Ascoviruses are large DNA viruses that infect insects, inducing a cellular pathology that resembles apoptosis but which differs by causing enormous cellular hypertrophy followed by cleavage of the cell into numerous viral vesicles for replication. Previous EM studies suggest that mitochondria are important for vesicle formation. Transcriptome analyses of Spodoptera frugiperda larvae infected with SfAV-1a showed that mitochondrial transcripts were similar to those from uninfected controls or increased slightly during vesicle formation, especially for ATP6, ATP8 synthase, and NADH dehydrogenase subunits. This pattern resembles that for chronic disease-inducing viruses, which conserve mitochondria, differing markedly from viruses causing short-term viral diseases, which degrade mitochondrial DNA. Though mitochondrial transcript increases were low, our results demonstrate that SfAV-1a alters host mitochondrial expression more than any other virus. Regarding innate immunity, although SfAV-1a destroys most fat body cells, certain immunity genes were highly upregulated (greater than 20-fold), suggesting that these transcripts may originate from other tissues.
KEYWORDS: ascovirus, mitochondrial genes, innate immunity genes, Toll, melanization, phagocytosis, dual transcriptome, cytoskeleton, dsDNA virus, innate immunity, membrane biogenesis, mitochondrial DNA transcription, vesicle formation
ABSTRACT
Ascoviruses are large, enveloped DNA viruses that induce remarkable changes in cellular architecture during which the cell is partitioned into numerous vesicles for viral replication. Previous studies have shown that these vesicles arise from a process resembling apoptosis yet which differs after nuclear lysis in that mitochondria are not degraded but are modified by the virus, changing in size, shape, and motility. Moreover, infection does not provoke an obvious innate immune response. Thus, we used in vivo RNA sequencing to determine whether infection by the Spodoptera frugiperda ascovirus 1a (SfAV-1a) modified expression of host mitochondrial, cytoskeletal, and innate immunity genes. We show that transcripts from many mitochondrial genes were similar to those from uninfected controls, whereas others increased slightly during vesicle formation, including those for ATP6, ATP8 synthase, and NADH dehydrogenase subunits, supporting electron microscopy (EM) data that these organelles were conserved for virus replication. Transcripts from 58 of 106 cytoskeletal genes studied increased or decreased more than 2-fold postinfection. More than half coded for mitochondrial motor proteins. Similar increases occurred for innate immunity transcripts and their negative regulators, including those for Toll, melanization, and phagocytosis pathways. However, those for many antimicrobial peptides, such as moricin, increased more than 20-fold. In addition, transcripts for gloverin-3, spod_x_tox, Hdd23, and lebocin, also antimicrobial, increased more than 20-fold. Interestingly, a phenoloxidase inhibitor transcript increased 12-fold, apparently to interfere with melanization. SfAV-1a destroys most fat body cells by 7 days postinfection, so innate immunity gene transcripts apparently occur in remaining cells in this tissue and possibly other major tissues, namely, epidermis and tracheal matrix.
IMPORTANCE Ascoviruses are large DNA viruses that infect insects, inducing a cellular pathology that resembles apoptosis but which differs by causing enormous cellular hypertrophy followed by cleavage of the cell into numerous viral vesicles for replication. Previous EM studies suggest that mitochondria are important for vesicle formation. Transcriptome analyses of Spodoptera frugiperda larvae infected with SfAV-1a showed that mitochondrial transcripts were similar to those from uninfected controls or increased slightly during vesicle formation, especially for ATP6, ATP8 synthase, and NADH dehydrogenase subunits. This pattern resembles that for chronic disease-inducing viruses, which conserve mitochondria, differing markedly from viruses causing short-term viral diseases, which degrade mitochondrial DNA. Though mitochondrial transcript increases were low, our results demonstrate that SfAV-1a alters host mitochondrial expression more than any other virus. Regarding innate immunity, although SfAV-1a destroys most fat body cells, certain immunity genes were highly upregulated (greater than 20-fold), suggesting that these transcripts may originate from other tissues.
INTRODUCTION
Ascoviruses are viruses of insects characterized by large enveloped virions with double-stranded DNA (dsDNA) genomes ranging from 150 to 180 kbp, primarily infecting larvae of the lepidopteran family Noctuidae (1). Unlike most insect viruses, ascoviruses are unusual in that they are not easily transmitted when fed to host larvae. Instead, various studies show that transmission by female endoparasitic wasps during oviposition is highly effective. Females acquire virions on their ovipositor while laying eggs in infected hosts and then mechanically transmit these to healthy larvae during the next oviposition event. Infection results in a chronic disease that significantly retards development, with larvae often living 4 to 6 weeks prior to death, compared to only 2 weeks for normal growth resulting in pupation. Interestingly, although causing a chronic disease, a few days after infection these viruses produce millions of spherical virion-containing vesicles, referred to as viral vesicles, 2 to 15 μm in diameter, that accumulate in the hemolymph (blood), turning it from translucent green to milky white (1–3).
While this combination of characteristics easily differentiates ascoviruses from all other viruses, their most unique feature is the remarkable changes in cell architecture that result in the formation of viral vesicles. The early stages of viral vesicle morphogenesis resemble apoptosis, with invagination of the nuclear membrane followed by lysis of the nucleus (1–6). In fact, the ascovirus type species, Spodoptera frugiperda ascovirus 1a (SfAV-1a), is the only known virus that encodes a functional executioner caspase, which is synthesized in vitro 9 h postinfection (4) and observed in vivo 24 h postinfection (7). However, the cell does not die as in a typical apoptotic pathway; rather, the cell undergoes significant hypertrophy (∼10 times the original cell size) and divides along cleavage planes into numerous vesicles. Simultaneously, the mitochondria change in shape, divide, and migrate under viral control, accumulating along the cleavage planes, the place where vesicle membranes are synthesized and assembled (1, 3).
The Spodoptera frugiperda mitochondrial genome consists of 15,365 bp (GenBank accession no. KM362176.1; described in reference 8) and has the typical gene order for mitochondrial genomes of noctuid larvae (8, 9), consisting of genes coding for 13 proteins, 22 tRNAs, and 2 rRNAs. The accumulation of dense aggregations of mitochondria during ascovirus infection at sites of membrane synthesis and plasmalemma invagination, along with several lipid-metabolizing enzymes encoded by all ascoviruses (10), provides evidence that these organelles are reorganized to provide energy for virus replication and vesicle formation. Thus, one objective of the present study was to investigate this possibility by examining host mitochondrial genome expression along with that of cytoskeletal genes possibly involved in moving mitochondria to sites of membrane synthesis in fat body tissue infected with SfAV-1a.
In addition, very little is known about host responses at the molecular level to the chronic disease caused by ascoviruses, especially given the circulation of viral vesicles, apparently with viral surface proteins, in the hemolymph for weeks. SfAV-1a primarily infects the fat body, and within a week of infection, this tissue is almost completely destroyed (2). This raises the question of whether the host’s innate immunity pathways, such as Toll, Imd, melanization, and phagocytosis, respond to ascovirus infection. Thus, our second objective was to use in vivo larval RNA sequencing (RNA-Seq) to determine whether these innate immune pathways responded in Spodoptera frugiperda third instars infected with SfAV-1a. Previous evidence for any innate host response to ascovirus infection is limited to conservation of an RNase III endonuclease in all ascovirus species associated with virus evasion of the host RNA interference (RNAi) machinery (11) and a recent transcriptome study of Heliothis virescens ascovirus (HvAV) 3h during infection in Spodoptera exigua larvae (12). The latter study showed activation of certain innate immune gene pathways, such as Toll-like receptor signaling and JAK/STAT.
Here, we show that during the first week of SfAV-1a infection, genes of the S. frugiperda mitochondrial genome were expressed at levels comparable to those in the healthy larvae or slightly higher during vesicle formation, especially genes for ATP synthase and NADH dehydrogenase subunits. Only 58 of 106 genes studied changed by 2-fold or more postinfection compared to the controls. Of these, 32 were identified as coding for kinesin-, dynein-, and myosin-related proteins. Regarding host innate immunity, SfAV-1a triggered upregulation of important antimicrobial genes as the infection advanced and of several key humoral and cellular genes of the Toll, melanization, and phagocytosis pathways. However, we also detected simultaneous increases of certain negative regulators of these pathways, suggesting that this might moderate the host immune response, prolonging the larval life span and thereby extending ascovirus transmission.
RESULTS
Host mitochondrial genome transcriptome pattern.
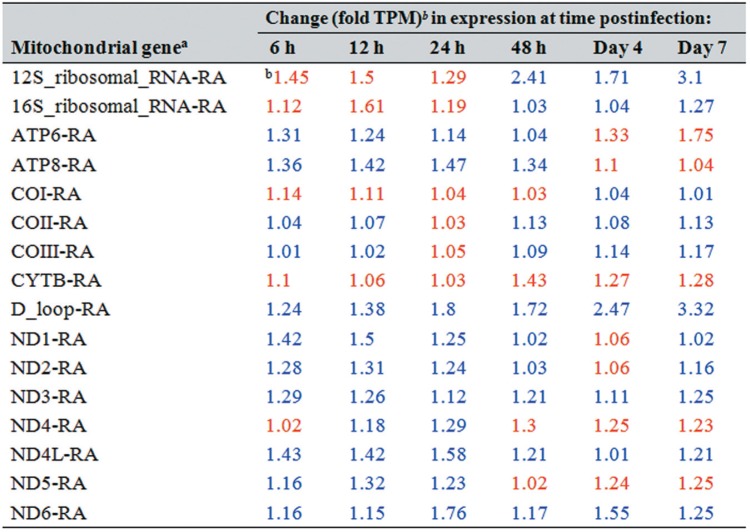
Ascoviruses have significant and unusual effects on mitochondrial shape, size, and distribution within infected cells regardless of the tissue, especially where mitochondria aggregate along cellular cleavage planes as viral vesicle-delimiting membranes form (3). These observations imply that mitochondria are conserved rather than destroyed after the infection and likely provide energy for this de novo membrane synthesis. Our transcriptome data support these observations. For example, both ATP synthase subunits (ATP6 and ATP8, i.e., mitochondrial complex V) were upregulated, especially during the vesicle formation period from 6 to 48 h postinfection (hpi). The upregulation level ranged from 1.04- to 1.47-fold (Table 1). At days 4 and 7 postinfection (p.i.), transcription for both subunits was downregulated (decreases ranged from 1.04- to 1.75-fold). In addition, the increases were typically lower than 2-fold (ranging from 1.01- to 1.76-fold) for transcripts of NADH dehydrogenase subunits 3, 4L, and 6 at all time points postinfection and similarly for subunits 1 and 2 at all time points except for day 4 (Table 1; Fig. 1). NADH dehydrogenase subunits 4 and 5 showed a slight upregulation early in the infection and then, beginning at 48 hpi, decreased at all time points by 1.02 to 1.3-fold. Therefore, the expression of ATP synthase and NADH dehydrogenase genes in infected larvae is best described as varying between stability and upregulation during viral vesicle formation. In contrast, mitochondrial 12S and 16S rRNA transcripts were upregulated late in the infection. In case of 12S rRNA, the increase was greater than 2-fold at 48 h (2.41-fold) and by day 7 p.i. (3.1-fold) (Table 1; Fig. 1), implying active mitochondrial translation after vesicle formation.
TABLE 1.
Changes in Spodoptera frugiperda third-instar larval mitochondrial gene expression levels at different time points after infection with Spodoptera frugiperda ascovirus 1a
The Spodoptera frugiperda mitochondrial genome used in this study is available under accession no. KM362176.1 (8). tRNAs are not listed in this table. ND1 to -6 refer to NADH dehydrogenase subunits, and COI to -III refer to cytochrome c oxidase subunits.
Blue and red type refer to upregulated and downregulated genes, respectively.
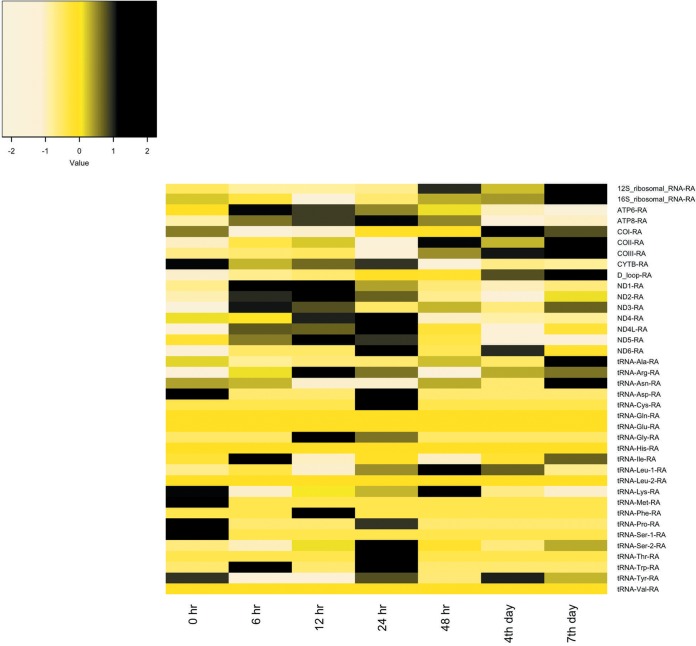
FIG 1.
Heat map representation of mitochondrial genome expression in Spodoptera frugiperda third-instar larvae infected with Spodoptera frugiperda ascovirus 1a. Mitochondrial gene expression is for the 0-h uninfected control compared with time points from 6 h to 7 days postinfection. The heat map colors represent the z-scale of the transcripts per million (TPM) value of replicate expression level averages at each time point. The Spodoptera frugiperda mitochondrial genome used for mapping RNA-Seq reads is available under accession no. KM362176.1 (8). ND1 to -6 refer to NADH dehydrogenase subunits, and COI to -III refer to cytochrome c oxidase subunits.
With respect to cytochrome c oxidase subunits, COII and COIII transcript levels increased, but by less than 2-fold at all times after infection compared to control levels, except for at 24 hpi, at which time expression showed a slight decrease (Table 1; Fig. 1). Alternatively, COI transcription decreased by 1.03- to 1.14-fold at almost all time points postinfection. Moreover, S. frugiperda mitochondrial cytochrome b (complex III) transcripts decreased by 1.03- to 1.43-fold. Most tRNA expression patterns fluctuated at the time points tested (Fig. 1). Thus, although expression of mitochondrial genes was upregulated during vesicle formation, the change was not statistically significant.
Cytoskeleton gene transcription pattern.
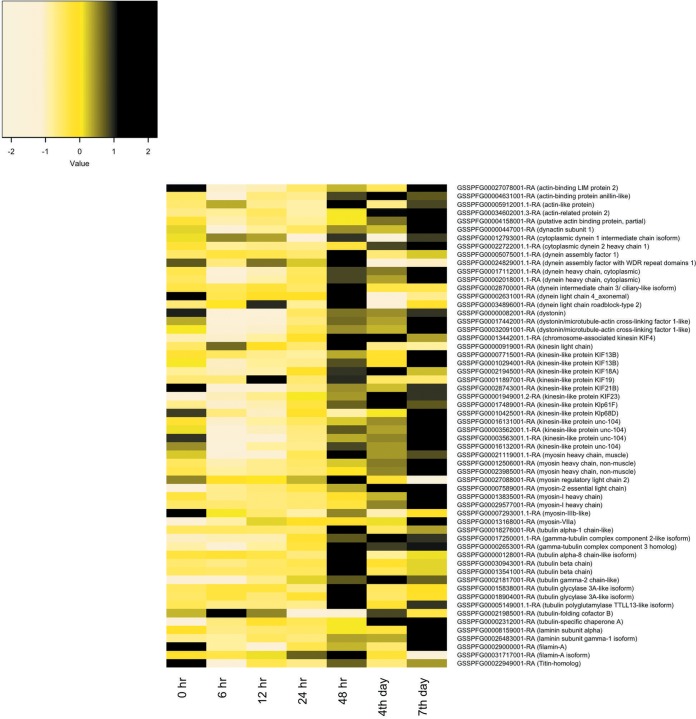
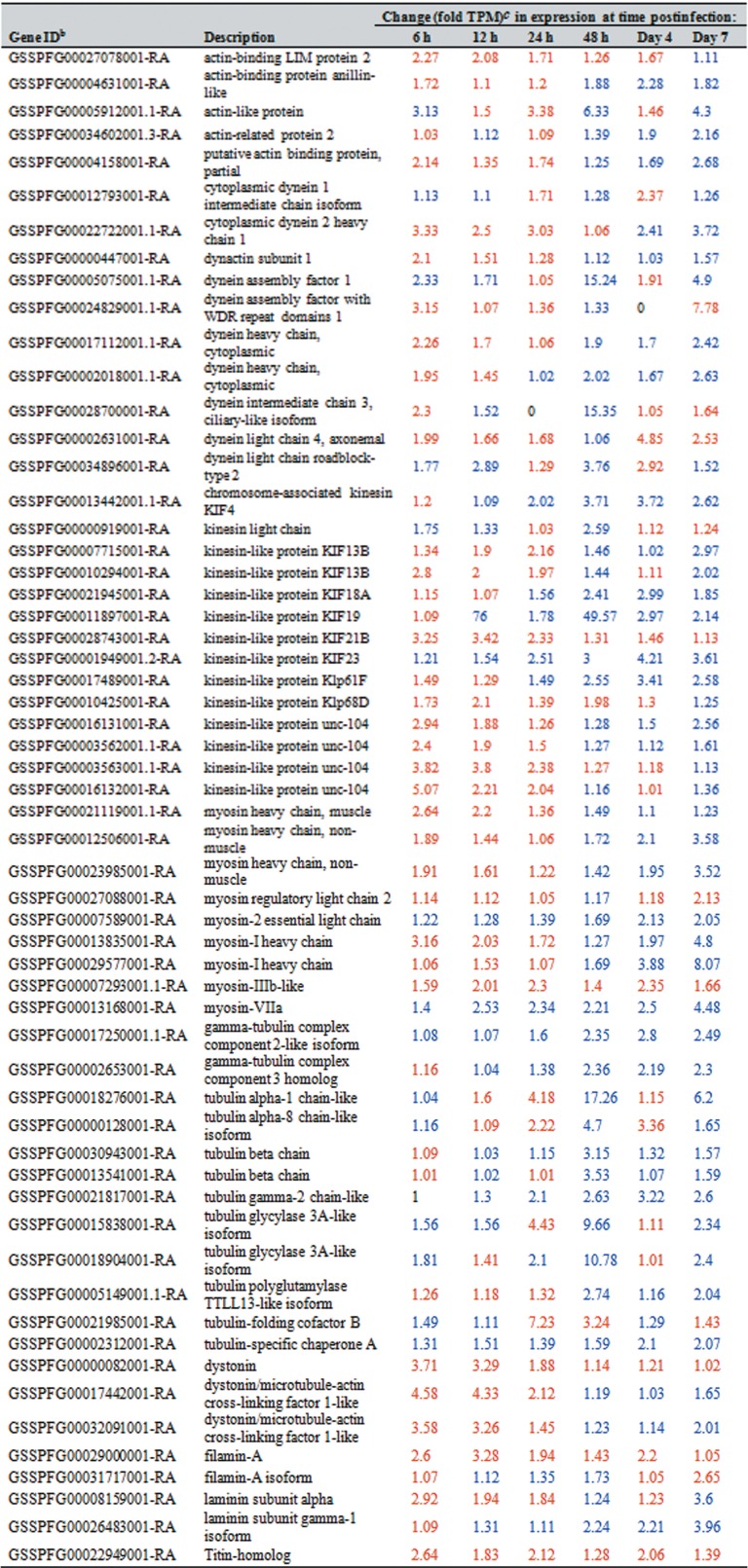
Of the 106 cytoskeleton genes studied, i.e., those coding for actin, actin-related proteins, tubulin, dynein, kinesin, lamin, filamin, myosin, and profilin, 58 changed postinfection by 2-fold or more (Fig. 2). Overall, most of these genes were downregulated from 6 to 24 hpi. However, beginning at 48 hpi through to the day 7 p.i. expression of many of these genes increased. Specifically, the expression of 48, 33, and 46 cytoskeleton genes increased at days 2, 4, and 7 p.i., respectively (Table 2; Fig. 2). Thirty-two of the 58 genes coded for proteins related to kinesin, dynein, and myosin. Kinesin and dynein are known as the main motors of the mitochondria, and myosin is associated with mitochondrial movement over short distances.
FIG 2.
Heat map representation of cytoskeleton gene expression in Spodoptera frugiperda third-instar larvae infected with Spodoptera frugiperda ascovirus 1a. Expression patterns are for genes changing by 2-fold or more at any time point postinfection. The heat map color scale represents the z-scale of the transcripts per million (TPM) value of replicate expression level averages at each time point. The S. frugiperda gene identifications can be accessed through the SfruDB Information system (http://bipaa.genouest.org/is/lepidodb/spodoptera_frugiperda/) (67).
TABLE 2.
Changes in Spodoptera frugiperda third-instar larval cytoskeleton gene levels at different time points after infection with Spodoptera frugiperda ascovirus 1aa
Only genes whose expression changed by 2-fold or more (at any time point postinfection) are included.
Gene IDs used in this study can be accessed through the SfruDB Information system (http://bipaa.genouest.org/is/lepidodb/spodoptera_frugiperda/) (67).
Blue and red type refer to upregulated and downregulated genes, respectively.
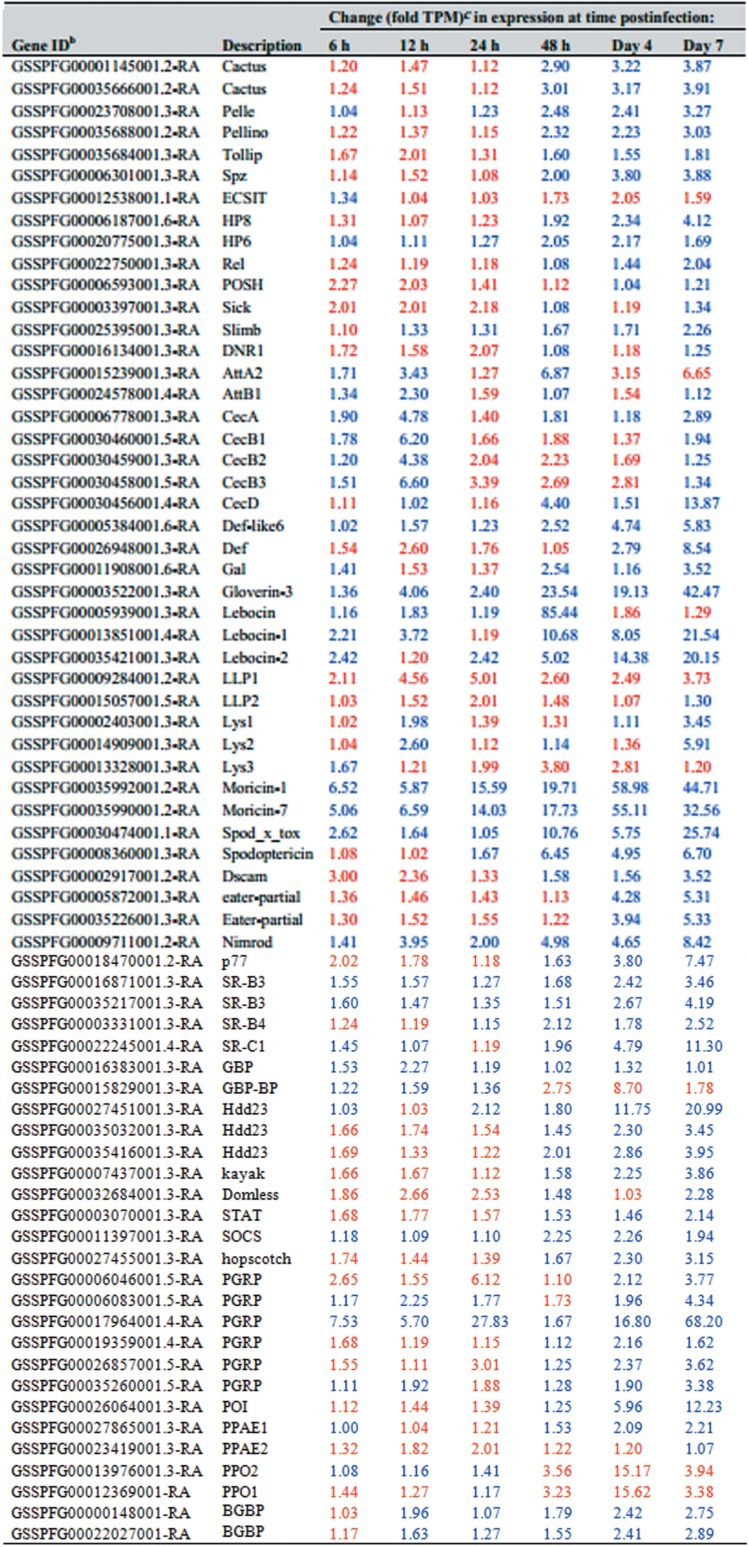
Innate immunity gene transcription patterns.
We quantified expression of the primary innate immunity genes identified in the S. frugiperda transcriptome and genome and characterized the results as local, humoral, or cellular, based on the structural characterization of the Drosophila immune system (13). As discussed below, major innate immunity genes were either upregulated or downregulated during ascovirus infection (Table 3; Fig. 3).
TABLE 3.
Changes in Spodoptera frugiperda third-instar larval innate immunity gene levels at different time points after infection with Spodoptera frugiperda ascovirus 1aa
Only genes whose expression changed by 2-fold or more (at any time point postinfection) are included.
The Spodoptera frugiperda innate immunity genes used in this study are listed in references 67 and 68. Gene identifications can be accessed through the SfruDB Information system (http://bipaa.genouest.org/is/lepidodb/spodoptera_frugiperda/).
Blue and red type refer to upregulated and downregulated genes, respectively.
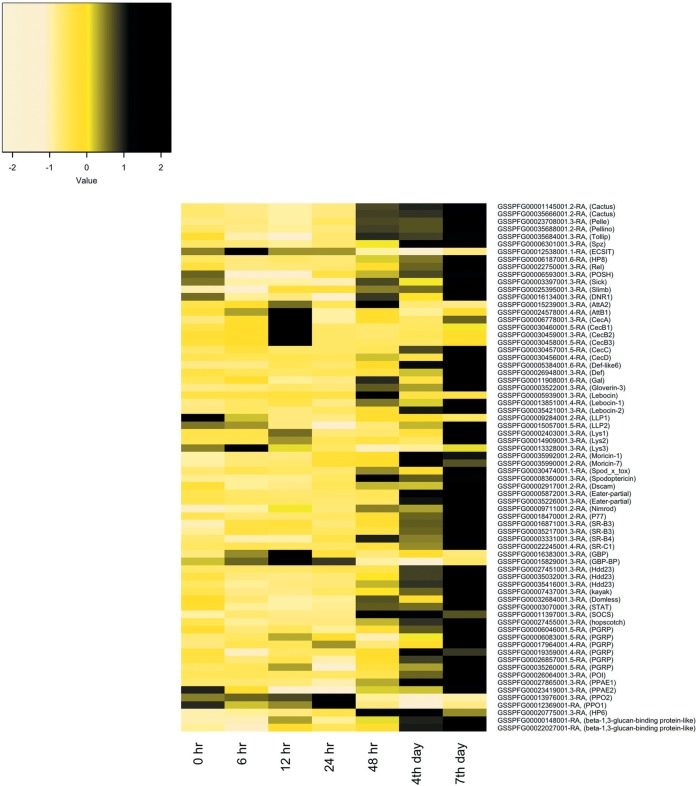
FIG 3.
Heat map representation of changes in innate immunity gene transcripts per million (TPM) in Spodoptera frugiperda third-instar larvae infected with Spodoptera frugiperda ascovirus 1a. Expression patterns are for genes changing by 2-fold or more at any time point postinfection. The heat map color scale represents the z-scale of the TPM value of replicate expression level averages at each time point. The S. frugiperda innate immunity gene identifications can be accessed through the SfruDB Information system (http://bipaa.genouest.org/is/lepidodb/spodoptera_frugiperda/) (67).
(i) Local immunity: AMPs. The local immune response is the first line of defense against foreign materials and is typically provoked by microbial pathogens. Activation of this response leads to the production of many antimicrobial peptides (AMPs), limiting the spread of pathogens from wounds or those that enter via the midgut. During SfAV-1a infection, we found upregulation by 2-fold or more by 6 and/or 12 hpi of genes for attacin A2 and B1, cecropins (A, B1, B2, and B3), gloverin-3, lebocin (1 and 2), Lys (2), moricin (1 and 7), and spod_x_tox (Fig. 3; Table 3). In addition, cecropin C (GSSPFG00030457001.5-RA) increased from 0 transcripts per million (TPM) at 0 h to more than 1 TPM at 6 h and was even higher by the days 4 and 7 p.i. (by 1.23, 5.83, and 11.08 TPM, respectively). Some of these, namely, gloverin-3, spod_x_tox, lebocin-1, and lebocin-2, achieved higher levels at later stages, increasing on day 7 p.i. by 42-fold, 25-fold, 21-fold, and 20–fold, respectively. Only moricin-1 and moricin-7 maintained an expression level higher than 2-fold, and sometimes markedly higher, at all times postinfection (ranging from about 6- to 59-fold and 5- to 55-fold, respectively), indicating their importance to S. frugiperda immunity and survival. Interestingly, lebocin (GSSPFG00005939001.3-RA) showed a sudden high upregulation (85-fold) at 48 hpi. Alternatively, certain other AMPs were downregulated by more than 2-fold at specific time points, including attacin (A2), cecropin (B2 and B3), defensin, Lys3, and LLP2. Only lysozyme LLP1 was downregulated (about 2- to 5-fold) by more than 2-fold at all the time points postinfection.
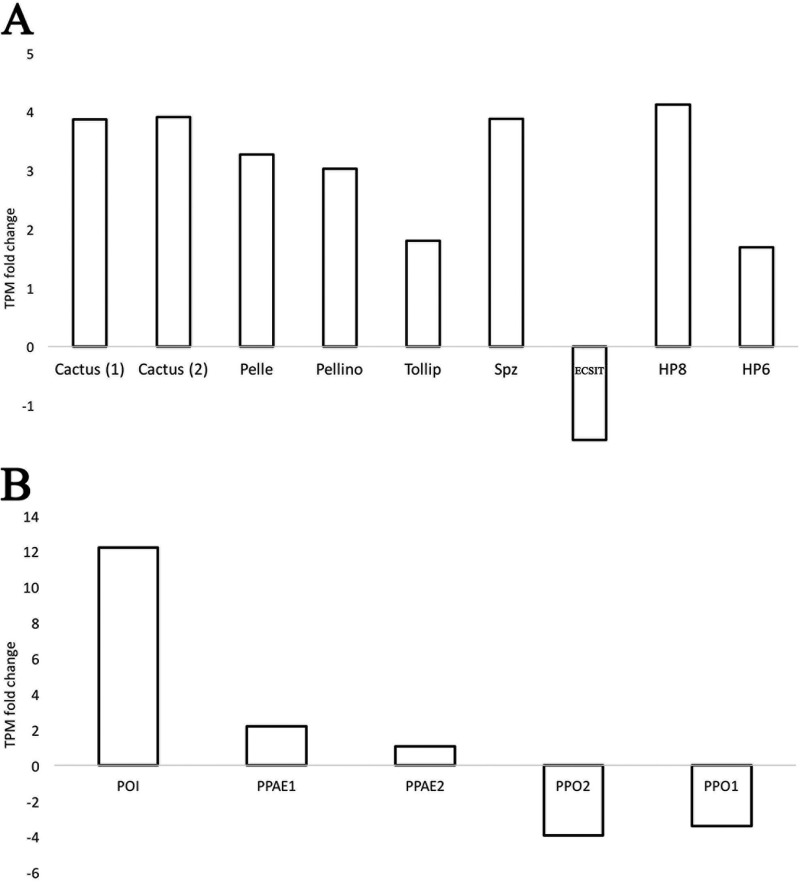
(ii) Humoral immunity: immune signaling pathways. (a) Toll pathway. Most of the S. frugiperda Toll pathway genes were upregulated in infected larvae by 2-fold or more, especially by 48 hpi or day 4 p.i., specifically, genes for Pelle, Pellino, Spätzle, HP8 (Spätzle-processing enzyme), and HP6 (serine protease persephone-like) and two Cactus genes (Table 3; Fig. 4). Cactus is a negative regulator of the Toll pathway (14). Interestingly, both Cactus genes were upregulated from day 2 and remained upregulated by 2-fold or more until day 7 p.i. (Fig. 4). For Cactus genes, the increases ranged from 2.9- to 3.9-fold. Tollip increased beginning at day 2 but by less than 2-fold. Finally, only an ECSIT gene was downregulated, and at almost all time points postinfection, ranging from 1.03- to 2.05-fold (Table 3).
FIG 4.
Upregulated and downregulated innate immunity genes of third-instar Spodoptera frugiperda infected with Spodoptera frugiperda ascovirus 1a. Toll pathway (A)- and phenoloxidase system (PO) and melanization cascade (B)-associated gene transcripts per million (TPM) levels at 7 days postinfection are shown. Expression level changes were calculated by comparing levels at day 7 postinfection with the 0-h control (uninfected) expression level. Cactus and phenoloxidase inhibitor (POI) are negative regulators of the Toll pathway and PO system, respectively. Cactus (1) refers to GSSPFG00035666001.2-RA and Cactus (2) refers to GSSPFG00001145001.2-RA (gene identifications for the Spodoptera frugiperda genome [67]).
(b) Imd and JNK pathways.
Although we noticed alterations in expression of most immune deficiency (Imd) genes, the level of up- or downregulation was less than 2-fold. Of the 17 S. frugiperda Imd pathway genes and negative regulators analyzed, only an NF-κB-like factor Relish gene and four Imd negative regulators showed a change of 2-fold or slightly more at any time after infection. The negative regulators were POSH, Slimb, DNR1, and Sick (Table 3). Only one Jun N-terminal kinase (JNK) pathway gene, that for transcription factor Kayak, changed by 2-fold or more. Kayak (GSSPFG00007437001.3-RA) was upregulated over 2-fold late in the infection (Fig. 3).
(c) JAK/STAT pathway.
The SOCS (a negative Janus kinase/signal transducer and activator of transcription [JAK/STAT] regulator) was upregulated at all times postinfection. STAT, hopscotch, and Domless were downregulated early during infection; however, they showed upregulation by 2.14-fold, 3.15-fold, and 2.28-fold at day 7 p.i., respectively, demonstrating activation of some JAK/STAT genes (Table 3; Fig. 3).
(d) Melanization.
Both prophenoloxidase subunit 1 (PPO1) and prophenoloxidase subunit 2 (PPO2) genes were downregulated markedly by day 4 p.i. (Table 3). Alternatively, a phenoloxidase (PO) inhibitor (POI) was upregulated, by about 6-fold at day 4 p.i. and by about 12-fold at day 7 p.i. (Fig. 4).
(iii) Cellular immunity.
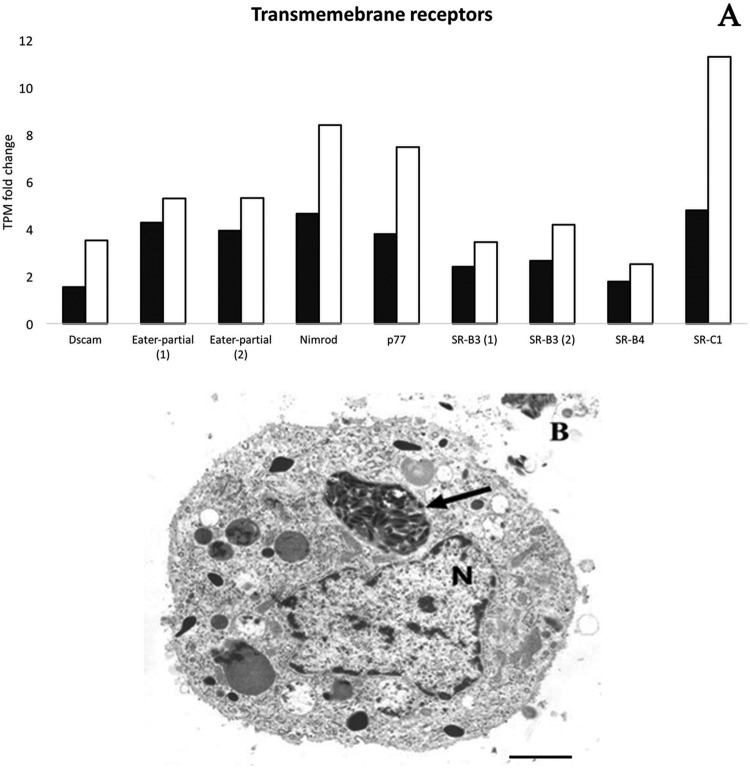
The RNA-Seq results (Table 3; Fig. 5) and the occurrence, though rare, of viral vesicles in the cytoplasm (Fig. 5B) of hemocytes provide support for the activation of the cellular immunity. SfAV overwhelmingly replicates in fat body tissue, and viral vesicles never appear in infected cells until hours after nuclear lysis. Thus, the presence of a viral vesicle packed with virions provides evidence that at least some hemocytes recognize these as foreign material. The marked reduction in prophenoloxidase transcripts and some AMPs, probably due to the loss of healthy fat body tissue, may also result in greater cellular immunity. However, the only cellular immunity we observed was increases in the phagocytosis pathway.
FIG 5.
Upregulation of Spodoptera frugiperda third-instar larvae transmembrane receptor genes after infection with Spodoptera frugiperda ascovirus 1a. These transmembrane receptor proteins are known to be associated with phagocytosis. (A) Transmembrane receptor gene transcripts per million (TPM) fold changes at day 4 (black bars) and day 7 (white bars) postinfection. Eater-partial (1) refers to GSSPFG00035226001.3-RA, and Eater-partial (2) refers to GSSPFG00005872001.3-RA (gene identifications for the S. frugiperda genome [67]). SR-B3 (1) refers to GSSPFG00016871001.3-RA, and SR-B3 (2) refers to GSSPFG00035217001.3-RA (gene identifications for the same genome). (B) Hemocyte with an ascovirus viral vesicle containing virions (arrow). Note that the nucleus (N) is intact, indicating an engulfed vesicle. Bar, 1.5 μm.
Phagocytosis.
Many pattern recognition receptors (PRRs) associated with phagocytosis were upregulated by SfAV-1a infection. The Nimrod and two Scavenger receptors (SR-B3) were upregulated at all time points postinfection. A DSCAM, two Eater genes, and Scavenger receptors (SR-B4 and SR-C1) were also upregulated, but late in infection (Table 3; Fig. 5). In addition, six peptidoglycan recognition proteins (PGRPs) were upregulated late during infection (Table 3).
Other genes associated with immunity.
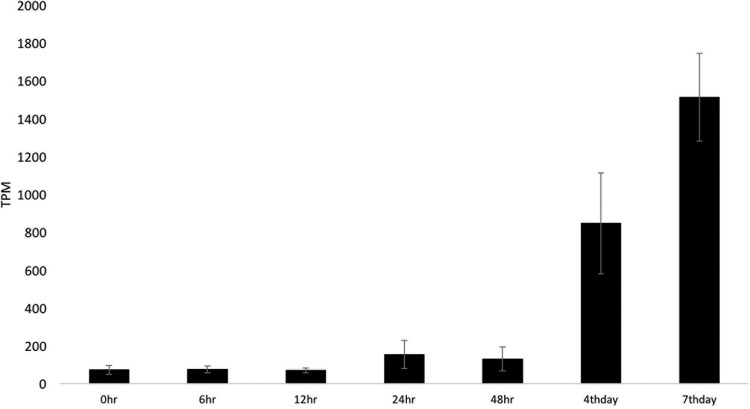
The innate immunity gene Hdd23-like GSSPFG00027451001.3-RA and two homologs (GSSPFG00035416001.3-RA and GSSPFG00035032001.3-RA) were upregulated, beginning at about 24 or 48 hpi. At day 7 p.i., expression of these increased by 21-, 3.95-, and 3.45-fold, respectively (Table 3; Fig. 6).
FIG 6.
Expression of the Spodoptera frugiperda Hdd23 gene from 0 h to 7 days postinfection with Spodoptera frugiperda ascovirus 1a. The high expression level is shown in transcripts per million (TPM). Error bars represent the standard deviation.
DISCUSSION
Studies of ascovirus cytopathology, especially ultrastructural investigations of the remarkable change in cell architecture that results in the formation of viral vesicles, suggest a very complex interaction between these viruses and their lepidopteran hosts. For example, changes in the shape of mitochondria and their movement to sites where the delimiting membrane of viral vesicles form suggest that these organelles are required for synthesis of these membranes (1, 15). Moreover, previous studies suggest that ascoviruses alter protein levels involved in innate immunity (2, 7, 12), for example, the Toll-like receptor and JAK/STAT pathway proteins (12). Moreover, the persistence of viral vesicles for 4 to 6 weeks or more while circulating in the hemolymph also suggests that the levels of some innate immunity proteins decrease. In the present study, therefore, we extended our previous in vivo investigations of the SfAV-1a transcriptome in infected S. frugiperda third instars (7) by focusing on host mitochondrial, cytoskeletal, and innate immunity gene expression patterns. As discussed below, we found that the expression of most mitochondrial genes in infected larvae was similar to that in controls, though several were upregulated during vesicle formation. In addition, we found upregulation of some host and viral mitochondrial motor genes and key host innate immune pathway genes.
The mitochondrial transcriptome of S. frugiperda third-instar larvae was stable prior to infection with SfAV-1a, but then postinfection typically increased less than 2-fold. Interestingly, transcript upregulation temporally correlated with the stage of vesicle formation, showing that mitochondria were conserved rather being destroyed after nuclear lysis, as occurs in apoptosis. Thus, our data support previous ultrastructural studies indicating that mitochondria participated in viral vesicle membrane formation (1, 3). In general, there are very few studies that examine mitochondrial genome expression in insects infected with viruses, or even in other eukaryotic species, with the exception of vertebrate animals or cell lines infected with mammalian viruses. Thus, it is not possible to make direct comparisons. There is, however, some evidence in at least one case that mitochondria play a role in human cells infected with cytomegalovirus. This virus typically causes a chronic infection, but chloramphenicol treatment significantly inhibited mitochondrial translation and virus replication, indicating that mitochondria directly enhanced viral replication (16). In other cases, for example, herpes simplex virus 1 (HSV-1), HSV-2, and Epstein-Barr virus (EBV), the role of mitochondria in viral replication appears to be minimal, if any, as mitochondrial DNA is typically degraded rapidly (17). In addition, in vertebrate cells such as the Vero cell line infected with African swine fever virus or chicken embryo fibroblasts infected with Frog virus 3, microtubules participate in moving mitochondria close to the virogenic stroma (18–20), apparently to make virus replication more efficient. Alternatively, in HSV-1, mitochondrial DNA is degraded by a virus-encoded mitochondrion-targeted nuclease (17, 21). Thus, unique among all viruses, the chronic nature of ascovirus infection and the use of ascovirus vesicles for virus replication and virion assembly help explain the need for maintaining mitochondrial function for many days, if not weeks, as the infection spreads throughout the fat body tissue.
The changes in mitochondrial shape and size during ascovirus infection may be partially explained by the RNA-Seq data as an outcome of expression level alterations of complex I (NADH dehydrogenases), complex III (cytochrome b), complex IV (cytochrome c oxidases), and complex V (ATP6 and ATP8 of ATP synthases), given that these subunits are embedded in the inner mitochondrial membrane. However, these changes alone most likely do not account for the radical changes in cell architecture characteristic of ascovirus infection. Furthermore, it is unlikely that mitochondrial genes carried in the nuclear genome are involved in these changes, because the nucleus and nuclear DNA are degraded early during cell infection. It is more likely, therefore, that these marked changes in the size, shape, and movement of mitochondria are due to several viral genes that contain putative sequences targeting the encoded proteins to mitochondria, identified in the previous study of the SfAV-1a transcriptome (7).
Whereas the changes in mitochondria are significant during ascovirus infections, based on biochemical studies it is known that several mammalian viral proteins manipulate these organelles (reviewed in references 18 and 22). For example, viral proteins can affect regulation of mitochondrial membrane potential (23), alter the intracellular distribution of mitochondria (19), “hijack” mitochondrial proteins to enable their genomes to enter the nucleus (24), and deplete mitochondrial DNA (25). It is also possible that other complex insect viruses, such as baculoviruses and entomopoxviruses, directly manipulate the mitochondrial transcriptome, but none has been identified. In general, more transcriptome studies are needed to determine which viruses are capable of manipulating mitochondria for virus replication and the mechanisms underlying these virus-directed changes. Such studies could be important to human health because it is now known that several neurological diseases of aging are due to mutations in mitochondrial proteins encoded in either the mitochondrial or nuclear genome (26–28).
For cytoskeletal genes, we found that the expression of only 58 of 106 genes studied was altered more than 2-fold postinfection. More than half of those were genes that coded for proteins related to kinesin, dynein, and myosin (Table 2). Kinesin and dynein are known as the main motors of the mitochondria. Specifically, kinesin is associated with antegrade and dynein with retrograde movement. For example, in Drosophila, cytoplasmic dynein was associated with retrograde mitochondrial transport. Myosins are associated with mitochondrial movement, however, only for short distances (29, 30). Moreover, a virus structural gene known as a dynein-like b chain (ORF084), which is conserved in Trichoplusia ni ascovirus (TnAV) and HvAV species, was identified recently as a late SfAV-1a gene, for which expression begins by 12 hpi and peaks at 24 hpi (4.14 log2 reads per kilobase per million [RPKM]) (7). The overall expression pattern of the 58 genes initially involves downregulation from 6 to 24 hpi. However, beginning at 48 hpi through to day 7 p.i., many of the genes were upregulated. Initial downregulation of genes was reported recently by Yu et al. (12), who found that most of the differentially expressed genes in S. exigua larvae infected with HvAV were downregulated from 6 to 12 hpi. However, they reported that subsequently, during the formation of viral vesicles (12 to 72 hpi), unigenes were upregulated. Analysis of the most abundant differentially expressed genes demonstrated that host genes associated with regulation of actin cytoskeletal genes were upregulated as follows: 4 genes at 6 h, 23 at 12 h, 27 at 72 h, and 50 by the day 7 postinfection (12). Chen et al. (31) have also reported initial downregulation of host genes for the Autographa californica multiple nucleopolyhedrovirus (AcMNPV) baculovirus, which attacks lepidopteran larvae. However, in their 6- to 48-hpi infection period in the Trichoplusia ni cell line, they reported a decrease through the remainder of the infection cycle.
Previous studies of lepidopteran larvae infected with ascoviruses reported the accumulation of high concentrations of viral vesicles in the hemolymph, which turns this tissue milky white. These studies also showed that viral vesicles continued to be produced and to circulate in the hemolymph for weeks, thereby favoring transmission of the virus by parasitic wasps (1, 2). Other than the change in color, however, there were no obvious signs of an innate immune response such as melanization of the vesicles or phagocytosis by hemocytes as they circulate in the hemolymph. Moreover, the long-term survival of infected larvae for transmission might favor an immune response to prevent them from succumbing to infections by other pathogens. This raises the question of whether the larval host is capable of recognizing the vesicles as foreign. Based on a recent transcriptome study, in S. exigua larvae infected with Heliothis virescens ascovirus (HvAV) 3h, genes for the Toll-like receptor signaling and JAK/STAT pathways along with other genes involved with immunity were upregulated upon infection (12).
The insect innate immune system is classified as being local, humoral, or cellular, based on the structural characterization of the Drosophila immune system (13). Interestingly, the local immunity represented in antimicrobial peptides such as attacin, cecropin, moricin, defensin, gloverin, gallerimycin, and lebocin, all known to be effective in combination against a wide range of bacterial, fungal, and viral pathogens that infect lepidopteran larvae (32–36), was upregulated either consistently or at different time points postinfection. Thus, they can be understood as a systemic antimicrobial defense induced by wounding of the insect cuticle (37, 38), which our infection process may have triggered, as it mimicked the type of wound made by a wasp ovipositor. Therefore, the induction of these antimicrobial peptides might prevent opportunistic pathogens from competing for host resources during infection. With respect to lysozymes, upregulation of Lys1 and Lys2 and downregulation of lysozyme-like LLP1, LLP2, and Lys3 at almost all the time points postinfection appear to be contradictory. However, these enzymes are known to be associated with roles other than peptidoglycan attack, for example, melanization (39).
Induction of the Toll pathway is initiated by interactions between pathogen-associated molecular patterns (PAMPs) and host pattern recognition receptors (PRRs). Gram-positive bacterial and fungal cell wall components, for example, are the main inducers of the Toll cascade (40). Whereas it has not been determined how the NF-κB-dependent pathways (Toll and Imd) are induced by a virus infection, these pathways have nevertheless been shown to limit insect virus infections. Specifically, the Toll pathway and cellular immunity limit Drosophila X virus (41), and the Imd pathway and cellular immunity, also in Drosophila, limit cricket paralysis virus (42). Alternatively, it has been shown that certain Toll-dorsal pathway genes, specifically, pelle, tube, and cactus, are involved in development in Drosophila dorsal-ventral patterning, a difference that distinguishes the invertebrate Toll pathway from that of mammals (43–46). Therefore, the upregulation of Toll genes after SfAV-1a infection may be both an immune response and to modify larval development. Of interest regarding the latter, development is significantly retarded during ascovirus disease compared to that of healthy larvae (3). Moreover, we detected the upregulation of some JAK/STAT-associated genes (SOCS, STAT, Domless, and hopscotch) either consistently or late in the infection (Table 3). Interestingly, the unpaired cytokine (Upd3) or the JAK/STAT pathway activator was not detected in the S. frugiperda transcriptome. Similarly, the Drosophila extracellular cytokine unpaired3 (Upd3) ortholog was missing in Manduca sexta and Tribolium castaneum (47, 48), implying the involvement of an undefined JAK/STAT pathway-activating cytokine that replaces Upd3 in these insects. The insect JAK/STAT is like the Toll pathway in terms of being associated with immunity and development (46, 48). The upregulation of Toll and JAK/STAT genes after SfAV-1a infection agrees with the S. exigua response to HvAV 3h (12).
The low variation in expression levels of Imd genes is not surprising because there is evidence that expression of this pathway can be deleterious to insects. For example, when the Imd pathway in Drosophila was downregulated, i.e., suppressed by a small fly protein known as Diedel, flies lived longer. When the die gene was mutated, the life span of the flies was reduced by about 10 days, which is significantly different from that of wild-type flies, which typically live for 70 days (49). Interestingly, the Imd-associated genes that varied (increased or decreased) 2-fold or more were an NF-κB-like factor Relish gene and genes for four negative regulators (POSH, Sick, Slimb, and DNR1) (Table 3). Of relevance here is that SfAV-1a synthesizes high levels of a Diedel homolog (ORF121) during infection, and thus its function may be to suppress the Imd pathway. If so, this would help explain the longevity of larvae infected with ascoviruses as well as the minor variations in expression levels of Imd pathway genes. Alternatively, the expression levels of Imd genes may be tightly controlled by mechanisms not yet identified during virus infections. Overall, the upregulation of the Toll pathway negative regulator of Cactus in combination with the highly expressed ascovirus virokine Diedel (ORF121), which apparently acts as an Imd suppressor (7), may ultimately help explain at the molecular level how SfAV-1a controls expression of two of the most important signaling innate immunity pathways.
In lepidopteran insects, the oenocytoids are the hemocytes associated with the production of melanization cascade enzymes. The antiviral and antibacterial effect of the melanin cascade-associated compounds has been demonstrated in many recent studies (50–52). Therefore, the phenoloxidase pathway gene alterations indicate the activation of the melanization cascade against SfAV-1a; however, the high upregulation of the host phenoloxidase inhibitor (∼12-fold by day 7 p.i.) may decrease the effectiveness of this pathway against this virus. The dedication of a specific viral protein to inhibit the PO is also reported for the viral protein WSSV453 produced by the white spot syndrome virus, which inhibits the melanization cascade in the shrimp Penaeus monodon by interacting with and preventing activation of pro PO-activating enzyme 2 (PmproPPAE2) (53). Whether ascovirus genomes possess a melanization inhibitor is not known, but ascoviruses have large DNA genomes and synthesize many proteins with unknown functions.
The SfAV-1a tissue tropism is primarily restricted to the S. frugiperda fat body. This tissue is completely destroyed by day 12 p.i., with dense accumulations of viral vesicles (108/ml) in the hemolymph by day 7 p.i. (15). Although the fat body, with functions similar to those of the mammalian liver, is not essential for larval survival, it does regulate many aspects of insect growth, development, and metamorphosis (53, 54). Very importantly, it also has primary responsibility for innate immunity responses (55). Interestingly, we detected many PRRs associated with phagocytosis upregulation by SfAV-1a infection, for example, Nimrod and Scavenger receptors, DSCAM, and Eater. For example, many PRRs are associated with phagocytic events (56–58) against bacteria, plasmodia (59), and (recently shown) viruses. For instance, recently the role of the class A (60) and C (61) scavenger receptors in virus phagocytosis was confirmed. The class C receptor in the shrimp Marsupenaeus japonicus is efficient in inducing hemocyte phagocytosis of white spot syndrome virus, thereby preventing systemic infection (61). Phagocytosis is a cellular immune response that takes place by either circulating or immobile hemocytes. In lepidopterans, the plasmatocytes are responsible for this process (52, 62). Each phagocyte can internalize and degrade numerous invading pathogens (63). Therefore, given that the fat body is almost completely destroyed by SfAV-1a infection by day 7, it is expected that humoral immunity would be significantly reduced as the infection increases. Indeed, Fig. 5B illustrates an S. frugiperda hemocyte that has engulfed an SfAV viral vesicle, indicating at least some recognition of foreign material by host cells.
Finally, we detected the high upregulation of an Hdd23 gene or an immune-associated protein, by ∼21-fold at day 7 p.i. The Hdd23 gene was identified in the fall webworm Hyphantria cunea as an early acute-phase immunity-associated protein upregulated continuously after a bacterial challenge (64). However, a protein that shares sequence similarity with Hdd23 (known as stress-responsive peptide [SRP]) was characterized recently in Helicoverpa armigera and found to be associated with prophenoloxidase and nodulation activation (65). The high upregulation of Hdd23-like proteins (Fig. 6) shows that this is in response to SfAV-1a infection, even though they apparently are largely ineffective against this virus. Thus, as in the case of the antimicrobial peptides, given the response to bacterial challenge in H. cunea and activation of nodule formation in H. armigera, the function of these proteins may be to inhibit bacterial infections during the prolonged larval stage caused by ascovirus infection.
In conclusion, the evolutionary success of the chronic characteristic of disease caused by ascoviruses in their lepidopteran larval hosts may reflect their unique ability to conserve mitochondrial functions while keeping the balance between the different innate immunity cascade inducers and inhibitor/negative regulators that are expressed simultaneously. The transcriptome data show that some of these processes are controlled by viral genes, such as the preservation of mitochondrial functions, whereas other are likely direct innate immune responses due directly to cuticular damage due to oviposition attempts. Studies of many vertebrate viruses over the past decade, especially of mammalian viruses, have identified many proteins involved in manipulating mitochondria and inducing innate immune responses. Ascoviruses have large genomes, and the functions of many genes remain unknown. Thus, it is possible that several of these, if not more, are involved in the direct manipulation of mitochondria and innate immunity, suggesting that a wealth of new information may be derived by transcriptome studies of additional ascoviruses and other large DNA insect viruses.
An important dilemma that remains to be resolved for ascoviruses is the discrepancy between the ultrastructural data suggesting that mitochondria play a major role in cell hypertrophy, membrane synthesis, and vesicle formation and the transcriptome data from two studies that suggest unremarkable changes in the upregulation of mitochondrial gene expression. This may be due to the normalization of transcriptome data per unit of tissue and possibly the release of nascent viral vesicles as the anucleate cell cleaves. Another enigma is how mitochondria continue to function after nuclear DNA is cleaved, considering that mitochondrial functions depend on numerous genes carried in the nuclear genome.
MATERIALS AND METHODS
Virus infection and disease development in infected larvae.
Spodoptera frugiperda ascovirus 1a (SfAV-1a), strain Sf82-126 (66), was used to inoculate Spodoptera frugiperda third-instar larvae. Larvae were grown on an artificial diet (Benzon Research, Carlisle, PA) under ambient laboratory conditions (22°C). To mimic infection by parasitic wasps, virus inoculation was performed manually using a minutin pin dipped in a suspension of viral vesicles (1 × 108/ml), which was then inserted briefly through the posterior abdominal cuticle into the fat body. Disease development was confirmed by examining a small drop of hemolymph to ensure it was opaque white and then by viewing the drop by phase-contrast microscopy for the presence of high concentrations of viral vesicles.
Extraction of total RNA from healthy and infected larvae.
Total RNA was isolated from healthy and SfAV-infected S. frugiperda larvae using TRIzol (Invitrogen, Life Technologies) following the protocol provided by the manufacturer. RNA was isolated from 50 to 100 mg of tissue by mechanical homogenization of the larval body in 1 ml TRIzol. RNA samples were isolated at 0 h (uninfected control larvae) and for SfAV-infected larvae at 6, 12, 24, and 48 h and 4 and 7 days postinfection (p.i.). Total RNA was extracted from three biological replicates at each time point. Three replicates were included in the calculations of transcripts per million (TPM) (described below).
DNase treatment and RNA purification.
To purify the isolated RNA and remove any DNA contamination, the instructions from the RNA Clean and Concentrator TM-5 kit manufacturer (Zymo Research) were followed, combined with the RNase-free DNase set (Qiagen). RNA quantity and quality were determined using a Thermo Scientific NanoDrop ND-2000c spectrophotometer.
Enrichment of mRNA, RNA-Seq library preparation, and sequencing.
RNA-Seq library preparation and sequencing were previously described in detail (7). Briefly, mRNA was enriched using the NEBNext poly(A) mRNA magnetic isolation module kit (New England BioLabs), followed by library preparation using the NEBNext Ultra Directional RNA library preparation kit (New England BioLabs) for Illumina sequencing. For sequencing, the HiSeq2500 Illumina and NextSeq500 sequencers were used (Noel T. Keen Genomics Core Facility at the UC Riverside Institute for Integrative Genome Biology).
Bioinformatic analysis and TPM calculations.
For quantification of S. frugiperda mitochondrial gene expression, we used the published S. frugiperda mitochondrial genome (8) (available through NCBI, accession no. KM362176.1) for transcript locations. In this reference genome, two transcripts are annotated at multiple loci but with identical transcript names: tRNA-Leu and tRNA-Ser. For clarity, we refer to these transcripts as tRNA-Leu-1 (located on the reference mitochondrial genome at positions 2992 to 3059), tRNA-Leu-2 (positions 12714 to 12783), tRNA-Ser-1 (positions 6164 to 6233), and tRNA-Ser-2 (positions 11690 to 11756). For quantification of innate immunity and cytoskeleton genes, RNA-Seq reads were mapped to the recently published Spodoptera frugiperda corn variant genome (67). The assembled genome can be accessed through the European Bioinformatics Institute (EMBL-EBI) (accession no. PRJEB13110). The innate immunity and cytoskeleton gene IDs listed in this study can be accessed through the SfruDB Information system (http://bipaa.genouest.org/is/lepidodb/spodoptera_frugiperda/). For innate immunity genes, we included representatives (97 genes) for every innate immunity pathway identified in the Spodoptera frugiperda transcriptome (Sf_TR2012b) and listed in reference 68. Thus, genes for Toll, Imd, JAK/STAT, JNK, phenoloxidase system (PO), extracellular signal transduction and cytokines, antimicrobial peptides (AMPs), transmembrane receptors, peptidoglycan recognition protein (PGRP), and the Gram-negative bacterium-binding protein (GNBP) were included. For cytoskeleton genes, we included 106 genes in the analysis. These included representatives for actin, actin-related proteins, tubulin, dynein, kinesin, lamin, filamin, myosin, and profilin. Transcript expression estimates for these genes were estimated as transcripts per million (TPM) using the RSEM program (69).
Determination of Spodoptera frugiperda genes upregulated and downregulated postinfection.
After TPM quantification for each S. frugiperda mitochondrial, cytoskeletal, or innate immunity gene, the genes were separated into two classes, upregulated or downregulated, based on the increase or decrease in expression compared to the 0-h control. Upregulated genes were defined as those for which transcription increased 2-fold or more postinfection (at any time point postinfection) and which had a basal transcription level equal to or greater than 1 TPM at 0 h or any time postinfection. Similarly, downregulated genes were those for which transcription decreased by equal to or greater than 2-fold (at any time point postinfection) and which had a basal transcription level equal to or greater than 1 TPM at 0 h or any time postinfection.
EM.
The transmission electron microscopy (EM) imaging was done following protocols described previously (1, 70). Briefly, for tissue fixation, 3% glutaraldehyde and 1% OsO4 buffers were used, followed by tissue embedding in Epon-Araldite. Ultrathin sections were cut on a Sorvall Ultrathin Microtome and examined in a Philips electron microscope.
Data availability.
The Spodoptera frugiperda RNA-Seq data and TPM values for mitochondrial, cytoskeletal, and innate immunity genes obtained in this study can be accessed through the NCBI Gene Expression Omnibus (GEO) series accession number GSE114901; samples are located in files GSM3154237 to GSM3154250 and GSM4259495 to GSM4259501.
ACKNOWLEDGMENT
Heba A. H. Zaghloul was funded by the Fulbright Program, sponsored by the U.S. Department of State’s Bureau of Educational and Cultural Affairs and administered by AMIDEAST in Egypt, her home country.
REFERENCES
- 1.Federici BA. 1983. Enveloped double stranded DNA insect virus with novel structure and cytopathology. Proc Natl Acad Sci U S A 80:7664–7668. doi: 10.1073/pnas.80.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govindarajan R, Federici BA. 1990. Ascovirus infectivity and effects of infection on the growth and development of noctuid larvae. J Invertebr Pathol 56:291–299. doi: 10.1016/0022-2011(90)90115-m. [DOI] [PubMed] [Google Scholar]
- 3.Federici BA, Bideshi DK, Tan Y, Spears T, Bigot Y. 2009. Ascoviruses: superb manipulators of apoptosis for viral replication and transmission. Curr Top Microbiol Immunol 328:171–196. doi: 10.1007/978-3-540-68618-7_5. [DOI] [PubMed] [Google Scholar]
- 4.Bideshi DK, Tan Y, Bigot Y, Federici BA. 2005. A viral caspase contributes to modified apoptosis for virus transmission. Genes Dev 19:1416–1421. doi: 10.1101/gad.1300205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asgari S. 2007. A caspase-like gene from the Heliothis virescens ascovirus (HvAV3e) is not involved in apoptosis but is essential for virus replication. Virus Res 128:99–105. doi: 10.1016/j.virusres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Bigot Y, Asgari S, Bideshi DK, Cheng X, Federici BA, Renault S. 2012. Family Ascoviridae, p 147–152. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses, 3rd ed Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 7.Zaghloul HAH, Hice R, Arensburger P, Federici BA. 2017. Transcriptome analysis of the Spodoptera frugiperda ascovirus in vivo provides insights into how its apoptosis inhibitors and caspase promote increased synthesis of viral vesicles and virion progeny. J Virol 91:e00874-17. doi: 10.1128/JVI.00874-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q-N, Chai X-Y, Bian D-D, Ge B-M, Zhou C-L, Tang B-P. 2016. The complete mitochondrial genome of fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Genes Genom 38:205–216. doi: 10.1007/s13258-015-0346-6. [DOI] [Google Scholar]
- 9.Babbucci M, Basso A, Scupola A, Patarnello T, Negrisolo E. 2014. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol Evol 6:3326–3343. doi: 10.1093/gbe/evu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bideshi DK, Bigot Y, Federici BA, Spears T. 2010. Ascoviruses, p 3–34. In Asgari S, Johnson KN (ed), Insect virology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 11.Hussain M, Abraham AM, Asgari S. 2010. An ascovirus-encoded RNase III autoregulates its expression and suppresses RNA interference-mediated gene silencing. J Virol 84:3624–3630. doi: 10.1128/JVI.02362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Li Z-Q, He L, Ou-Yang Y-Y, Li N, Huang G-H. 2018. Response analysis of host Spodoptera exigua larvae to infection by Heliothis virescens ascovirus 3h (HvAV-3h) via transcriptome. Sci Rep 8:5367. doi: 10.1038/s41598-018-23715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valanne S, Kallio J, Kleino A, Rämet M. 2012. Large-scale RNAi screens add both clarity and complexity to Drosophila NF-κB signaling. Dev Comp Immunol 37:9–18. doi: 10.1016/j.dci.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay SA, Wasserman SA. 2014. Conventional and non-conventional Drosophila toll signaling. Dev Comp Immunol 42:16e24–16e24. doi: 10.1016/j.dci.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federici BA, Govindarajan R. 1990. Comparative histology of three ascovirus isolates in larval noctuids. J Invertebr Pathol 56:300–311. doi: 10.1016/0022-2011(90)90116-N. [DOI] [PubMed] [Google Scholar]
- 16.Karniely S, Weekes MP, Antrobus R, Rorbach J, van Haute L, Umrania Y, Smith DL, Stanton RJ, Minczuk M, Lehner PJ, Sinclair JH. 2016. Human cytomegalovirus infection upregulates the mitochondrial transcription and translation machineries. mBio 7:e00029. doi: 10.1128/mBio.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duguay BA, Saffran HA, Ponomarev A, Duley SA, Eaton HE, Smiley JR. 2014. Elimination of mitochondrial DNA is not required for herpes simplex virus 1 replication. J Virol 88:2967–2976. doi: 10.1128/JVI.03129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand SK, Tikoo SK. 2013. Viruses as modulators of mitochondrial functions. Adv Virol 2013:738794. doi: 10.1155/2013/738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol 72:7583–7588. doi: 10.1128/JVI.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly DC. 1975. Frog virus 3 replication: electron microscope observations on the sequence of infection in chick embryo fibroblasts. J Gen Virol 26:71–86. doi: 10.1099/0022-1317-26-1-71. [DOI] [PubMed] [Google Scholar]
- 21.Claus C, Liebert UG. 2014. A renewed focus on the interplay between viruses and mitochondrial metabolism. Arch Virol 159:1267–1277. doi: 10.1007/s00705-013-1841-1. [DOI] [PubMed] [Google Scholar]
- 22.Khan M, Syed GH, Kim SJ, Siddiqui A. 2015. Mitochondrial dynamics and viral infections: a close nexus. Biochim Biophys Acta 1853:2822–2833. doi: 10.1016/j.bbamcr.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nudson WA, Rovnak J, Buechner M, Quackenbush SL. 2003. Walleye dermal sarcoma virus orf C is targeted to the mitochondria. J Gen Virol 84:375–381. doi: 10.1099/vir.0.18570-0. [DOI] [PubMed] [Google Scholar]
- 24.Matthews DA, Russell WC. 1998. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol 79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 25.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep 8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith DR. 2013. RNA-Seq data: a goldmine for organelle research. Brief Funct Genomics 12:454–456. doi: 10.1093/bfgp/els066. [DOI] [PubMed] [Google Scholar]
- 27.Hodgkinson A, Idaghdour Y, Gbeha E, Grenier JC, Hip-Ki E, Bruat V, Goulet JP, de Malliard T, Awadalla P. 2014. High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 344:413–415. doi: 10.1126/science.1251110. [DOI] [PubMed] [Google Scholar]
- 28.Tian Y, Smith DR. 2016. Recovering complete mitochondrial genome sequences from RNA-Seq: a case study of Polytomella non-photosynthetic green algae. Mol Phylogenet Evol 98:57–62. doi: 10.1016/j.ympev.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Pilling AD, Horiuchi D, Lively CM, Saxton WM. 2006. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell 17:2057–2068. doi: 10.1091/mbc.e05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M, Kalyanasundaram A, Zhu J. 2013. Structural and biomechanical basis of mitochondrial movement in eukaryotic cells. Int J Nanomedicine (Lond) 8:4033–4042. doi: 10.2147/IJN.S52132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y-R, Zhong S, Fei Z, Gao S, Zhang S, Li Z, Wang P, Blissard G. 2014. Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus. J Virol 88:13781–13797. doi: 10.1128/JVI.02243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara S, Yamakawa M. 1995. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J Biol Chem 270:29923–29927. doi: 10.1074/jbc.270.50.29923. [DOI] [PubMed] [Google Scholar]
- 33.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. 2008. The discovery and analysis of a diverged family of novel antifungal moricin-like peptides in the wax moth Galleria mellonella. Insect Biochem Mol Biol 38:201–212. doi: 10.1016/j.ibmb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Hultmark D, Engstrom A, Andersson K, Steiner H, Bennich H, Boman HG. 1983. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J 2:571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi H-Y, Chowdhury M, Huang Y-D, Yu X-Q. 2014. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol 98:5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badapanda C, Chikara SK. 2016. Lepidopteran antimicrobial peptides (AMPs): overview, regulation, modes of action, and therapeutic potentials of insect-derived AMPs, p 141–163. In Raman C, Goldsmith M, Agunbiade T (ed), Short views on insect genomics and proteomics. Entomology in focus, vol 4 Springer, Cham, Switzerland. [Google Scholar]
- 37.Brey PT, Lee WJ, Yamakawa M, Koizumi Y, Perrot S, François M, Ashida M. 1993. Role of the integument in insect immunity: epicuticular abrasion and induction of cecropin synthesis in cuticular epithelial cells. Proc Natl Acad Sci U S A 90:6275–6279. doi: 10.1073/pnas.90.13.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler- Joseph S, Michaut L, Reichhart J, Hoffmann JA. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J 17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao XJ, Ling E, Yu XQ. 2010. The role of lysozyme in the prophenoloxidase activation system of Manduca sexta: an in vitro approach. Dev Comp Immunol 34:264–271. doi: 10.1016/j.dci.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanji T, Hu X, Weber AN, Ip YT. 2007. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol 27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A 102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa A, Jan E, Sarnow P, Schneider DS. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 44.Belvin MP, Anderson KV. 1996. A conserved signaling pathway: the Drosophila Toll-dorsal pathway. Annu Rev Cell Dev Biol 12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 45.Hultmark D. 2003. Drosophila immunity: paths and patterns. Curr Opin Immunol 15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, Cherry S. 2014. Viruses and antiviral immunity in Drosophila. Dev Comp Immunol 42:67–84. doi: 10.1016/j.dci.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. 2007. Comparative genome analysis of the Tribolium immune system. Genome Biol 8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao X, He Y, Hu Y, Wang Y, Chen YR, Bryant B, Clem RJ, Schwartz LM, Blissard G, Jiang H. 2015. The immune signaling pathways of Manduca sexta. Insect Biochem Mol Biol 62:64–74. doi: 10.1016/j.ibmb.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamiable O, Kellenberger C, Kemp C, Troxler L, Pelte N, Boutros M, Marques JT, Daeffler L, Hoffmann JA, Roussel A, Imler JL. 2016. Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc Natl Acad Sci U S A 113:698–703. doi: 10.1073/pnas.1516122113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao P, Lu Z, Strand MR, Jiang H. 2011. Antiviral, anti-parasitic, and cytotoxic effects of 5,6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochem Mol Biol 41:645–652. doi: 10.1016/j.ibmb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charoensapsri W, Amparyup P, Suriyachan C, Tassanakajon A. 2014. Melanization reaction products of shrimp display antimicrobial properties against their major bacterial and fungal pathogens. Dev Comp Immunol 47:150–159. doi: 10.1016/j.dci.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Sutthangkul J, Amparyup P, Eum JH, Strand MR, Tassanakajon A. 2017. Anti-melanization mechanism of the white spot syndrome viral protein, WSSV453, via interaction with shrimp proPO-activating enzyme, PmproPPAE2. J Gen Virol 98:769–778. doi: 10.1099/jgv.0.000729. [DOI] [PubMed] [Google Scholar]
- 53.Mirth CK, Riddiford LM. 2007. Size assessment and growth control: how adult size is determined in insects. Bioessays 29:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- 54.Hoshizaki DK. 2005. Fat-cell development, p 315–345. In Gilbert LI, Iatrou K, Gill S (ed), Complete molecular insect science, vol 2 Elsevier, Berlin, Germany. [Google Scholar]
- 55.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol 7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 56.Mukhopadhyay S, Gordon S. 2004. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiol 209:39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Ando I. 2007. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol 17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 58.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 59.Dong Y, Taylor HE, Dimopoulos G. 2006. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol 4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dansako H, Yamane D, Welsch C, McGivern DR, Hu F, Kato N, Lemon SM. 2013. Class A scavenger receptor 1 (MSR1) restricts hepatitis C virus replication by mediating toll-like receptor 3 recognition of viral RNAs produced in neighboring cells. PLoS Pathog 9:e1003345. doi: 10.1371/journal.ppat.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang M, Shi X, Yang H, Sun J, Xu L, Wang X, Zhao X, Wang J. 2016. Scavenger receptor C mediates phagocytosis of white spot syndrome virus and restricts virus proliferation in shrimp. PLoS Pathog 12:e1006127. doi: 10.1371/journal.ppat.1006127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Honti V, Csordas G, Kurucz E, Markus R, Ando I. 2014. The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev Comp Immunol 42:47e56–47e56. doi: 10.1016/j.dci.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Oliver JD, Dusty Loy J, Parikh G, Bartholomay L. 2011. Comparative analysis of hemocyte phagocytosis between six species of arthropods as measured by flow cytometry. J Invertebr Pathol 108:126e130. doi: 10.1016/j.jip.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Shin SW, Park SS, Park DS, Kim MG, Kim SC, Brey PT, Park HY. 1998. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem Mol Biol 28:827–837. doi: 10.1016/s0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- 65.Qiao C, Li J, Wei XH, Wang JL, Wang YF, Liu XS. 2014. SRP gene is required for Helicoverpa armigera prophenoloxidase activation and nodulation response. Dev Comp Immunol 44:94–99. doi: 10.1016/j.dci.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 66.Bigot Y. 2011. Genus ascovirus, p 73–78. In Tidona C, Darai G (ed), Springer index of viruses, 2nd ed Springer, Heidelberg, Germany. [Google Scholar]
- 67.Gouin A, Bretaudeau A, Nam K, Gimenez S, Aury J-M, Duvic B, Hilliou F, Durand N, Montagné N, Darboux I, Kuwar S, Chertemps T, Siaussat D, Bretschneider A, Moné Y, Ahn S-J, Hänniger S, Grenet A-SG, Neunemann D, Maumus F, Luyten I, Labadie K, Xu W, Koutroumpa F, Escoubas J-M, Llopis A, Maïbèche-Coisne M, Salasc F, Tomar A, Anderson AR, Khan SA, Dumas P, Orsucci M, Guy J, Belser C, Alberti A, Noel B, Couloux A, Mercier J, Nidelet S, Dubois E, Liu N-Y, Boulogne I, Mirabeau O, Le Goff G, Gordon K, Oakeshott J, Consoli FL, Volkoff A-N, Fescemyer HW, Marden JH, Luthe DS, Herrero S, Heckel DG, Wincker P, Kergoat GJ, Amselem J, Quesneville H, Groot AT, Jacquin-Joly E, Nègre N, Lemaitre C, Legeai F, d'Alençon E, Fournier P. 2017. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep 7:11816. doi: 10.1038/s41598-017-10461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Legeai F, Gimenez S, Duvic B, Escoubas J-M, Gosselin Grenet A-S, Blanc F, Cousserans F, Séninet I, Bretaudeau A, Mutuel D, Girard P-A, Monsempes C, Magdelenat G, Hilliou F, Feyereisen R, Ogliastro M, Volkoff A-N, Jacquin-Joly E, d'Alençon E, Nègre N, Fournier P. 2014. Establishment and analysis of a reference transcriptome for Spodoptera frugiperda. BMC Genomics 15:704. doi: 10.1186/1471-2164-15-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Federici BA. 1982. A new type of insect pathogen in larvae of the clover cutworm, Scotogramma trifolii. J Lnvertebr Pathol 40:41–54. doi: 10.1016/0022-2011(82)90035-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Spodoptera frugiperda RNA-Seq data and TPM values for mitochondrial, cytoskeletal, and innate immunity genes obtained in this study can be accessed through the NCBI Gene Expression Omnibus (GEO) series accession number GSE114901; samples are located in files GSM3154237 to GSM3154250 and GSM4259495 to GSM4259501.