Flaviviruses and other arthropod-transmitted viruses represent a widespread global health problem, with limited treatment options currently available. Thus, a better understanding of the cellular requirements for their infection is needed. Both DENV and ZIKV rely on expansion of the endoplasmic reticulum (ER) and the induction of autophagy to establish productive infections. However, little is known regarding the interplay between the requirements for autophagy initiation during infection and the mechanisms used by these viruses to avoid clearance through the autophagic pathway. Our study highlights the importance of the host factor BPIFB3 in regulating flavivirus replication and further confirms that the RETREG1-dependent reticulophagy pathway is antiviral to both DENV and ZIKV.
KEYWORDS: BPIFB3, autophagy, flavivirus, reticulophagy, viral membrane manipulation, viral replication
ABSTRACT
Flaviviruses, including dengue virus (DENV) and Zika virus (ZIKV), rely heavily on the availability of endoplasmic reticulum (ER) membranes throughout their life cycle, and degradation of ER membranes restricts flavivirus replication. Accordingly, DENV and ZIKV restrict ER turnover by protease-mediated cleavage of reticulophagy regulator 1 (RETREG1), also known as FAM134B, an autophagy receptor responsible for targeted ER sheet degradation. Given that the induction of autophagy may play an important role in flavivirus replication, the antiviral role of RETREG1 suggests that specialized autophagic pathways may have differential effects on the flavivirus life cycle. We previously identified BPI fold-containing family B member 3 (BPIFB3) as a regulator of autophagy that negatively controls enterovirus replication. Here, we show that in contrast to enteroviruses, BPIFB3 functions as a positive regulator of DENV and ZIKV infection and that its RNA interference-mediated silencing inhibits the formation of viral replication organelles. Mechanistically, we show that depletion of BPIFB3 enhances RETREG1-dependent reticulophagy, leading to enhanced ER turnover and the suppression of viral replication. Consistent with this, the antiviral effects of BPIFB3 depletion can be reversed by RETREG1 silencing, suggesting a specific role for BPIFB3 in regulating ER turnover. These studies define BPIFB3 as a required host factor for both DENV and ZIKV replication and further contribute to our understanding of the requirements for autophagy during flavivirus infection.
IMPORTANCE Flaviviruses and other arthropod-transmitted viruses represent a widespread global health problem, with limited treatment options currently available. Thus, a better understanding of the cellular requirements for their infection is needed. Both DENV and ZIKV rely on expansion of the endoplasmic reticulum (ER) and the induction of autophagy to establish productive infections. However, little is known regarding the interplay between the requirements for autophagy initiation during infection and the mechanisms used by these viruses to avoid clearance through the autophagic pathway. Our study highlights the importance of the host factor BPIFB3 in regulating flavivirus replication and further confirms that the RETREG1-dependent reticulophagy pathway is antiviral to both DENV and ZIKV.
INTRODUCTION
Flaviviruses, which include dengue virus (DENV) and Zika virus (ZIKV), are enveloped, positive-sense RNA viruses that replicate exclusively in association with endoplasmic reticulum (ER) membranes of infected cells (1, 2). Upon entry and uncoating, the flaviviral genome is directly translated as a single polyprotein and embedded in the ER. Following processing of the polyprotein by viral and host proteases, the nonstructural proteins induce expansion of the ER and the formation of viral replication organelles (ROs) (3–6). ROs function to sequester replication machinery within membrane-bound compartments and thus provide a high concentration of host and viral replication factors, while isolating replication intermediates from clearance by the innate immune system (7, 8).
Autophagy is an intrinsic cellular pathway that not only functions in cellular maintenance but also plays an important role in the clearance of intracellular pathogens (9, 10). In the absence of infection, macroautophagy, referred to here as autophagy, functions to degrade bulk cytoplasmic contents or damaged organelles by fusion with the lysosome, referred to as autophagic flux (11, 12). Autophagy can thus target many aspects of flavivirus infection, including direct clearance of viral particles and clearance of viral replication factories within the ER (13–15). However, the full relationship between flaviviruses and autophagy remains unclear. Evidence suggests that DENV and ZIKV induce autophagy as a mechanism to promote viral infection (16, 17). Specifically, DENV infection induces the degradation of lipid droplets by autophagy, in a process termed lipophagy, to allow increased energy during infection (18, 19). In contrast, we previously demonstrated that degradation of the ER through selective autophagy (in a process referred to as reticulophagy) restricts flavivirus replication, likely through the targeting of ROs for degradation (15). These studies, combined with the evidence that numerous autophagy components are required for flavivirus membrane manipulation and spread, suggest not only that individual autophagy pathways play a unique role during flavivirus infection but also that the timing of autophagy induction is important (20).
We previously identified bactericidal/permeability increasing protein (BPI) fold-containing family B member 3 (BPIFB3) as a regulator of infection with coxsackievirus group B (CVB), a positive-sense RNA virus belonging to the genus Enterovirus, through its negative regulation of a noncanonical form of autophagy (21). Similar to flaviviruses, CVB relies on the availability of intracellular membranes to establish replication compartments; however, the source of these cellular membranes is variable (22). In this study, we determined whether BPIFB3 was more broadly involved in regulating RNA virus replication, specifically focusing on flaviviruses because of their close association with the ER. Here, we show that in contrast to CVB, BPIFB3 expression is required for the replication of two flaviviruses, Dengue virus (DENV) and Zika virus (ZIKV). Moreover, we show that BPIFB3 regulates ER membrane morphology and that loss of BPIFB3 prevents the establishment of ER-localized flavivirus ROs. Our data provide evidence that BPIFB3 silencing increases the level of ER turnover through RETREG1-dependent reticulophagy and that inhibition of this pathway by silencing of RETREG1 expression restores flavivirus infection. Our study thus demonstrates the differential control of flavivirus and enterovirus replication by BPIFB3 and further suggests that reticulophagy is anti-flaviviral. These data also define a more specific role for BPIFB3 in regulating reticulophagy and provides insights into the cellular function of this protein.
RESULTS
BPIFB3 is required for efficient flavivirus infection.
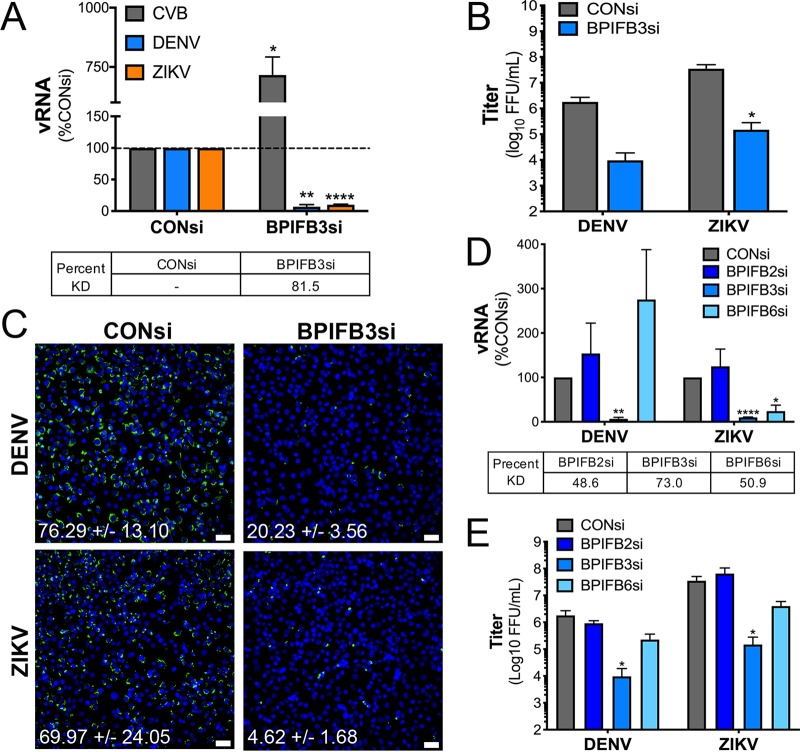
We previously identified BPIFB3 as a negative regulator of coxsackievirus group B (CVB) replication through the restriction of autophagy (21). To determine if BPIFB3 functions more broadly to regulate the replication of other RNA viruses, we assessed the impact of BPIFB3 silencing on flavivirus replication. For these studies, we utilized human brain microvascular endothelial cells (HBMEC), an immortalized cell line model of the blood-brain barrier microvasculature (23). HBMEC were transfected with a small interfering RNA (siRNA)-targeting BPIFB3 (BPIFB3si) or a scrambled control siRNA (CONsi) and infected with DENV or ZIKV at a multiplicity of infection (MOI) of 1 for 48 h or with CVB for 16 h. Infection was quantified by reverse transcription-quantitative PCR (RT-qPCR) for DENV, ZIKV, and CVB viral RNA (vRNA) (Fig. 1A) or by fluorescent focus unit assays (FFU) for DENV and ZIKV (Fig. 1B). In contrast to CVB, BPIFB3 silencing significantly restricted both DENV and ZIKV infection, resulting in an approximately 90% reduction in vRNA and a 100-fold decrease in infectious-particle production (Fig. 1A and B). Consistent with this, we observed a stark decrease in staining of double-stranded vRNA, a replication intermediate of DENV and ZIKV, in BPIFB3si-transfected cells infected with DENV and ZIKV compared to that in CONsi-transfected cells (Fig. 1C).
FIG 1.
BPIFB3 is specifically required for both DENV and ZIKV infection. (A) Infection levels of CVB, DENV, and ZIKV determined by RT-qPCR. Data are presented as percent fold change from CONsi-transfected cells. Knockdown (KD) efficiency for BPIFB3 was determined by RT-qPCR and is shown as percent KD compared to CONsi. (B) Titration (by fluorescence focus unit assay) of DENV and ZIKV infectious-particle production from HBMEC from panel A. (C) Immunofluorescence microscopy for dsRNA (green), a replication intermediate, in CONsi- or BPIFB3si-transfected HBMEC. The average percent infected cells with the standard deviation is listed below each image. Bar, 50 μm. (D) RT-qPCR for vRNA infection levels of DENV and ZIKV in cells depleted of BPIFB2, BPIFB3, or BPIFB6. Percent KD compared to CONsi is shown for each BPIFBsi. (E) Titration of DENV and ZIKV by FFU assay. Student's t test was performed to determine statistical significance (*, P < 0.05; **, P < 0.01; ****, P < 0.0001).
To determine whether other members of the BPIFB family are also required for flavivirus replication, we tested the effects of RNA interference (RNAi)-mediated silencing of the other ER-localized BPIFB proteins, including BPIFB2 and BPIFB6. We showed previously that in contrast to BPIFB3, BPIFB6 is a proviral host factor for CVB, and other enterovirus, replication (24). We found that unlike BPIFB3 silencing, silencing of BPIFB2 did not significantly impact DENV and ZIKV, while silencing of BPIFB6 restricted ZIKV replication and infectious-particle production while having no significant effect on DENV (Fig. 1D and E). Given that we previously showed that BPIFB6 regulates Golgi complex morphology, this discrepancy between the effects of BPIFB6 silencing on DENV and ZIKV may be indicative of distinct trafficking pathways utilized by these viruses (24). In all studies, knockdown efficiency was confirmed by RT-qPCR (Fig. 1A and D). These data point to a specific role for BPIFB3 in flavivirus replication.
BPIFB3 depletion restricts flavivirus infections at an early stage of replication.
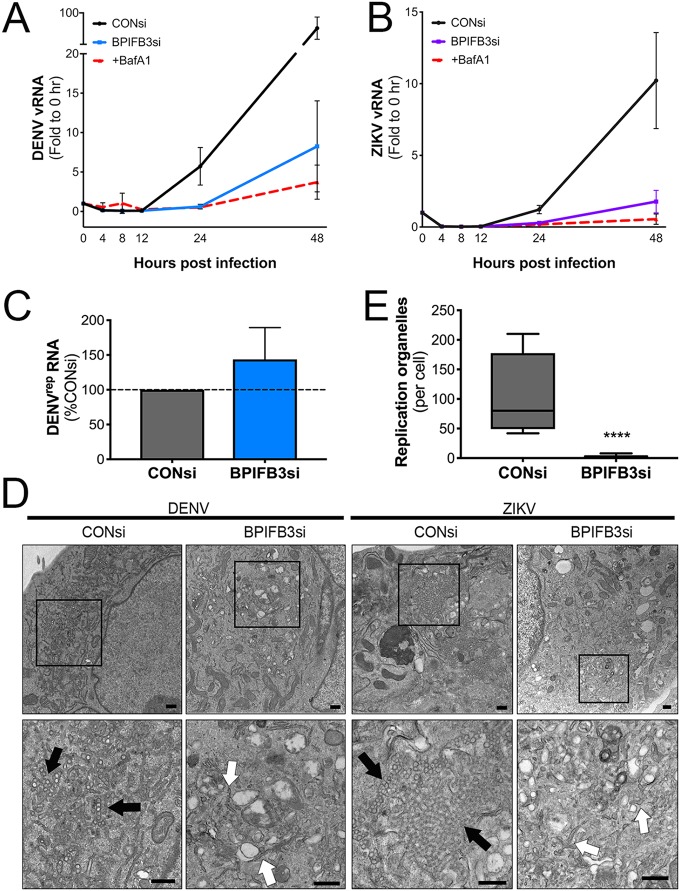
To define which step(s) of flavivirus replication was facilitated by BPIFB3 expression, we assessed the effects of BPIFB3 silencing on viral RNA (vRNA) replication and membrane rearrangement. To address the effects of BPIFB3 expression on vRNA replication, we performed multistep growth curves with DENV and ZIKV (MOI of 1) in CONsi- and BPIFB3si-transfected HBMEC and in HBMEC treated with bafilomycin A1 (BafA1) to prevent viral entry (Fig. 2A and B). In CONsi-transfected HBMEC, we detected increased vRNA between 12 and 24 h postinfection (hpi). In contrast, levels of vRNA remained very low in BPIFB3si-transfected cells and were comparable to those in BafA1-treated cells at 24 hpi (Fig. 2A and B). However, by 48 hpi, vRNA in BPIFB3si-transfected cells had a slight increase compared to that in BafA1-treated cells. These data suggest that BPIFB3 facilitates flavivirus infection at an early stage after entry, at or prior to genome replication. To confirm this, we utilized HBMEC stably propagating a DENV subgenomic replicon (HBMECrep) that expresses the full seven nonstructural proteins, allowing the replication of replicon RNA in membrane-bound replication organelles similar to viral infection (25). We found that silencing of BPIFB3 in HBMECrep had no effect on replicon RNA levels 48 h after BPIFB3 depletion (Fig. 2C), suggesting that BPIFB3si restriction may occur before replication of the viral genome. This lack of effect on replicon RNA can likely be attributed to HBMECrep cells’ having already undergone extensive ER remodeling to establish and maintain replicon expression. This could further suggest that BPIFB3 is required for the initial stages of ER remodeling.
FIG 2.
BPIFB3 depletion restricts flavivirus infection prior to genome replication. (A) RT-qPCR analysis of vRNA from multistep growth curves for DENV (A) and ZIKV (B) performed using an MOI of 1. Data are presented as fold change relative to CONsi at the 0-h time point. (C) DENV replicon RNA levels determined by RT-qPCR in response to BPIFB3 depletion, presented as percentages relative to CONsi transfection. (D) TEM of HBMEC transfected with CONsi or BPIFB3si and infected with DENV or ZIKV. Black arrows indicate viral replication organelles, and white arrows indicate ER membranes lacking signs of viral replication. Bars, 2 μm (top) and 500 nm (bottom). (E) Quantification of ZIKV replication organelles per cell in TEM images in panel D. Student's t test was performed to determine statistical significance (****, P < 0.0001).
Prior to vRNA replication, flaviviruses induce large-scale expansion and remodeling of the endoplasmic reticulum in order to allow the formation of ROs. To determine if BPIFB3 depletion had an effect on these membrane remodeling stages, we performed transmission electron microscopy (TEM) in CONsi- or BPIFB3si-transfected HBMEC infected with either DENV or ZIKV. Depletion of BPIFB3 prior to DENV or ZIKV infection significantly inhibited the formation of ER-bound ROs (Fig. 2D and E). Additionally, BPIFB3 silencing inhibited other hallmarks of flaviviral membrane manipulation (Fig. 2D, arrows), including the formation of convoluted membranes frequently seen adjacent to ROs that serve as sites of lipid synthesis.
BPIFB3 depletion restricts flavivirus replication independent of innate immune activation and viral binding.
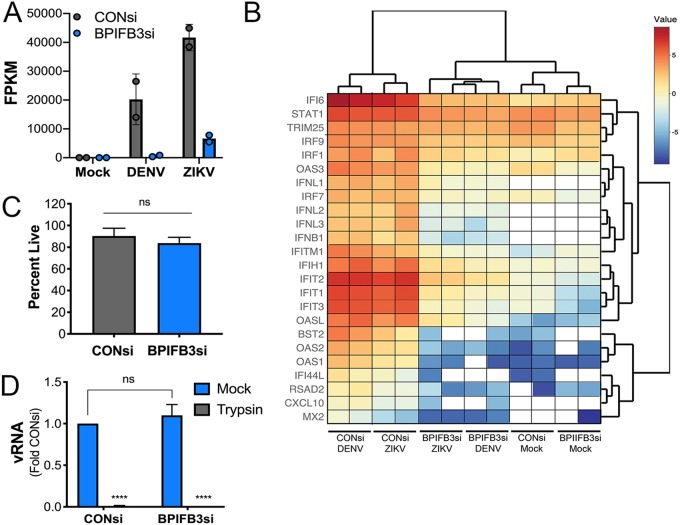
To eliminate any impact of BPIFB3 on innate immune signaling, which could inhibit flaviviral replication, we performed whole-genome transcriptome sequencing (RNA-seq) on CONsi- and BPIFB3si-transfected HBMEC in mock-, DENV-, or ZIKV-infected cells. Consistent with our RT-qPCR-based approaches, RNA-seq confirmed the very low levels of flaviviral vRNA in BPIFB3si-transfected cells, with numbers of viral fragments per kilobase of transcript per million mapped reads (FPKM) being significantly lower in BPIFB3-depleted cells (Fig. 3A). Consistent with this, we did not detect any induction of innate immune signaling, as indicated by the lack of changes in the expression of type I interferon (beta interferon [IFN-β]) or type III IFN (IFN-γ), or in the induction of interferon-stimulated genes (ISGs) (Fig. 3B). As expected, we observed robust induction of IFNs and ISGs in CONsi-transfected HBMEC infected with both DENV and ZIKV (Fig. 3B).
FIG 3.
BPIFB3-dependent regulation of flavivirus infection is not caused by enhanced innate immune signaling or impairment of viral binding. (A) RNA-seq showing viral FPKM values for DENV- and ZIKV-infected CONsi- or BPIFB3si-treated HBMEC. Mock-infected cells for both had zero reads for both DENV and ZIKV. (B) RNA-seq analysis of mock-, DENV-, or ZIKV-infected CONsi or BPIFB3si HBMEC showing interferon and ISG mRNA levels from ln(RPKM) values; white indicates no reads for the specified sample. (C) Trypan blue stain for cell viability in CONsi and BPIFB3si HBMEC. (D) ZIKV binding assay in CONsi and BPIFB3si HBMEC, quantified by RT-qPCR for vRNA. Cells were incubated either in normal media (mock) or in media with trypsin added to inhibit ZIKV binding as a control. Data are normalized to results obtained with mock CONsi transfection.
We next confirmed that the decreases in flavivirus replication were not due to decreases in cell viability or in the ability of viral particles to bind to the cell surface. We found that there were no differences in the extent of cell viability between CONsi- and BPIFB3si-transfected cells, as assessed by trypan blue staining (Fig. 3C). In addition, there were no differences in the levels of viral binding, as assessed by RT-qPCR of vRNA bound to cells (Fig. 3D).
Flavivirus infection of BPIFB3-depleted cells induces aberrant ER morphology.
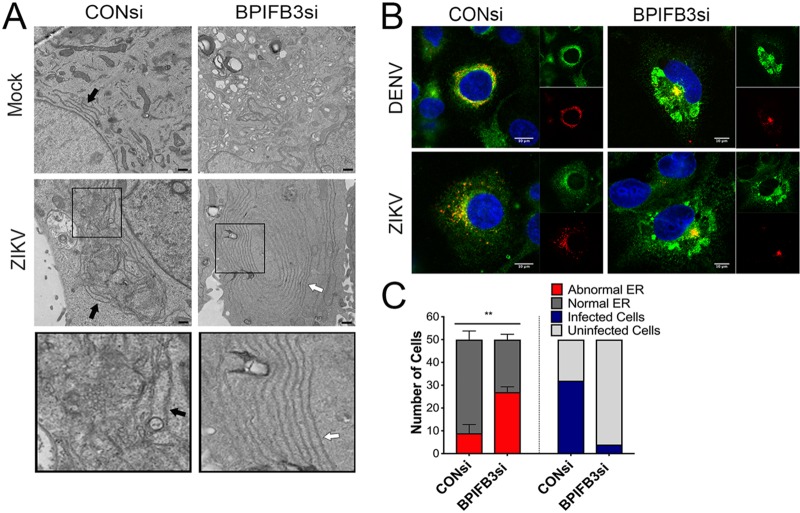
We next performed transmission electron microscopy (TEM) to determine the impact of BPIFB3 silencing on ER morphology with ultrastructural detail. These studies revealed remarkable effects of BPIFB3 silencing on ER morphology that were exaggerated in cells infected with DENV or ZIKV, including ER “whorls” and stacked membranes (Fig. 4A), which have been associated with microautophagy in yeasts but remain largely uncharacterized in mammalian cells (26, 27). To further characterize these changes in ER morphology, we performed immunofluorescence imaging of HBMEC for the ER sheet marker Climp63 and viral double-stranded RNA (dsRNA) to identify infected cells. In uninfected cells, ER sheets originate at the nuclear envelope and extend to the cell periphery in a uniform arrangement; however, during infection with DENV and ZIKV, ER sheets condense around the perimeter of the nucleus, where they colocalize with viral dsRNA (Fig. 4B), designating the location of viral membrane remodeling and replication organelle formation (15). In cases where viral replication was detected by dsRNA staining in BPIFB3-depleted cells, the ER sheet marker Climp63 showed aberrant rearrangements that did not colocalize with sites of viral replication (Fig. 4B). To assess how frequent these unique ER structures were during BPIFB3 depletion, we quantified abnormal ER morphology across 50 individual cells from ZIKV-infected CONsi- or BPIFB3si-treated HBMEC by TEM, as well as scoring them for visible replication organelles (Fig. 4C). BPIFB3-depleted cells infected with ZIKV showed significantly more instances of aberrant ER structures than CONsi-treated cells, which lacked traditional ER-localized viral replication organelles.
FIG 4.
BPIFB3 depletion induces aberrant ER phenotypes in response to flavivirus infection. (A) TEM images from CONsi- and BPIFB3si-transfected HBMEC infected with ZIKV (or mock-infected controls). Bars, 2 μm. Black arrows indicate normal ER morphology, and white arrows indicate examples of abnormal ER that are quantified in panel C. (B) Confocal microscopy from CONsi or BPIFB3si HBMEC infected with DENV or ZIKV (MOI = 1) and stained for dsRNA (red) and Climp63 (green) 48 h postinfection. Bars, 10 μm. (C) Abnormal ER expansion by TEM (A) from 50 cells was scored blindly as either normal or abnormal. The same cells were assessed for visible viral replication and scored as either infected or uninfected.
RETREG1 silencing restores flavivirus replication in BPIFB3si-transfected cells.
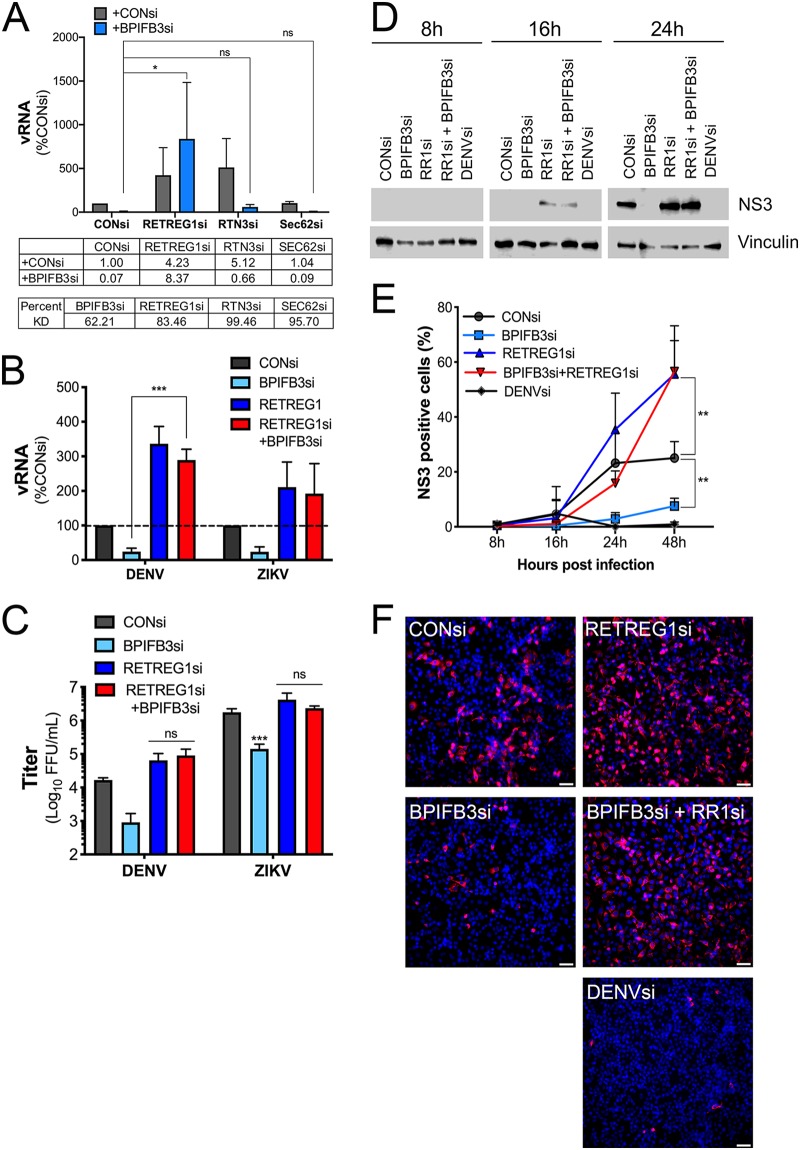
DENV and ZIKV are dependent on the availability of ER membranes to replicate, and we previously showed that reticulophagy functions as an antiviral pathway that limits the availability of these membranes (15). We next determined whether codepletion of RETREG1 and/or other reticulophagy receptors, including reticulon 3 (RTN3) and Sec62, restored flavivirus infection in BPIFB3si-transfected cells. We found that cosilencing of RETREG1, but not RTN3 or Sec62, reversed the inhibition of flavivirus infection in BPIFB3si-transfected cells, as determined by both qPCR for vRNA and FFU for viral titers (Fig. 5A to C). In contrast, silencing of RTN3 or Sec62 had no impact on BPIFB3si-mediated suppression of DENV infection (Fig. 5A). Knockdown efficiency for all reticulophagy receptors was confirmed by RT-qPCR (Fig. 5A).
FIG 5.
Inhibition of RETREG1 reticulophagy by RNAi restores flavivirus replication. (A) RT-qPCR of DENV infection of BPIFB3 depletion alone or with the reticulophagy receptor RETREG1, RTN3, or Sec62 in HBMEC. The mean fold change relative to CONsi for each knockdown is shown below the graph. Percent KD was determined by RT-qPCR and is shown for each siRNA. (B) RT-qPCR for DENV and ZIKV infection levels in cells depleted of BPIFB3 and RETREG1 alone or together. (C) Infectious particle production from BPIFB3- and RETREG1-depleted cells determined by FFU assay from panel B. (D) Western blot for DENV NS3 at 8, 16, and 24 h postinfection in BPIFB3- and RETREG1-depleted cells. (E) siRNA-transfected HBMEC infected with DENV were stained and imaged for NS3 at 8, 16, 24, and 48 h postinfection and quantified by automated imaging software to determine the percent NS3-positive cells per field. (F) Representative images from the assay whose results are presented in panel E.
To define the kinetics of RETREG1-mediated restoration of flavivirus infection in BPIFB3si-transfected cells, we examined the production of the DENV nonstructural protein 3 (NS3) by immunoblotting at various times postinfection. Consistent with our findings for vRNA, silencing of BPIFB3 significantly inhibited the production of NS3 at 8, 16, and 24 hpi (Fig. 5D). Moreover, RETREG1si transfection enhanced the production of NS3 at 16 hpi compared to that in CONsi-transfected cells (Fig. 5D, middle). Cosilencing of RETREG1 with BPIFB3 reversed the inhibition of NS3 production and enhanced the production of NS3 by 16 hpi in BPIFB3si-transfected cells (Fig. 5D, middle). We confirmed these findings by immunofluorescence microscopy for NS3 followed by quantitative image analysis; representative images for NS3 staining are shown for the 48-h time point (Fig. 5E and F).
BPIFB3-depleted cells exhibit enhanced reticulophagy.
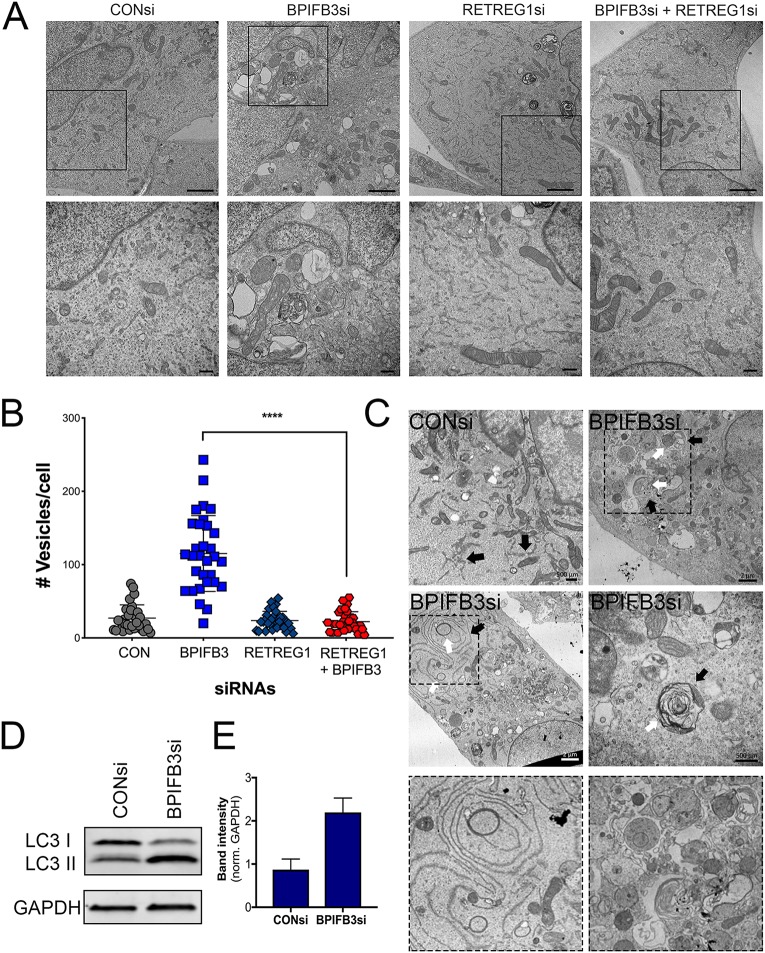
To mechanistically determine whether the reversal of flavivirus infection by RETREG1 codepletion corresponded with a reversal in the autophagy phenotype associated with BPIFB3 depletion, we performed TEM of HBMEC transfected with BPIFB3si or RETREG1si, alone or in combination, or with CONsi (Fig. 6A). Quantification of the number of vesicles under each condition demonstrated that codepletion of RETREG1 with BPIFB3 inhibited the increase in autophagy observed in BPIFB3si-transfected cells (Fig. 6B). Given that depletion of RETREG1 reversed the enhancement of autophagy and the inhibition of viral infection associated with BPIFB3 silencing, we determined whether BPIFB3-depleted cells exhibited increases in reticulophagy specifically. To test this, we performed TEM to directly examine the association of the ER with autophagosomes and lysosomes. TEM imaging confirmed that the ER is closely associated with autophagosomes during BPIFB3 depletion (Fig. 6C, arrows). Last, we confirmed that BPIFB3 silencing enhanced overall levels of autophagosome formation by immunoblotting for the autophagy adapter protein, LC3. In agreement with our prior studies (21), BPIFB3 depletion increased the autophagy-committed form of LC3 II (Fig. 6D and E). Taken together, these data suggest that BPIFB3 is required to facilitate flavivirus replication due to its regulation of RETREG1-dependent reticulophagy.
FIG 6.
RETREG1 depletion inhibits BPFB3si induced autophagy. (A) TEM of HBMEC depleted of BPIFB3 or RETREG1 or both. (Top) Total cell morphology. Bars, 2 μm. Black boxes indicate regions magnified in the bottom panel to show the ER membrane and vesicle morphology. (B) Quantification of total number of vesicles (autophagosomes, lysosomes, and enlarged endosomes/amphisomes) per cell from TEM images in panel A. (C) TEM images of CONsi or BPIFB3si HBMEC showing ER membranes closely associated with autophagosomes. (D) Western blot of siRNA-transfected HBMEC for the autophagy protein LC3 and the GAPDH as a loading control, quantified in panel E.
DISCUSSION
The success of flavivirus infection depends on the cooperation of numerous cellular organelles and pathways that function to produce progeny virions, specifically relying on host membranes throughout their life cycles. Here, we show that BPIFB3 is required for DENV and ZIKV infection by regulating the availability of ER membranes for viral remodeling of the ER. Our data show that BPIFB3 depletion enhances ER sheet turnover by RETREG1-mediated reticulophagy. These findings not only define the role of specific autophagic pathways in the regulation of flavivirus infection but also identify BPIFB3 as a novel regulator of RETREG1-specific forms of reticulophagy.
Unlike other RNA viruses, flaviviruses depend solely on ER-derived membranes for their replication. The viral genome is delivered to the rough ER following entry and uncoating, where translation of viral proteins induces expansion of the ER. Of the seven nonstructural proteins, the majority remain associated with the ER throughout the life cycle, where they function in viral replication, membrane remodeling, and inactivation of reticulophagy and ER stress pathways (5–7, 15, 28, 29). While it has been suggested that the virally encoded nonstructural proteins NS1, NS4A, and NS4B are involved in membrane manipulation during DENV infection, little is known regarding host factors essential for this process. Currently, only three host factors have been implicated in membrane expansion, including fatty acid synthase (FASN), RETREG1, and reticulon 3.1A (RTN3.1A). FASN is recruited to sites of replication organelle formation by the DENV protease NS3, demonstrating that increased lipid synthesis is important for membrane remodeling (18). Additionally, both DENV and ZIKV inhibit ER degradation by cleaving the RETREG1 reticulophagy receptor, allowing an accumulation of ER membranes (15). Lastly, RTN3.1A localizes to viral replication organelles to facilitate proper membrane curvature; however, it does not interact with DENV or ZIKV NS4A during membrane remodeling (30). The work presented here further confirms that degradation of the ER is an antiviral process and suggests BPIFB3 as a new host cell factor that regulates ER turnover. RNAi-mediated silencing of BPIFB3 leads to enhanced levels of reticulophagy, which decreases the availability of ER membranes for flavivirus replication. Concurrent depletion of RETREG1 with BPIFB3 overcomes this defect, demonstrating that the antiviral effects of BPIFB3 depletion are specific to RETREG1-mediated reticulophagy.
One method proposed to promote membrane expansion during flavivirus infection is the induction of autophagy (29). However, our data demonstrate that enhanced levels of reticulophagy, particularly early during infection, inhibit membrane remodeling and replication organelle formation. Recent work has identified a number of ER-specific autophagy pathways that differ by the receptor used to target cargo to autophagosomes (12, 31, 32). However, it remains unclear whether these pathways are regulated by the same machinery that controls canonical macroautophagy. The growing diversity in the various forms of autophagy further complicates our understanding of the relationship between viral infection and this pathway, as certain forms of autophagy may differentially regulate viral replication at various stages of the viral life cycle. The work presented here, in combination with our previous work characterizing BPIFB3 as a negative regulator of CVB infection, demonstrates the unique requirements for autophagy between different RNA virus families. In contrast to the unclear role for distinct autophagic pathways in flavivirus infection, CVB benefits from autophagy induction, as it uses autophagosomes and other cytoplasmic vesicles for replication organelle formation. Importantly, CVB inhibits fusion of the autophagosome with the lysosome, which enhances the number of cytoplasmic vesicles and prevents the degradation of viral replication machinery (33–35). Conversely, it has not been demonstrated whether flaviviruses have developed strategies to avoid clearance through the macroautophagy pathway, similarly to CVB and other enteroviruses. While the induction of autophagy during flavivirus infection has been implicated in enhancing viral replication (36), the precise timing of induction may have distinct effects on the viral life cycle. Furthermore, the ability to specifically activate one form of autophagy while inhibiting others may be essential for successful flavivirus infection. The distinction between membrane manipulation during CVB infection and flavivirus infection explains the differential effects of BPIFB3 in regulating these unique viruses and further suggests that increased flux through autophagy is detrimental to flavivirus replication.
The BPIFB family of proteins were initially named and identified because of their homology to the bactericidal/permeability-increasing (BPI) protein, a secreted antimicrobial protein that functions through binding to lipopolysaccharide (LPS) (37–39). Despite the high degree of predicted structural homology, BPIFB3 localizes to the ER and is not secreted (21). Of the other members of the family, BPIFB2 and BPIFB6 are also ER localized; however, neither appears to regulate autophagy (24) or flavivirus infection. BPIFB proteins contain two BPI folds demonstrated to have lipid binding properties. Unlike other BPIFB proteins, the first BPI domain (BPI1) of BPIFB3 lacks the ability to bind lipids, while BPI2 is capable of binding phosphatidic acid, phosphatidylserine, cardiolipin, and other lipid molecules (24). Of the related proteins, BPIFB6 is the only protein to be characterized, and it has been demonstrated to regulate secretory trafficking and Golgi complex morphology (24). Together with the data presented here, this suggests that a possible unifying function of these proteins is to regulate sites of vesicle trafficking; however, the mechanism by which they do so remains unclear. BPIFB3 overexpression has been associated with inhibition of autophagy, while here we show that its depletion specifically enhances RETREG1 reticulophagy but not other reticulophagy pathways universally. In comparison, BPIFB6 depletion results in Golgi complex dispersal and a disruption of retrograde and anterograde trafficking (24). This alludes to a possible mechanism whereby BPIFB3 and BPIFB6 expression is associated with decreased vesicle trafficking to the autophagic and secretory pathways, respectively, while loss of expression leads to enhanced vesicle trafficking originating in the ER. Importantly, expression of BPIFB3 is remarkably low, and we are unable to detect endogenous protein by either Western blotting or immunofluorescence assays. Despite its low expression, depletion of BPIFB3 elicits a dramatic phenotype in cells, suggesting an essential role in regulating the morphology of the cellular membrane network. This is consistent with other ER structural proteins that drastically effect membrane morphology at very low levels of endogenous expression (40). Their potential roles in vesicle trafficking have important implications for the ability of these proteins to impact the trafficking and spread of a variety of viruses. However, further characterization is required to delineate the different methods by which viruses are trafficked during infection.
The relationship between flavivirus infection and the autophagic pathway is likely to be complex. While the initiation of autophagy and that of lipophagy have been demonstrated as proviral pathways (17, 19, 29, 36), flux through the autophagic pathway and reticulophagy are antiviral (15, 16, 41). Thus, further characterization of the role of specific autophagic pathways in the regulation of flavivirus infection is needed to understand and develop new mechanisms to control infection.
MATERIALS AND METHODS
Cells and viruses.
Human brain microvascular endothelial cells (HBMEC) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 10% Nu-Serum, 1× nonessential amino acids, 1× minimum essential medium vitamins, 1% sodium pyruvate, and 1% antibiotic. Human bone osteosarcoma U2OS (ATCC HTB-96) and Vero (ATCC CCL-81) cells were grown in Dulbecco’s minimal essential medium (DMEM) with 10% FBS and 1% antibiotic. Development of DENV replicon HBMECs using constructs provided by Theodore Pierson (NIH/NIAID) was described previously (42). Aedes albopictus midgut C6/36 cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotic at 28°C in a 5% CO2 atmosphere. Cell viability was assessed by trypan blue staining at a 1:1 ratio and analyzed on a Bio-Rad TC20 automated cell counter.
DENV2 16881 and ZIKV Paraiba/2015 (provided by David Watkins, University of Miami) were propagated in C6/36 and Vero cells, respectively (43). Titers were determined by fluorescent focus unit assay as previously described, using recombinant anti-double-stranded RNA monoclonal antibody (provided by Abraham Brass, University of Massachusetts) (44). Propagation and titration of CVB3 (RD) were described previously (45). All experiments measuring infection levels were performed using a multiplicity of infection (MOI) of 1 for 16 h (CVB) or 48 h (DENV and ZIKV) unless stated otherwise, and infection was quantified by RT-qPCR or fluorescent focus unit assay.
siRNAs, plasmids, and transfections.
Characterization of siRNAs targeting BPIFB3, BPIFB2, BPIFB6, and RETREG1 (FAM134B) was described previously (15, 21). Sequences of siRNAs targeting RTN3 and Sec62 are CCACUCAGUCCCAUUCCAUtt for RTN3 and GAAGGAUGAGAAAUCUGAAtt for Sec62. All siRNAs, including the scrambled control (CONsi), were purchased from Sigma. Efficiency of knockdown was determined by RT-qPCR for each siRNA target. siRNAs were reverse transfected at 25 nM in to HBMEC using Dharmafect 1, and then either cells were infected or RNA was collected 48 h posttransfection.
Development of green fluorescent protein (GFP)-tagged RETREG1 and RETREG1mutLIR have been described elsewhere (15). Plasmids were transfected into HBMEC cells using Lipofectamine 3000 according to the manufacturer’s protocol and fixed for fluorescence microscopy or infected at 48 h posttransfection.
RNA extraction, cDNA synthesis, and RT-qPCR.
RNA was isolated using the GenElute total RNA miniprep kit from Sigma according to the kit protocol. RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) with 1 μg of RNA per sample. RT-qPCR was performed using IQ SYBR green SuperMix (Bio-Rad) in a Bio-Rad CFX96 Touch real-time PCR detection system. A modified threshold cycle (ΔCT) method was used to calculate gene expression using human actin for normalization. Primer sequences for actin, DENV, ZIKV, CVB, BPIFB3, and RETREG1 have been described previously (15, 46).
Virus binding assay.
HBMEC were transfected with an siRNA targeting BPIFB3 or a nontargeting control siRNA. Approximately 48 h posttransfection, cells were washed with phosphate-buffered saline (PBS), and ZIKV (MOI = 10) was adsorbed for 1 h at 16°C. Following adsorption, cells were washed extensively with PBS and directly lysed for RNA extraction (referred to here as “mock”) or incubated with trypsin (3 min), washed with PBS, and lysed for RNA extraction. Following RNA extraction, cDNA synthesis, and qPCR, the data were analyzed using the ΔΔCT method and normalized to mock CONsi.
RNA-seq.
Total RNA was isolated as described above, and RNA sequencing was performed as previously described (25). Analysis of RNA-seq data sets was performed using CLC Genomics 11 (Qiagen) to process and map sequences to the human genome (hg19) or the appropriate viral genome to calculate viral fragments per kilobase of transcript per million mapped reads (FPKM) values. Differentially expressed genes were identified using the DeSeq2 package in R with a significance cutoff of 0.001 and a fold change cutoff of 2 (47). Gene set enrichment analysis (GSEA) and manual sorting were used to identify pathways or specific transcripts differentially regulated. Heat maps were generated using MeViewer software based on ln(RPKM) (where RPKM is reads per kilobase million) values.
Antibodies.
The mouse monoclonal anti-V5 epitope tag was purchased from Invitrogen (R960-25). Rabbit polyclonal antibody against CKAP4 (16686-1-AP) was purchased from ProteinTech. Rabbit polyclonal antibodies to DENV NS3 (GTX124252) and ZIKV NS4B (GTX133311) were purchased from GeneTex. Recombinant mouse monoclonal anti-dsRNA was provided by Abraham Brass (University of Massachusetts). Alexa Fluor-conjugated secondary antibodies were purchased from Invitrogen.
Immunofluorescence and electron microscopy.
Immunofluorescence microscopy was performed on cells grown in 8-well chamber slides (Millipore Sigma), fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton. In some cases, cells were fixed in ice-cold methanol. Primary antibodies were incubated in PBS with cells for 1 h, followed by staining with Alexa Fluor-conjugated secondary antibodies for 30 min. Slides were mounted with coverslips using Vectashield containing 4′,6-diamino-2-phenylindole (DAPI). Imaging was performed on an Olympus IX83 inverted microscope (wide-field microscopy) or a Zeiss LSM 710 confocal microscope. All image quantification was performed using ImageJ/FIJI. Pixel intensity measurements were performed using isolated channels on individual cells with the region-of-interest (ROI) manager. Data are presented as mean pixel intensity, normalized to cell area. Quantification of fluorescent puncta was performed manually, counting the number of ER-localized vesicles alone or those colocalized with the indicated marker. Preparation of samples for TEM was done by the Center for Biologic Imaging (University of Pittsburgh) as previously described (46). Imaging was performed on a JEOL 1011 transmission electron microscope. Quantification of TEM images was performed manually.
Statistical analyses.
All analyses were performed using GraphPad Prism. Experiments were performed at least three times. Student's t test, two-way analysis of variance (ANOVA), and one-way ANOVA were used where indicated. Analysis of fluorescence microscopy data was done using a nonparametric Kruskal-Wallis test. Data are presented as means ± standard deviations, with specific P values presented in the figure legends.
Data availability.
RNA sequencing files have been deposited to the NCBI Sequence Read Archive (SRA) and can be located under the accession number PRJNA606334.
ACKNOWLEDGMENTS
We thank Abraham Brass (University of Massachusetts) for providing anti-dsRNA antibody and Theodore Pierson (NIH/NIAID) for providing DENV replicon constructs.
This project was supported by NIH R01-AI081759 (C.B.C.), NIH T32-AI049820 (A.S.E.), a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award (C.B.C), and the Children’s Hospital of Pittsburgh of the UPMC Health System (C.B.C.).
REFERENCES
- 1.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufusi PH, Kelley JF, Yanagihara R, Nerurkar VR. 2014. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One 9:e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 5.Peña J, Harris E. 2012. Early dengue virus protein synthesis induces extensive rearrangement of the endoplasmic reticulum independent of the UPR and SREBP-2 pathway. PLoS One 7:e38202. doi: 10.1371/journal.pone.0038202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 8.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkegaard K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr Top Microbiol Immunol 335:323–333. doi: 10.1007/978-3-642-00302-8_16. [DOI] [PubMed] [Google Scholar]
- 10.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. 2016. Mammalian autophagy: how does it work?. Annu Rev Biochem 85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 12.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hübner CA, Dikic I. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 13.Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, Green DR, Blander JM. 2017. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171:809–823.e13. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stolz A, Grumati P. 2019. The various shades of ER‐phagy. FEBS J 286:4642–4649. doi: 10.1111/febs.15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennemann NJ, Coyne CB. 2017. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13:322–332. doi: 10.1080/15548627.2016.1265192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metz P, Chiramel A, Chatel-Chaix L, Alvisi G, Bankhead P, Mora-Rodríguez R, Long G, Hamacher-Brady A, Brady NR, Bartenschlager R. 2015. Dengue virus inhibition of autophagic flux and dependency of viral replication on proteasomal degradation of the autophagy receptor p62. J Virol 89:8026–8041. doi: 10.1128/JVI.00787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. 2016. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heaton NS, Perera R, Berger KL, Khadka S, LaCount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan TX, Randall G. 2017. Dengue virus activates the AMP kinase-mTOR axis to stimulate a proviral lipophagy. J Virol 91:e02020-16. doi: 10.1128/JVI.02020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abernathy E, Mateo R, Majzoub K, van Buuren N, Bird SW, Carette JE, Kirkegaard K. 2019. Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLoS Biol 17:e2006926. doi: 10.1371/journal.pbio.2006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delorme-Axford E, Morosky S, Bomberger J, Stolz DB, Jackson WT, Coyne CB. 2014. BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. mBio 5:e02147-14. doi: 10.1128/mBio.02147-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. 2010. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol 84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stins MF, Badger J, Sik Kim K. 2001. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb Pathog 30:19–28. doi: 10.1006/mpat.2000.0406. [DOI] [PubMed] [Google Scholar]
- 24.Morosky S, Lennemann NJ, Coyne CB. 2016. BPIFB6 regulates secretory pathway trafficking and enterovirus replication. J Virol 90:5098–5107. doi: 10.1128/JVI.00170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques E, Cherry S, Sadovsky Y, Coyne CB. 2016. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuck S, Gallagher CM, Walter P. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dykstra KM, Pokusa JE, Suhan J, Lee TH. 2010. Yip1A structures the mammalian endoplasmic reticulum. Mol Biol Cell 21:1556–1568. doi: 10.1091/mbc.e09-12-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol 80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datan E, Roy SG, Germain G, Zali N, McLean JE, Golshan G, Harbajan S, Lockshin RA, Zakeri Z. 2016. Dengue-induced autophagy, virus replication and protection from cell death require ER stress (PERK) pathway activation. Cell Death Dis 7:e2127. doi: 10.1038/cddis.2015.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. 2017. The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep 21:1639–1654. doi: 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 31.Grumati P, Morozzi G, Hölper S, Mari M, Harwardt M-L, Yan R, Müller S, Reggiori F, Heilemann M, Dikic I. 2017. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6:e25555. doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Soldà T, D'Antuono R, Raimondi A, Jung M, Melnyk A, Schorr S, Schreiber A, Simonelli L, Varani L, Wilson-Zbinden C, Zerbe O, Hofmann K, Peter M, Quadroni M, Zimmermann R, Molinari M. 2016. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 33.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris KG, Morosky SA, Drummond CG, Patel M, Kim C, Stolz DB, Bergelson JM, Cherry S, Coyne CB. 2015. RIP3 regulates autophagy and promotes coxsackievirus B3 infection of intestinal epithelial cells. Cell Host Microbe 18:221–232. doi: 10.1016/j.chom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H, Zhai X, Chen Y, Wang R, Lin L, Chen S, Wang T, Zhong X, Wu X, Wang Y, Zhang F, Zhao W, Zhong Z. 2016. Protein 2B of coxsackievirus B3 induces autophagy relying on its transmembrane hydrophobic sequences. Viruses 8:131. doi: 10.3390/v8050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Yuan J, Kirkegaard K. 2013. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol 87:1312–1321. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bingle CD, Craven CJ. 2002. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum Mol Genet 11:937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- 38.Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. 2004. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci 13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bingle CD, Seal RL, Craven CJ. 2011. Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily. Biochem Soc Trans 39:977–983. doi: 10.1042/BST0390977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Tukachinsky H, Romano FB, Rapoport TA. 2016. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife 5:e18605. doi: 10.7554/eLife.18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang YT, Wan SW, Lu YT, Yao JH, Lin CF, Hsu LJ, Brown MG, Marshall JS, Anderson R, Lin YS. 2014. Autophagy facilitates antibody-enhanced dengue virus infection in human pre-basophil/mast cells. PLoS One 9:e110655. doi: 10.1371/journal.pone.0110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bramley JC, Drummond CG, Lennemann NJ, Good CA, Kim KS, Coyne CB. 2017. A three-dimensional cell culture system to model RNA virus infections at the blood-brain barrier. mSphere 2:e00206-17. doi: 10.1128/mSphere.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medina F, Medina JF, Colon C, Vergne E, Santiago GA, Munoz-Jordan JL. 2012. Dengue virus: isolation, propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15:Unit 15D.2. doi: 10.1002/9780471729259.mc15d02s27. [DOI] [PubMed] [Google Scholar]
- 44.Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. 2006. Quantitation of flaviviruses by fluorescent focus assay. J Virol Methods 134:183–189. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Coyne CB, Bergelson JM. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 46.Delorme-Axford E, Donker RB, Mouillet J-F, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, Sadovsky Y, Coyne CB. 2013. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A 110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing files have been deposited to the NCBI Sequence Read Archive (SRA) and can be located under the accession number PRJNA606334.