Rotaviruses are significant human pathogens that result in diarrhea, dehydration, and deaths in many children around the world. Rotavirus vaccines have suboptimal efficacy in low- to middle-income countries, where the burden of the diseases is the most severe. With the ultimate goal of improving current vaccines, we aim to better understand how rotavirus interacts with the host innate immune system in the small intestine. Here, we demonstrate that interferon-activated RNase L signaling blocks rotavirus replication in a strain-specific manner. In addition, virus-encoded VP3 antagonizes RNase L activity both in vitro and in vivo. These studies highlight an ever-evolving arms race between antiviral factors and viral pathogens and provide a new means of targeted attenuation for next-generation rotavirus vaccine design.
KEYWORDS: gastrointestinal infection, innate immunity, interferons, rotavirus, virus-host interactions
ABSTRACT
Our understanding of how rotavirus (RV) subverts host innate immune signaling has greatly increased over the past decade. However, the relative contribution of each virus-encoded innate immune antagonist has not been fully studied in the context of RV infection in vivo. Here, we present both in vitro and in vivo evidence that the host interferon (IFN)-inducible 2′-5′-oligoadenylate synthetase (OAS) and RNase L pathway effectively suppresses the replication of heterologous RV strains. VP3 from homologous RVs relies on its 2′-5′-phosphodiesterase (PDE) domain to counteract RNase L-mediated antiviral signaling. Using an RV reverse-genetics system, we show that compared to the parental strain, VP3 PDE mutant RVs replicated at low levels in the small intestine and were shed less in the feces of wild-type mice, and such defects were rescued in Rnasel−/− suckling mice. Collectively, these findings highlight an important role of VP3 in promoting viral replication and pathogenesis in vivo in addition to its well-characterized function as the viral RNA-capping enzyme.
IMPORTANCE Rotaviruses are significant human pathogens that result in diarrhea, dehydration, and deaths in many children around the world. Rotavirus vaccines have suboptimal efficacy in low- to middle-income countries, where the burden of the diseases is the most severe. With the ultimate goal of improving current vaccines, we aim to better understand how rotavirus interacts with the host innate immune system in the small intestine. Here, we demonstrate that interferon-activated RNase L signaling blocks rotavirus replication in a strain-specific manner. In addition, virus-encoded VP3 antagonizes RNase L activity both in vitro and in vivo. These studies highlight an ever-evolving arms race between antiviral factors and viral pathogens and provide a new means of targeted attenuation for next-generation rotavirus vaccine design.
INTRODUCTION
Despite the availability of several safe and effective vaccines, rotaviruses (RVs) remain a major human pathogen. On a global scale, RVs are the leading etiological agent of severe, acute gastroenteritis and dehydrating diarrhea in infants and young children, resulting in over 200,000 deaths annually (1, 2). Although symptomatic RV infections occur commonly in the young of almost all mammalian species (3), the replication of heterologous RV strains (strains not native to the host species from which they are isolated) is greatly restricted by the host innate immune system in the small intestine (4–7). Attenuation of heterologous RVs in humans serves as the genetic basis for attenuation of several currently licensed vaccines (8, 9). It was previously shown by our laboratory and others that both type I and type III interferon (IFN)-mediated antiviral signaling partially underlies this host range restriction (HRR) in a suckling mouse model (10–13). However, the precise antirotaviral effectors activated by host IFNs remain incompletely understood.
Members of the Reoviridae family, RVs have a triple-layered icosahedral capsid and an 11-segmented double-stranded RNA (dsRNA) genome (2, 14). Like many segmented-genome viruses, they can undergo gene reassortment at a high frequency during mixed infections (15). We previously used in vitro mixed infection followed by plaque purification to generate a reassortant library between a homologous murine RV EW strain (native to the mouse species) and a heterologous simian RV RRV strain (7). A set of genetic studies linked several murine RV genes to an efficient intestinal replication and spread phenotype in vivo (7). Using this system, we identified murine RV genes encoding NSP1 along with a constellation of VP3, NSP2, and NSP3 that seemed to work in concert to promote intestinal replication in a host-specific manner (7). The precise roles of these viral proteins in promoting RV replication are not yet defined. With the advent of a tractable RV reverse-genetics system (16), we now have the ability to more specifically dissect and examine these individual genes and their functions in RV replication at a molecular level.
In this study, we address a key gap in our knowledge of RV-host interactions. What are the IFN-associated antiviral effectors? And of the RV genes that are different between homologous and heterologous RVs, what viral factors, if any, confer resistance to host IFN signaling? Here, we show that the IFN-inducible 2′-5′-oligoadenylate synthetase (OAS)-RNase L pathway, recently shown to be targeted by the RV VP3 2′-5′-phosphodiesterase (PDE) domain (17), suppresses RV replication in a virus strain- and host species-specific manner both in vitro and in vivo. Using a set of molecular tools, including the recently described RV reverse-genetics system, we demonstrate, for the first time, that VP3’s PDE activity functionally antagonizes RNase L signaling and is critical for successful RV replication and pathogenesis in vivo.
RESULTS
RV VP3 confers viral resistance to IFN signaling in an RV strain-specific manner.
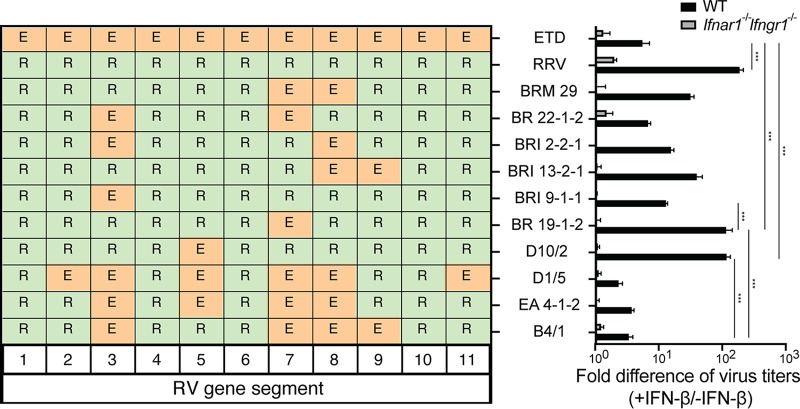
To uncover the roles of specific RV genes and their protein products in resisting IFN-mediated antiviral factors, we first examined the replication of homologous or heterologous RVs in mock- or IFN-β-treated murine embryonic fibroblasts (MEFs). The replication of the simian RV RRV strain, measured by a focus-forming unit (FFU) assay, was significantly reduced (∼130-fold) in IFN-β-stimulated MEFs compared to wild-type (WT) MEFs (Fig. 1). In contrast, the replication of the tissue culture-adapted murine RV ETD strain was minimally affected by IFN pretreatment (Fig. 1), suggesting that distinct RV strains responded differently to IFN-β-stimulated cellular signaling. To identify the specific RV gene segment(s) in the murine RV that was responsible for the IFN-β resistance phenotype, we took advantage of a collection of reassortants that we previously isolated from murine RV- and simian RV-coinfected suckling mice (7). Importantly, all the reassortant RVs that contain gene 3 (which encodes VP3) derived from the murine RV EW strain background, including BR 22-1-2, BRI 2-2-1, BRI 9-1-1, D1/5, and EA 4-1-2, exhibited reduced insensitivity to exogenous IFN signaling (Fig. 1) (<15-fold). However, reassortant RVs that encode VP3 derived from the simian RV RRV strain, including BRM 29, BR 13-2-1, BR 19-1-2, and D10/2, were highly sensitive (>30-fold) to IFN treatment (Fig. 1). Gene 8, which encodes NSP2, albeit to a lesser degree, also contributed to this phenotype. Therefore, RV’s ability to counteract exogenous IFN activity and replicate efficiently cosegregates with VP3 and NSP2 originating from the murine parental virus. As expected, IFN-β had no effect on RV replication in MEFs isolated from Ifnar1−/− Ifngr1−/− mice that lack the type I IFN receptor (Fig. 1). Collectively, using MEFs and RV genetics, we demonstrate that RV strains responded differently to exogenous IFN treatment and that the murine RV gene encoding VP3 cosegregated with enhanced virus replication in IFN-treated MEFs in a strain-specific manner.
FIG 1.
Murine RV VP3 confers resistance to IFN antiviral activity in MEFs. WT and Ifnar1−/− Ifngr1−/− MEFs were pretreated with or without 1,000 U/ml murine IFN-β for 24 h and then infected with the indicated RV strains (MOI = 1). At 24 h postinfection (hpi), virus-infected cells were harvested, and virus yields were determined by a focus-forming unit (FFU) assay. Fold differences in virus titers were calculated as follows: yield of WT or Ifnar1−/− Ifngr1−/− MEFs with IFN treatment (FFU per milliliter)/yield of MEFs without IFN treatment. The fold differences in virus titers presented are the means of quadruple data points. R represents viral genes from the simian RV RRV strain, and E represents viral genes from the murine RV EW strain.
The OAS-RNase L pathway restricts heterologous RV replication in mice.
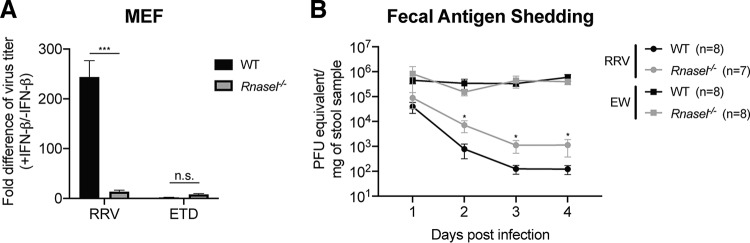
It was previously shown that a C-terminal PDE domain of RV VP3 enabled this domain to degrade 2′-5′-oligoadenylate (2-5A) and thus prevent RNase L activation (17). However, in those studies, the PDE activity of VP3 was examined as a truncated protein, not in the context of full-length RV VP3, and not in the context of an actual RV infection, either in vitro or in vivo. In order to directly dissect the physiological relevance of the OAS-RNase L pathway for actual RV replication in vitro or in vivo, we first isolated MEFs from WT or Rnasel knockout (KO) mice. Consistent with the data described above (Fig. 1), we found that IFN-β stimulation significantly reduced simian RV but not murine RV replication in WT MEFs (Fig. 2A). More importantly, simian RV became resistant to exogenous IFN treatment in Rnasel−/− MEFs, suggesting that the OAS-RNase L pathway may account for the majority of the IFN-mediated suppression of replication of the simian RV in MEFs from WT mice.
FIG 2.
RNase L signaling inhibits simian RV replication in vitro and in vivo. (A) WT and Rnasel−/− MEFs were pretreated with or without 1,000 U/ml murine IFN-β for 24 h and then infected with simian RRV or murine ETD RV at an MOI of 1 for 24 h. Virus yields were determined by a focus-forming unit assay. Fold differences in virus titers were calculated as follows: yield of WT or Rnasel−/− MEFs with IFN treatment (FFU per milliliter)/yield of MEFs without IFN treatment. The fold differences in virus titers presented are the means of quadruple data points. n.s., not significant. (B) Five-day-old WT and Rnasel−/− pups were orally inoculated with simian RRV (107 PFU) or murine EW (104 DD50). Fecal specimens were collected on the indicated days postinfection and subjected to a real-time PCR-based assay measuring RV gene NSP5 levels with standard curves to determine infectious virus particles per gram of stool sample. The numbers of mice in each group are indicated in parentheses.
Next, we examined whether the OAS-RNase L pathway also plays a role in controlling RV replication in vivo using a well-established suckling mouse model (18, 19). To this end, we perorally inoculated 5-day-old WT and Rnasel−/− suckling mice with either the simian RV RRV strain (107 FFU) or the murine RV EW strain (104 50% diarrhea doses [DD50]) and monitored shedding of viral RNA in the feces over a 4-day time course. The amount of virus for each strain was determined based on data from previous publications (6, 7, 10, 12, 19–21). For RRV infection, we detected significantly more (∼10-fold) infectious RVs in stool samples harvested from Rnasel−/− mice than in those from the WT littermates at 2, 3, and 4 days postinfection (dpi) (Fig. 2B). In line with our in vitro results, levels of EW shedding were comparable in the WT and Rnasel−/− groups (Fig. 2B). Taken together, these results indicate that the OAS-RNase L pathway plays a significant role in restricting the replication of heterologous RV both in vitro and in vivo.
The OAS-RNase L pathway restricts the replication of certain RV strains in human cells.
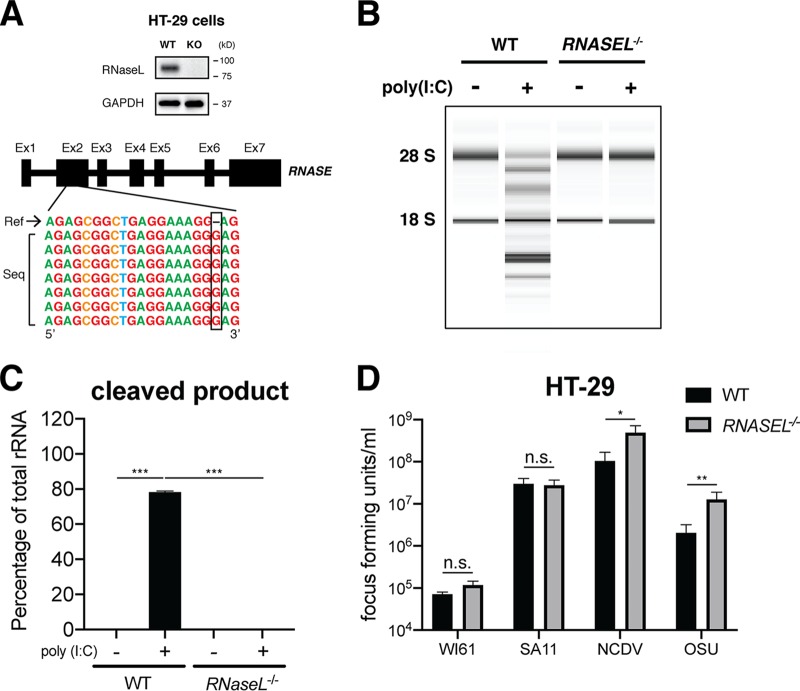
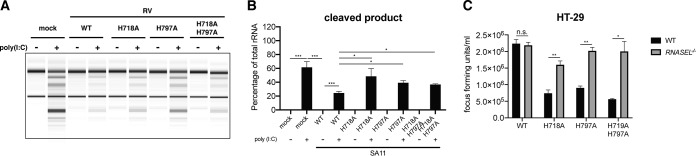
To facilitate a more mechanistic study of OAS-RNase L signaling in human intestinal cells in vitro, we utilized CRISPR-Cas9 to genetically delete RNASEL in HT-29 cells, a human intestinal epithelial cell (IEC) line commonly used for RV studies because of its small intestinal features (20, 22). RNase L depletion was confirmed by both Western blotting and Sanger sequencing (Fig. 3A). RNase L-deficient HT-29 cells were fully viable and did not exhibit altered cell proliferation or metabolic activities (data not shown). We also used an rRNA cleavage-based assay (23) to confirm the loss of function of the OAS-RNase L pathway in RNASEL−/− HT-29 cells. Specifically, following the transfection of cells with poly(I·C), a dsRNA mimetic that activates the OAS enzymes (24–26), rRNA, represented as the 18S and 28S bands, was prominently degraded in WT HT-29 cells into smaller digested fragments (Fig. 3B and C). In contrast, RNASEL−/− HT-29 cells remained completely resistant to poly(I·C)-induced rRNA degradation (Fig. 3B and C).
FIG 3.
RNase L depletion facilitates strain-specific RV replication in HT-29 cells. (A) Single clonal RNASEL−/− HT-29 cells were generated by CRISPR-Cas9 (detailed in Materials and Methods) and validated by Western blotting (top) and Sanger sequencing (bottom). (B) WT and RNASEL−/− HT-29 cells were transfected with 4.0 μg/ml poly(I·C) for 9 h, and total RNA was extracted and resolved by an RNA chip assay. The positions of 18S and 28S rRNAs are indicated. (C) Quantification of intact 18S and 28S rRNA bands and RNase L cleavage products in panel B. The data presented are the arithmetic means ± standard deviations from three independent experiments. (D) WT and RNASEL−/− HT-29 cells were infected with the indicated RV stains (MOI = 1) for 24 h. Virus yields were determined by a focus-forming unit assay. The virus growth ratio was calculated as follows: yield of RNASEL−/− (FFU per milliliter)/yield of WT (FFU per milliliter) HT-29 cells. The ratios presented are the means of quadruple data points.
Paralleling our findings in WT and Rnasel-deficient MEFs (Fig. 2A), our findings in human HT-29 cells showed a virus strain-specific and partial-host-range phenotype (Fig. 2A). The replication of WI61, a human RV strain, and SA11, a simian RV strain, was not affected by the loss of RNase L signaling (Fig. 3D). However, virus titers of more distantly related RVs, including the bovine RV NCDV strain and the porcine RV OSU strain, were enhanced by approximately 5- to 7-fold in RNASEL−/− cells compared to those in WT HT-29 cells (Fig. 3D), further highlighting RNase L as an RV strain-specific inhibitory mechanism.
Intact PDE activity is required for OAS-RNase L inhibition by RV VP3.
In order to study the role of PDE activity in the full-length VP3 protein, we used site-directed mutagenesis to specifically replace the two key catalytic histidine (H) residues at positions 718 and 797 (17) with arginine (R), either individually or in combination. All green fluorescent protein (GFP)-tagged VP3 proteins, including the WT, the single mutants H718R and H797R, and a double mutant, H718R/H797R, were equally expressed when transfected into HEK293 cells (Fig. 4A). We also found by fluorescence imaging that the mutant proteins were expressed at comparable levels and that the specific PDE site mutations did not alter the subcellular localization of VP3 (Fig. 4B). Compared to the diffuse GFP signal in the cytoplasm and nucleus, WT and all three mutant VP3s colocalized with cytochrome C oxidase IV (COXIV), a well-characterized mitochondrial marker, as previously reported (20) (Fig. 4B). Given that the VP3 PDE domain alone inhibited RNase L-mediated rRNA degradation (17), we next tested the ability to block RNase L activation by different VP3 constructs. Using the previously established rRNA degradation assay (Fig. 3B), we noticed that compared to mock transfection or the GFP empty vector control, full-length VP3 significantly inhibited poly(I·C)-induced 18S and 28S cleavage (Fig. 4C and D). However, mutation of either H site to R abolished VP3’s ability to inhibit RNase L signaling (Fig. 4C and D), suggesting that the PDE activity is essential for RNase L inhibition by VP3.
FIG 4.
VP3 PDE activity is required for inhibiting the OAS-RNase L pathway in the full-length VP3 protein. (A) HEK293 cells were transfected with the GFP empty vector or GFP-tagged WT and VP3 mutants from the simian RV RRV strain. At 24 h, cell lysates were harvested and either immunoprecipitated with an anti-GFP antibody and probed for GFP in a Western blot (top) or directly examined by Western blotting for GAPDH (bottom). (B) HEK293 cells were transfected with the GFP empty vector or the GFP-tagged WT and VP3 mutants from the simian RV RRV strain. At 24 hpt, cells were fixed with 4% paraformaldehyde, stained for COXIV (mitochondrial marker [mito]), and imaged with a Zeiss LSM710 confocal microscope. Inset images are labeled with red boxes in the merged channel. Yellow signals, indicative of VP3 and COXIV colocalization, are pointed out with white arrows. (C) HEK293 cells were transfected with GFP and the indicated RRV VP3 WT and mutants for 24 h and transfected with 1 μg/ml poly(I·C) for another 6 h. Total RNA was then extracted and resolved by an RNA chip assay. (D) Quantification of intact 18S and 28S rRNA bands and RNase L cleavage products in panel C. The arithmetic means ± standard deviations from three independent experiments are shown.
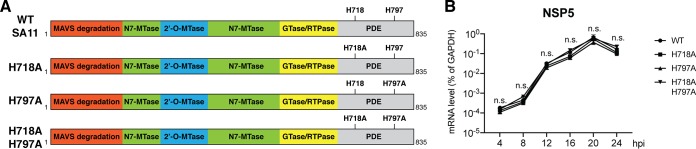
Next, we examined the relative contribution of VP3 PDE antagonism of the RNase L pathway to RV replication. To this end, we utilized the recently described RV reverse-genetics system (16) to rescue recombinant RVs that encode either WT or mutant VP3s. We successfully recovered four recombinant viruses: the parental simian RV SA11 strain and three mutant RVs with key VP3 H residues mutated to alanine (A), creating the single mutants H718A and H797A and a double mutant, H718A/H797A (Fig. 5A). The ability of these RVs to replicate in MA104 cells, an IFN-defective monkey kidney cell line most frequently used for RV propagation (27), was not compromised by any of the VP3 mutations at any time point examined (Fig. 5B). We also used plaque assays to quantify the yield of infectious RVs (WT SA11 and three VP3 mutants) from infected MA104 cells and observed no replication differences (data not shown). The mutant RVs were genetically stable and did not revert back to WT VP3 sequences during serial passage in MA104 cells (data not shown).
FIG 5.
VP3 PDE mutations minimally affect RV replication in MA104 cells. (A) Schematic diagram of the recombinant simian RV SA11 strain rescued using reverse genetics. MTase, methyltransferase; RTPase, RNA 5′-triphosphatase. (B) MA104 cells were infected by recombinant WT or VP3 mutant RVs at an MOI of 1. At the indicated time points, total RNA was harvested, and the RV gene NSP5 level was measured by reverse transcription-quantitative PCR (RT-qPCR) and normalized to the value for GAPDH.
We hypothesized that this lack of attenuation might be due to a relatively weak IFN-RNase L response in MA104 cells. Therefore, we proceeded to examine the capacity of WT and VP3 mutant RVs to block RNase L activation in IFN-signaling-competent HT-29 cells. Interestingly, there was no rRNA degradation observed in RV infection alone without poly(I·C) stimulation. We reason that this could be due to an alternative mechanism of RV-mediated RNase L inhibition, independent of the PDE activity (28). Consistent with our hypothesis, poly(I·C)-induced rRNA degradation was significantly blocked by WT VP3 RV (Fig. 6A and B). In contrast, poly(I·C) stimulation-mediated rRNA degradation was enhanced after the H-to-A mutations, in particular at the H797 site (Fig. 6A and B). As a consequence, these VP3 PDE mutant RVs also replicated less efficiently than the WT recombinant virus in WT HT-29 cells (Fig. 6C). Importantly, such a defect was rescued in RNASEL−/− HT-29 cells, suggesting that the RNase L pathway accounts for the attenuated replication phenotypes of these VP3 mutant RVs (Fig. 6C).
FIG 6.
RV VP3 PDE activity contributes to viral replication in HT-29 cells. (A) WT HT-29 cells were transfected with 4 μg/ml of poly(I·C) for 1 h and subsequently infected with recombinant WT or VP3 mutant RVs at an MOI of 10 for 9 h. Total RNA was extracted and resolved by an RNA chip assay. (B) Quantification of intact 18S and 28S rRNA bands and RNase L cleavage products in panel A. The arithmetic means ± standard deviations from three independent experiments are shown. (C) WT and RNASEL−/− HT-29 cells were infected with recombinant WT or VP3 mutant RVs at an MOI of 1 for 24 h. Virus particles in the supernatant were titrated by an FFU assay.
VP3 PDE activity promotes intestinal RV replication in vivo.
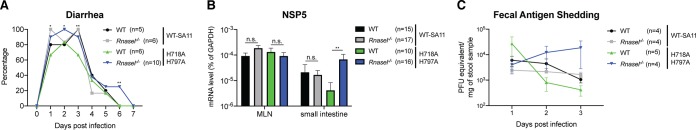
Finally, to examine the pathophysiological relevance of RNase L-mediated RV inhibition and VP3 PDE-dependent RNase L counteraction, we turned to an in vivo mouse model (18, 19) and studied the replication and pathogenesis of WT and VP3 mutant RVs in WT C57BL/6 and Rnasel−/− mice on the same genetic background. Five-day-old suckling pups were orally inoculated with an equal dose of 106 PFU of the recombinant WT SA11 or the VP3 double mutant H718A/H797A viruses. We monitored the induction of diarrhea and intraintestinal replication by standard diarrhea scores (29) and real-time PCR assays (19), respectively.
In C57BL/6 mice, WT SA11 had slightly more diarrhea occurrence than the VP3 mutant virus at 1 and 3 dpi (Fig. 7A). In Rnasel−/− mice, we observed overall increases in diarrhea development for both viruses at 1, 2, 3, and 6 dpi (Fig. 7A). Altogether, these results suggest that RNase L at least partially regulates RV-related disease severity and that encoding a functional copy of VP3 confers small yet significant advantages over viruses with VP3 PDE point mutations.
FIG 7.
RV VP3 PDE activity is important for viral replication in vivo. (A) Five-day-old WT and Rnasel−/− pups were orally inoculated with 106 PFU of WT recombinant SA11 or SA11 encoding H718A/H797A mutated VP3. Diarrhea was monitored from days 1 to 7 postinfection. (B) Mesenteric lymph node (MLN) and small intestinal tissues from infected mice were collected at 3 days postinfection. Vial gene NSP5 expression was measured by RT-qPCR and normalized to that of GAPDH. (C) Stool samples were harvested from infected mice at 1, 2, and 3 days postinfection, and viral RNA levels were measured by RT-qPCR and converted to PFU counts based on the standard curve. The numbers of mice in each group are indicated in parentheses.
Vis-à-vis the viral load within tissues, we quantified virus replication levels in all four groups at 3 dpi. We did not observe detectable numbers of RV RNA copies in the liver, biliary tract, or blood (data not shown). In addition, almost identical levels of WT and H718A/H797A VP3 SA11 were found in the mesenteric lymph node (MLN), the immune cell compartment of the gastrointestinal tract, in C57BL/6 mice (30). In contrast, we detected significantly lower (1-log) levels of replication of the VP3 mutant virus than of WT SA11 in the small intestinal tissues (Fig. 7B), suggesting that the effect of VP3 on enhancing RV replication via its PDE domain is likely to be a small bowel epithelium-specific phenotype. Furthermore, H718A/H797A VP3 SA11 replication was enhanced by at least 10-fold in the small intestinal epithelial cells of Rnasel−/− mice (Fig. 7B), comparably to that of WT SA11, suggesting that once the restriction pressure is lifted, having VP3 PDE activity does not confer further replication advantages in the gut. Finally, the amount of H718A/H797A mutant SA11 in fecal specimens was decreased compared to that of the WT VP3 virus in WT mice at 2 and 3 dpi, and such attenuation was again rescued in the absence of Rnasel (Fig. 7C). Collectively, using CRISPR-Cas9 KO cells, animal models, and genetic tools (including natural reassortants and the reverse-genetics system), we demonstrated that in the small intestine, OAS-RNase L signaling plays an important role in the restriction of RVs in a virus strain-specific manner.
DISCUSSION
The levels of RV replication and virulence (in this model system, diarrhea) are strongly influenced by virus strains, cell types, and host species. Viral genetic factors controlling these phenotypes are likely to be multigenic. Here, we examined the molecular mechanisms underlying differential RV resistance to exogenous IFN signaling. Using a naturally derived reassortant RV library (7), we pinpointed RV’s ability to antagonize the effects of exogenous type I IFN to gene segment 3, which encodes the RNA-capping enzyme VP3 (Fig. 1). We then determined that in both human HT-29 cells and suckling mice in vivo, the OAS-RNase L pathway is key to mediating IFN’s suppression of selected heterologous RVs, and it is counteracted by the presence of a homologous RV VP3 (Fig. 2 to 4). We next demonstrated, using the recently established reverse-genetics system (16), that RVs encoding VP3 PDE mutants fail to inhibit RNase L and replicate poorly in HT-29 cells and in mouse intestine (Fig. 6 and 7). In conclusion, we used a multipronged approach to directly demonstrate that VP3-encoded PDE activity can counteract RNase L signaling within the context of RV infection and is important for promoting RV replication and pathogenesis in vivo.
Cell specificity becomes an important aspect of studying innate immunity, as many pattern recognition receptors and antiviral molecules are expressed in a highly specific manner in certain cells and tissues (31). In our study of the OAS-RNase L system, we also observed such differential activation in MA104 versus HT-29 cells (Fig. 5B and Fig. 6C). In fact, a cell-type-specific activation of OAS-RNase L was previously reported for coronavirus infections (32–36). This is particularly important in light of the recent 2019 novel coronavirus (SARS-CoV-2) outbreak (37). It would be interesting to examine whether this new bat coronavirus exhibits PDE activity to inhibit RNase L function in a cell-type-dependent manner, paving the way for future targeted attenuation in vaccine design.
Our data demonstrating that point mutations within the PDE domain of VP3 lead to reduced RV replication in HT-29 cells are particularly intriguing (Fig. 6). Given that the human RV vaccines now in use are all based on live attenuation (8), we predict that targeted mutations of VP3 PDE activity would create recombinant viruses that are less virulent and potentially safer. Theoretically, with the additional targeted VP3 attenuation, a higher dose of RV vaccine could be administered to enhance immunogenicity without causing diarrhea or other adverse effects. Testing of this hypothesis would require the recovery of a fully recombinant human RV using the reverse-genetics system, which would ideally then be tested in a primary human intestinal enteroid-hematopoietic coculture system. The other alternative is to further investigate the role of VP3 and PDE mutations in the background of a completely infectious and virulent murine RV. Such a recombinant viral construct will enable studies of adaptive immune responses both during the initial infection and using a subsequent challenge model to examine vaccine efficacy.
Another interesting observation made in this study is that in vivo, heterologous simian RV replication is suppressed by RNase L signaling in the intestinal epithelium but not in the MLNs (Fig. 7). Cell-type-specific RNase L activity has previously been reported (35, 36). This is also reminiscent of our previous findings with heterologous RV infections in Mavs−/− mice (20). We reason that this could be due to distinct IFN responses in epithelial cells compared to hematopoietic cells (38). It is plausible that immune cells in MLNs have other non-IFN-mediated antiviral signaling pathways (i.e., Toll-like receptors or the inflammasome, etc.) to compensate for the loss of RNase L. It is also possible that other IFN-stimulated genes besides OASs are at play in this immune compartment, whereas OAS-RNase L accounts for the majority of IFN’s antiviral activity in epithelial cells. Using a now readily available and highly tractable reverse-genetics system, we report the recovery of the first VP3 mutant RVs and demonstrate that intact PDE activity is required for optimal virus replication in vitro and in vivo. Although much work remains, the present study represents an important additional step toward the mechanistic understanding of the complex and multigenic nature of RV-IFN signaling interactions.
MATERIALS AND METHODS
Cells and viruses.
The African green monkey MA104 kidney epithelial cell line was cultured in complete M199 medium. The human embryonic fibroblast HEK293 cell line was cultured in complete Dulbecco’s modified Eagle’s medium (DMEM). The human colonic epithelial HT-29 cell line was cultured in complete advanced DMEM–F-12 medium. Human and animal RV strains, including human WI61, simian RRV and SA11, bovine NCDV, porcine OSU, murine EW and ETD (a tissue culture-adapted strain of the EDIM-derived EW strain), and reassortants between RRV and EW, were used in this study. EW was propagated in suckling pups as previously described (39). The other RV strains were propagated and titers were determined by plaque assays in MA104 cells (40).
Mice and RV infection.
Wild-type 129sv, C57BL/6, and Ifnar1−/− Ifngr1−/− mice were originally purchased from the Jackson Laboratory and maintained as individual in-house breeding colonies. Rnasel−/− mice were purchased from the Jackson Laboratory and were originally developed by, and provided with permission of, Robert H. Silverman (41). Isolation of MEFs from different mouse backgrounds was performed as described previously (42). MEF infection and virus titration were performed as described previously (7). Five-day-old WT and Rnasel−/− pups were orally inoculated with simian RRV (107 PFU), murine EW (104 DD50), or the recombinant simian RV SA11 strain (106 PFU) rescued using the reverse genetic method (16). Fecal specimens were collected on the indicated days postinfection and subjected to a real-time PCR-based assay measuring RV gene NSP5 levels with standard curves to determine infectious virus particles per gram of stool sample, as described previously (29, 43). All mice were housed at the Veterinary Medical Unit of the Palo Alto VA Health Care System. All animal studies were approved by the Stanford Institutional Animal Care Committee.
Generation of recombinant RVs by the reverse-genetics method.
The recombinant simian RV SA11 strain was rescued as described previously using the reverse genetic method (16, 44). BHK-T7 cells were seeded into 6-well plates for 48 h and then transfected with the plasmids pT7-VP1SA11, pT7-VP2SA11, pT7-VP3SA11, pT7-VP4SA11, pT7-VP6SA11, pT7-VP7SA11, pT7-NSP1SA11, pT7-NSP2SA11, pT7-NSP3SA11, pT7-NSP4SA11, and pT7-NSP5SA11 to generate SA11 WT virus. We generated pT7-VP3H718ASA11, pT7-VP3H797ASA11, and pT7-VP3H718AH797ASA11 using site-directed mutagenesis. At 24 h posttransfection (hpt), we replaced the medium with serum-free DMEM. After another 24 h, we removed the medium, seeded 105 cells/well of MA104 cells in serum-free DMEM supplemented with trypsin (0.5 μg/ml), and incubated the cells for 72 h. MA104 cells were infected by recombinant WT or VP3 mutant RVs at a multiplicity of infection (MOI) of 1 to quantify replication kinetics.
Generation of CRISPR-Cas9 knockout cells.
Single clonal RNASEL knockout HT-29 cells were generated using transient transfection of a PX458 vector (Addgene plasmid 48138) that expresses Cas9 and single guide RNA (sgRNA) against RNASEL. Green fluorescent protein (GFP)-positive cells were single-cell sorted into 96-well plates at 48 h posttransfection using a BD Aria II instrument, allowed to grow into colonies, and screened by Western blotting and Sanger sequencing for knockout single clones as previously described (22).
RNase L-mediated rRNA cleavage assay.
HEK293 cells in 24-well plates were transfected with RRV WT and PDE mutant VP3 using Lipofectamine 3000. At 24 hpt, cells were transfected with 1 μg/ml of poly(I·C) for 6 h. For HT-29 cells, 4 μg/ml of poly(I·C) was used for 1 h, and the cells were then infected with recombinant WT or VP3 mutant RVs at an MOI of 10 for 9 h. Total RNA was extracted using a Thermo Fisher PureLink RNA minikit and resolved by an RNA chip assay. The cleavage products of 18S and 28S rRNAs were quantitated using the Bioanalyzer system as described previously (28). For quantification, the areas below the curves of the regions corresponding to 18S and 28S and those between 18S and 28S were calculated from the electropherogram, and their sum was taken as 100%. The fraction represented by each band was then calculated.
Western blotting.
Cell lysates were harvested in radioimmunoprecipitation assay (RIPA) buffer supplemented with a protease inhibitor cocktail and a phosphatase inhibitor. Proteins were resolved by SDS-PAGE and analyzed as described previously (38) using antibodies for RNase L (catalog number aa139-167; LifeSpan) (1:1,000), GFP (catalog number A11122; Thermo Fisher) (1:1,000), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog number 60004-1; Proteintech) (1:1,000). Secondary antibodies were anti-rabbit (catalog number 7074; CST) (1:5,000) or anti-mouse (catalog number 7076; CST) (1:5,000) immunoglobulin G-horseradish peroxidase-linked antibodies. Protein bands were visualized with the Clarity ECL substrate, Amersham Hyperfilm, and a Structurix X-ray film processor.
Fluorescence imaging.
HEK293 cells in 8-well chamber slides were transfected with GFP-tagged WT or PDE mutant VP3 and fixed with 4% paraformaldehyde at 24 h, as previously described (22). Samples were then stained with a primary antibody against COXIV (catalog number 4850; Cell Signaling) and DAPI (4′,6-diamidino-2-phenylindole) (catalog number P36962; Thermo Fisher) and imaged with a Zeiss LSM 710 confocal microscope. Images were analyzed using Volocity v5.2.
Statistical analysis.
Data in the bar graphs are displayed as means ± standard errors of the means (SEM). Statistical significance of the data in Fig. 2A, Fig. 3D, and Fig. 6C was calculated by Student’s t test, and statistical significance of the data in Fig. 2B, Fig. 3C, Fig. 4D, Fig. 5B, Fig. 6B, and Fig. 7A and C was calculated by analysis of variance (ANOVA) using Prism 8 (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). Statistics in Fig. 1 were calculated using single-factor ANOVA and post hoc pairwise comparison. All experiments, unless otherwise noted, were repeated at least three times.
ACKNOWLEDGMENTS
We are grateful to Yize Li and Susan Weiss (University of Pennsylvania) for useful discussions. We thank all members of the Greenberg lab for their input.
This work is supported by National Institutes of Health (NIH) grants R01 AI125249 and U19 AI116484 and by VA Merit grant GRH0022 awarded to H.B.G., NIH grants K99/R00 AI135031 and R01 AI150796 and an early career award from the Thrasher Research Fund to S.D., and NIH grants R01 AI104887 and R01 AI135922 to R.H.S.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization–Coordinated Global Rotavirus Surveillance Network. 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 2.Parashar UD, Gibson CJ, Bresee JS, Glass RI. 2006. Rotavirus and severe childhood diarrhea. Emerg Infect Dis 12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blutt SE, Conner ME. 2007. Rotavirus: to the gut and beyond! Curr Opin Gastroenterol 23:39–43. doi: 10.1097/MOG.0b013e328011829d. [DOI] [PubMed] [Google Scholar]
- 4.Ward LA, Rosen BI, Yuan L, Saif LJ. 1996. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J Gen Virol 77(Part 7):1431–1441. doi: 10.1099/0022-1317-77-7-1431. [DOI] [PubMed] [Google Scholar]
- 5.Ramig RF. 1988. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb Pathog 4:189–202. doi: 10.1016/0882-4010(88)90069-1. [DOI] [PubMed] [Google Scholar]
- 6.Feng N, Sen A, Wolf M, Vo P, Hoshino Y, Greenberg HB. 2011. Roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J Virol 85:2686–2694. doi: 10.1128/JVI.02408-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng N, Yasukawa LL, Sen A, Greenberg HB. 2013. Permissive replication of homologous murine rotavirus in the mouse intestine is primarily regulated by VP4 and NSP1. J Virol 87:8307–8316. doi: 10.1128/JVI.00619-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciarlet M, Schodel F. 2009. Development of a rotavirus vaccine: clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 27(Suppl 6):G72–G81. doi: 10.1016/j.vaccine.2009.09.107. [DOI] [PubMed] [Google Scholar]
- 9.Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 10.Lin JD, Feng N, Sen A, Balan M, Tseng HC, McElrath C, Smirnov SV, Peng J, Yasukawa LL, Durbin RK, Durbin JE, Greenberg HB, Kotenko SV. 2016. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog 12:e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vancott JL, McNeal MM, Choi AH, Ward RL. 2003. The role of interferons in rotavirus infections and protection. J Interferon Cytokine Res 23:163–170. doi: 10.1089/107999003321532501. [DOI] [PubMed] [Google Scholar]
- 12.Feng N, Kim B, Fenaux M, Nguyen H, Vo P, Omary MB, Greenberg HB. 2008. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J Virol 82:7578–7590. doi: 10.1128/JVI.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angel J, Franco MA, Greenberg HB, Bass D. 1999. Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J Interferon Cytokine Res 19:655–659. doi: 10.1089/107999099313802. [DOI] [PubMed] [Google Scholar]
- 14.Pesavento JB, Crawford SE, Estes MK, Prasad BV. 2006. Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol 309:189–219. doi: 10.1007/3-540-30773-7_7. [DOI] [PubMed] [Google Scholar]
- 15.McDonald SM, Matthijnssens J, McAllen JK, Hine E, Overton L, Wang S, Lemey P, Zeller M, Van Ranst M, Spiro DJ, Patton JT. 2009. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog 5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci U S A 110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng N, Franco MA, Greenberg HB. 1997. Murine model of rotavirus infection. Adv Exp Med Biol 412:233–240. doi: 10.1007/978-1-4899-1828-4_35. [DOI] [PubMed] [Google Scholar]
- 19.Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li HB, Wang G, Lei X, de Zoete MR, Zhao J, Zheng Y, Chen H, Zhao Y, Jurado KA, Feng N, Shan L, Kluger Y, Lu J, Abraham C, Fikrig E, Greenberg HB, Flavell RA. 2017. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546:667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding S, Zhu S, Ren L, Feng N, Song Y, Ge X, Li B, Flavell RA, Greenberg HB. 2018. Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. Elife 7:e39494. doi: 10.7554/eLife.39494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenaux M, Cuadras MA, Feng N, Jaimes M, Greenberg HB. 2006. Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J Virol 80:5219–5232. doi: 10.1128/JVI.02664-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding S, Diep J, Feng N, Ren L, Li B, Ooi YS, Wang X, Brulois KF, Yasukawa LL, Li X, Kuo CJ, Solomon DA, Carette JE, Greenberg HB. 2018. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat Commun 9:1485. doi: 10.1038/s41467-018-03782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Banerjee S, Wang Y, Goldstein SA, Dong B, Gaughan C, Silverman RH, Weiss SR. 2016. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci U S A 113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem 270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding S, Mooney N, Li B, Kelly MR, Feng N, Loktev AV, Sen A, Patton JT, Jackson PK, Greenberg HB. 2016. Comparative proteomics reveals strain-specific beta-TrCP degradation via rotavirus NSP1 hijacking a host Cullin-3-Rbx1 complex. PLoS Pathog 12:e1005929. doi: 10.1371/journal.ppat.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez-Tacuba L, Rojas M, Arias CF, López S. 2015. Rotavirus controls activation of the 2′-5′-oligoadenylate synthetase/RNase L pathway using at least two distinct mechanisms. J Virol 89:12145–12153. doi: 10.1128/JVI.01874-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Ding S, Feng N, Mooney N, Ooi YS, Ren L, Diep J, Kelly MR, Yasukawa LL, Patton JT, Yamazaki H, Shirao T, Jackson PK, Greenberg HB. 2017. Drebrin restricts rotavirus entry by inhibiting dynamin-mediated endocytosis. Proc Natl Acad Sci U S A 114:E3642–E3651. doi: 10.1073/pnas.1619266114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spahn TW, Kucharzik T. 2004. Modulating the intestinal immune system: the role of lymphotoxin and GALT organs. Gut 53:456–465. doi: 10.1136/gut.2003.023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. 2014. Cell-type-specific effects of RNase L on viral induction of beta interferon. mBio 5:e00856-14. doi: 10.1128/mBio.00856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birdwell LD, Zalinger ZB, Li Y, Wright PW, Elliott R, Rose KM, Silverman RH, Weiss SR. 2016. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J Virol 90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Dong B, Wei Z, Silverman RH, Weiss SR. 2019. Activation of RNase L in Egyptian rousette bat-derived RoNi/7 cells is dependent primarily on OAS3 and independent of MAVS signaling. mBio 10:e02414-19. doi: 10.1128/mBio.02414-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Weiss SR. 2016. Antagonism of RNase L is required for murine coronavirus replication in Kupffer cells and liver sinusoidal endothelial cells but not in hepatocytes. J Virol 90:9826–9832. doi: 10.1128/JVI.01423-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L, Birdwell LD, Wu A, Elliott R, Rose KM, Phillips JM, Li Y, Grinspan J, Silverman RH, Weiss SR. 2013. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol 87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Horby PW, Hayden FG, Gao GF. 2020. A novel coronavirus outbreak of global health concern. Lancet 395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolen CR, Ding S, Robek MD, Kleinstein SH. 2014. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludert JE, Feng N, Yu JH, Broome RL, Hoshino Y, Greenberg HB. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J Virol 70:487–493. doi: 10.1128/JVI.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren L, Ding S, Song Y, Li B, Ramanathan M, Co J, Amieva MR, Khavari PA, Greenberg HB. 2019. Profiling of rotavirus 3′UTR-binding proteins reveals the ATP synthase subunit ATP5B as a host factor that supports late-stage virus replication. J Biol Chem 294:5993–6006. doi: 10.1074/jbc.RA118.006004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J 16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng N, Sen A, Nguyen H, Vo P, Hoshino Y, Deal EM, Greenberg HB. 2009. Variation in antagonism of the interferon response to rotavirus NSP1 results in differential infectivity in mouse embryonic fibroblasts. J Virol 83:6987–6994. doi: 10.1128/JVI.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sen A, Rothenberg ME, Mukherjee G, Feng N, Kalisky T, Nair N, Johnstone IM, Clarke MF, Greenberg HB. 2012. Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc Natl Acad Sci U S A 109:20667–20672. doi: 10.1073/pnas.1212188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komoto S, Kanai Y, Fukuda S, Kugita M, Kawagishi T, Ito N, Sugiyama M, Matsuura Y, Kobayashi T, Taniguchi K. 2017. Reverse genetics system demonstrates that rotavirus nonstructural protein NSP6 is not essential for viral replication in cell culture. J Virol 91:e00695-17. doi: 10.1128/JVI.00695-17. [DOI] [PMC free article] [PubMed] [Google Scholar]