels of ZO-1, occludin, and claudin-5 were detected by western blot. The results indicated that the expression levels of ZO-1, occludin, and claudin-5 were significantly increased in the LINC00673 OE group, whereas they were significantly decreased in shLINC00673 group, compared with the vector and shNC groups (Figure 2F). Furthermore, immunofluorescence analysis revealed that ZO-1, occludin, and claudin-5 were significantly decreased and exhibited discontinuously distribution in the shLINC00673 group (Figure 2G).
RBFOX1 Targeted LINC00673 and Reduced LINC00673 Stability
It was demonstrated that the differential expression of LINC00673 exists in the RBFOX1-depleted GECs. Also, qRT-PCR further determined that LINC00673 expression was decreased in the RBFOX1 OE group, compared with the vector group. Additionally, LINC00673 expression was increased in the shRBFOX1 group, compared with the shNC group (Figure 3A). Subsequently, the RNA immunoprecipitation (RIP) assay showed that LINC00673 was enriched in the RBFOX1 immunoprecipitates compared with the immunoglobulin G (IgG) immunoprecipitates (Figure 3B). RNA pull-down assays further demonstrated that LINC00673 bound with RBFOX1, instead of LINC00673-mutant (Mut) and anti-sense RNA (Figure 3C). Hence, RBFOX1 directly bound to LINC00673 and negatively regulated LINC00673 expression in GECs. Given the possibility that some RBPs regulate lncRNA stability,23 we analyzed the half-life of LINC00673 in RBFOX1 OE GECs. Newly synthesized RNA was labeled and isolated using the Click-iT Nascent RNA capture kit, and further qRT-PCR results suggested that the nascent LINC00673 was unaffected in the RBFOX1 OE group and vector group (Figure 3D). Then, we evaluated LINC00673 stability with RBFOX1 gain or loss in GECs under the action of actinomycin D (2 μg/mL). The results indicated that the half-life of LINC00673 is 5.5 h in the RBFOX1 OE group, while the half-life of LINC00673 is 8 h in the vector group (Figure 3E). Thus, RBFOX1 negatively regulated the expression level of LINC00673 through inhibiting LINC00673 stability.
Figure 3.
Upregulation of RBFOX1 Destabilized LINC00673, and LINC00673 Was Involved in RBFOX1-Mediated Regulation on BTB Permeability
(A) Effects of RBFOX1 overexpression or inhibition on LINC00673 expression by a qRT-PCR assay. Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the vector group; #p < 0.05, ##p < 0.01 versus the shNC group. (B) Enrichment of LINC00673 in RBFOX1 immunoprecipitates was measured by a RIP assay. Data represent means ± SD (n = 3, each). ∗∗p < 0.01 versus the anti-normal IgG group. (C) RBFOX1 and GAPDH protein levels in immunoprecipitation with LINC00673 RNA were evaluated by western blots. The expression levels of RBFOX1 and GAPDH proteins are shown. (D) The Click-iT Nascent RNA capture kit (Life Technologies) was used to label and capture newly synthesized RNA, and nascent LINC00673 was analyzed using qRT-PCR. (E) GECs after RBFOX1 overexpression were exposed to actinomycin D (2 μg/mL), and total RNA was separated at the indicated times and subjected to qRT-PCR to assess the half-life of LINC00673 RNA. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the vector group. RBFOX1 and LINC00673 were co-transfected in GECs. (F and G) Effects of RBFOX1 upregulation and LINC00673 knockdown or RBFOX1 knockdown and LINC00673 upregulation on TEER values (F) and HRP flux (G). (H) Effects of RBFOX1 upregulation and LINC00673 knockdown or RBFOX1 knockdown and LINC00673 upregulation on ZO-1, occludin, and claudin-5 expression levels as determined by western blot. Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the RBFOX1 OE+LINC00673 OE group; #p < 0.05, ##p < 0.01 versus the shRBFOX1+shLINC00673 group; ▲p < 0.05, ▲▲p < 0.01 versus the control group.
LINC00673 Was Involved in RBFOX1-Regulated BTB Permeability
To clarify whether LINC00673 was involved in RBFOX1-mediated regulation of BTB permeability as well as the expression of ZO-1, occludin, and claudin-5, we performed a co-transfection assay. As shown in Figures 3F and 3G, TEER values were decreased more in the RBFOX1 OE+shLINC00673 group than those in the RBFOX1 OE+LINC00673 OE group. However, HRP flux was increased more in the RBFOX1 OE+shLINC00673 group than that in RBFOX1 OE+LINC00673 OE group. Moreover, TEER values were increased in the shRBFOX1+LINC00673 OE group, whereas HRP flux decreased, compared with the shRBFOX1+LINC00673 OE group. The expression levels of ZO-1, occludin, and claudin-5 were decreased more in the RBFOX1 OE+shLINC00673 group than those in the RBFOX1 OE+LINC00673 OE group. Additionally, these levels were increased in the shRBFOX1+LINC00673 OE group, compared with the shRBFOX1+shLINC00673 group (Figure 3H). These results clarified that RBFOX1 increased BTB permeability by inhibiting LINC00673 expression.
MAFF Was a Target of LINC00673, and LINC00673 Regulated MAFF Expression by the Staufen1 (STAU1)-Mediated mRNA Decay (SMD) Process
The regulation of protein-coding genes occurs in a series of processes in their product messenger RNAs (mRNAs), indicating both synthesis and decay. Accordingly, many human diseases are due to aberrancies in the regulation of mRNA half-life.24,25 In the present study, we found that the Alu element exists in LINC00673 and MAFF mRNA 3′ UTRs by searching RepeatMasker and IntaRNA bioinformatics tools, and LINC00673 bound to MAFF mRNA 3′ UTR in a sequence-specific manner with their Alu element (Figure 4A). A subsequent dual-luciferase assay demonstrated that the luciferase activity is significantly reduced in the MAFF-3′ UTR-wild-type (WT)+LINC00673 OE group compared with the MAFF-3′ UTR-WT+vector group. However, luciferase activity was not affected in the control group and MAFF-3′ UTR-Mut+LINC00673 OE group (Figure 4B). Given that STAU1 was bound to the 3′ UTR of a number of mRNAs so as to reduce their half-life in UPF1-dependent mechanisms,26,27 we performed a RIP assay. The results showed that LINC00673 and MAFF mRNA were enriched in STAU1 immunoprecipitates compared with IgG immunoprecipitates. However, LINC00673 knockdown decreased the expression of LINC00673 and MAFF mRNA in STAU1 immunoprecipitates (Figure 4C). Subsequently, qRT-PCR showed that the half-life of MAFF mRNA was 6 h in LINC00673 OE group and 9 h in the control group, whereas the half-life of MAFF mRNA was 12.5 h in the shSTAU1 group and shUPF1 group (Figure 4D). Furthermore, western blot demonstrated that the expression levels of MAFF mRNA and protein were decreased in the LINC00673 OE+shNC group, but increased in the vector+shSTAU1 group, compared with the vector+shNC group. Notably, the downregulation of STAU1 significantly reversed the LINC00673 upregulation-induced decrease in MAFF mRNA and protein expression (Figures 4E and 4F). These results demonstrated that LINC00673 promoted MAFF degradation through the SMD process in GECs.
Figure 4.
LINC00673 Targets MAFF and Negatively Regulated MAFF through the SMD Pathway, as well as MAFF Involved in LINC00673-Mediated Regulation in Promoting BTB Permeability
(A) The putative LINC00673 binding site in MAFF 3′ UTR (MAFF 3′ UTR-WT) and the mutant sequence (MAFF 3′ UTR-Mut) are illustrated. (B) Relative luciferase activity in HEK293T cells co-transfected with MAFF-3′ UTR-WT or MAFF 3′ UTR-Mut and LINC00673 OE or vector. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the MAFF-3′ UTR-WT+vector group. (C) The enrichment of LINC00673 and MAFF mRNA in GECs and LINC00673-deficient GECs was measured by a RIP assay. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the anti-IgG group; #p < 0.05 versus the shLINC00673 group. (D) After overexpression of LINC00673, knockdown of STAU1, and knockdown of UPF1, GECs were treated with actinomycin D (2 μg/mL), and total RNA was isolated at the indicated times and subjected to qRT-PCR to assess the half-life of MAFF mRNA. Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the control group. LINC00673 and STAU1 co-transfected in GECs are shown. (E) Expression levels of MAFF mRNA were detected by qRT-PCR. (F) Expression levels of MAFF protein were detected by western blot. Data represent mean ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the vector+shNC group. MAFF was co-transfected with LINC00673 in GECs. (G and H) TEER (G) and HRP (H) were detected. (I) The expression of ZO-1, occludin, and claudin-5 was detected by western blot. Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the LINC00673 OE+MAFF OE group; #p < 0.05, ##p < 0.01 versus the shLINC00673+shMAFFgroup; ▲p < 0.05, ▲▲p < 0.01 versus the control group.
MAFF Was Engaged in LINC00673-Mediated Regulation of BTB Permeability
To investigate whether MAFF was involved in LINC00673-mediated regulation of BTB function, we performed co-transfection assays. TEER values were increased in the LINC00673 OE+shMAFF group, whereas HRP flux was decreased, in comparison with the LINC00673 OE+shMAFF group. Additionally, TEER values were decreased in the shLINC00673+MAFF OE group, whereas HRP flux was increased, in comparison with the shLINC00673+MAFF OE group (Figures 4G and 4H). Meanwhile, the expression levels of ZO-1, occludin, and claudin-5 were increased in LINC00673 OE+shMAFF group, compared with the LINC00673 OE+MAFF OE group. However, the expression of ZO-1, occludin, and claudin-5 were decreased in the shLINC00673+MAFF OE group, compared with the shLINC00673+shMAFF group (Figure 4I). These results demonstrated that LINC00673 increased BTB permeability by increased MAFF expression.
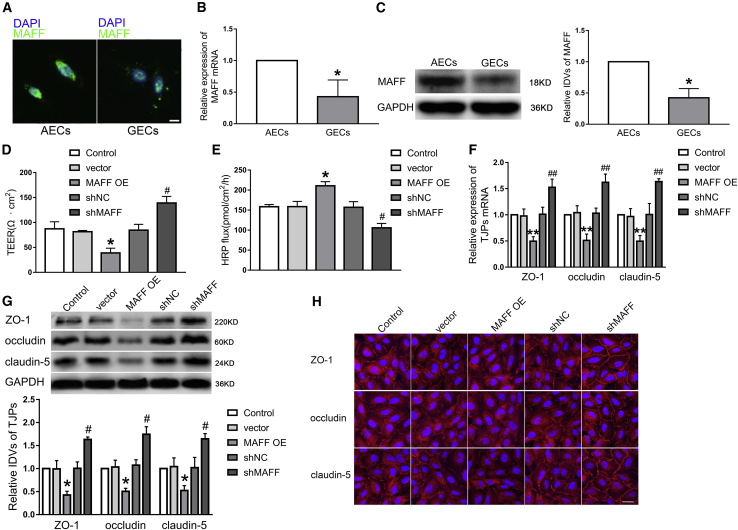
MAFF Was Downregulated in GECs, and Upregulated MAFF Increased the Permeability of the BTB and Reduced the Expression of the Tight Junction-Related Protein in GECs
After confirming that MAFF was a target of LINC00673, we further detected the location and expression of MAFF in AECs and GECs. A FISH assay revealed that MAFF was located in both the nucleus and cytoplasm in cells, and expression was lower in GECs than that in AECs (Figure 5A). Also, qRT-PCR and western blot results showed that the expression of MAFF mRNA and protein was downregulated in GECs, compared with AECs (Figures 5B and 5C). To clarify the functional roles of MAFF in BTB permeability, the stable MAFF overexpression or MAFF silencing of GEC lines was established, respectively. TEER values decreased whereas HRP flux increased in the MAFF OE group compared with the vector group; however, opposite results were found in the shMAFF group (Figures 5D and 5E). Subsequently, qRT-PCR and western blots suggested that the expression levels of ZO-1, occludin, and claudin-5 decreased in the MAFF OE group, compared with those in the vector group. However, the expression levels of ZO-1, occludin, and claudin-5 were increased in the shMAFF group compared with the shNC group (Figures 5F and 5G). Furthermore, immunofluorescence analysis revealed that ZO-1, occludin, and claudin-5 were significantly decreased and exhibited discontinuous distribution in the MAFF OE group (Figure 5H)
Figure 5.
MAFF Was Lowly Expressed in GECs, and Overexpression of MAFF Increased BTB Permeability
(A) FISH was applied to detect expression and location of MAFF in AECs and GECs (green, MAFF; blue, DAPI nuclear staining). Scale bar represents 20 μm. (B) The expression levels of MAFF mRNA in AECs and GECs were detected by a qRT-PCR assay. (C) The expression levels of MAFF protein in AECs and GECs were detected by a western bolt assay. Data represent means ± SD (n = 3, each). ∗p < 0.05 versus the AECs group. (D and E) Effects of MAFF upregulation or knockdown on TEER values (D) and HRP flux (E). (F and G) Effects of MAFF upregulation or knockdown on ZO-1, occludin, and claudin-5 mRNA and protein expression levels determined by qRT-PCR (F) and western blot (G). Data represent means ± SD (n = 3, each). ∗p < 0.05, ∗∗p < 0.01 versus the vector group; #p < 0.05, ##p < 0.01 versus the shNC group. (H) Effects of MAFF upregulation or knockdown on ZO-1, occludin, and claudin-5 expression levels and distribution as determined by immunofluorescence staining (n = 3, each). ZO-1, occludin, and claudin-5 (red) were labeled with secondary antibodies anti-ZO-1, anti-occludin, and anti-claudin-5, respectively, and nuclei (blue) were labeled with DAPI. Scale bar represents 30 μm.
MAFF Bound to the Tight Junction-Related Proteins ZO-1, Occludin, and Claudin-5 and Inhibited Promoter Activities
Having confirmed that MAFF overexpression reduced the expression of the tight junction-related proteins ZO-1, occludin, and claudin-5 at the mRNA and protein levels, we hypothesized that MAFF may directly regulate the expression of tight junction proteins at the transcriptional level. In the present study, the sequences of ZO-1, occludin, and claudin-5 promoters were selected using the bioinformatics database DBTSS. At the same time, the positions of the transcription start site (TSSs) of ZO-1, occludin, and claudin-5 were predicted by DBTSS. Moreover, we identified the sites to which MAFF bound by analyzing the DNA sequence 2,000 bp upstream of the TSS and its 200-bp downstream sequence. Then, dual-luciferase reporter gene analysis and chromatin immunoprecipitation (ChIP) were used to verify the activity and binding effects of MAFF combined with the ZO-1, occludin, and claudin-5 promoter-binding sites, respectively. As shown in Figures 6A and 6C, the activities of the ZO-1, occludin, and claudin-5 promoters were significantly reduced in the pEX3 MAFF group, compared with those in the pEX3 empty vector group. Also, a series of DNA fragments was constructed and ligated into the pGL3 basic vector. Wild-type and deletion constructs and putative MAFF binding sites were indicated. Upon co-transfection with pEX3-MAFF, ZO-1 promoter activity was downregulated by the −683 site region (Figure 6A), occludin promoter activity was downregulated by the +100 site region (Figure 6B), and claudin-5 promoter activity was downregulated by the −462 site region, but there was no significant change by the −789 site region (Figure 6C). Finally, ChIP assays were performed to determine whether MAFF directly binds the promoters of ZO-1, occludin, and claudin-5 in GECs. The putative MAFF binding sites were indicated. As an NC, PCR was conducted to amplify the 2,000-bp upstream region of the putative MAFF binding site that was not expected to associate with MAFF. The results demonstrated that there was an association of MAFF with the putative binding site PCR2 of ZO-1, the putative binding site PCR2 of occludin, and the putative binding site PCR3 of claudin-5. There was no association of MAFF with all of the control regions (Figures 6D–6F). The above results indicate that MAFF bound to the promoter region of ZO-1, occludin, and claudin-5 and inhibited its transcription activity.
Figure 6.
MAFF Bound to the Promoters of Tight Junction-Related Proteins and Transcriptionally Depressed ZO-1, Occludin, and Claudin-5 Expression in GECs, and RBFOX1 Overexpression Combined with LINC0067 Knockdown Promoted the Delivery of Dox as well as Apoptosis of Glioma Cells
(A–C) Schematic depictions of the different reporter plasmids and the relative luciferase activity for ZO-1 (A), occludin (B), and claudin-5 (C). The y bar shows the deletion positions on the promoter fragments. The x bar shows the reporter vector activity after normalization with the cotransfected reference vector (pRL-TK), and relative to the activity of the pEX3 empty vector, the activity of which was set to 1. Data represent means ± SD (n = 3, each). ∗p < 0.05. (D–F) Schematic representations of the human ZO-1 (D), occludin (E), and claudin-5 (F) promoter region 2,000 bp upstream of the transcription start sites (TSSs), which were designated as +1. ChIP PCR products for putative MAFF binding sites and an upstream region not supposed to associate with MAFF are depicted with bold lines. Dashed lines represent the primers used for each PCR. GECs were utilized to conduct ChIP assays. PCR was conducted with the resulting precipitated DNA. Images make up independent experiments (n = 3). (G) RBFOX1 and LINC00673 were co-transfected in GECs. Flow cytometry analysis of U251 cells in the lower chamber with Dox (20 μM) for 12 h is shown. Data represent mean ± SD (n = 3, each). ∗∗p < 0.01 versus the control group; ##p < 0.01 versus the Dox group; &&p < 0.01 versus the RBFOX1 OE+Dox group; ▪▪p < 0.01 versus the shLINC00673+Dox group.
RBFOX1 Overexpression Combined with LINC00673 Knockdown Promoted Doxorubicin Delivery and Glioma Cell Apoptosis
To maximize the therapeutic efficacy of brain glioma and minimize the side effects, anti-tumor drugs should be penetrated through the BTB and then targeted to brain carcinoma cells. Doxorubicin (Dox) is an anthracycline antibiotic used for tumor treatment that inhibits the synthesis, replication, and transcription of DNA to impair the cell cycle.28 A recent study showed that Dox could successfully treat malignant glioma.29 In the present study, we have stable transfected RBFOX1 overexpression, LINC00673 knockdown, and co-transfected RBFOX1 overexpressing and LINC00673 knockdown in endothelial cells, respectively. Then in vitro BTB models were established with U251. Then, on the 4th day of the establishment of the BTB, 20 μM Dox was added into the upper compartment of the Transwell system. 12 h later, U251 cells in the lower chamber were selected and double stained with annexin V-phycoerythrin (PE)/7-aminoactinomycin D (7AAD). To verify the effects of transfection of RBFOX1 overexpression or LINC00673 knockdown alone and RBFOX1 overexpression and LINC00673 knockdown co-transfection of BTB permeability to promote Dox delivery, flow cytometry was performed. The results showed that compared with the Dox group, the RBFOX1 OE+Dox group and shLINC00673+Dox group increased the apoptosis of U251 cells, respectively. Notably, RBFOX1 overexpression combined with LINC00673 knockdown exhibited a high apoptosis rate (Figure 6G). In addition, compared with the carboplatin group, the RBFOX1 OE+carboplatin group and the shLINC00673+carboplatin group increased the apoptosis of U251 glioma cells, respectively (Figure S2). Collectively, the combination of RBFOX1 overexpression and LINC00673 knockdown promoted Dox delivery and glioma cell apoptosis.
Discussion
Endothelial cells are an immortalized human cerebral microvascular endothelial cell line that recapitulates most properties of the blood-brain barrier (BBB) in situ. Also, this approach is often used as a BBB model in vitro.30 The in vitro BTB models established by co-culturing cells simulate high TEER, restricted permeability across the monolayer, and changes of the morphological characteristics of endothelial cells in vivo. Thus, this approach has been utilized to detect the nature of BTB cellular interactions occurring in vivo. Based on the BTB models in vitro, TEER values and HRP flux are essential indices used to evaluate BTB permeability. In the present study, we selected these two indices to determine the effects of genetic changes on BTB permeability.
Human RBFOX1 is the first identified as an interactive partner of ataxin-2. Abnormal expression of RBFOX1 in the central nervous system promotes human neurodevelopmental disorders.31 RBFOX1 exhibits a low expression level in the brain of autistic patients.32 Knockdown of RBFOX1 in the mouse central nervous system leads to the susceptibility of increased seizures.33 Studies have also shown that RBFOX1 is related to the development of some tumors. The expression level of RBFOX1 protein in breast cancer tissues is significantly reduced.34 The presence of RBFOX1 gene deletion mutations in colon cancer is positively related to the development of colon cancer.35 RBFOX1 is downregulated in gastric cancer and human mesothelioma.36,37 RBFOX1 is a tissue-specific alternative splicing factor of malignant glioma. RBFOX1 inhibits the expression of exon 6a and 6b of the tumor suppressor TPM1, and promotes the occurrence of glioma.7,38 In this study, we demonstrated the regulation of RBFOX1 in brain vascular endothelial cells and enriched the current knowledge on RBFOX1. Drugs cross the BTB by two pathways: the caveolin-mediated transcellular pathway, and the opening of tight junction paracellular pathways. Tight junction proteins consisting of cytosolic scaffold proteins Zos and transmembrane proteins occludins and claudins, among others, are responsible for the structural integrity of the BBB and the regulation of BBB permeability.39 For the BTB, the expression levels of tight junction proteins, including ZO-1, occludin, and claudin-5, are significantly downregulated when BTB permeability is increased.40 Our data demonstrated that the expression levels of tight junction proteins ZO-1, occludin, and claudin-5 were decreased following RBFOX1 overexpression, indicating that upregulated RBFOX1 increased BTB permeability by opening tight junctions through paracellular pathways.
Aberrantly expressed lncRNAs are vital regulatory factors in glioma blood vessels. A number of studies indicate that lncRNAs are demonstrated to be necessary to regulate BTB permeability and glioblastoma multiforme (GBM) angiogenesis.20,41 Meanwhile, several studies have reported that lncRNA LINC00673 is involved in the progression of tumors and considered as a potential novel biomarker for early diagnosis of lung cancer.42 In this regard, it is urgent to ascertain the underlying mechanism of LINC00673 dysregulation in tumors. It has been reported that LINC00673 is triggered by the transcription factor SP1 and directly targets LSD1 and EZH2, and that it acts as a scaffold to inhibit the expression of KLF2 and LATS2, thereby promoting the growth and invasion of gastric cancer cells.11 In the present study, we screened out several lncRNAs after downregulation of RBFOX1 in GECs, and finally we verified the LINC00673. Subsequently, the expression and location of LINC00673 in GECs, as well as the detailed function of LINC00673, were further detected. Our results indicated that knockdown of LINC00673 reduced ZO-1, occludin, and claudin-5 expression, destroyed the integrity of the BTB, and increased the permeability of the BTB. Meanwhile, restoration of RBFOX1 dramatically reduced the LINC00673 expression and increased BTB permeability. Accordingly, whether there is a supervisory relationship between RBFOX1 and LINC00673 has attracted our attention. Recent studies have demonstrated that RBPs affect biological function via regulating RNA in a variety of ways. RBP NFAT5 positively regulates the expression of lncRNA SBF2-AS1 and promotes GBM angiogenesis through the miR-338-3p/EGFL7 pathway.43 HuR promotes tongue cancer by increasing the stability of lnc-Sox5.44 Also, HuR enhances the stability of lncRNA NEAT1, inhibits the expression of miR-124-3p, and promotes ovarian cancer.45 In this study, our results indicated that the RBP RBFOX1 plays a functional role in regulating the permeability of the BTB by decreasing the stability of LINC00673. Furthermore, accumulating evidence shows that RBFOX1 acts in the regulation of the stability of target genes. RBFOX1 downregulation results in a decrease of mRNA stability by encoding the synaptic transmission protein MARK2 and induces the occurrence of Alzheimer’s disease.46
MAFF is deregulated in various cells and plays a crucial role in various physiological and pathological processes, such as emergency signals, hematopoiesis, central nervous system function, and tumorigenesis.47 Additionally, MAFF expression is decreased in endothelial progenitor cells derived from the circulating bone marrow of patients with type I diabetes, which inhibits cardiovascular production and tissue regeneration.16 Our data demonstrated that MAFF was downregulated in GECs and located in both cytoplasm and nucleus. Overexpression of MAFF reduced the expression of tight junction-related proteins ZO-1, occludin, and claudin-5 and increased the permeability of the BTB.
It was predicted that the transcription factor MAFF mRNA 3′ UTR processed a binding site for LINC00673 by using the bioinformatics software IntaRNA, and we further verified the fact of binding between LINC00673 and MAFF-3′ UTR. Moreover, inhibition of LINC00673 markedly prolonged the half-life of MAFF mRNA. Furthermore, the silencing of LINC00673 increased the permeability of the BTB by elevating MAFF mRNA stability. There are numerous ways in which lncRNAs exert biological effects. Accumulated evidence has indicated that lncRNAs directly regulate the expression of downstream target genes at the transcriptional and post-transcriptional levels through the “sponging” of miRNAs or by altering the stability of RNAs.48 Additionally, a study has reported that lncRNAs could regulate the stability of target genes, for example, lncRNA AC132217.4, increasing the stability of IGF2 mRNA to accelerate the metastasis of oral squamous cell carcinoma by binding to IGF2 3′ UTR.49
SMD is one of the means of regulation of mRNA degradation in mammalian cells.26 In the SMD process, the double-stranded binding protein STAU1 targets the STAU1 binding site (SBS) formed by lncRNAs with target gene mRNAs via their Alu elements to recruit ATP-dependent helical UPF1, resulting in the degradation of the target mRNA.50 In this study, we found that LINC00673 overexpression destabilized MAFF mRNA, whereas STAU1 or UPF1 knockdown increased the stabilization of MAFF mRNA. Furthermore, STAU1 or UPF1 knockdown rescued MAFF mRNA and protein expression downregulated by LINC00673 overexpression. MAFF and LINC00673 were co-transfected, and MAFF knockdown significantly reversed the effect of promoting BTB permeability by LINC006673 alone knockdown. Thereby, LINC00673 knockdown increased the BTB permeability and decreased the expression of ZO-1, occludin, and claudin-5 by reducing MAFF mRNA degradation in an SMD manner. In line with our results, recent studies also demonstrated that the SMD pathway plays an important role in the regulation of varieties of biological effects. For example, during the process of adipogenesis, the abundance of PNRC2 or STAU1 was increased and UPF1 was hyperphosphorylated, and hyperphosphorylated UPF1 forms a stable complex with PNRC2, resulting in KLF2 mRNA destabilization and promoting the process of adipogenesis.51
Accumulating studies have demonstrated that transcription factors regulate the target genes through binding to their promoters. A recent study has suggested that the transcription factor Twist1, regulated by transforming growth factor β (TGF-β) through SMD, binds to the SNCG promoter, which promotes hepatocellular carcinoma cell migration and invasion.52 Furthermore, MAFF leads to perinatal lethality through targeting the US2 motif of the oxytocin receptor (OTR) promoter specifically, activating US2 transcriptional activity and promoting the OTR expression.53 In the tumor microenvironment, the transcription inhibitor MAFF binds to mutP53, inhibits SIL-1Ra promoter activity, and facilitates the progression of malignancy.54 In the present study, we demonstrated that MAFF bound to the promoter of ZO-1, occludin, and claudin-5 and inhibited their transcriptions.
Owing to the partially intact BTB, the treatment of brain glioma remains a great challenge. The BTB limits most chemotherapeutics in their delivery to tumors. Different methods, including focused ultrasound (FUS), leukotriene C4, and bradykinin, have been used and have had promising outcomes. However, these methods have been either invasive, non-targeted, or required the function of new drugs.55 In the present study, RBFOX1 overexpression increased BTB permeability, and LINC00673 knockdown increased BTB permeability, and these results revealed the methods of opening the BTB from the molecular level. Recent research reported that diphtheria toxin (DT)270–326 significantly increased the permeability of the BTB in vitro, and the combination of DT270–326 with Dox significantly enhanced the loss of cell viability and the apoptosis of glioma cells.56 In the present study, we demonstrated that RBFOX1 overexpression combined with LINC00673 knockdown promoted Dox delivery across the BTB. In conclusion, the present study for the first time reveals the significance of the interaction among RBP RBFOX1, lncRNA-LINC00673, and transcription factor MAFF in the regulation of BTB permeability, and the underlying mechanisms presented. The results of the study not only provide a new theoretical and experimental basis for functional regulation of BTB, but they also provide new ideas for the comprehensive treatment against glioma.
Materials and Methods
Clinical Specimens
All human glioma specimens and NBTs were obtained from the Department of Neurosurgery of Shengjing Hospital, China Medical University. All specimens were immediately frozen and preserved in liquid nitrogen. Two experienced clinical pathologists classified glioma specimens into two grades, that is, LGG (WHO grade I–II, n = 5) and HGG (WHO grade III–IV, n = 5), according to the WHO classification. Specimens of NBTs (n = 5) were used as the NC. All participants signed and provided informed consent, and this study was approved by the Institutional Review Board of Shengjing Hospital of China Medical University.
LCM
Tissue specimens were frozen and sectioned at 10-μm thickness using a microtome/cryostat (Microm International, Walldorf, Germany). Then, LCM was performed as previously described.57 Sections of vessels in glioma (or normal brain) were stained by fluorescent dye-tagged lectin, Ulex europaeus agglutinin I (UEA-I) (Vector Laboratories, Burlington, ON, Canada), according to the manufacturer’s protocol. After that, LCM was conducted by using the ArcturusXT microdissection instrument. The respectively captured microvessels were transferred into CapSure LCM caps (Applied Biosystems, Foster City, CA, USA) and further processed for RNA isolation.
Cell Lines and Cultures
The human cerebral microvascular endothelial cell line hCMEC/D3, marked as endothelial cells, was kindly provided by Dr. P.O. Couraud (Institut Cochin, Paris, France). The hCMEC/D3 cells (passage 30–35) were cultured in flasks or plates (Corning Life Sciences, Corning, NY, USA). Culture medium contained endothelial basal medium (EBM-2) (Lonza, Walkersville, MD, USA) coated with 150 μg/mL Cultrex rat collagen I (R&D Systems, Minneapolis, MN, USA). Normal human astrocytes (NHAs) were purchased from Science Cell Research Laboratories (Carlsbad, CA, USA) and maintained in astrocyte medium (Science Cell Research Laboratories, Carlsbad, CA, USA). The human glioma cell line U251MG and the human embryonic kidney 293T (HEK293T) cell line were purchased from the Shanghai Institutes for Biological Sciences Cell Resource Center and maintained in Dulbecco’s modified Eagle’s medium (DMEM) of high glucose with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Paisley, UK). All of the cells were cultured in a humidified atmosphere containing 37°C and 5% CO2.
Establishment of In Vitro BTB (or BBB) Models
The in vitro BTB or BBB models were established by co-culturing endothelial cells and U251 or endothelial cells and NHAs, as we described previously.58 Briefly, for in vitro BTB models, 2 × 104 U251 cells were seeded in six-well plates. When U251 cells were confluent at 2 × 105 per well, endothelial cells were seeded on the upper chamber of a six-well with Transwell inserts coated with Cultrex rat collagen I. Then, the GECs, endothelial cells co-cultured with U251 cells, were obtained in vitro after 4 days, with medium replacement every 2 days. For the in vitro BBB model, endothelial cells were co-cultured with NHAs using the same approach described above. The AECs, modeling the endothelial cell phenotype of the normal brain, were obtained after co-culturing astrocytes for 4 days.
Real-Time PCR Assay
Total RNAs were separated from AECs and GECs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as described by the manufacturer. Then, we detected the concentration and quality of RNAs by using a NanoDrop spectrophotometer (ND-100, Thermo Scientific, Waltham, MA, USA). A One-Step SYBR PrimeScript RT-PCR kit (Perfect Real Time, Takara Bio) was used to performed the following gene expression assays: RBFOX1 (NM_001308117.1), LINC00673 (NR_036488.1), MAFF (NM_001161572.1), ZO-1 (NM_001301025.3), occludin (NM_001205254.2), and claudin-5 (NM_001130861.1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the endogenous control. The relative expression values were calculated using the relative quantification (2−ΔΔCt) method. Primers used in present study are shown in Table S1.
Western Blot Analysis
Total proteins were isolated with radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Jiangsu, China). Then, equal amounts of protein samples were separated by SDS-PAGE and electrophoretically transferred onto polyvinylidene fluoride (PVDF) membranes. To avoid non-specific binding, membranes were blocked with 5% nonfat milk for 2 h at room temperature. Subsequently, membranes were incubated with primary antibodies: RBFOX1 (1:1,000, Proteintech, USA, 22647-1-AP), MAFF (1:1,000, Proteintech, USA, 12771-1-AP), GAPDH (1:10,000, Proteintech, USA, 60004-1-Ig), ZO-1 (1:500, Life Technologies, Frederick, MD, USA, 61-7300), occludin (1:1,000, Proteintech, USA, 27260-1-AP), claudin-5 (1:300, Life Technologies, Frederick, MD, USA, 35-2500) at 4°C overnight. Then, after washing, the membranes were incubated with HRP-conjugated secondary antibodies for 2 h at room temperature. After washing three times with Tris-buffered saline with Tween 20 (TBST), immunoblots were visualized by an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotechnology, Dallas, TX, USA) and scanned using the ChemiImager 5500 v2.03 software (Alpha Innotech, San Leandro, CA, USA). The integrated density values were calculated through FluorChem 2.0 software.
Immunofluorescence Assays
The endothelial cells on insert filters were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.3% Triton X-100 for 10 min at room temperature (ZO-1 and claudin-5) or fixed with methanol for 10 min at −20°C (occludin). After blocking with 5% bovine serum album for 2 h, the cells were incubated with primary antibodies for ZO-1 (1:50; Life Technologies), occludin (1:50; Abcam), and claudin-5 (1:50; Life Technologies) at 4°C overnight, respectively. Then, cells were incubated with Alexa Fluor 555-labeled goat anti-mouse IgG and anti-rabbit IgG (1:500; Beyotime Institute of Biotechnology, Hangzhou, Jiangsu, China) for 2 h at room temperature. DAPI (4′,6′-diamidino-2-phenylindole) was applied at 5 min to the samples to visualize cell nuclei. Staining was visualized using confocal microscopy (Olympus, JP, FV-1000).
Cell Transfection
The short hairpin RNAs (shRNAs) directly against the human RBFOX1 gene, MAFF gene, and STAU1 (NM_001037328.2), or UPF1 (NM_001297549.1) were constructed into the GV248/puro (GeneChem, Shanghai, China) vector (shRBFOX1, shMAFF, shSTAU1, and shUPF1), respectively. Additionally, human LINC00673 (NR_036488.1) shRNA was reconstructed in the pGPU6/Neo (GenePharma, Shanghai, China) vector (shLINC00673). Empty vectors were used as their NC (shNC). Full-length RBFOX1-pcDNA3.1/Hypro(+) vector (RBFOX1 OE) and full-length MAFF-pcDNA3.1/Hypro(+) vector (MAFF OE) were purchased from GenScript (Piscataway, NJ, USA). Meanwhile, full-length LINC00673-pIRES2 vector (LINC00673 OE) was purchased from GenePharma (Shanghai, China), and its empty vector was used as the NC (vector). We seeded endothelial cells in 24-well plates and then transfected them using LTX and PLUS reagent (Invitrogen, USA) until confluence reached ∼80%. Stable cell lines were established via Geneticin (G418; Sigma-Aldrich, St. Louis, MO, USA), puromycin (BioFroxx, Germany), and hygromycin (Solarbio, China) selection. The sequences of shRBFOX1, shLINC00673, shMAFF, shSTAU1, shUPF1, and NC are shown in Table S2. The transfection efficiencies of RBFOX1, MAFF, and LINC00673 are shown in Figures S1A–S1C.
TEER Measurements
A Millicell-ERS instrument (Millipore, Billerica, MA, USA) was used to measure the integrity of the BTB after in vitro BTB models were established. Before measuring the TEER values, co-culture inserts were placed in room temperature for 30 min. TEER values were measured directly after the medium exchange. Then, the background electrical resistance was subtracted before final resistances were calculated (Ω·cm2).
HRP Flux Assays
The in vitro BTB permeability was determined by detecting HRP (44 kDa) permeability. After the in vitro BTB models were established, 1 mL of serum-free EBM-2 medium containing 10 μg/mL HRP was added to the upper compartment of inserts. In addition, 2 mL of complete medium was added to the well. After incubation at 37°C for 1 h, 5 μL of culture medium was gathered from the lower compartment. Then, samples were identified using the tetramethylbenzidine colorimetry approach. Using a spectrophotometer, absorbance was measured at 370 nm. Finally, the HRP flux was expressed as pmol passed per cm2 surface area per hour.
Click-iT Captures Nascent RNA
A Click-iT Nascent RNA Capture kit (Invitrogen) was used to label and isolate newly synthesized RNA, as previously reported.59 Briefly, we used 5-ethynyl uridine (EU) to label the nascent RNA, which was then isolated using streptavidin magnetic beads.
ChIP Assay
A SimpleChip Enzymatic Chromatin IP kit (Cell Signaling Technology, Danvers, MA, USA) was applied for the ChIP assay, following the manufacturer’s instructions. Primers used for ChIP PCR are given in Table S3.
Dual-Luciferase Reporter Assay
The putative binding sequence of MAFF in LINC00673 and the mutant sequence were synthesized and cloned into the pmirGLO Dual-Luciferase vector (Promega, Madison, WI, USA). Wild-type pmirGLO-MAFF or MAFF mutant reporter plasmid and shLINC00673 or shNC were co-transfected into HEK293T cells. Luciferase activity was analyzed by the Dual-Luciferase reporter assay system (Promega), which was recorded as the ratio of firefly luciferase activity to luciferase activity.
For the putative binding site of MAFF in ZO-1, occludin, and the claudin-5 promoter region, we identified the predictor using the Dual-Luciferase reporter assay. The fragments of ZO-1, occludin, and claudin-5 promoters were amplified from human geneomic DNA, and then construct the sequence into the pGL3-Basic vector (Promega). Human full-length MAFF was constructed into the pEX3 vector (GenePharma). The luciferase activity of these binding sites was measured as above.
FISH
We conducted a FISH assay as previously described. Then, we characterized LINC00673 and MAFF rearrangement in GECs. A LINC00673 probe (red-labeled; Exiqon, Copenhagen, Denmark) and MAFF probe (green-labeled; BioSense, Guangzhou, China) were used.
Human lncRNA Microarrays
For lncRNA analysis, sample preparation and microarray hybridization were performed by KangChen Bio-tech (Shanghai, China).
RIP Assay
RIP was assayed using a Magna RBP immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Whole cells were lysed by RNA lysis buffer containing RBFOX1 and STAU1 antibody (Proteintech, China). Normal mouse IgG (Millipore, USA) served as an NC. Furthermore, co-precipitated RNAs were extracted and RNAs were purified and examined by quantitative real-time PCR.
RNA Pull-Down
The Pierce magnetic RNA-protein pull-down kit (Thermo Fisher Scientific) was conformed to examine the interaction between RBFOX1 and LINC00673 according to the manufacturer’s protocols. Furthermore, we co-incubated biotin-labeled LINC00673 or antisense RNA with protein extract of GECs and magnetic beads. After washing, the bead compound was boiled in SDS buffer, and the retrieved protein was detected by western blot with GAPDH as the control.
Analysis of Apoptosis by Flow Cytometry
The in vitro BTB models were established successfully. After cells were confluent to monolayer, GECs were treated with doxorubicin (20 μM) for 12 h. Untreated cells used as a control group. An annexin V-PE/7AAD staining apoptosis detection kit (SouthernBiotech, Birmingham, AL, USA) was used to analyze apoptosis, according to the manufacturer’s instructions. The cells acquired were detected by flow cytometry (FACScan, BD Biosciences, San Jose, CA, USA) and analyzed by CellQuest 3.0 software.
Statistical Analysis
Data are presented as the means ± standard deviation (SD), and statistical analysis was performed with GraphPad Prism7 (GraphPad, La Jolla, CA, USA). Statistical analysis was performed using Student’s t test and one-way ANOVA. The difference was considered statistically significant when p < 0.05.
Author Contributions
YX.X. and YH.L. contributed to conceived and designed the project. SY.S., D.W., Y.L. and J.M. contributed to performed most of the experiments. J.Z., LB. L., H. C. and T.M. did the statistics analysis. SY.S., CQ. Y. and XB.L. wrote the manuscript. LN.Z., P. A and Z. L. did western blotting experiments. YX.X. and YH.L. contributed to the manuscript revision. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work is supported by grants from the Natural Science Foundation of China (81872503, 81872073 and 81602726), China Postdoctoral Science Foundation (2019M661172), Liaoning Science and Technology Plan Project (No. 2017225020, 2015225007), Project of Key Laboratory of Neuro-oncology in Liaoning Province (112-2400017005), special developmental project guided by central government of Liaoning Province (No. 2017011553-301).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.03.014.
Supplemental Information
References
- 1.Gravina G.L., Mancini A., Marampon F., Colapietro A., Delle Monache S., Sferra R., Vitale F., Richardson P.J., Patient L., Burbidge S., Festuccia C. The brain-penetrating CXCR4 antagonist, PRX177561, increases the antitumor effects of bevacizumab and sunitinib in preclinical models of human glioblastoma. J. Hematol. Oncol. 2017;10:5. doi: 10.1186/s13045-016-0377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendricks B.K., Cohen-Gadol A.A., Miller J.C. Novel delivery methods bypassing the blood-brain and blood-tumor barriers. Neurosurg. Focus. 2015;38:E10. doi: 10.3171/2015.1.FOCUS14767. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack B.L., Lai L., Northrop J.L., Justice M.J., Hirschi K.K. Visceral endoderm function is regulated by quaking and required for vascular development. Genesis. 2006;44:93–104. doi: 10.1002/gene.20189. [DOI] [PubMed] [Google Scholar]
- 4.Ceolotto G., De Kreutzenberg S.V., Cattelan A., Fabricio A.S., Squarcina E., Gion M., Semplicini A., Fadini G.P., Avogaro A. Sirtuin 1 stabilization by HuR represses TNF-α- and glucose-induced E-selectin release and endothelial cell adhesiveness in vitro: relevance to human metabolic syndrome. Clin. Sci. (Lond.) 2014;127:449–461. doi: 10.1042/CS20130439. [DOI] [PubMed] [Google Scholar]
- 5.Conboy J.G. Developmental regulation of RNA processing by Rbfox proteins. Wiley Interdiscip. Rev. RNA. 2017;8:e1398. doi: 10.1002/wrna.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carreira-Rosario A., Bhargava V., Hillebrand J., Kollipara R.K., Ramaswami M., Buszczak M. Repression of pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev. Cell. 2016;36:562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J., Ho A.L., Yuan L., Hu B., Hua S., Hwang S.S., Zhang J., Hu T., Zheng H., Gan B. Neutralization of terminal differentiation in gliomagenesis. Proc. Natl. Acad. Sci. USA. 2013;110:14520–14527. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H., Xu Q., Liu F., Ye X., Wang J., Meng X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J. Thorac. Oncol. 2015;10:645–654. doi: 10.1097/JTO.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 10.Arnes L., Liu Z., Wang J., Maurer C., Sagalovskiy I., Sanchez-Martin M., Bommakanti N., Garofalo D.C., Balderes D.A., Sussel L. Comprehensive characterisation of compartment-specific long non-coding RNAs associated with pancreatic ductal adenocarcinoma. Gut. 2019;68:499–511. doi: 10.1136/gutjnl-2017-314353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M., Hou J., Wang Y., Xie M., Wei C., Nie F., Wang Z., Sun M. Long Noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Massrieh W., Derjuga A., Doualla-Bell F., Ku C.Y., Sanborn B.M., Blank V. Regulation of the MAFF transcription factor by proinflammatory cytokines in myometrial cells. Biol. Reprod. 2006;74:699–705. doi: 10.1095/biolreprod.105.045450. [DOI] [PubMed] [Google Scholar]
- 13.Blank V., Andrews N.C. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen T., Huang H.C., Pickett C.B. Transcriptional regulation of the antioxidant response element: activation by Nrf2 and repression by MafK. J. Biol. Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 15.Kimura T., Ivell R., Rust W., Mizumoto Y., Ogita K., Kusui C., Matsumura Y., Azuma C., Murata Y. Molecular cloning of a human MafF homologue, which specifically binds to the oxytocin receptor gene in term myometrium. Biochem. Biophys. Res. Commun. 1999;264:86–92. doi: 10.1006/bbrc.1999.1487. [DOI] [PubMed] [Google Scholar]
- 16.van Oostrom O., de Kleijn D.P., Fledderus J.O., Pescatori M., Stubbs A., Tuinenburg A., Lim S.K., Verhaar M.C. Folic acid supplementation normalizes the endothelial progenitor cell transcriptome of patients with type 1 diabetes: a case-control pilot study. Cardiovasc. Diabetol. 2009;8:47. doi: 10.1186/1475-2840-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Y.T., Qin L.J., Qin X., Xu F. The molecular mechanism of dexamethasone-mediated effect on the blood-brain tumor barrier permeability in a rat brain tumor model. Neurosci. Lett. 2009;452:114–118. doi: 10.1016/j.neulet.2008.12.047. [DOI] [PubMed] [Google Scholar]
- 18.Balda M.S., Flores-Maldonado C., Cereijido M., Matter K. Multiple domains of occludin are involved in the regulation of paracellular permeability. J. Cell. Biochem. 2000;78:85–96. [PubMed] [Google Scholar]
- 19.Komarova Y., Malik A.B. Regulation of endothelial permeability via paracellular and transcellular transport pathways. Annu. Rev. Physiol. 2010;72:463–493. doi: 10.1146/annurev-physiol-021909-135833. [DOI] [PubMed] [Google Scholar]
- 20.Guo J., Cai H., Zheng J., Liu X., Liu Y., Ma J., Que Z., Gong W., Gao Y., Tao W., Xue Y. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Ferrè F., Colantoni A., Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief. Bioinform. 2016;17:106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao S., Zheng J., Liu X., Liu Y., Ruan X., Ma J., Liu L., Wang D., Yang C., Cai H. FXR1 promotes the malignant biological behavior of glioma cells via stabilizing MIR17HG. J. Exp. Clin. Cancer Res. 2019;38:37. doi: 10.1186/s13046-018-0991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Xue Y., Liu X., Zheng J., Shen S., Yang C., Chen J., Li Z., Liu L., Ma J. ZRANB2/SNHG20/FOXK1 axis regulates vasculogenic mimicry formation in glioma. J. Exp. Clin. Cancer Res. 2019;38:68. doi: 10.1186/s13046-019-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamin D., Moroni C. mRNA stability and cancer: an emerging link? Expert Opin. Biol. Ther. 2007;7:1515–1529. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X.J., Wu J., Shi L., Li X.X., Zhu L., Sun X., Qian J.Y., Wang Y., Wei J.F., Ding Q. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. J. Exp. Clin. Cancer Res. 2017;36:149. doi: 10.1186/s13046-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park E., Maquat L.E. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2013;4:423–435. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damas N.D., Marcatti M., Côme C., Christensen L.L., Nielsen M.M., Baumgartner R., Gylling H.M., Maglieri G., Rundsten C.F., Seemann S.E. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F., Teves S.S., Kemp C.J., Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upadhyay K.K., Bhatt A.N., Mishra A.K., Dwarakanath B.S., Jain S., Schatz C., Le Meins J.F., Farooque A., Chandraiah G., Jain A.K. The intracellular drug delivery and anti tumor activity of doxorubicin loaded poly(γ-benzyl l-glutamate)-b-hyaluronan polymersomes. Biomaterials. 2010;31:2882–2892. doi: 10.1016/j.biomaterials.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 30.Luissint A.C., Federici C., Guillonneau F., Chrétien F., Camoin L., Glacial F., Ganeshamoorthy K., Couraud P.O. Guanine nucleotide-binding protein Gαi2: a new partner of claudin-5 that regulates tight junction integrity in human brain endothelial cells. J. Cereb. Blood Flow Metab. 2012;32:860–873. doi: 10.1038/jcbfm.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.A., Tang Z.Z., Black D.L. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin C.L., Duvall J.A., Ilkin Y., Simon J.S., Arreaza M.G., Wilkes K., Alvarez-Retuerto A., Whichello A., Powell C.M., Rao K. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 33.Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.H., Shiue L., Ares M., Jr., Mody I., Black D.L. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge X., Yamamoto S., Tsutsumi S., Midorikawa Y., Ihara S., Wang S.M., Aburatani H. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Sengupta N., Yau C., Sakthianandeswaren A., Mouradov D., Gibbs P., Suraweera N., Cazier J.B., Polanco-Echeverry G., Ghosh A., Thaha M. Analysis of colorectal cancers in British Bangladeshi identifies early onset, frequent mucinous histotype and a high prevalence of RBFOX1 deletion. Mol. Cancer. 2013;12:1. doi: 10.1186/1476-4598-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada M., Kanai F., Tanaka Y., Sanada M., Nannya Y., Tateishi K., Ohta M., Asaoka Y., Seto M., Imazeki F. Prognostic significance of genetic alterations detected by high-density single nucleotide polymorphism array in gastric cancer. Cancer Sci. 2010;101:1261–1269. doi: 10.1111/j.1349-7006.2010.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klorin G., Rozenblum E., Glebov O., Walker R.L., Park Y., Meltzer P.S., Kirsch I.R., Kaye F.J., Roschke A.V. Integrated high-resolution array CGH and SKY analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines. Cancer Genet. 2013;206:191–205. doi: 10.1016/j.cancergen.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung H.C., Baggerly K.A., Tsavachidis S., Bachinski L.L., Neubauer V.L., Nixon T.J., Aldape K.D., Cote G.J., Krahe R. Global analysis of aberrant pre-mRNA splicing in glioblastoma using exon expression arrays. BMC Genomics. 2008;9:216. doi: 10.1186/1471-2164-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung D., Manias J.L., Stewart D.J., Nag S. Decreased junctional adhesion molecule-A expression during blood-brain barrier breakdown. Acta Neuropathol. 2008;115:635–642. doi: 10.1007/s00401-008-0364-4. [DOI] [PubMed] [Google Scholar]
- 40.Liu L.B., Xue Y.X., Liu Y.H., Wang Y.B. Bradykinin increases blood-tumor barrier permeability by down-regulating the expression levels of ZO-1, occludin, and claudin-5 and rearranging actin cytoskeleton. J. Neurosci. Res. 2008;86:1153–1168. doi: 10.1002/jnr.21558. [DOI] [PubMed] [Google Scholar]
- 41.Yu H., Xue Y., Wang P., Liu X., Ma J., Zheng J., Li Z., Li Z., Cai H., Liu Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6:e303. doi: 10.1038/oncsis.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Q., Yu Y., Li N., Jing W., Zhou H., Qiu S., Liang C., Yu M., Tu J. Identification of long non-coding RNA 00312 and 00673 in human NSCLC tissues. Mol. Med. Rep. 2017;16:4721–4729. doi: 10.3892/mmr.2017.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H., Zheng J., Liu X., Xue Y., Shen S., Zhao L., Li Z., Liu Y. Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338-3p-mediated EGFL7 expression change. Front. Mol. Neurosci. 2017;10:301. doi: 10.3389/fnmol.2017.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Ye S., Wang J., Gu Z., Zhang Y., Zhang C., Ma X. HuR stabilizes lnc-Sox5 mRNA to promote tongue carcinogenesis. Biochemistry (Mosc.) 2017;82:438–445. doi: 10.1134/S0006297917040046. [DOI] [PubMed] [Google Scholar]
- 45.Chai Y., Liu J., Zhang Z., Liu L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016;5:1588–1598. doi: 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alkallas R., Fish L., Goodarzi H., Najafabadi H.S. Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat. Commun. 2017;8:909. doi: 10.1038/s41467-017-00867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 48.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., Ma C., Zhang L., Li N., Zhang X., He J., He R., Shao M., Wang J., Kang L., Han C. LncRNAAC132217.4, a KLF8-regulated long non-coding RNA, facilitates oral squamous cell carcinoma metastasis by upregulating IGF2 expression. Cancer Lett. 2017;407:45–56. doi: 10.1016/j.canlet.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho H., Kim K.M., Han S., Choe J., Park S.G., Choi S.S., Kim Y.K. Staufen1-mediated mRNA decay functions in adipogenesis. Mol. Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Shao T., Song P., Hua H., Zhang H., Sun X., Kong Q., Wang J., Luo T., Jiang Y. Gamma synuclein is a novel Twist1 target that promotes TGF-β-induced cancer cell migration and invasion. Cell Death Dis. 2018;9:625. doi: 10.1038/s41419-018-0657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye X., Li Y., Huang Q., Yu Y., Yuan H., Wang P., Wan D., Gu J., Huo K., Li Y.Y., Lu H. The novel human gene MIP functions as a co-activator of hMafF. Arch. Biochem. Biophys. 2006;449:87–93. doi: 10.1016/j.abb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Ubertini V., Norelli G., D’Arcangelo D., Gurtner A., Cesareo E., Baldari S., Gentileschi M.P., Piaggio G., Nisticò P., Soddu S. Mutant p53 gains new function in promoting inflammatory signals by repression of the secreted interleukin-1 receptor antagonist. Oncogene. 2015;34:2493–2504. doi: 10.1038/onc.2014.191. [DOI] [PubMed] [Google Scholar]
- 55.Groothuis D.R. The blood-brain and blood-tumor barriers: a review of strategies for increasing drug delivery. Neuro-oncol. 2000;2:45–59. doi: 10.1093/neuonc/2.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y., Wang P., Liu Y.H., Shang X.L., Chen L.Y., Xue Y.X. DT270–326, a truncated diphtheria toxin, increases blood-tumor barrier permeability by upregulating the expression of caveolin-1. CNS Neurosci. Ther. 2016;22:477–487. doi: 10.1111/cns.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W., Mojsilovic-Petrovic J., Andrade M.F., Zhang H., Ball M., Stanimirovic D.B. The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. FASEB J. 2003;17:2085–2087. doi: 10.1096/fj.02-1131fje. [DOI] [PubMed] [Google Scholar]
- 58.Ma J., Wang P., Liu Y., Zhao L., Li Z., Xue Y. Krüppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5. J. Cell. Physiol. 2014;229:916–926. doi: 10.1002/jcp.24523. [DOI] [PubMed] [Google Scholar]
- 59.Li C., Li S., Kong D.H., Meng X., Zong Z.H., Liu B.Q., Guan Y., Du Z.X., Wang H.Q. BAG3 is upregulated by c-Jun and stabilizes JunD. Biochim. Biophys. Acta. 2013;1833:3346–3354. doi: 10.1016/j.bbamcr.2013.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.