Abstract
The prognosis of breast cancer brain metastasis (BCBM) is extremely poor due to its resistance to conventional therapy. Elucidation of the molecular mechanisms of BCBM could contribute to the development of new therapeutic targets. In this study, we isolated RNA samples from primary breast cancer or BCBM, and then performed mRNA profiling. We determined that SOX2 is associated with the occurrence of BCBM and could be a predictor of BCBM. High levels of SOX2 were significantly associated with decreasing BCBM-free survival in patients. Overexpression of SOX2 in breast cancer cells enhanced cancer cell adhesion to brain microvascular endothelial cells, transendothelial migration, and in vitro blood-brain barrier (BBB) migration, whereas silencing SOX2 inhibited these events. SOX2 can increase cancer cell migration and BBB permeability by upregulating FSCN1 and HBEGF, thereby promoting BBB migration of breast cancer cells. Moreover, high levels of FSCN1 and HBEGF were significantly associated with reducing BCBM-free survival in breast cancer patients. Further study indicated that SOX2 mediates the expression of HBEGF and FSCN1 by activating AKT and β-catenin signaling pathways. Additionally, in vivo experiments showed that SOX2 promotes the development of BCBM. This study demonstrated that SOX2 promotes BCBM by upregulating the expression of FSCN1 and HBEGF.
Keywords: breast cancer, brain metastasis, SOX2, FSCN1, HBEGF
Graphical Abstract
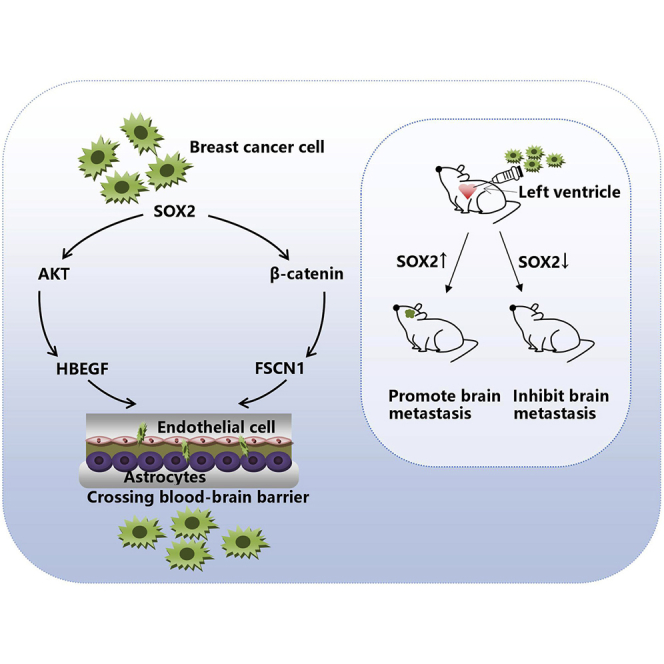
Xiao et al. identify that SOX2 is associated with the occurrence of breast cancer brain metastasis after microarray screening and extended sample validation. They demonstrate that SOX2 upregulates the expression of HBEGF and FSCN1 by activating the β-catenin and AKT signaling pathways, which together promote breast cancer brain metastasis.
Introduction
Breast cancer is the most common malignant tumor in women. The main cause of death of breast cancer is distant metastasis. Common distant metastatic sites of breast cancer include the bone, the lung, the liver, and the brain.1 Patients with breast cancer brain metastasis (BCBM) have an incidence of approximately 15% and the worst prognosis.2 HER2-positive and triple-negative breast cancer are the high-risk subtypes of brain metastasis, and the incidence of brain metastasis in these two subtypes can be as high as 30%–50%. The median survival time of patients with BCBM is only 7.5–15 months.3,4 Thus, BCBM is a serious threat to the life and health of patients.
The control of extracranial diseases has improved with the advancement of breast cancer treatment strategies. The incidence of BCBM continues to increase with prolonged patient survival.5 In metastatic brain tumors, the incidence of breast cancer-derived brain metastasis ranks second only to that of lung cancer.6 Brain metastasis is a major problem in breast cancer because the therapeutic effect of BCBM is very unsatisfactory.1 Therefore, it is urgently needed to understand the molecular mechanisms of how metastatic cancer cells penetrate, colonize, and grow in the brain. This understanding will help identify new targets for the treatment of BCBM.
The blood-brain barrier (BBB) consists of tightly connected brain microvascular endothelial cells (BMECs) and astrocytes, which can limit circulating tumor cells into the brain parenchyma.7,8 Studies on an animal model of brain metastases have indicated that circulating metastatic cancer cells first adhere to BMECs and then cross the BBB through multiple steps, which is a critical event in brain metastasis.9 The interaction between BMECs and breast cancer cells is regulated by multiple molecules. For example, αB-crystallin can increase the adhesion of breast cancer cells to BMECs.10 Animal experiments have shown that cyclooxygenase-2 (COX-2), the epidermal growth factor receptor (EGFR) ligand HBEGF, and the α-2,6-sialyltransferase ST6GALNAC5 can increase the ability of tumor cells to cross the BBB.11
Previous studies have suggested that brain metastatic tumors are very different from their primary tumors in terms of gene mutations and expression.12,13 In addition, some studies have shown that therapeutically induced mutations can be present in metastatic tumors rather than in primary tumors.14,15 Therefore, we explored the possible molecular mechanisms of BCBM by analyzing differences in gene expression between primary breast cancer and its brain metastasis. We screened differentially expressed genes between BCBM and paired primary tumors. We found that SOX2 is a major gene in BCBM that is upregulated compared with that in primary tumors. Importantly, high levels of SOX2 were significantly associated with brain metastasis-free survival (BMFS) in breast cancer patients. We demonstrated that SOX2 upregulates the expression of two essential genes, HBEGF and FSCN1, that induce brain metastasis by activating the β-catenin and AKT signaling pathways, which together promote increased permeability of the BBB and tumor cell infiltration in the brain.
Results
Overexpression of SOX2 in BCBM
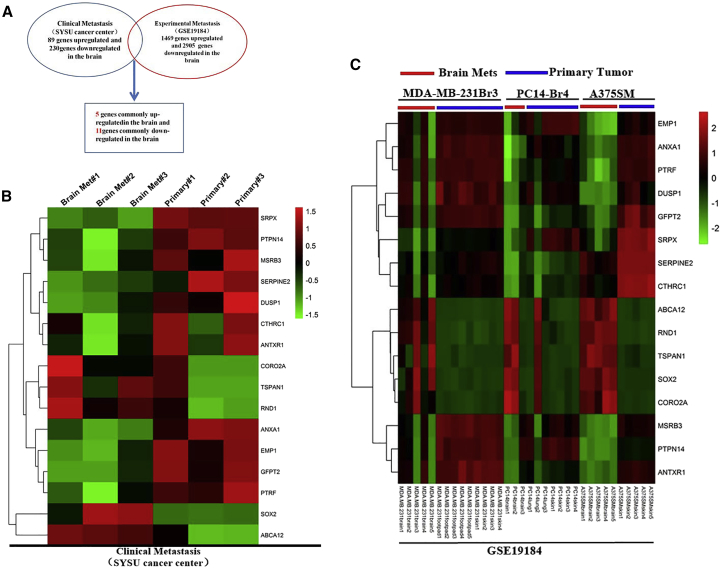
To identify genes that specifically participate in the progression of BCBM, we obtained clinical specimens of primary breast cancer and metastatic lesions in the brain. Primary breast cancer and brain metastasis were confirmed by pathological examination. We isolated total RNA only from the tumor area to avoid potential mix with normal tissue. Affymetrix 3′IVT expression profile chips were used to analyze total RNA and then normalized to Robust Multichip Average (RMA). Our analysis showed that the expression of 89 genes was significantly upregulated, whereas 230 genes were downregulated in BCBM (fold change ≥ 2; p < 0.05; false discovery rate [FDR] < 0.05; Figure 1A). Gene Ontology (GO) enrichment analysis (Figure S1A) showed that the functional enrichment of differential genes is mainly in extracellular matrix structural components, collagen, Wnt protein, Platelet Derived Endothelial Growth Factor (PDEGF), fibronectin, and the like. Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway analysis (Figure S1B) suggested that differential genes were enriched in malignant tumor-associated signaling pathways, focal adhesions, and extracellular matrix receptor binding. To obtain insights into how disseminated tumor cells acquire the essential features of successful brain growth from primary sites, we analyzed the public gene expression profiles of brain-specific metastases in nude mice injected with various human cancer cells (MDA-MB-231 cells for breast cancer, PC14 cells for lung cancer, and A375SM cells for melanoma).
Figure 1.
Schematic of the Microarray Analysis
(A) Compared with primary breast tumors (Sun Yat-sen University Cancer Center [SYSUCC]), patients’ brain metastases showed differential gene expression, of which 89 genes were significantly upregulated. Cancer cells were injected into immunodeficient mice to produce in situ primary tumors (MDA-MB-231 cells for breast tumors, PC14 for lung tumors, and A375SM for melanoma) and experimental brain metastases (all three cell lines). Brain metastases derived from these three cancer cell lines showed 1,469 commonly upregulated genes compared with their respective primary tumors (GEO: GSE19184). SOX2 is one of the only five commonly increased genes in the brain metastases of two datasets. (B) Heatmap showing the expression of 5 commonly upregulated and 11 commonly downregulated genes in clinical brain metastases (SYSUCC data). (C) Heatmap showing the expression of 5 commonly upregulated and 11 commonly downregulated genes in experimental brain metastases (GEO: GSE19184 data).
As shown in the screening flowchart in Figure 1A, we first extracted the GEO: GSE19184 data from the GEO database. The GSE19184 data contain differentially expressed gene profiles between brain metastasis and primary tumors of various human malignancies in animal models. We analyzed the differentially expressed genes in our (Sun Yat-sen University Cancer Center [SYSUCC]) cohort and the GEO: GSE19184 cohort, and identified the genes that were significantly different in these two cohorts. Finally, we found 5 upregulated genes (SOX2, ABCA12, RND1, CORO2A, and TSPAN1; Figure S2A) and 11 downregulated genes (ANXA1, EMP1, GFPT2, SRPX, PTPN14, MSRB3, DUSP1, SERPINE2, CTHRC1, ANTXR1, and PTRF; Figure S2B). Figures 1B and 1C shows differential expression of 16 common differentially expressed genes in the clinical brain metastasis group (SYSUCC) and the experimental brain metastasis group (GEO: GSE19184), respectively.
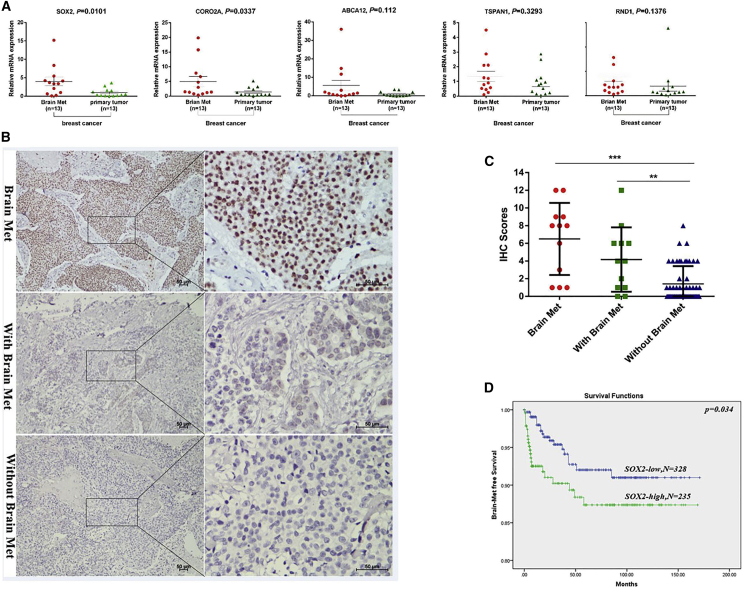
We first performed quantitative real-time polymerase chain (PCR) reaction on five genes that were upregulated in both groups to verify the microarray results. The validation results showed that only SOX2 and CORO2A were significantly higher in brain metastasis than in the primary tumor (Figure 2A). The oncogenic function of SOX2 has been well documented, and its role in cancer metastasis has been reported in several cancer types. Considering that SOX2 is a gene related to tumor metastasis and the difference of its expression is more significant than that of CORO2A, in this study, we mainly explored the role and mechanism of SOX2 in the brain metastasis of breast cancer. In addition, we used immunohistochemistry (IHC) to verify the protein levels of SOX2 in brain lesions, primary tumors with brain metastasis, and primary tumors without brain metastasis. As shown in Figure 2B, the level of SOX2 was indeed higher in the brain metastasis lesion than in the primary lesion, which confirms the results of qPCR. Moreover, the expression level of SOX2 was significantly increased in primary tumors with brain metastasis compared with breast tumors without brain metastasis (Figure 2C). To examine whether SOX2 is associated with brain metastasis in the clinical setting, we performed a meta-analysis of SOX2 expression in patients with or without BCBM using a combination of existing databases (GEO: GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020). We found that high levels of SOX2 were significantly associated with poor BMFS in breast cancer patients (Figure 2D). Our results strongly suggest that SOX2 plays an important role in brain metastasis, and that the expression of SOX2 can be used as a biomarker for predicting the risk for brain metastasis in breast cancer patients.
Figure 2.
SOX2 Expression Is Upregulated in Brain Metastasis of Breast Cancer and Is Associated with Brain Metastasis
(A) Quantitative real-time PCR verified the mRNA expression of five upregulated genes (SOX2, ABCA12, RND1, CORO2A, and TSPAN1). (B) Immunohistochemistry on the expression of SOX2 and the evaluation of the intensity of SOX2 staining in brain metastases, primary tumors with brain metastases, and primary tumors without brain metastasis. (C) IHC scores of brain metastasis tissues and breast primary tissues. ∗∗p < 0.01. (D) Kaplan-Meier analysis of brain metastasis-free survival in 710 breast cancer patients using a combined GEO database (GEO: GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020). Patients were divided into two groups according to the state of expression of SOX2 in the primary tumor.
Overexpression of SOX2 Promotes Breast Cancer Progression
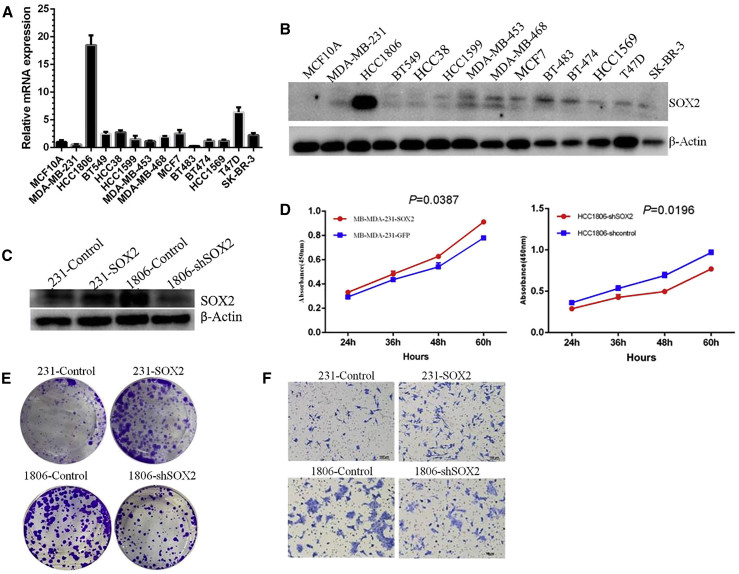
To examine the role of SOX2 in breast cancer progression, we first examined the mRNA and protein levels of SOX2 in established human breast cancer cell lines and found that the HCC1806 cell line showed high basal SOX2 expression, whereas the MDA-MB-231 cell line showed low basal SOX2 expression (Figures 3A and 3B). To investigate the function of SOX2 in breast cancer progression, we selected HCC1806 and MDA-MB-231 cells. We first established two modulated cell lines: 231-SOX2 and 1806-shSOX2. Western blotting confirmed the overexpression of SOX2 in 231 cells and the knockdown of SOX2 in the 1806 cell line (Figure 3C). Cell Counting Kit-8 (CCK-8) and plate cloning experiments suggested that overexpression of SOX2 increased the proliferative potential of 231 cells, whereas knockdown of SOX2 expression in 1806 cells reduced their cell proliferation activity (Figures 3D and 3E). Transwell invasion assays (Figure 3F) demonstrated that overexpression of SOX2 promoted migration and invasion of breast cancer cells, and knockdown of SOX2 expression inhibited these events.
Figure 3.
SOX2 Promotes the Proliferation and Invasion of Breast Cancer Cells In Vitro
(A) qPCR assay on the mRNA expression of SOX2 in a group of breast cancer cells and MCF10A epithelial cells. (B) Western blot analysis on the protein expression of SOX2 in a group of breast cancer cells and MCF10A epithelial cells. (C) Western blot analysis of SOX2 overexpression in 231 cells and SOX2 knockdown by small hairpin RNA (shRNA) in 1806 cells. (D) CCK-8 assays on the proliferation of 231-SOX2 cells, 1806-shSOX2 cells, and control cells. (E) Colony formation assays on the proliferation of 231-SOX2 cells, 1806-shSOX2 cells, and control cells. (F) Transwell assays for the invasive ability of 231-SOX2 cells, 1806-shSOX2 cells, and control cells.
SOX2 Promotes Adhesion of Breast Cancer Cells to the Brain Endothelium, Transendothelial Migration, and In Vitro BBB Migration
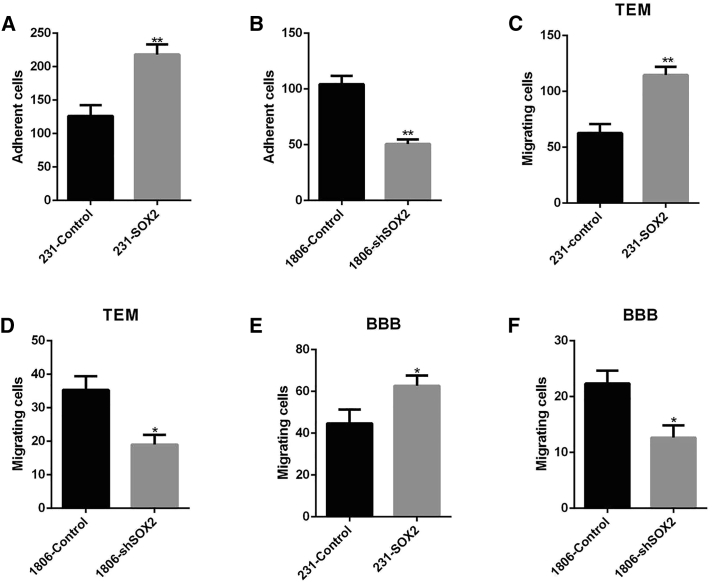
Crossing the BBB is a key step in brain metastasis. In 231 cells, we observed that SOX2 overexpression significantly increased the adhesion of 231 cells to HBMECs (Figure 4A), enhanced transendothelial migration through human brain microvascular endothelial cells (HBMECs) (Figure 4B), and enhanced BBB migration in vitro (Figure 4C). Compared with 1806-control cells, 1806-shSOX2 cells showed significantly attenuated adhesion to HBMECs (Figure 4D), trans-cerebral vascular endothelial migration (Figure 4E), and in vitro BBB migration (Figure 4F). Taken together, these findings indicated that SOX2 promotes the adhesion of breast cancer cells to HBMECs, transendothelial migration, and migration across the BBB in vitro.
Figure 4.
SOX2 Overexpression in Breast Cancer Cells, Adhesion to Brain Endothelium, Transendothelial Migration, and In Vitro BBB Migration
(A) Adhesion of 231-SOX2 and 231-Control to HBMECs. Cells were seeded on confluent HBMECs for 2 h, and attached EGFP-positive cells were scored (mean ± SEM; n = 3; ∗∗p < 0.01). (B) Adhesion of 1806-shSOX2 and 1806-Control to HBMECs. Cells were seeded on confluent HBMECs for 2 h, and attached EGFP-positive cells were scored (mean ± SEM; n = 3; ∗∗p < 0.01). (C) Transendothelial migration (TEM) of 231-SOX2 and 231-Control cells at 24 h (mean ± SEM; n = 3; ∗∗p < 0.01). (D) Transendothelial migration (TEM) of 1806-shSOX2 and 1806-Control cells at 24 h (mean ± SEM; n = 3; ∗p < 0.05). (E) In vitro blood-brain barrier (BBB) migration. The 231-SOX2 and 231-Control cells transferred by two cell layers (HBMECs and astrocytes) were scored at 48 h (mean ± SEM; n = 3; ∗p < 0.05). (F) In vitro blood-brain barrier (BBB) migration. The 1806-shSOX2 and 1806-Control cells transferred by two cell layers (HBMECs and astrocytes) were scored at 48 h (mean ± SEM; n = 3; ∗p < 0.05).
SOX2 Enhances the Expression of the FSCN1 and HBEGF Genes
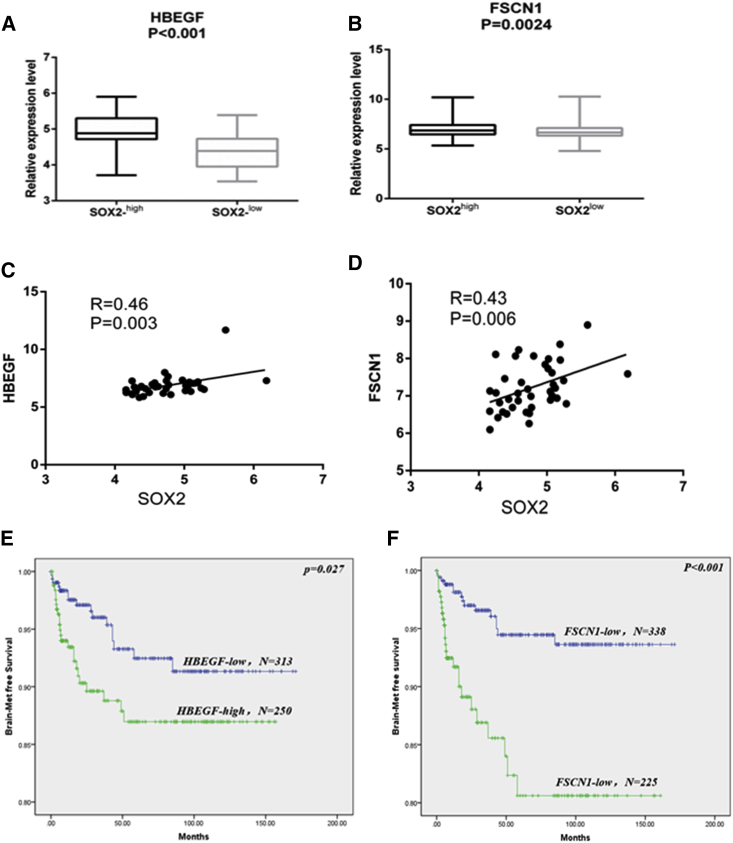
Bos et al.16 defined 17 genes associated with the BM signature (GEO: GSE12237). The GEO: GSE12237 data included 47 breast cancer samples with BM. These 17 genes include 13 upregulated genes (ANGPTL4, PLOD2, COL13A1, PTGS2, PELI1, MMP1, B4GALT6, HBEGF, CSF3, RGC32, LTBP1, FSCN1, and LAMA4) and 4 downregulated genes (TNFSF10, RARRES3, SCNN1A, and LAMA4). To further investigate how SOX2 regulates BM in breast cancer, we first investigated the association of SOX2 and these 17 BCBM-associated genes. In the GEO database, we found that two genes (HBEGF and FSCN1) showed the strongest correlation with SOX2 expression levels (Figures 5A and 5B). Correlation analysis showed that HBEGF or FSCN1 was significantly positively correlated with SOX2 in brain metastasis (Figures 5C and 5D).
Figure 5.
The Expression of SOX2 in Brain Metastases Is Associated with the Expression of the Brain Metastatic Genes HBEGF and FSCN1
(A) HBEGF was significantly increased in lesions with high SOX2 expression. (B) FSCN1 was significantly increased in lesions with high SOX2 expression. (C) Significant correlation between HBEGF and SOX2 expression in brain metastases. (D) Significant correlation between FSCN1 and SOX2 expression in brain metastases. (E) Kaplan-Meier analysis showed that HBEGF expression was significantly associated with brain metastasis-free survival. (F) Kaplan-Meier analysis showed FSCN1 expression significantly associated with brain metastasis-free survival.
In addition, when we used meta-data (GEO: GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020) for BMFS of breast cancer patients, we found that high expression of HBEGF and FSCN1 was significantly associated with BMFS (Figures 5E and 5F). These results indicated that FSCN1 and HBEGF can also effectively predict BMFS.
FSCN1 and HBEGF Are Overexpressed in Brain Metastatic Tumors and Breast Tumors with Breast Metastasis
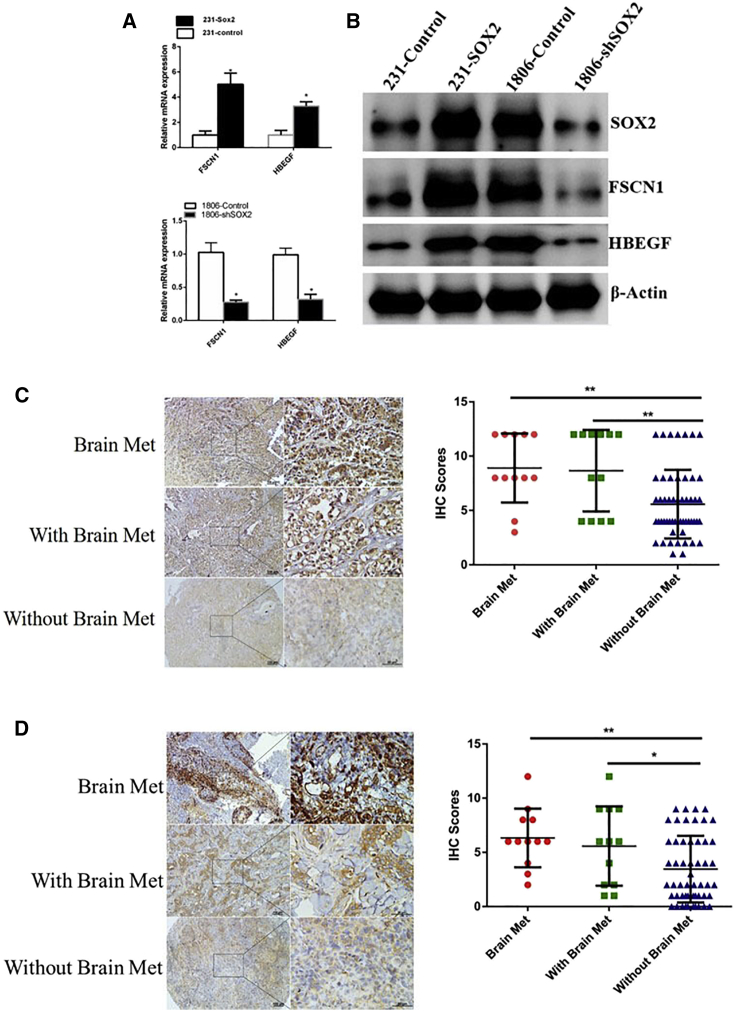
We found that overexpression of SOX2 in 231 cells also upregulated the expression of HBEGF and FSCN1, whereas knockdown of SOX2 in 1806 cells reduced the expression of these two genes (Figures 6A and 6B). This result indicated that SOX2 may regulate HBEGF and FSCN1 expression. To further investigate the relationship between the expression of FSCN1 and HBEGF and BCBM in the clinical setting, we performed immunohistochemical analysis of FSCN1 and HBEGF in breast tumors and brain metastasis. As shown in Figures 6C and 6D, the expression levels of FSCN1 and HBEGF in brain metastasis and primary lesions with brain metastasis were significantly higher than those of breast cancer patients without brain metastasis. Therefore, HBEGF and FSCN1 can be used as biomarkers for predicting the risk for brain metastasis in breast cancer patients.
Figure 6.
SOX2 Promotes Brain Metastasis by Regulating HBEGF and FSCN1
(A) Modulation of SOX2 expression in 231 or 1806 cells resulted in changes in the mRNA levels of HBEGF and FSCN1. (B) Modulation of SOX2 expression in 231 or 1806 cells resulted in changes in the protein levels of HBEGF and FSCN1. (C) Representative immunohistochemical (IHC) staining images and scores of HBEGF expressed in brain metastases, primary lesions with brain metastases, and primary lesions without brain metastasis. ∗∗p < 0.01. (D) Representative IHC staining images and scores of FSCN expressed in brain metastases, primary lesions with brain metastases, and primary lesions without brain metastasis. ∗∗p < 0.01, ∗p < 0.05.
SOX2 Upregulates the Expression of FSCN1 and HBEGF by Activating the β-Catenin and AKT Signaling Pathways
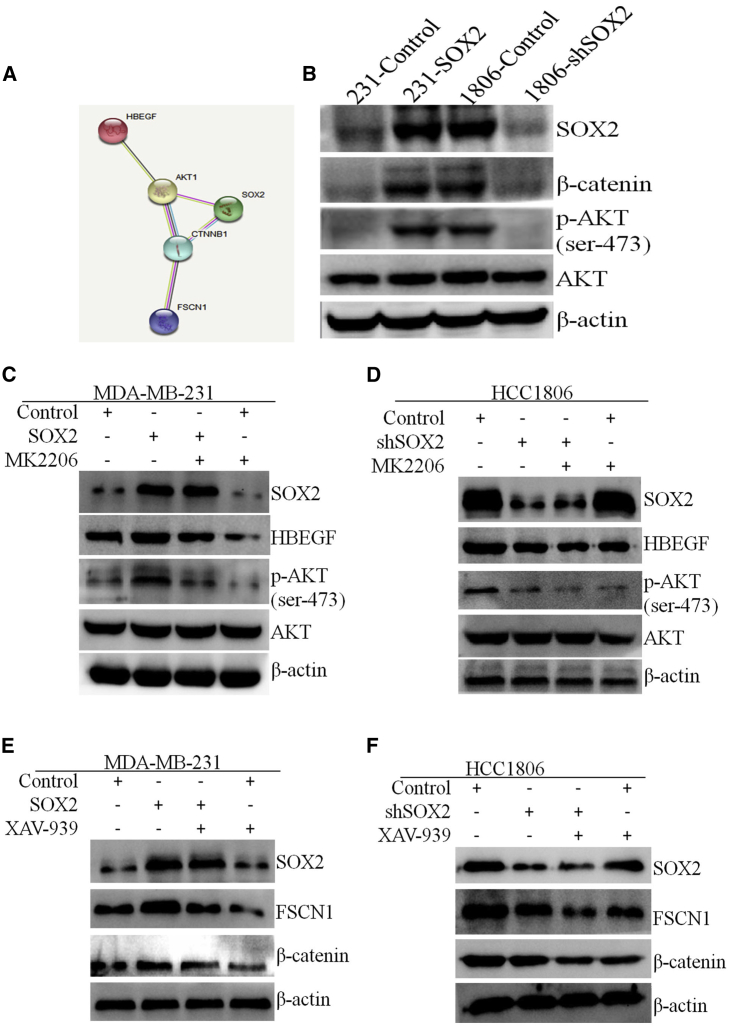
Several studies have reported that Wnt/β-catenin and PI3K/AKT signaling pathways are involved in breast cancer metastasis. Some studies have shown that SOX2 promotes the progression of malignant tumors through Wnt/β-catenin and PI3K/AKT signaling pathways.17, 18, 19, 20, 21 Consistent with these findings, overexpression of SOX2 in 231 cells induced activation of AKT and β-catenin, whereas knockdown of SOX2 in 1806 cells inhibited the AKT and β-catenin signaling pathways (Figure 7B). Based on the STRING database (https://string-db.org), SOX2 can regulate FSCN1 and HBEGF by activating the β-catenin and AKT pathways (Figure 7A). To further investigate the role of SOX2 and AKT signaling in the regulation of HBEGF expression, we used the AKT inhibitor MK2206 to treat breast cancer cells with different levels of SOX2 to observe the effect of MK2206 on SOX2, AKT, and HBEGF. Western blot analysis showed that MK2206 treatment reduced the expression of p-AKT and HBEGF in cells that overexpressed SOX2, whereas the total AKT level did not change significantly (Figures 7C and 7D). Similarly, to observe the role of SOX2 and β-catenin signaling pathways in the regulation of FSCN1 expression, we used XAV-939, a β-catenin inhibitor, to treat breast cancer cells with different levels of SOX2 expression. Western blot analysis showed that XAV-939 reduced the expression of β-catenin and FSCN1 in cells that overexpressed SOX2, but there was no significant change in SOX2 expression (Figures 7E and 7F).
Figure 7.
SOX2 Regulates the Up-Expression of FSCN1 and HBEGF by Activating the AKT and β-Catenin Signaling Pathways
(A) The protein interaction network in the STRING database (https://string-db.org) shows the relationship among SOX2, AKT, β-catenin, FSCN1, and HBEGF. (B) AKT and β-catenin expression in 231-SOX2 and 1806-shSOX2 and the respective control cells. (C) Detection of HBEGF and AKT expression in 231 cells after 72 h of treatment with SOX2 and/or MK2206. (D) Detection of HBEGF and AKT expression in 1806 cells after 72 h of treatment with SOX2 and/or MK2206. (E) FSCN1 and β-catenin expression in 231 cells after 72 h of treatment with SOX2 and/or XAV-939. (F) FSCN1 and β-catenin expression in 1806 cells after 72 h of treatment with SOX2 and/or XAV-939.
SOX2 Promotes BCBM In Vivo
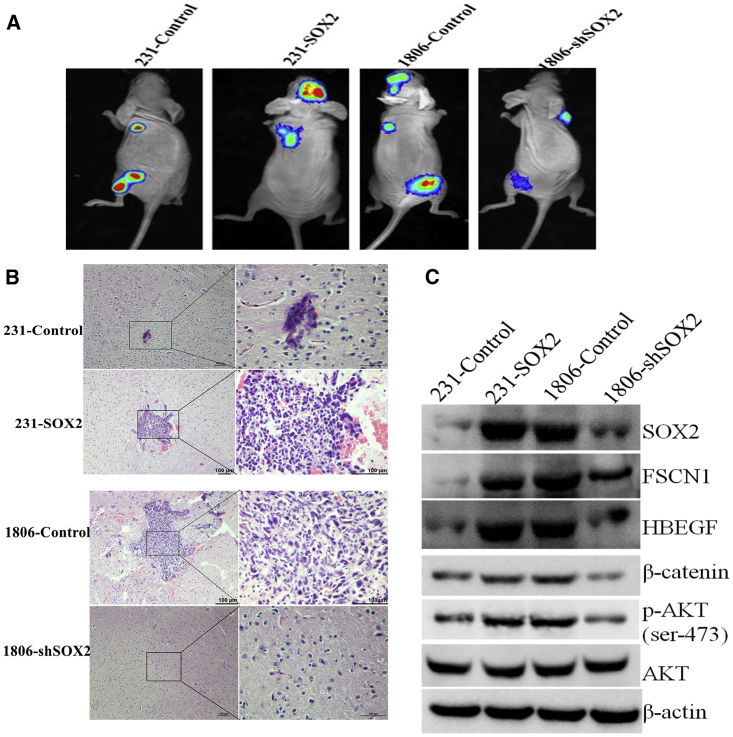
To explore the potential role of SOX2 in brain metastasis in vivo, we injected 231-SOX2 or 1806-shSOX2 cells and their respective control cells into the left ventricle of nude mice and then monitored the growth of metastatic tumors in the brain. After 6 weeks of injection of 231-SOX2 cells, metastatic tumor growth in the brain was observed in four out of five mice. However, only one of the five mice injected with 231-control cells showed BM (Figures 8A and 8B; Figure S3). After 6 weeks of injection of 1806-shSOX2 cells into the left ventricle, no metastatic tumor growth in the brain was observed in any of the five mice. In contrast, brain metastasis (Figures 8A and 8B) was observed in three of five mice in which the left ventricle was injected with 1806-control cells. In addition, western blot analysis showed that the levels of p-AKT, β-catenin, HBEGF, and FSCN1 were higher in tumors that overexpressed SOX2, whereas the levels of p-AKT, β-catenin, HBEGF, and FSCN1 were lower in tumors with low SOX2 expression (Figure 8C).
Figure 8.
In Vivo Study of SOX2 Promotion of Brain Metastasis in Breast Cancer
(A) Bioluminescence images of brain metastatic lesions from representative mice of each experimental group. (B) H&E staining images of brain metastatic lesions from representative mice of each experimental group. (C) Western blot images of SOX2, FSCN1, HBEGF, β-catenin, p-AKT, AKT, and β-actin in brain metastatic lesions from representative mice of each experimental group.
Discussion
BCBM is particularly fatal, and effective treatment is still lacking due to insufficient information about its underlying mechanisms. The process of brain metastasis is highly complex and involves multiple steps. Unlike other organ metastases, cancer cells that metastasize to the brain must cross the BBB. SOX2, an important embryonic stem cell transcription factor, has been reported to be associated with invasive tumor phenotypes in breast cancer, melanoma, and other cancers.11,22, 23, 24 In this study, we demonstrated that SOX2 is a regulator of BCBM and may be involved in the process of metastatic breast cancer cells crossing the BBB.
As one of the key transcription factors that regulate stem cell characteristics, SOX2 plays an important role in the development of the central nervous system (CNS).25,26 Lee et al.26 compared the gene expression profiles of 20 primary breast cancer tissues (with recurrence involved the brain) and 41 brain metastatic tumors, and found that SOX2 was upregulated in brain metastasis. However, the researchers did not validate the expression of SOX2 or explore the role of SOX2 in the brain metastasis of breast cancer. SOX2 plays an important role in the maintenance of embryonic and adult stem cells, as well as cancer stem cells and metastasis. Previous studies have demonstrated the important role of SOX2 in transcriptional regulation, cell proliferation, and migration, suggesting the potential role of SOX2 in promoting tumor progression.27,28 Consistent with previous studies, this study confirmed the clinical significance of SOX2 as an independent prognostic marker for breast cancer patients. More interestingly, patients with high expression of SOX2 in primary breast tumors have shorter BMFS, which indicates that SOX2 may be used to identify patients at high risk for brain metastases. Functional experiments showed that SOX2 overexpression can strongly promote the invasion and metastasis, whereas SOX2 knockdown can effectively inhibit the invasion and metastasis of breast cancer. These results revealed the role of SOX2 in promoting breast cancer progression and participating in BCBM.
Under normal circumstances, the BBB protects the brain from tumor cells. Successfully crossing the BBB is considered the first critical step in BCBM. We observed that overexpression of SOX2 promoted breast cancer cells across the in vitro model of BBB and in vivo BCBM, whereas inhibition of SOX2 expression had the opposite effect. These results strongly suggested that SOX2 promotes BBB penetration. However, the specific role of SOX2 in metastatic cancer cells across the BBB is unclear. Bos et al.16 previously identified 17 genes as brain metastatic signatures. By using an existing cohort database that included 47 brain metastatic breast cancer patient samples, we found that two of these genes (FSCN1 and HBEGF) were significantly associated with SOX2 expression. Furthermore, when we used the existing data in the GEO database to examine patient survival, we found that the expression of FSCN1 and HBEGF was significantly associated with BMFS. These results indicated that HBEGF and FSCN1 play important roles in BCBM. We also examined the expression of HBEGF and FSCN1 in clinical samples of brain metastasis lesions and primary tumors by IHC, and found that HBEGF and FSCN1 were significantly upregulated in brain metastasis and in primary lesions with brain metastasis. HBEGF and FSCN1 are involved in the pathogenesis of breast cancer metastasis and are characteristic genes of brain metastasis. Specifically, HBEGF was reported to link breast cancer cells with brain infiltration and is a mediator of cancer cells passing through the BBB. Increased expression of HBEGF in ER (estrogen receptor)-negative breast tumors is associated with increased metastasis of distant organs and faster recurrence of disease after primary tumor resection.29 HBEGF is also associated with the progression of many other solid tumors, including glioblastoma, neuroblastoma, and ovarian cancer.30, 31, 32 FSCN1 has been reported to support the invasion and metastasis of various cancer cells.33, 34, 35 In this study, HBEGF and FSCN1 were also shown to be potential targets for SOX2 in breast cancer. Ectopic expression of SOX2 upregulated HBEGF and FSCN1 expression, and SOX2 knockdown reduced the expression of HBEGF and FSCN1.
This study has some limitations that should be acknowledged. First, the number of tissue specimens used for gene expression profiling and subsequent verification is relatively small. More samples are needed to verify the findings of this study in the future. Second, we do not yet know the cause of the increased expression of SOX2 in breast cancer brain metastases. It would be interesting to find out the upstream regulation of SOX2 in future research. Third, although we found multiple genes that were differentially expressed between brain metastases and primary foci, the research focused only on SOX2. The role and mechanism of other differentially expressed genes, such as CORO2A, need to be further verified and studied. CORO2A is a member of the actin-binding protein Coronin family, and the interaction between this family member and F-actin plays a role in the regulation of the actin cytoskeleton. The function of CORO2A in tumors has not been determined. Fourth, although this study found that SOX2 can regulate the expression of HBEGF and FSCN1, the specific regulatory mechanism is not clear enough. Last, but not least, based on our results, we are temporarily unable to determine whether SOX2 plays a regulatory role in other organ metastases. Therefore, the role of SOX2 in the metastasis of other organs is unclear.
In summary, this is the first report showing the association between SOX2 and brain metastasis in breast cancer. This study indicated that SOX2 is a potential biomarker for indicating brain metastasis. Because patients with overexpression of SOX2 might be at high risk for brain metastases, these patients are most likely to benefit from brain metastasis prevention measures, such as prophylactic cranial irradiation.
Materials and Methods
Patient Samples
Brain metastatic tumor tissues and paired primary breast cancer tissues were obtained from the Sun Yat-sen University Cancer Center (SYSUCC). Patients were diagnosed and admitted to the Sun Yat-sen University Cancer Center from 2000 to 2014. Three pairs of RNAlater (Invitrogen, Thermo Fisher Scientific)-preserved brain metastases and primary breast cancer tissues were used for Affymetrix mRNA expression profiling. Thirteen pairs of RNAlater-preserved brain metastases and breast cancer tissues were used for quantitative real-time PCR detection. A total of 77 formalin-fixed, paraffin-embedded (FFPE) tissues (12 brain metastases, 12 breast tumors with brain metastases, and 53 breast tumors without brain metastases) were used for IHC detection. Of these tumors, 34 were HR+ (HER2−), 24 were HER2+, and 16 were triple-negative tumors. The subtypes of the remaining three tumors were unknown. All patients signed an informed consent form. This study was approved by the Research Ethics Committee of SYSUCC. The research methodology is in accordance with the standards set by the Declaration of Helsinki.
Microarray Analysis
Three pairs of breast tumors and brain metastatic tumor tissue samples from patients were analyzed using the Affymetrix 3′IVT expression profiling chip. Affymetrix mRNA expression profiling was performed by extracting the total RNA of the sample to be detected for in vitro amplification and biotin labeling. The labeling procedure was performed using the Ambion #1792 cRNA amplification labeling kit. With total RNA, T7 Oligo(dT) Primer containing the T7 promoter sequence was used as a primer, and first-strand cDNA was synthesized using First Strand Enzyme Mix. The double-stranded DNA was synthesized by converting the RNA strand in the DNA-RNA hybrid into second-strand cDNA using Second Strand Enzyme Mix. Using second-strand cDNA as a template, cRNA was synthesized with T7 Enzyme Mix, and biotin was incorporated. The cRNA was purified using magnetic beads to remove impurities such as salts and enzymes, and then the cRNA was quantified. The cRNA was fragmented into a size suitable for hybridization. Finally, chip hybridization, cleaning, staining, and scanning were sequentially performed. AGCC software (Affymetrix GeneChip Command Console Software) was used to save the scanned image of the chip as a .DAT file for analysis. A fold change >2 and p <0.05 in the microarray data were defined as significant differential expression. We further confirmed the microarray results of 13 genes in 13 paired brain metastases and breast cancer tissues by quantitative real-time PCR.
Cell Culture and Treatment
All cell lines, including a normal breast epithelial cell line (MCF-10A), human breast cancer cell lines (MDA-MB-231, HCC1806, BT549, HCC38, HCC1599, MDA-MB-453, MDA-MB-468, MCF-7, BT483, BT474, HCC1569, T47D, and SK-BR-3), and a human embryonic kidney 293T cell line (HEK293T cells), were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were passaged in our laboratory for less than 6 months, maintained according to the supplier’s instructions, and identified for mycoplasma infection and authenticity by DNA fingerprinting prior to use.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). cDNA was synthesized using the PrimeScript RT kit (TaKaRa Bio, Dalian, China), and the total RNA for cDNA synthesis was adjusted to 100–1,000 ng according to the kit’s instructions. The quantitative real-time PCR assays were performed using All-in-One mRNA qRT-PCR Assay Kit (GeneCopoeia, Rockville, MD, USA). The mRNA expression levels were normalized relative to the endogenous β-actin control. The relative expression rate of genes in tumor tissues and cells was calculated by the −ΔCt method. Data are shown by means ± SEM of three independent experiments. Primer information is provided in Table S1.
Western Blot Analysis
Proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer with protease inhibitor and quantified by a BCA kit (Thermo, USA). Proteins in the lysate were separated by 10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membrane was incubated with 5% skim milk powder for 2 h at room temperature to block nonspecific binding. The membrane was then treated with antibody against SOX2 (1:1,000; Cell Signaling Technology [CST]), β-catenin (1:1,000; CST), AKT (1:1,000; CST), HBEGF (1:1,000; Santa Cruz), FSCN1 (1:1,000; Santa Cruz), and β-actin (1:1,000; CST) at 4°C overnight. Target proteins were visualized using secondary antibodies (1:5,000 dilution) and enhanced chemiluminescence (ECL) western blot detection reagents (ECL New England Biolabs, USA).
IHC Analyses
Tissues were formalin-fixed and paraffin-embedded (FFPE). IHC staining was performed according to the instructions provided by the Envision Immunohistochemistry Kit (Dako, Denmark). The tumor tissues were treated with monoclonal antibody against SOX2 (1:100; CST), polyclonal antibodies against HBEGF (1:200; Santa Cruz), and FSCN1 (1:200; Santa Cruz) at 4°C overnight. The IHC score was based on the proportion of immunostaining areas (0%, 0; 1%–25%, 1; 26%–50%, 2; 51%–75%, 3; and 76%–100%, 4) multiplied by the intensity of the staining (0, negative; 1, weak; 2, moderate; and 3, intense). The median score was selected as the cutoff value to define high and low expression.
Animal Experiments
For experimental metastasis assays, female BABL/c nude mice (7–8 weeks) were injected with 1 × 105 or 5 × 105 luciferase-labeled cancer cells in PBS into the left ventricle in a total volume of 100 μL. Six weeks later, the mice were euthanized, and the luciferase signal obtained from the brain metastatic nodules was detected by a small-animal in vivo imaging system (Bruke, MI, USA). Hematoxylin and eosin (H&E) staining was performed to confirm metastatic nodules. All animal experiments were conducted in accordance with the institutional standards guidelines of the Sun Yat-sen University Cancer Center.
Transendothelial Migration and In Vitro BBB Migration
Detection of transendothelial migration was performed according to methods described in previous literature.36 Transwell inserts (Corning-Costar) with 8-μm wells were coated with fibronectin (20 μg/mL) overnight. One day later, 50,000 cancer cells were added to each well. After 24 (231-EGFP) or 48 h (1806-EGFP), non-migrating cancer cells and HBMECs were removed, and EGFP-fluorescent cells were fixed in 10% formalin and scored in every 10× field (Leica MZ10F stereomicroscope). For the BBB assay, primary HBMECs were cocultured with human primary astrocytes on the opposite side of the Transwell insert as described. Twenty thousand primary human astrocytes were plated on the membrane surface and incubated at 37°C for 30 min. The insert was then inverted, as in the endothelial migration assay, and HBMECs were added and grown to confluence. One day after the cells reached confluence, 50,000 cancer cells were seeded on the upper chamber of the insert, and EGFP migration through the endothelial and astrocyte layers was observed in every 10× field at 48 (231-EGFP) or 72 h (1806-EGFP). Fluorescent cancer cells were scored.
CCK-8 Assay
Five thousand cancer cells were seeded in 96-well plates for 48 h. Cell proliferation was measured by the CCK-8 assay (Dojindo) every 24 h.
Statistical Analysis
All statistical analyses were performed using the SPSS 22.0 software (SPSS, Chicago, IL, USA). Data from at least three experiments were tested as mean ± SEM. Differences between groups were compared by Student’s t test. Spearman’s correlation test was used to assess correlations between parameters. Survival curves were calculated by the Kaplan-Meier method and compared with a log rank test. p < 0.05 was considered statistically significant.
Author Contributions
The design of this study was carried out by X.X., H.T., and W.X. Experiments and data analysis were performed on W.X., S.Z., and X.X. Clinical data analysis was performed by X.L., L.Z., and A.Y. Bioinformatics analysis was performed by W.X. and S.Z. The manuscript was written by W.X., S.Z., and X.X. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (grant 81772961 to H.T.; grant 81872152 to X.X.) and the Natural Science Foundation of Guangdong Province (grant 2019A1515011450 to H.T.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.03.001.
Contributor Information
Jian Wang, Email: wangjian2@sysucc.org.cn.
Hailin Tang, Email: tanghl@sysucc.org.cn.
Xiaoming Xie, Email: xiexm@sysucc.org.cn.
Supplemental Information
References
- 1.Pedrosa R.M.S.M., Mustafa D.A., Soffietti R., Kros J.M. Breast cancer brain metastasis: molecular mechanisms and directions for treatment. Neuro Oncol. 2018;20:1439–1449. doi: 10.1093/neuonc/noy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennecke H., Yerushalmi R., Woods R., Cheang M.C., Voduc D., Speers C.H., Nielsen T.O., Gelmon K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 3.Kim J.S., Kim K., Jung W., Shin K.H., Im S.A., Kim H.J., Kim Y.B., Chang J.S., Choi D.H., Park Y.H. Survival outcomes of breast cancer patients with brain metastases: A multicenter retrospective study in Korea. Breast. 2019;49:41–47. doi: 10.1016/j.breast.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasquier D., Darlix A., Louvel G., Fraisse J., Jacot W., Brain E., Petit A., Mouret-Reynier M.A., Goncalves A., Dalenc F. Treatment and outcomes in patients with central nervous system metastases from breast cancer in the real-life ESME MBC cohort. Eur J. Cancer. 2020;125:22–30. doi: 10.1016/j.ejca.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Verduin M., Zindler J.D., Martinussen H.M., Jansen R.L., Croes S., Hendriks L.E., Eekers D.B., Hoeben A. Use of Systemic Therapy Concurrent With Cranial Radiotherapy for Cerebral Metastases of Solid Tumors. Oncologist. 2017;22:222–235. doi: 10.1634/theoncologist.2016-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boire A., Brastianos P.K., Garzia L., Valiente M. Brain metastasis. Nat. Rev. Cancer. 2020;20:4–11. doi: 10.1038/s41568-019-0220-y. [DOI] [PubMed] [Google Scholar]
- 7.Avraham H.K., Jiang S., Fu Y., Nakshatri H., Ovadia H., Avraham S. Angiopoietin-2 mediates blood-brain barrier impairment and colonization of triple-negative breast cancer cells in brain. J. Pathol. 2014;232:369–381. doi: 10.1002/path.4304. [DOI] [PubMed] [Google Scholar]
- 8.Wang S., Liang K., Hu Q., Li P., Song J., Yang Y., Yao J., Mangala L.S., Li C., Yang W. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Invest. 2017;127:4498–4515. doi: 10.1172/JCI91553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kienast Y., von Baumgarten L., Fuhrmann M., Klinkert W.E., Goldbrunner R., Herms J., Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 10.Malin D., Strekalova E., Petrovic V., Deal A.M., Al Ahmad A., Adamo B., Miller C.R., Ugolkov A., Livasy C., Fritchie K. αB−crystallin: a novel regulator of breast cancer metastasis to the brain. Clin Cancer Res. 2014;20 doi: 10.1158/1078-0432.CCR-13-1255. 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng L., Hailin T., Cailu S., Jin W., Bo C., Xiaojia H., Xiaoqing P., Longzhong L. SOX2 Promotes Cell Proliferation and Metastasis in Triple Negative Breast Cancer. Front Pharmacol. 2018;9:942. doi: 10.3389/fphar.2018.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cacho-Díaz B., García-Botello D.R., Wegman-Ostrosky T., Reyes-Soto G., Ortiz-Sánchez E., Herrera-Montalvo L.A. Tumor microenvironment differences between primary tumor and brain metastases. J. Transl. Med. 2020;18:1. doi: 10.1186/s12967-019-02189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulsbergen A.F.C., Claes A., Kavouridis V.K., Ansaripour A., Nogarede C., Hughes M.E., Smith T.R., Brastianos P.K., Verhoeff J.J.C., Lin N.U., Broekman M.L.D. Subtype switching in breast cancer brain metastases: a multicenter analysis. Neuro Oncol. 2020 doi: 10.1093/neuonc/noaa013. Published online January 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyran M., Carbuccia N., Garnier S., Guille A., Adelaïde J., Finetti P., Toulzian J., Viens P., Tallet A., Goncalves A. A Comparison of DNA Mutation and Copy Number Profiles of Primary Breast Cancers and Paired Brain Metastases for Identifying Clinically Relevant Genetic Alterations in Brain Metastases. Cancers (Basel). 2019;11:e665. doi: 10.3390/cancers11050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson S.D., Zheng S., Xiu J., Zhou S., Khasraw M., Brastianos P.K., Kesari S., Hu J., Rudnick J., Salacz M.E. Profiles of brain metastases: Prioritization of therapeutic targets. Int. J. Cancer. 2018;143:3019–3026. doi: 10.1002/ijc.31624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos P.D., Zhang X.H., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A., Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S.J., Lee H.W., Kim H.R., Lee H., Shin C.H., Yun S.I., Lee D.H., Kim D.H., Kim K.K., Joo K.M., Kim H.H. Ubiquitin-specific protease 4 controls metastatic potential through β-catenin stabilization in brain metastatic lung adenocarcinoma. Sci. Rep. 2016;6:21596. doi: 10.1038/srep21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Yang J., Yu Q., Wu H., Liu B., Xiong H., Hu G., Zhao J., Yuan X., Liao Z. Associations between single-nucleotide polymorphisms in the PI3K-PTEN-AKT-mTOR pathway and increased risk of brain metastasis in patients with non-small cell lung cancer. Clin. Cancer Res. 2013;19:6252–6260. doi: 10.1158/1078-0432.CCR-13-1093. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen D.X., Chiang A.C., Zhang X.H., Kim J.Y., Kris M.G., Ladanyi M., Gerald W.L., Massagué J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilarinho S., Erson-Omay E.Z., Mitchell-Richards K., Cha C., Nelson-Williams C., Harmancı A.S., Yasuno K., Günel M., Taddei T.H. Exome analysis of the evolutionary path of hepatocellular adenoma-carcinoma transition, vascular invasion and brain dissemination. J. Hepatol. 2017;67:186–191. doi: 10.1016/j.jhep.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Rhun E., Bertrand N., Dumont A., Tresch E., Le Deley M.C., Mailliez A., Preusser M., Weller M., Revillion F., Bonneterre J. Identification of single nucleotide polymorphisms of the PI3K-AKT-mTOR pathway as a risk factor of central nervous system metastasis in metastatic breast cancer. Eur. J. Cancer. 2017;87:189–198. doi: 10.1016/j.ejca.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Ravindran Menon D., Luo Y., Arcaroli J.J., Liu S., KrishnanKutty L.N., Osborne D.G., Li Y., Samson J.M., Bagby S., Tan A.C. CDK1 Interacts with Sox2 and Promotes Tumor Initiation in Human Melanoma. Cancer Res. 2018;78:6561–6574. doi: 10.1158/0008-5472.CAN-18-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin C.M., Durinck S., Stawiski E.W., Poirier J.T., Modrusan Z., Shames D.S., Bergbower E.A., Guan Y., Shin J., Guillory J. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao S., Chang R.M., Yang M.Y., Lei X., Liu X., Gao W.B., Xiao J.L., Yang L.Y. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63:1256–1271. doi: 10.1002/hep.28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bani-Yaghoub M., Tremblay R.G., Lei J.X., Zhang D., Zurakowski B., Sandhu J.K., Smith B., Ribecco-Lutkiewicz M., Kennedy J., Walker P.R., Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Lee J.Y., Park K., Lee E., Ahn T., Jung H.H., Lim S.H., Hong M., Do I.G., Cho E.Y., Kim D.H. Gene Expression Profiling of Breast Cancer Brain Metastasis. Sci. Rep. 2016;6:28623. doi: 10.1038/srep28623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaal C.M., Bora-Singhal N., Kumar D.M., Chellappan S.P. Regulation of Sox2 and stemness by nicotine and electronic-cigarettes in non-small cell lung cancer. Mol. Cancer. 2018;17:149. doi: 10.1186/s12943-018-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keysar S.B., Le P.N., Miller B., Jackson B.C., Eagles J.R., Nieto C., Kim J., Tang B., Glogowska M.J., Morton J.J. Regulation of Head and Neck Squamous Cancer Stem Cells by PI3K and SOX2. J. Natl. Cancer Inst. 2017;109:djw189. doi: 10.1093/jnci/djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z.N., Sharma V.P., Beaty B.T., Roh-Johnson M., Peterson E.A., Van Rooijen N., Kenny P.A., Wiley H.S., Condeelis J.S., Segall J.E. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014;33:3784–3793. doi: 10.1038/onc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin C.H., Robinson J.P., Sonnen J.A., Welker A.E., Yu D.X., VanBrocklin M.W., Holmen S.L. HBEGF promotes gliomagenesis in the context of Ink4a/Arf and Pten loss. Oncogene. 2017;36:4610–4618. doi: 10.1038/onc.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaviglio A.L., Knelson E.H., Blobe G.C. Heparin-binding epidermal growth factor-like growth factor promotes neuroblastoma differentiation. FASEB J. 2017;31:1903–1915. doi: 10.1096/fj.201600828R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun S., Kwak Y., Nam S.K., Seo A.N., Oh H.K., Kim D.W., Kang S.B., Lee H.S. Ligand-Independent Epidermal Growth Factor Receptor Overexpression Correlates with Poor Prognosis in Colorectal Cancer. Cancer Res. Treat. 2018;50:1351–1361. doi: 10.4143/crt.2017.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Tian T., Li Z.-Y., Wang C.-Y., Deng R., Deng W.-Y., Yang A., Chen Y.-F., Li H. FSCN1 is an effective marker of poor prognosis and a potential therapeutic target in human tongue squamous cell carcinoma. Cell Death Dis. 2019;10:356. doi: 10.1038/s41419-019-1574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao W., Zhang C., Li W., Li H., Sang J., Zhao Q., Bo Y., Luo H., Zheng X., Lu Y. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol. Ther. 2019;27:365–379. doi: 10.1016/j.ymthe.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Zhang S., Pei M., Wu L., Liu Y., Li H., Lu J., Li X. FSCN1 Promotes Epithelial-Mesenchymal Transition Through Increasing Snail1 in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2018;49:1766–1777. doi: 10.1159/000493622. [DOI] [PubMed] [Google Scholar]
- 36.Xing F., Sharma S., Liu Y., Mo Y.-Y., Wu K., Zhang Y.-Y., Pochampally R., Martinez L.A., Lo H., Watabe K. miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-α. Oncogene. 2015;34:4890–4900. doi: 10.1038/onc.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.