Flavivirus infections are transmitted through mosquitos or ticks and lead to significant morbidity and mortality worldwide with a spectrum of manifestations. Infection with an emerging flavivirus, DTMUV, manifests with clinical symptoms that include lesions of the immune organs and neurological dysfunction, leading to heavy egg drop and causing serious harm to the duck industry in China, Thailand, Malaysia, and other Southeast Asian countries. Mosquito cells, bird cells, and mammalian cell lines are all susceptible to DTMUV infection. An in vivo study revealed that BALB/c mice and Kunming mice were susceptible to DTMUV after intracerebral inoculation. Moreover, there are no reports about DTMUV-related human disease, but antibodies against DTMUV and viral RNA were detected in serum samples of duck industry workers. This information implies that DTMUV has expanded its host range and may pose a threat to mammalian health. However, the pathogenesis of DTMUV is largely unclear. Our results show that NS2A strongly blocks the STING-induced signal transduction cascade by binding with STING, which subsequently blocks STING-STING binding and TBK1 phosphorylation. More importantly, the W164, Y167, or S361 residues in duSTING were identified as important interaction sites between STING and NS2A that are vital for NS2A-induced IFN production and effective phases of IFN response. Uncovering the mechanism by which DTMUV NS2A inhibits IFN in the cells of its natural hosts, ducks, will help us understand the role of NS2A in DTMUV pathogenicity.
KEYWORDS: duck Tembusu virus, NS2A, duSTING, duTBK1, immune inhibition, interaction sites
ABSTRACT
Duck Tembusu virus (DTMUV), which is similar to other mosquito-borne flaviviruses that replicate well in most mammalian cells, is an emerging pathogenic flavivirus that has caused epidemics in egg-laying and breeding waterfowl. Immune organ defects and neurological dysfunction are the main clinical symptoms of DTMUV infection. Preinfection with DTMUV makes the virus impervious to later interferon (IFN) treatment, revealing that DTMUV has evolved some strategies to defend against host IFN-dependent antiviral responses. Immune inhibition was further confirmed by screening for DTMUV-encoded proteins, which suggested that NS2A significantly inhibited IFN-β and IFN-stimulated response element (ISRE) promoter activity in a dose-dependent manner and facilitated reinfection with duck plague virus (DPV). DTMUV NS2A was able to inhibit duck retinoic acid-inducible gene-I (RIG-I)-, and melanoma differentiation-associated gene 5 (MDA5)-, mitochondrial-localized adaptor molecules (MAVS)-, stimulator of interferon genes (STING)-, and TANK-binding kinase 1 (TBK1)-induced IFN-β transcription, but not duck TBK1- and interferon regulatory factor 7 (IRF7)-mediated effective phases of IFN response. Furthermore, we found that NS2A competed with duTBK1 in binding to duck STING (duSTING), impaired duSTING-duSTING binding, and reduced duTBK1 phosphorylation, leading to the subsequent inhibition of IFN production. Importantly, we first identified that the W164A, Y167A, and S361A mutations in duSTING significantly impaired the NS2A-duSTING interaction, which is important for NS2A-induced IFN-β inhibition. Hence, our data demonstrated that DTMUV NS2A disrupts duSTING-dependent antiviral cellular defenses by binding with duSTING, which provides a novel mechanism by which DTMUV subverts host innate immune responses. The potential interaction sites between NS2A and duSTING may be the targets of future novel antiviral therapies and vaccine development.
IMPORTANCE Flavivirus infections are transmitted through mosquitos or ticks and lead to significant morbidity and mortality worldwide with a spectrum of manifestations. Infection with an emerging flavivirus, DTMUV, manifests with clinical symptoms that include lesions of the immune organs and neurological dysfunction, leading to heavy egg drop and causing serious harm to the duck industry in China, Thailand, Malaysia, and other Southeast Asian countries. Mosquito cells, bird cells, and mammalian cell lines are all susceptible to DTMUV infection. An in vivo study revealed that BALB/c mice and Kunming mice were susceptible to DTMUV after intracerebral inoculation. Moreover, there are no reports about DTMUV-related human disease, but antibodies against DTMUV and viral RNA were detected in serum samples of duck industry workers. This information implies that DTMUV has expanded its host range and may pose a threat to mammalian health. However, the pathogenesis of DTMUV is largely unclear. Our results show that NS2A strongly blocks the STING-induced signal transduction cascade by binding with STING, which subsequently blocks STING-STING binding and TBK1 phosphorylation. More importantly, the W164, Y167, or S361 residues in duSTING were identified as important interaction sites between STING and NS2A that are vital for NS2A-induced IFN production and effective phases of IFN response. Uncovering the mechanism by which DTMUV NS2A inhibits IFN in the cells of its natural hosts, ducks, will help us understand the role of NS2A in DTMUV pathogenicity.
INTRODUCTION
Tembusu virus (TMUV) is a member of the genus Flavivirus in the family Flaviviridae (1) that was first isolated in 1955 from Culex tritaeniorhynchus mosquitoes in Kuala Lumpur, Malaysia (2). Beginning in April 2010, an outbreak of duck TMUV (DTMUV) occurred in major duck-farming regions in China (3). The affected ducks exhibited lesions of the immune organs and neurological dysfunction, which caused significant economic losses in the duck industry (4, 5). A recent study revealed that DTMUV could infect multiple avian species and replicate well in a wide spectrum of mammalian cell lines (such as A549, BHK21, HeLa, Vero, and SH-SY5Y cells) and mosquito cell lines (such as C6/36 and Aedes albopictus cells) (6–8). Moreover, DTMUV exhibited pathogenicity in BALB/c and Kunming mice following intracerebral inoculation (9, 10). There are no reports about DTMUV-related human disease, but antibodies against DTMUV and viral RNA were detected in serum samples of duck industry workers (11). This information implies that DTMUV has expanded its host range and may pose a threat to mammalian health. DTMUV is an enveloped virus with a 10,990-bp genome of single-stranded, positive-sense RNA. The open reading frame (ORF) encodes a unique polyprotein precursor that is subsequently cleaved by cellular and viral proteases into three structural proteins (core, membrane, and envelope) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (2). Increasing numbers of studies have provided evidence that the structural proteins play a critical role in receptor binding, entry, and fusion during the viral life cycle and participate in the formation of viral particles, whereas the NS proteins are involved in viral RNA replication, virion assembly, and evasion of the host innate immune responses (12). However, the immunologic features of DTMUV NS proteins remain unknown.
Viruses rely on the host for protein synthesis and RNA replication; the host innate immune response is initiated as the first line of defense against viral infection. In brief, the incoming viruses are recognized by pattern recognition receptors (PRRs) such as retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) (13, 14) that subsequently trigger signaling cascades leading to the activation of interferons (IFNs) (15). Upon activation, RIG-I and MDA5 form a complex through recruiting mitochondrial-localized adaptor molecules (MAVS; also named virus-induced signaling adaptor [VISA], IPS-1, or Cardif), leading to the subsequent activation of TANK-binding kinase 1 (TBK1) and the transcription factors interferon regulatory factor 3/7 (IRF-3/7) and NF-κB. Upon translocation into the nucleus, IRF7/IRF3 binds to the beta interferon (IFN-β) promoter to activate its transcription and subsequent production of IFN-β, which, in turn, binds to its receptor on the cell surface and activates the JAK-STAT pathway to induce interferon-stimulated gene (ISG) expression (14). To combat recognition by the innate immune system and to replicate well in hosts, most viruses have developed sophisticated strategies to evade or subvert the host innate response. The NS proteins of flaviviruses have evolved various mechanisms to antagonize the type I IFN response. First, flaviviruses directly inhibit PRRs or their adaptor proteins to evade the host immune response (16, 17). Second, several flavivirus NS proteins subvert IFN-α/β induction by targeting critical signaling molecules downstream of several PRRs (18, 19). Third, flaviviruses also have two major ways to block the IFNAR-dependent signaling cascade: inhibition of JAK1 or tyrosine kinase 2 (TYK2) activity and targeting of the key transcription factors STAT1 and STAT2 (20–22).
Flavivirus NS2A is an approximately 22-kDa hydrophobic transmembrane protein (23). The topology of dengue virus (DENV) NS2A was studied recently, which showed that NS2A is composed of eight predicted transmembrane domains (pTMDs), including five integral transmembrane domains (TMD3, TMD4, and TMD6 to TMD8) that span the endoplasmic reticulum (ER) membrane (24). Kunjin virus (KUNV) NS2A colocalizes with viral double-stranded RNA (dsRNA) and interacts with the 3′ untranscribed region (UTR) of the viral RNA, suggesting that NS2A plays a critical role in viral RNA synthesis (25). Mutagenesis studies of NS2A have shown that the mutation of yellow fever virus (YFV) NS2A at amino acid 198 (K198S) blocked the production of infectious virus (26), the I59N mutation of NS2A reduced KUNV virion production (27), and the R84A mutation of NS2A disabled DENV-2 assembly (24). Collectively, these data indicate that NS2A plays an important role in the viral life cycle. Flavivirus NS2A has also been confirmed to antagonize the host immune response. DENV NS2A inhibited IFN-α/β signaling through the inhibition of the JAK-STAT signaling pathway by decreasing STAT1 phosphorylation (28). The A30P mutation of KUNV NS2A reduced the antagonistic effects on IFN-α/β signaling, attenuating KUNV virulence in mice (29). One previous study suggested that DTMUV NS1 disrupted the RIG-I-like receptor (RLR) pathway in HEK293 cells via targeting VISA (30). However, it remains to be determined whether immune inhibition exists in the cells of the DTMUV natural host, the duck. Understanding the mechanism by which DTMUV blocks the host antiviral response will help to uncover the mechanisms of flavivirus infection. Here, we found that NS2A inhibited RIG-I-mediated IFN expression signaling in a dose-dependent manner. DTMUV NS2A competitively bound to stimulator of interferon genes (STING) with TBK1, reducing TBK1 phosphorylation and suppressing the IFN production and effective phases of IFN response. Our findings offer important insights into how DTMUV establishes a mechanism to subvert the host innate immune response and suggest a role for NS2A in DTMUV pathogenicity. Additionally, the identified potential interaction sites might lead to the identification of new targets for vaccines and antiviral therapeutics.
RESULTS
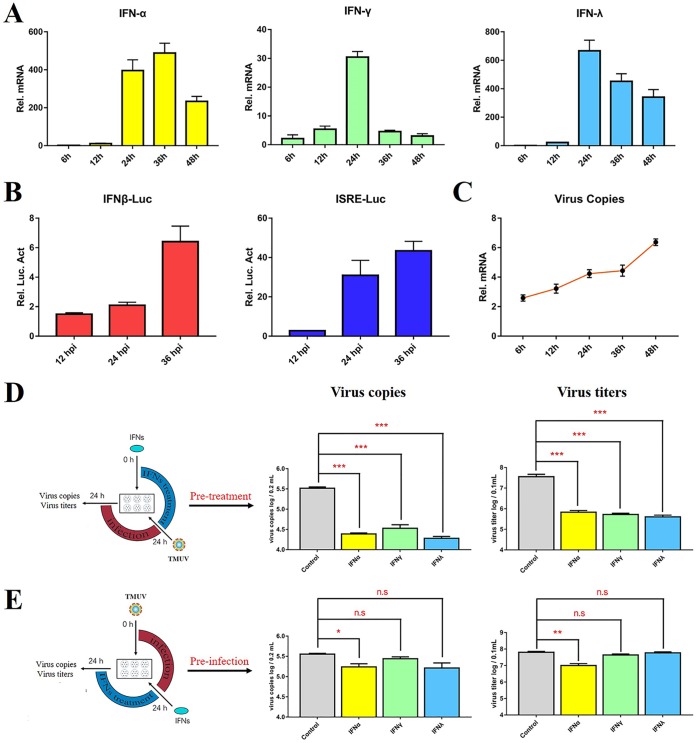
The antiviral effects of IFNs were abolished in cells preinfected with DTMUV.
To investigate the expression of avian IFNs in response to DTMUV infection, we infected duck embryo fibroblasts (DEFs) with DTMUV for multiple lengths of time and detected the expression of IFNs by reverse transcriptase quantitative PCR (RT-qPCR) and dual-luciferase reporter assays. DTMUV infection significantly upregulated the mRNA levels of duck IFNs (IFN-α, IFN-γ, and IFN-λ) and activated IFN-β/IFN-stimulated response element (ISRE)-luciferase (Luc) promoter signaling in DEFs (Fig. 1A to C). To understand the effects of IFN on DTMUV inhibition, the DEFs were pretreated with avian IFNs for 24 h and then infected with DTMUV, while other DEFs were preinfected with DTMUV for 24 h and then treated with IFN for 24 h. We found that IFN pretreatment for 24 h prior to infection significantly reduced the number of copies of DTMUV and the viral titers in DEFs. However, the antiviral effects of IFNs were abolished in the DTMUV preinfection group (Fig. 1E). These results demonstrated that DTMUV might interfere with the function of IFNs once viral replication is initiated. Therefore, we hypothesized the existence of a certain immune evasion stratagem by which DTMUV proteins abolish the IFN-dependent immune response.
FIG 1.
The antiviral effects of IFNs were abolished in cells preinfected with DTMUV. (A) The expression level of duIFN mRNA in DTMUV-infected DEFs. DEFs were treated with 100 μl DTMUV (1,000 TCID50) and harvested at 6, 12, 24, 36, and 48 hpi. Then, the transcription of duIFN (duIFN-α, duIFN-γ, and duIFN-λ) mRNA was detected by RT-qPCR. All results were normalized to those of duβ-actin. (B) IFN-β-Luc or ISRE-Luc promoter luciferase activity assay. The DTMUV-infected DEFs were cotransfected with the pGL3-IFN-β-Luc (or pGL4-ISRE-Luc) (400 ng/well) and pRL-TK plasmids (40 ng/well). At 12, 24, and 36 hpi, the cells were harvested, and luciferase activity was determined with a Dual-Glo luciferase assay system (Promega) and normalized on the basis of Renilla luciferase activities. All luciferase reporter assays were repeated three times. (C) Detection of virus copy numbers in DTMUV-infected DEFs after 12, 24, 36, and 48 h. (D, E) The effects of avian IFNs on DTMUV replication. The DEFs were pretreated with avian IFNs for 24 h and later infected with DTMUV for 24 h (D) or postinfected with DTMUV for 24 h and later treated with IFN for 24 h (E). The cells were harvested for virus copy number and titer detection. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
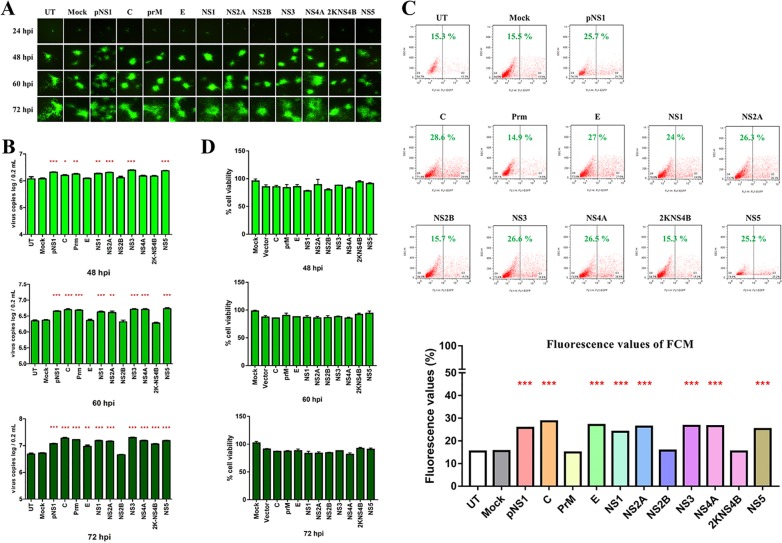
DTMUV proteins facilitate BAC-DPV-EGFP replication in vitro.
To determine the immune effects of each protein encoded by DTMUV, 10 genes (C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) were overexpressed in DEFs. The pCAGGS vector was used as a negative control, while the influenza A virus PR/8-NS1 plasmid (pNS1, a known IFN antagonist) served as a positive control (31). After 24 h of transfection, cells were infected with bacterial artificial chromosome (BAC)-DPV-enhanced green fluorescent protein (EGFP) (100 μl containing 100 50% tissue culture infective dose [TCID50] per well). At 24, 48, 60, and 72 h postinfection, cells were collected for further analysis of the viral copy number and fluorescence and for flow cytometry (FCM). The fluorescence values and viral copy numbers revealed that BAC-DPV-EGFP replication was highly enhanced in DTMUV C-, prM-, NS1-, NS2A-, NS3-, and NS5-overexpressing cells (Fig. 2). Based on the ability of DTMUV proteins to facilitate BAC-DPV-EGFP replication in transfected DEFs, we identified several potential DTMUV proteins (C, prM, NS1, NS2A, NS3, and NS5) that probably inhibited the IFN signaling pathway. Cell viability was measured by using a cell counting kit-8 (CCK-8) kit, suggesting that this inhibition was not a result of DTMUV protein-mediated cytotoxicity (Fig. 2D).
FIG 2.
DTMUV proteins facilitate BAC-DPV-EGFP replication in vitro. DEFs were transfected with pCAGGS plasmids expressing His-tagged DTMUV proteins (C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, or NS5) (800 ng/well), and the same dose of pCAGGS vector was used as a negative control, while pCAGGS-pNS1 served as a positive control. After 24 h of transfection, cells were infected with BAC-DPV-EGFP (100 μl containing 100 TCID50 per well). After infection, cells were collected for further fluorescence analysis at 24, 48, 60, and 72 hpi (A), the detection of viral copy numbers at 48, 60, and 72 hpi (B), and flow cytometry (FCM) at 60 hpi (C). (D) Cytotoxicity of DTMUV-derived expression plasmids in DEFs. The DEFs in 96-well plates were transiently transfected with each of DTMUV-derived expression plasmids (50 ng/well) and subsequently transfected with pRL-TK plasmid (5 ng/well), pGL3-IFN-β-Luc (50 ng/well), or pGL4-ISRE-Luc (50 ng/well). Similarly, the control group was transfected with 400 ng of pCAGGS vector, and the mock group did not transfect anything and served as a black control. At 48 h, 60 h, and 72 h posttransfection, CCK-8 reagent (10 μl) was added into each well for 2 h at 37°C. After that, the plates were evaluated at the 450-nm wavelength with a multidetection microplate reader. The results were expressed relative to black control cells, which were defined as 100 % viable. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
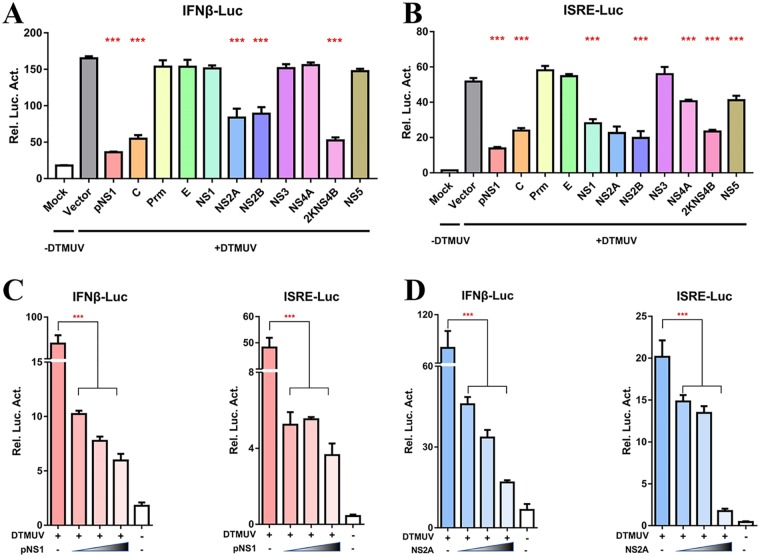
DTMUV NS2A inhibits the virus-induced IFN signaling pathway.
To further determine the antagonistic effects of each DTMUV protein on the RIG-I signaling pathway, 10 genes (C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, 2K-NS4B, and NS5) were coexpressed with a luciferase reporter in a reporter plasmid harboring the IFN-β and ISRE promoters. We found that the expression of the DTMUV C, NS2A, NS2B, and 2K-NS4B proteins significantly inhibited the DTMUV-triggered activation of IFN-β promoter activity, while DTMUV C, NS1, NS2A, NS2B, NS4A, and 2K-NS4B reduced the activity of the ISRE-Luc promoter (Fig. 3A and B). This enhancement was most prominent and significantly exceeded that observed in pNS1-, NS2A-, and NS2B-expressing cells. According to Fig. 2, among NS proteins, only the NS2A protein was identified as a potential IFN system antagonist based on the ability to facilitate BAC-DPV-EGFP replication. NS2B was investigated by our lab fellow (32), and 2KNS4B was chosen as individual study of its evasion mechanism on host immune response (data not shown). Therefore, in this study, we focus on the NS2A protein to further characterize the mechanism evolved by DTMUV to antagonize the host antiviral immune response. We found NS2A significantly inhibited the virus-induced activation of the IFN-β and ISRE promoters in a dose-dependent manner, suggesting that NS2A acts as an antagonist of IFN induction and effective phases of IFN response (Fig. 3D). The pNS1 plasmid significantly inhibited the virus-induced activation of the IFN-β and ISRE promoters in a dose-dependent manner and was used as a positive control (Fig. 3C). These results suggested that the expression of DTMUV NS2A leads the suppression of host IFN signaling.
FIG 3.
DTMUV NS2A inhibits the virus-induced IFN signaling pathway. (A, B) DEFs were transiently transfected with each of DTMUV-derived expression plasmids (400 ng/well) and subsequently transfected with pRL-TK plasmid (40 ng/well), pGL3-IFN-β-Luc (400 ng/well), or pGL4-ISRE-Luc (400 ng/well). At 24 h posttransfection, cells were infected with DTMUV (25 μl containing 100 TCID50 per well), and the IFN-β/ISRE-Luc activity was measured at 36 h postinfection. (C, D) Dose-dependent analysis of the inhibition of DTMUV-mediated IFN-β/ISRE promoter activity by both pNS1 and NS2A. DEFs were transiently transfected with the indicated amount of pNS1 and NS2A plasmid (100 ng, 200 ng, or 400 ng/well) and subsequently transfected with pRL-TK plasmid (40 ng/well), pGL3-IFN-β-Luc (400 ng/well), or pGL4-ISRE-Luc (400 ng/well). At 24 h posttransfection, cells were infected with DTMUV (25 μl containing 100 TCID50 per well); the IFN-β/ISRE-Luc activity was measured at 36 h postinfection. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
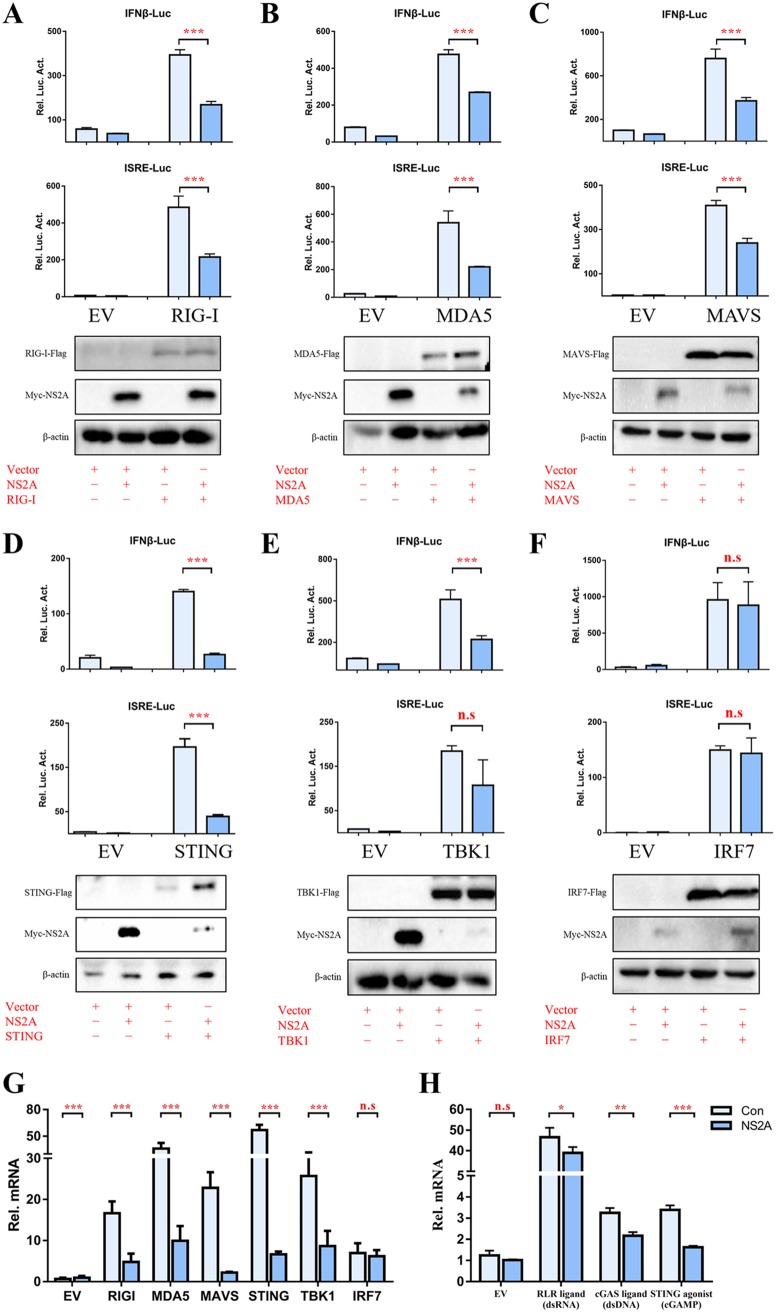
DTMUV NS2A inhibits RIG-I-, MAD5-, MAVS- and STING-mediated IFN induction.
To determine the molecular target of NS2A in the IFN induction signaling pathway, plasmids containing key molecules (duck [duRIG-I], duMDA5, duMAVS, duSTING, duTBK1, and duIRF7) were coexpressed with NS2A and the luciferase reporter produced from a reporter plasmid harboring the IFN-β and ISRE promoters. Analysis of the luciferase activities at 24 h posttransfection showed that the expression of NS2A significantly inhibited RIG-I-, MDA5-, MAVS-, and STING-induced IFN-β and ISRE promoter activation (Fig. 4A to E). NS2A only inhibited TBK1-induced IFN-β promoter activation but not inhibited ISRE promoter activation. Conversely, the induction of IFN-β and ISRE promoter activities were not affected by the coexpression of IRF7 and NS2A (Fig. 4F), which suggested that NS2A likely inhibits the IFN signaling pathway at the TBK1 step. The cells were harvested to detect duIFN-β mRNA levels by RT-qPCR. The expression of DTMUV NS2A inhibited the duIFN-β mRNA expression level induced by the components, except for IRF7 (Fig. 4G). Moreover, NS2A significantly disrupts RLR ligand (dsRNA)-, cyclic GMP-AMP (cGAMP) synthase (cGAS) ligand (dsDNA)-, and STING agonist (cGAMP)-activated IFN-β transcription in DEFs (Fig. 4H), suggesting that their common downstream molecule STING might be the target of NS2A to inhibit IFN-β induction.
FIG 4.
DTMUV NS2A inhibits RIG-I-, MAD5-, MAVS-, and STING-mediated IFN induction. (A to F) DEFs were transiently transfected with each of the above components (400 ng/well) and NS2A (400 ng/well) and subsequently transfected with pRL-TK plasmid (40 ng/well), pGL3-IFN-β-Luc, or pGL4-ISRE-Luc (400 ng/well). At 24 h posttransfection, the IFN-β/ISRE-Luc activity was measured. (G) DEFs were transiently transfected with each of the pCAGGS plasmids expressing Flag-tagged components (400 ng/well) and His-tagged NS2A (400 ng/well). After 24 h of transfection, cells were collected and treated with 1 ml RNAiso Plus reagent for the detection of duIFN-β mRNA by RT-qPCR. (H) DEFs were transiently cotransfected with RLR ligand (dsRNA) (1 μg/well), cGAS ligand (dsDNA) (3 μg/well), or STING agonist (cGAMP) (10 μg/well) with His-tagged NS2A (400 ng/well). After 24 h posttransfection, cells were collected and treated with 1 ml RNAiso Plus reagent for the detection of duIFN-β mRNA by RT-qPCR, and protein expression levels were determined by Western blot analysis. All results were normalized to those of duβ-actin. All results were normalized to those of duβ-actin. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
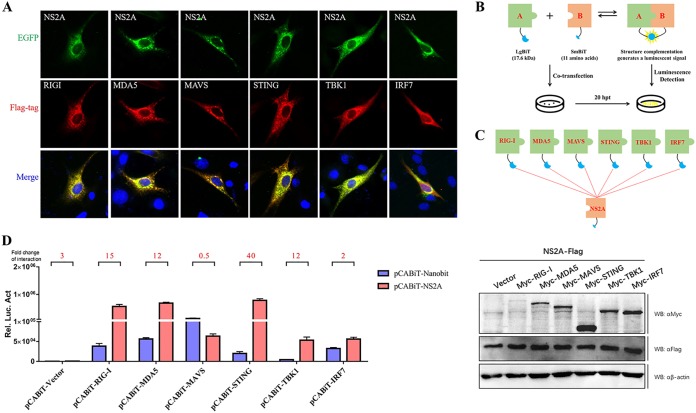
DTMUV NS2A interacts with STING.
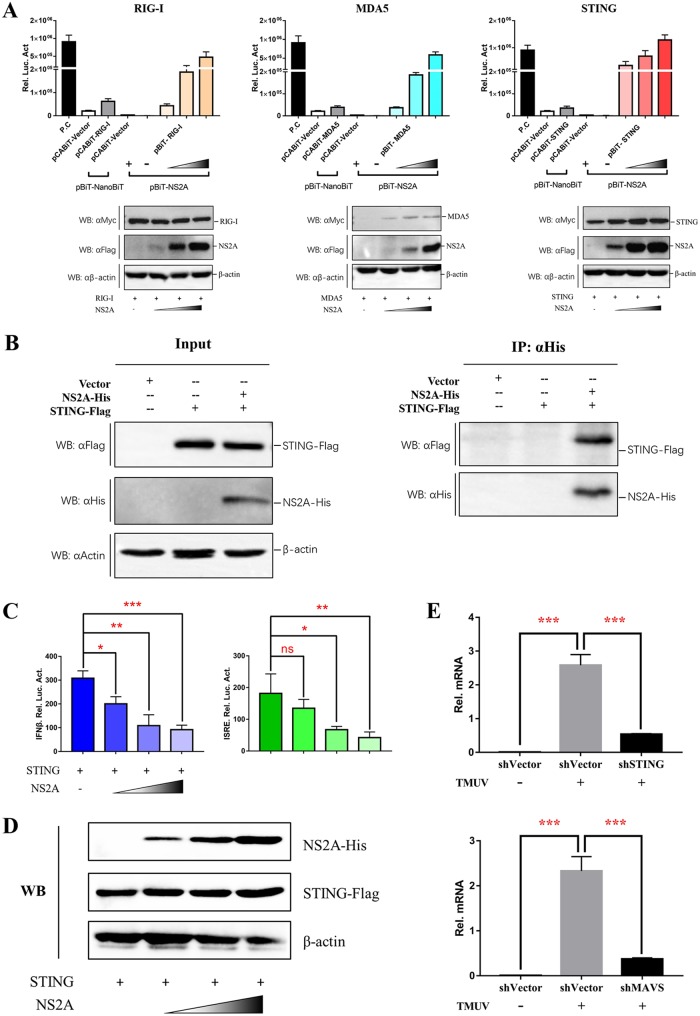
To determine the molecular interaction of NS2A in the IFN induction signaling pathway, the duRIG-I, duMDA5, duMAVS, duSTING, duTBK1, or duIRF7 plasmids were coexpressed with NS2A, and indirect immunofluorescence assays (IFAs) were performed. As shown in Fig. 5A, DTMUV NS2A colocalized with the upstream components of duSTING (containing duSTING, duRIGI, duMAD5, and duMAVS). Then, the RIG-I, MDA5, MAVS, STING, TBK1, or IRF7 plasmids with a Large Binary Technology (LgBiT) tag were coexpressed with NS2A with a Small BiT (SmBiT) tag, and NanoLuc Binary Technology (NanoBiT) protein-protein interaction (PPI) assays were performed (Fig. 5B and C). The luciferase activities were measured at 20 h posttransfection. Interestingly, we found that NS2A could interact with RIG-I, MDA5, and STING (Fig. 5D). Moreover, NS2A could interact with STING even at every low dose according to a dose-dependent assay, while NS2A could interact with RIG-I and MDA5 only at high doses (Fig. 6A). These results were further confirmed by a coimmunoprecipitation (co-IP) assay (Fig. 6B). In addition, we observed that NS2A strongly and significantly suppressed the duSTING-mediated activation of the IFN-β, IFN-β-Luc, and ISRE-Luc promoters in a dose-dependent manner (Fig. 6C). To exclude the effects of increasing doses of NS2A plasmid on STING expression, the protein levels of duck STING and NS2A were detected by Western blotting. With increasing doses of NS2A, STING expression was constant (Fig. 6D). Moreover, to demonstrate the essential role of STING in DTMUV activation of IFN-β transcription in the infected cells, we have performed additional knockdown experiments, suggested that the induction of IFN-β mRNA expression by TMUV infection was significantly reduced following knocking down STING or MAVS expression in DEFs (Fig. 6E). Collectively, these results revealed that DTMUV NS2A inhibited the duSTING-mediated activation of IFN-β/ISRE-Luc promoter activity via directly interacting with STING.
FIG 5.
DTMUV NS2A inhibits RIG-I-, MAD5-, MAVS-, and STING-mediated IFN induction. (A) BHK-21 cells were cotransfected with each of the components (pCAGGS-RIG-I, MDA5, MAVS, STING, TBK1, and IRF7-Flag) (400 ng/well) and pCAGGS-NS2A-His (400 ng/well). At 24 h posttransfection, cells were washed with cold PBS three times and subsequently fixed in 4% paraformaldehyde overnight at 4°C for IFA. (B, C) Schematics of the NanoBiT assay and plasmid construction. (D) DEFs were cotransfected with pCABiT-NS2A-SmBiT-Flag (400 ng/well) and plasmids encoding each of the components (pCABiT-RIG-I, MDA5, MAVS, STING, TBK1, and IRF7-LgBiT-Myc) (400 ng/well), and the luciferase activities were measured at 20 h posttransfection. Luminescence was measured at a user-defined time point or continuously for up to 2 h. According to the operational instructions, luminescence 10-fold higher than that of the negative control indicated a specific PPI (protein-protein interaction). Protein expression levels were determined by Western blot analysis. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 6.
DTMUV NS2A binds with STING. (A) Dose-dependent analysis of candidate components that interact with NS2A. DEFs were transiently transfected with the indicated amount of pCABiT-NS2A-SmBiT-Flag plasmid (100 ng, 200 ng, or 400 ng/well), along with 400 ng/well of pCABiT-RIG-I, MDA5, or STING-LgBiT-Myc plasmid, respectively. The luciferase activities were measured at 20 h posttransfection, and protein expression levels were determined by Western blot analysis. (B) Coimmunoprecipitation of NS2A and STING. DEF cells were cotransfected with pCAGGS-NS2A-His (800 ng/well) and pCAGGS-STING-Flag (800 ng/well). At 24 h posttransfection, cells were lysed in Pierce IP lysis buffer (Thermo Fisher), and whole-cell extracts (WCEs) were loaded as input. WCEs were incubated with 10 μg of the indicated antibody and 1 mg of SureBeads Protein G. Finally, the precipitates were fractionated by SDS-PAGE, and Western blotting was performed with the appropriate antibody. (C, D) Dose-dependent analysis of the inhibition of STING-mediated IFN-β-Luc/ISRE-Luc promoter activity by NS2A. DEFs were cotransfected with indicated amount of pCAGGS-NS2A-His plasmid (100 ng, 200 ng, or 400 ng/well) and pCAGGS-STING-Flag (400 ng/well) and subsequently transfected with pRL-TK plasmid (40 ng/well), pGL3-IFN-β-Luc (400 ng/well), or pGL4-ISRE-Luc (400 ng/well); the IFN-β/ISRE-Luc activity was measured at 36 h postinfection. Protein expression levels were determined by Western blot analysis. (E) DEFs were transfected with shSTING (800 ng/well), shMAVS (800 ng/well), or NC-shRNA (800 ng/well), respectively. After 24 h transfection, cells were infected with TMUV (25 μl containing 100 TCID50 per well) for 24 h; the virus copies were detected by RT-qPCR. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
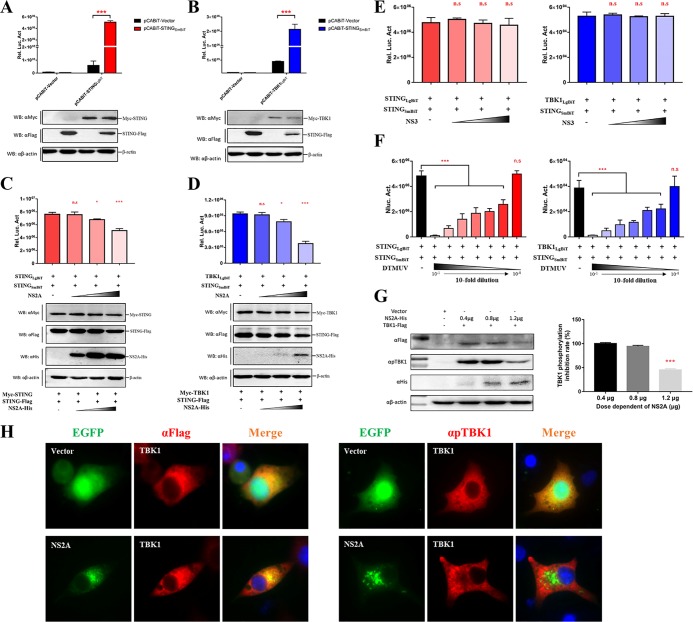
NS2A impairs the STING-STING interaction and the STING-TBK1 interaction, which reduces the phosphorylation of TBK1.
Following the dimerization of STING, TBK1 and IRF3 can be recruited to the C-terminal cytoplasmic domain of STING to form a complex that facilitates TBK1 phosphorylation, subsequently leading to the activation of the IFN-β response. To confirm the dimerization of STING and the STING-TBK1 complex, STING with an SmBiT tag was cotransfected with STING with an LgBiT tag or TBK1 with an LgBiT tag. According to the results of a NanoBiT PPI assay, we found that STING could interact with itself to form a dimer (Fig. 7A) and also interacted with TBK1 (Fig. 7B). To determine the effects of NS2A on the formation of STING-STING complex and STING-TBK1 complex, NS2A was cotransfected with STING with a SmBiT tag and STING with a LgBiT tag or STING with a SmBiT tag and TBK1 with a LgBiT tag. NS2A significantly impaired the formation of the STING-STING complex and the STING-TBK1 complex in a dose-dependent manner (Fig. 7C and D). As additional controls, NS3 had no inhibitory activity on the STING-STING complex and the STING-TBK1 complex in a dose-dependent manner. (Fig. 7E). Consistently, DTMUV infection could also disrupt the STING-STING and STING-TBK1 interaction in infected cells (Fig. 7F). Furthermore, the phosphorylation of TBK1 was reduced in cells expressing a high level of NS2A, as shown by Western blotting and IFA (Fig. 7G and H). The Western blotting results were analyzed by ImageJ software, and we found that TBK1 phosphorylation was inhibited by 55% in cells transfected with 1.2 μg of NS2A plasmid (Fig. 7G) compared with TBK1 phosphorylation in cells transfected with 0 μg of NS2A plasmid. These results indicated that the interaction between NS2A and STING inhibited the recruitment of TBK1 to STING, which subsequently reduced the phosphorylation of TBK1, leading to the inhibition of the IFN-β signaling pathway. Thus, NS2A inhibits the IFN-β signaling pathway by interacting with STING, which disrupts the formation of the STING-STING complex and the STING-TBK1 complex.
FIG 7.
NS2A impairs the formation of the STING-STING interaction and the STING-TBK1 interaction, which reduces the phosphorylation of TBK1. (A, B) The interaction between STING-STING and STING-TBK1. pCABiT-STING-SmBiT-Flag (400 ng/well) was cotransfected with pCABiT-STING-LgBiT-Myc or pCABiT-TBK1-LgBiT-Myc (400 ng/well) into DEF cells for 20 h. The luminescence was detected, and protein expression was measured by Western blot analysis. (C, D) NS2A competitively inhibits the interactions of STING-STING and STING-TBK1. pCABiT-STING-SmBiT-Flag (400 ng/well) was cotransfected with pCABiT-STING-LgBiT-Myc or pCABiT-TBK1-LgBiT-Myc (400 ng/well) into DEF cells in the presence of pCAGGS-NS2A-His at different doses (100 ng, 200 ng, or 400 ng/well). After 20 h of transfection, the luminescence was detected, and protein expression was measured by Western blot analysis. (E) NS3 has no inhibitory activity on STING-STING or STING-TBK1 interaction. pCABiT-STING-SmBiT-Flag (400 ng/well) was cotransfected with pCABiT-STING-LgBiT-Myc or pCABiT-TBK1-LgBiT-Myc (400 ng/well) into DEF cells in the presence of pCAGGS-NS3-His at different doses (100 ng, 200 ng, or 400 ng/well). After 20 h of transfection, the luminescence was detected. (F) DTMUV infection disrupts the STING-STING and STING-TBK1 interaction in infected cells. DEFs were infected with several dilutions of DTMUV (10-fold serially diluted in DMEM from 10−1 to 10−8; 100 μl/well) for 24 h; the control group was treated with 100 μl DMEM. Subsequently, pCABiT-STING-SmBiT-Flag (400 ng/well) was cotransfected with pCABiT-STING-LgBiT-Myc or pCABiT-TBK1-LgBiT-Myc (400 ng/well) into DEF. After 20 h of transfection, the luminescence was detected. (G, H) NS2A suppresses the phosphorylation of TBK1. pCAGGS-TBK1-Flag (1,200 ng/well) was cotransfected with different doses of pCAGGS-NS2A-His (400 ng, 800 ng, or 1,200 ng/well) into DEF cells for 24 h. (G) The protein expression levels were determined by Western blot analysis with the phospho-TBK1/NAK (Ser172) rabbit MAb (Cell Signaling Technology, USA), and the results were quantified by ImageJ software. pCAGGS-TBK1-Flag (1,200 ng/well) was cotransfected with pEGFP-NS2A or pEGFP vector (1,200 ng/well) into BHK-21 cells. (H) At 24 h posttransfection, cells were washed with cold PBS three times and subsequently fixed in 4% paraformaldehyde overnight at 4°C for IFA with the phospho-TBK1/NAK (Ser172) rabbit MAb. All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *,P < 0.05; **, P < 0.01; ***, P < 0.001.
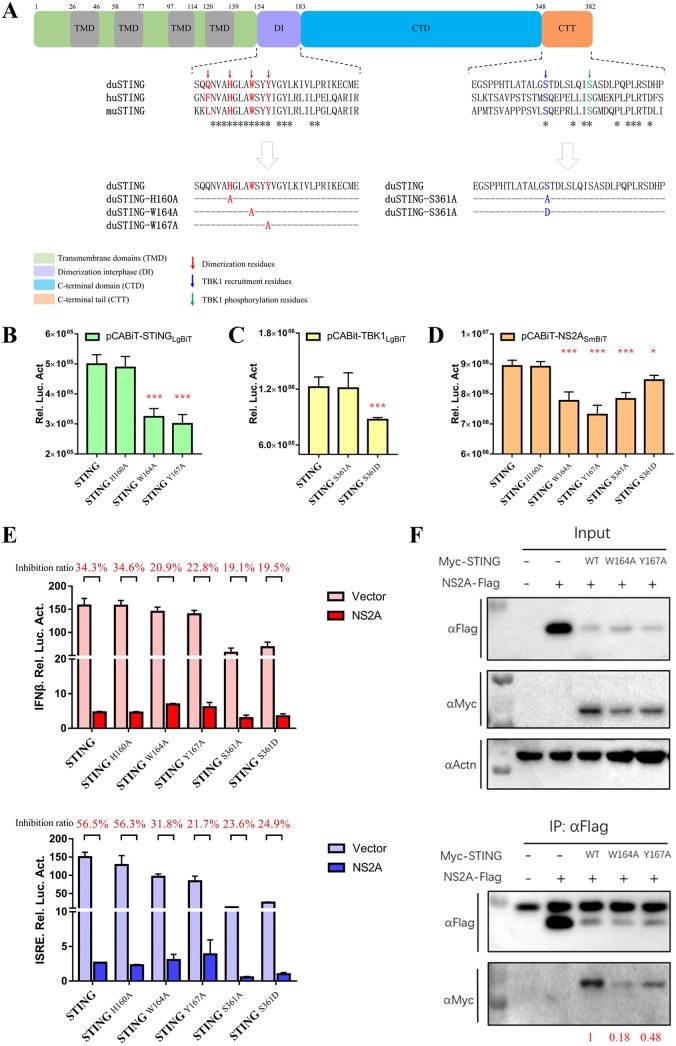
STING dimerization and phosphorylation are critical for its interactions with NS2A and subsequent IFN induction.
According to amino acid sequence alignment of duSTING, human STING (huSTING), and mouse STING (muSTING), the potential key dimerization sites (H160, W164, and Y167) and phosphorylated residues (S361) are mostly conserved (Fig. 8A). To understand the molecular mechanisms of the interaction between NS2A and the STING-TBK1 complex, STING mutants were constructed as follows. Alanine mutations were made in the dimerization residues (H160A, W164A, and Y167A) of duSTING and the phosphorylated STING residues (S361A and S361D; changing both residues into Asp residues could create a continuous negatively charged surface to mimic the phosphorylation of serine residues) (33), and these mutants were introduced into the pCABiT-SmBiT vector fused with a Flag tag or the pCABiT-LgBiT vector fused with a Myc tag, respectively (Fig. 8A). As shown in Fig. 8B, we found the W164A and Y167A mutations in STING significantly reduced duSTING homodimerization. Moreover, the S361D mutation significantly impaired the ability of STING to recruit TBK1, while the S361A mutation did not (Fig. 8C). To determine whether the dimerization and phosphorylation of STING are essential for the NS2A-STING interaction, DEF cells were cotransfected with STING mutants with an LgBiT tag and NS2A with an SmBiT tag. According to the NanoBiT PPI assay, we found that the STING W164A, Y167A, and S361A mutations significantly affected the NS2A-STING interaction, which suggested that duSTING homodimerization is important for the NS2A-STING interaction (Fig. 8B and C).
FIG 8.
The dimerization and phosphorylation sites of STING are critical for its interactions with NS2A and subsequent IFN induction. (A) Sequence analysis of duSTING protein functional domains and multiple alignment of duSTING (GenBank accession number XP_027323921.1), huSTING (GenBank accession number NP_938023), and muSTING (GenBank accession number NP_082537). (B) Interaction of STING with STING dimerization-related mutants. DEF cells were cotransfected with pCABiT-STING-LgBiT-Myc (50 ng/well) and pCABiT-STING-SmBiT-Flag mutants (H160A, W164A, and Y167A) (50 ng/well). After 20 h of transfection, the luminescence was detected. (C) Interaction of TBK1 with STING phosphorylation-related mutants. DEF cells were cotransfected with pCABiT-TBK1-LgBiT-Myc (50 ng/well) and pCABiT-STING-SmBiT-Flag mutants (S361A and S361D) (50 ng/well). After 20 h of transfection, the luminescence was detected. (D) Interaction of NS2A with STING dimerization- and phosphorylation-related mutants. DEF cells were cotransfected with pCABiT-NS2A-SmBiT-Flag (50 ng/well) and pCABiT-STING-LgBiT-Myc mutants (H160A, W164A, Y167A, S361A, and S361D) (50 ng/well). After 20 h of transfection, the luminescence was detected. (E) The inhibitory effects of NS2A on STING mutants-mediated IFN-β/ISRE-Luc reporter activities. DEF cells were cotransfected with pCAGGS-NS2A-His (50 ng/well) and pCAGGS-STING-Flag mutants (H160A, W164A, Y167A, S361A, and S361D) (50 ng/well) and subsequently transfected with pRL-TK plasmid (5 ng/well), pGL3-IFN-β-Luc, or pGL4-ISRE-Luc (50 ng/well). At 24 h posttransfection, the IFN-β/ISRE-Luc activity was measured. The inhibition ratio was determined using the following formula: inhibition ratio (%) = (1-IFN-β/ISRE-Luc reporter value of NS2A transfected / IFN-β/ISRE-Luc reporter value of vector transfected) × 100%. (F) Coimmunoprecipitation of NS2A and STING mutants (H160A, W164A, and Y167A). DEF cells were cotransfected with pCAGGS-NS2A-His and pCAGGS-STING-Flag mutants (H160A, W164A, and Y167A). At 24 h posttransfection, cells were lysed in Pierce IP lysis buffer (Thermo Fisher), and whole-cell extracts (WCEs) were loaded as input. WCEs were incubated with 10 μg of the indicated antibody and 1 mg of SureBeads Protein G. Finally, The protein expression levels were determined by Western blot analysis, and the results were quantified by ImageJ software (represented with red). All data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The dephosphorylation (S361A) of STING had no effort on the TBK1-STING interaction, while its autophosphorylation (S361D) indeed significantly reduced the TBK1-STING complex. However, the dephosphorylation (S361A) of STING significantly reduced the NS2A-STING interaction. Importantly, STING autophosphorylation reduced the inhibition of the interaction between dephosphorylated STING (S361A) and NS2A (Fig. 8D). Additionally, the mutation of the phosphorylation site in STING, residue S361, significantly abolished its ability to induce IFN-β and ISRE-Luc promoter activity, and the inhibitory effects of NS2A on the W164A, Y167A, S361A, and S361D STING mutants mediated IFN-β/ISRE-Luc promoter activity, which was decreased compared to the effects on WT STING-mediated promoter activity (Fig. 8E). A subsequent co-IP assay was performed to confirmed that W164A and Y167A mutations in STING significantly impaired the NS2A-STING interaction (Fig. 8F). These results demonstrated that the dimerization residues of STING (W164 and Y167) are critical for the formation of the STING-NS2A complex, leading to the inhibition of the STING mutant-mediated IFN-β signaling pathway by NS2A. Similarly, the phosphorylation site of STING (S361) was important for the interaction between NS2A and STING, and the dephosphorylation of STING impaired the STING-NS2A interaction and IFN induction. These results also indicated that the binding of NS2A to STING may be dependent on the phosphorylation of STING at S361, thus impairing the recruitment of TBK1 by STING.
DISCUSSION
Once invading viral pathogens are recognized by RLRs, the host initiates the activation of the type I IFN response against viral infection. Consequently, most viruses evoke efficient strategies beneficial to their survival to avoid the type I IFN response of the host. Accumulating research has been conducted on the coevolution between flaviviruses and their hosts. Some flaviviruses are sensed by pattern recognition receptors, such as TLRs and RLRs, leading to the initiation of the IFN response (34); however, flaviviruses have developed several strategies to combat the host innate immune response (35). As an emerging avian flavivirus, DTMUV was reported to initiate the IFN response in cells infected with DTMUV to induce the expression of IFN-regulated genes or ISGs (36). In our previous in vivo and in vitro studies, infection by DTMUV triggered a series of immune responses to maintain control of virus invasion and replication (37, 38), and goose IFNs could effectively reduce the replication of DTMUV due to downstream antiviral genes, such as goMx and goOASL (39). However, the immune characteristics of DTMUV nonstructural proteins remain largely unknown.
Mechanistically, our data showed that DTMUV NS2A could promote the replication of DPV-GFP in vitro and suppress virus-mediated IFN-β and ISRE promoter activity. Accumulating evidence has identified several strategies by which flavivirus NS2A evades the innate immune system. NS2A from DENV-2 was first found to act as an interferon antagonist in 2003. DENV-2 NS2A overexpression enhanced the replication of NDV-GFP in chick embryo fibroblast (CEF) and A549 cells, indicating that NS2A may be a potential DENV-2 anti-IFN protein. Reporter assays were performed to show that NS2A has the ability to block both IFN induction by viral infection and IFN-stimulated signal transduction (28). A recent report indicated that DENV-1/2/4 NS2A inhibited IFN-β production by blocking TBK1 phosphorylation and activation (18). Research on the Kunjin strain of West Nile virus (WNV) (WNVKUN) has shown that NS2A inhibits STAT2 translocation to the nucleus, hence blocking IFN-α signaling (40). Moreover, in WNVKUN NS2A, the A30P mutation was shown to be responsible for the suppression of IFN-β transcription both in vitro and in vivo, leading to attenuated virulence in mice (41). ZIKV NS2A, NS2B, and NS4B antagonize IFN-β production through blocked TBK1 phosphorylation (42). Although flavivirus NS2A was reported to inhibit IFN production by targeting TBK1 or STAT2, research on how NS2A targets components and the interaction sites between NS2A and these components has not been carried out. Here, based on IFN-β/ISRE promoter activity, NS2A significantly inhibited RIG-I-, MDA5-, MAVS-, and STING-induced IFN-β and ISRE promoter activation, whereas NS2A only inhibited TBK1-induced IFN-β promoter activation but did not inhibit ISRE promoter activation; we speculate that the binding of NS2A to duck STING may cause structural changes of STING protein or unexpected steric effects, which subsequently affect TBK1 and STING interaction. Moreover, NS2A significantly disrupts RLR ligand-, cGAS ligand-, and STING agonist-activated IFN-β transcription in DEFs, suggesting that their common downstream molecule STING might be the target of NS2A to inhibit IFN-β induction. Subsequently, according to the results of IFA, NanoBiT, and co-IP assays, we showed that NS2A directly interacts with duSTING and therefore reduces STING-mediated IFN-β/ISRE promoter activity, but Wang et al. (30) observed DTMUV NS1, not other nonstructural proteins, inhibited type I IFN expression by targeting VISA in HEK293 cells. The reason of the discrepancy is that it is possible that the different species of cells were used. Additionally, our recent study suggested the cleavage of STING by DTMUV NS2B3 to impaired IFN-β induction (32). It is very possible that different DTMUV proteins target the same cellular components through different strategies to antagonize IFN-β production.
However, we also noticed that the association of NS2A with RIG-I or MDA5 may be an indirect or spatial interaction. Indeed, both RIG-I and MDA5 activate MAVS through caspase-recruitment domain (CARD)-CARD interactions, which is essential for the activation of downstream type I IFN production (43). Additionally, STING directly interacts with RIG-I, MAVS, and TBK1, which activate interferon regulatory factor (IRF) signaling (44). This may be the reason for the indirect interaction of NS2A with other molecules (such as RIG-I and MDA5 in our experiments). NS2A interacted with STING and inhibited subsequent promoter activity, which was not due to a reduction in the expression level of STING. Subsequently, we confirmed the formation of the STING-STING complex, STING-TBK1 complex, and the phosphorylation of TBK1 were significantly affected by NS2A. A physical interaction between STING and TBK1 is essential for facilitating the recruitment of IRF3/IRF7 and the activation of type I IFN signaling. Therefore, we speculate that the binding of NS2A to duSTING may cause structural changes in the duSTING protein, which subsequently cannot bind to itself or to duTBK1. Alternatively, the binding sites between NS2A and duSTING may overlap or be near the sites for STING-STING interaction and the STING-TBK1 interaction, which may affect STING homodimer and the STING-TBK1 heterodimer formation. However, the molecular mechanism by which NS2A interferes with the STING homodimer and the STING-TBK1 heterodimer requires further investigation.
Previous reports have also shown that huSTING and muSTING are dimeric, which is mediated by the region containing residues 152 and 173 (45). Once activated, the recruitment of TBK1 is dependent on the phosphorylation of huSTING at S358, which is mediated by TBK1 (44). In our study, we found that the predicted dimerization sites (H160, W164, and Y167) and STING phosphorylation site (S361) are almost conserved among duSTING, huSTING, and muSTING. Therefore, we constructed H160A, W164A, Y167A, S361A, and S361D STING mutants. Both the W164A and Y167A mutations could affect STING-STING binding and its interaction with NS2A, leading to the subsequent inhibition of IFN-β/ISRE promoter activity, which may affect STING homodimer formation. The S361A mutation of STING abolished its ability to induce IFN-β/ISRE promoter activity but did not affect the recruitment of TBK1. It is possible that the phosphorylation sites of duSTING are not the same sites responsible for the recruitment of duTBK1. Indeed, a recent study revealed that the huSTING C-terminal tail (amino acids [aa] 370 to 380) is the huTBK1 binding site (46). Therefore, we concluded that steric hindrance on the surface of the STING S361D mutant significantly diminished the binding between STING and TBK1. However, the steric hindrance of STING S361D only moderately affected the interaction between NS2A and STING. We propose that the formation of the duSTING dimer is important for the NS2A-STING interaction. We hypothesize that the oligomerization and phosphorylation of STING change the protein structure compared with the structure of the STING monomer, which might make STING spatially accessible to NS2A.
In conclusion, our results uncovered a novel mechanism by which DTMUV NS2A subverts host innate immune responses. NS2A significantly inhibited IFN-β and ISRE promoter activity in a dose-dependent manner and facilitated DPV infection. We found that NS2A competes with TBK1 in binding to STING, impairs STING-STING binding, and reduces the phosphorylation of TBK1, leading to the subsequent inhibition of IFN production. Importantly, we identified that the W164A, Y167A, and S361A STING mutations obviously impaired the NS2A-STING interaction, which is important for NS2A-induced immune inhibition. Hence, our data demonstrate that DTMUV NS2A disrupted STING-dependent antiviral cellular defenses by binding to STING.
MATERIALS AND METHODS
Ethics statement.
The animal studies were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (no. XF2014-18) and followed the National Institutes of Health guidelines for the performance of animal experiments.
Cells, viruses and antibodies.
Duck embryo fibroblasts (DEFs) and BHK-21 cells were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco Life Technologies, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Gibco Life Technologies, Shanghai, China). All cells were cultured in 6-well or 12-well plates at 37°C in 5% CO2. The BAC-DPV-EGFP virus employed in this study was constructed by our research center. The duck Tembusu virus (DTMUV) CQW1 strain (GenBank accession number KM233707) was isolated by our laboratory (47) with a measured viral titer of 6.3 × 10−6 TCID50/100 μl, as reported previously (38). Antibodies (Abs) against Flag, His, Myc, and β-actin were purchased from TransGen Biotech (Beijing, China). Phospho-TBK1/NAK (Ser172) rabbit MAb was purchased from Cell Signaling Technology (CST, USA). The RLR ligand (dsRNA), cGAS ligand (dsDNA), and STING agonist (cGAMP) were purchased from InvivoGen (USA).
Plasmid construction.
The sequences of the structural (C, P, and E) and nonstructural (NS) (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) viral proteins were amplified from the DTMUV CQW1 strain genome and cloned into the pCAGGS expression vector with a His tag at the C terminus using standard molecular biology techniques. The duck RIG-I, MDA5, MAVS, TBK1, IRF7, and STING genes were cloned into the pCAGGS expression vector with a Flag tag at the C terminus. The DTMUV NS2A gene was cloned into pCABiT-SmBiT with a Flag tag, and each immune component (duRIG-I, duMDA5, duMAVS, duTBK1, duIRF7, and duSTING) was cloned into pCABiT-LgBiT with a Myc tag for NanoLuc Binary Technology (NanoBiT) assay. duSTING was also cloned into pCABiT-SmBiT with a Flag tag. Moreover, duSTING mutants (STINGW160A, STINGH164A, and STINGY167A) were constructed and cloned into pCABiT-LgBiT with a Myc tag, while other duSTING mutants (STINGS361A and STINGS361D) were constructed and cloned into both pCABiT-LgBiT with a Myc tag and pCABiT-SmBiT with a Flag tag. Plasmid containing influenza A virus PR/8-NS1 was donated by the Shanghai Veterinary Research Institute.
Real-time RT-PCR.
Total RNA was isolated from selected tissues using RNAiso Plus reagent. The quantity of the RNA in each sample was determined using a NanoDrop 2000 (Thermo, Waltham, MA, USA), and RT-PCR was performed on each sample using a 5× All-In-One RT master mix reagent kit in accordance with the manufacturer’s instructions (Applied Biological Materials, Richmond, BC, Canada). Finally, the cDNAs were stored at −80°C until use. qPCR was used to detect the expression of duIFN-α, duIFN-γ, and duIFN-λ in the samples. qPCR was performed using the Bio-Rad CFX96 real time detection system (Bio-Rad, USA). Threshold cycle (CT) values were normalized to those of the housekeeping genes duβ-actin or goGAPDH, and the relative expression levels of each target gene were calculated with the comparative CT (2−ΔΔCT) method. The real-time PCR conditions and protocols have been previously described (48).
Viral infection and IFN treatment.
The recombinant plasmids pcDNA3.1 (+)-goIFN-α, pcDNA3.1 (+)-goIFN-γ, and pcDNA3.1 (+)-goIFN-λ were transfected into BHK-21 cells. Cell lysates from BHK-21 cells were harvested at 24 h posttransfection and clarified by centrifugation at 500 × g for 10 min after being frozen and thawed three times. Then, DEFs were pretreated with 100 μl goIFN-α, goIFN-γ, and goIFN-λ. After 24 h of treatment, the cells were infected with 400 μl DTMUV (at 100 TCID50) for 24 h. In addition, some cells were preinfected with DTMUV for 24 h and treated with IFNs 24 h later. All cells were collected for the detection of viral copy numbers and viral titers. Samples (200 μl) were extracted with a nucleic acid extraction kit (Tiangen, Shanghai, China), and then the DTMUV copy numbers were detected by RT-qPCR using special primers based on the DTMUV-E gene. Subsequently, DTMUV titers in DEFs were determined by an endpoint dilution assay, and the results were analyzed using the Reed-Muench method (TCID50).
Transfection and infection of DEF cells with BAC-DPV-EGFP.
DEFs were transfected with each of the 10 DTMUV-derived plasmids; the pCAGGS vector was used as a negative control, while pCAGGS-pNS1 (influenza A virus NS1, a known IFN antagonist) served as a positive control. After 24 h of transfection, cells were infected with BAC-DPV-EGFP (100 μl containing 100 TCID50 per well). At 48, 60, and 72 h postinfection, the cells were collected to analyze viral copy numbers and fluorescence and for flow cytometry (FCM).
Indirect immunofluorescence assay.
Transfected BHK-21 cells were washed three times with cold phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde overnight at 4°C. After three washes with PBS containing 0.1% Tween 20 (PBST) for 5 min each, the cells were permeabilized with 0.2% Triton X-100 for 30 min at 4°C and blocked with 5% bull serum albumin (BSA) in PBS for 1 h at room temperature. The cells were subsequently washed three times with PBST and incubated with primary antibody (diluted 1:2,000) for 2 h at room temperature in 1% BSA. Following three washes with PBST, the cells were incubated with a secondary antibody (diluted 1:5,000) for 1 h at room temperature in 1% BSA and then incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Finally, the coverslips were washed extensively and fixed onto slides. Fluorescence images were taken with a fluorescence microscope (Bio-Rad, USA).
Luciferase reporter assay.
The pGL3-IFN-β-Luc expression plasmid was constructed with the sequence of the duck IFN-β promoter region (GenBank accession number KM032183.1). The commercialized pGL4-ISRE-Luc expression plasmid was purchased from Promega (Madison, WI, USA). First, DEFs were seeded onto a 48-well plate and transiently transfected with pGL3-IFN-β-Luc (400 ng/well) or pGL4-ISRE-Luc (400 ng/well). Then, the cells were transfected with a pRL-TK plasmid (40 ng/well) (Promega, Madison, WI, USA), which acted as an internal control to normalize the transfection efficiency. Twenty-four hours later, the cells were challenged with 100 μl DTMUV (1,000 TCID50). At 12, 24, 36, and 48 hpi, the cells were harvested for luciferase assays. The luciferase activities were determined with a Dual-Glo luciferase assay system (Promega) and normalized based on the Renilla luciferase activity.
NanoBiT protein-protein interaction assay.
NanoBiT is a complementation reporter designed for the quantitative investigation of protein interaction dynamics under relevant physiological conditions. The NanoBiT assay is a two-subunit system based on NanoLuc luciferase that can be used for the intracellular detection of PPIs. The Large BiT (LgBiT; 17.6 kDa) and Small BiT (SmBiT; 11 amino acids) subunits are fused to proteins of interest (Fig. 1), and when expressed, PPI brings the subunits into close proximity to form a functional enzyme that generates a bright, luminescent signal (Fig. 5B). Two proteins that potentially interact were fused to the LgBiT or SmBiT subunit and transfected into DEF cells for 20 h. Protein kinase cAMP-activated catalytic subunit alpha (PRKACA-SmBiT) and protein kinase cAMP-dependent type II regulatory subunit alpha (PRKAR2A-LgBiT) were used as positive controls, whereas the LgBiT fusion plasmid coexpressed with HaloTag-SmBiT was used as a negative control. Then, the medium was aspirated and replaced with 100 μl of buffered Opti-MEM I reduced serum medium (Life Technologies; catalog no. 11058), 25 μl of Nano-Glo live cell reagent was added to each well, and the plate was gently mixed by hand or with an orbital shaker (15 s at 300 to 500 rpm). Finally, the luminescence was measured at a user-defined time point or continuously for up to 2 h. Importantly, if the signal from the unknown PPI pair was less than 10-fold higher than that of the negative control, this potentially indicated a nonspecific interaction between the fusion partners.
Coimmunoprecipitation and Western blot analysis.
Transfected DEF cells were lysed in Pierce IP lysis buffer (Thermo Fisher) and incubated on ice for 5 min with periodic mixing. Then, the lysate was transferred to a microcentrifuge tube and centrifuged at ∼13,000 × g for 10 min at 4°C to pellet the cellular debris, and the supernatant was collected to determine the protein concentration and for further analysis. From each sample, 0.5 ml of cell lysate was incubated with 10 μg of the indicated antibody and 1 mg of SureBeads Protein G (Thermo Fisher) at room temperature for 2 h. The SureBeads were washed 3 times with 1 ml PBST and centrifuged at ∼13,000 × g for 1 min. Finally, the precipitates were fractionated by SDS-PAGE, and Western blotting was performed with the appropriate antibody.
RNA interference.
Short hairpin RNA (shRNA) targeting duSTING and duMAVS was chemically synthesized by GenePharma (China). The target sequences were inserted to the pGPU6 plasmid to generate pGPU6-shSTING (shSTING) and pGPU6-shMAVS (shMAVS). A nontargeting shRNA (NC-shRNA) was synthesized as a negative control. The efficiencies of the shRNA were measured by RT-qPCR.
Statistical analysis.
Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). The differences between values were evaluated by Student's t test. A P value of < 0.05 indicated statistical significance, and all values are expressed as the mean ± standard error of the mean (SEM).
Data availability.
Data for huSTING and muSTING are available in GenBank under accession numbers NP_938023 and NP_082537, respectively.
ACKNOWLEDGMENTS
This work was funded by grants from the National Key Research and Development Program of China (2017YFD0500800), Sichuan-International Joint Research for Science and Technology (2018HH0098), the China Agricultural Research System (CARS-42-17), the Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and Integration and Demonstration of Key Technologies for Goose Industrial Chain in Sichuan Province (2018NZ0005).
REFERENCES
- 1.Yan Z, Shen H, Wang Z, Lin W, Xie Q, Bi Y, Chen F. 2017. Isolation and characterization of a novel Tembusu virus circulating in Muscovy ducks in South China. Transbound Emerg Dis 64:e15–e17. doi: 10.1111/tbed.12525. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Chen S, Mahalingam S, Wang M, Cheng A. 2017. An updated review of avian-origin Tembusu virus: a newly emerging avian flavivirus. J Gen Virol 98:2413–2420. doi: 10.1099/jgv.0.000908. [DOI] [PubMed] [Google Scholar]
- 3.Su J, Li S, Hu X, Yu X, Wang Y, Liu P, Lu X, Zhang G, Hu X, Liu D, Li X, Su W, Lu H, Mok NS, Wang P, Wang M, Tian K, Gao GF. 2011. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One 6:e18106. doi: 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P, Lu H, Li S, Moureau G, Deng YQ, Wang Y, Zhang L, Jiang T, de Lamballerie X, Qin CF, Gould EA, Su J, Gao GF. 2012. Genomic and antigenic characterization of the newly emerging Chinese duck egg-drop syndrome flavivirus: genomic comparison with Tembusu and Sitiawan viruses. J Gen Virol 93:2158–2170. doi: 10.1099/vir.0.043554-0. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Lu H, Li S, Wu Y, Gao GF, Su J. 2013. Duck egg drop syndrome virus: an emerging Tembusu-related flavivirus in China. Sci China Life Sci 56:701–710. doi: 10.1007/s11427-013-4515-z. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Diao Y, Chen H, Ou Q, Liu X, Gao X, Yu C, Wang L. 2015. Isolation and genetic characterization of a Tembusu virus strain isolated from mosquitoes in Shandong, China. Transbound Emerg Dis 62:209–216. doi: 10.1111/tbed.12111. [DOI] [PubMed] [Google Scholar]
- 7.Wang HJ, Li XF, Liu L, Xu YP, Ye Q, Deng YQ, Huang XY, Zhao H, Qin ED, Shi PY, Gao GF, Qin CF. 2016. The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammalian interferon antiviral signaling. J Virol 90:6538–6548. doi: 10.1128/JVI.00197-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Guinn ML, Turell MJ, Kengluecha A, Jaichapor B, Kankaew P, Miller RS, Endy TP, Jones JW, Coleman RE, Lee JS. 2013. Field detection of Tembusu virus in western Thailand by RT-PCR and vector competence determination of select culex mosquitoes for transmission of the virus. Am J Trop Med Hyg 89:1023–1028. doi: 10.4269/ajtmh.13-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Li X, Zhang L, Wang Y, Yu X, Tian K, Su W, Han B, Su J. 2013. Duck Tembusu virus exhibits neurovirulence in BALB/c mice. Virol J 10:260. doi: 10.1186/1743-422X-10-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ti J, Zhang M, Li Z, Li X, Diao Y. 2016. Duck Tembusu virus exhibits pathogenicity to Kunming mice by intracerebral inoculation. Front Microbiol 7:190. doi: 10.3389/fmicb.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Gao X, Diao Y, Feng Q, Chen H, Liu X, Ge P, Yu C. 2013. Tembusu virus in human, China. Transbound Emerg Dis 60:193–196. doi: 10.1111/tbed.12085. [DOI] [PubMed] [Google Scholar]
- 12.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 13.Saito T, Gale M Jr. 2008. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med 205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loo YM, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalrymple NA, Cimica V, Mackow ER. 2015. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio 6:e00553-15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Chen Z, Li Y, Zhao Z, He W, Zohaib A, Song Y, Deng C, Zhang B, Chen H, Cao S. 2017. Japanese encephalitis virus NS5 inhibits type I Interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-kappaB. J Virol 91:e00039-17. doi: 10.1128/JVI.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JML, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez C, tenOever BR, García-Sastre A. 2014. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol 80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers TJ, McCourt DW, Rice CM. 1989. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 26.Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol 76:4773–4784. doi: 10.1128/jvi.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J Virol 82:4731–4741. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz-Jordan JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melian EB, Edmonds JH, Nagasaki TK, Hinzman E, Floden N, Khromykh AA. 2013. West Nile virus NS2A protein facilitates virus-induced apoptosis independently of interferon response. J Gen Virol 94:308–313. doi: 10.1099/vir.0.047076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Lei CQ, Ji Y, Zhou H, Ren Y, Peng Q, Zeng Y, Jia Y, Ge J, Zhong B, Li Y, Wei J, Shu HB, Zhu Q. 2016. Duck Tembusu virus nonstructural protein 1 antagonizes IFN-beta signaling pathways by targeting VISA. J Immunol 197:4704–4713. doi: 10.4049/jimmunol.1502317. [DOI] [PubMed] [Google Scholar]
- 31.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu Z, Zhang W, Wu Y, Wang T, Wu S, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Merits A, Chen S, Cheng A. 2019. Binding of the duck Tembusu virus protease to STING Is mediated by NS2B and is crucial for STING cleavage and for impaired induction of IFN-beta. J Immunol 203:3374–3385. doi: 10.4049/jimmunol.1900956. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M Jr. 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol 82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gack MU, Diamond MS. 2016. Innate immune escape by dengue and West Nile viruses. Curr Opin Virol 20:119–128. doi: 10.1016/j.coviro.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Wang Y, Li R, Liu J, Zhang J, Cai Y, Liu S, Chai T, Wei L. 2015. Immune responses of ducks infected with duck Tembusu virus. Front Microbiol 6:425. doi: 10.3389/fmicb.2015.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Liu M, Sun K, Yang Q, Wu Y, Zhao X, Cheng A. 2018. Duck stimulator of interferon genes plays an important role in host anti-duck plague virus infection through an IFN-dependent signalling pathway. Cytokine 102:191–199. doi: 10.1016/j.cyto.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Zhou H, Chen S, Wang M, Jia R, Zhu D, Liu M, Liu F, Yang Q, Wu Y, Sun K, Chen X, Jing B, Cheng A. 2016. Antigen distribution of TMUV and GPV are coincident with the expression profiles of CD8alpha-positive cells and goose IFNgamma. Sci Rep 6:25545. doi: 10.1038/srep25545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Zhang W, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Liu M, Sun K, Yang Q, Wu Y, Chen X, Cheng A. 2017. Goose Mx and OASL play vital roles in the antiviral effects of type I, II, and III interferon against newly emerging avian flavivirus. Front Immunol 8:1006. doi: 10.3389/fimmu.2017.01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol 79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol 80:2396–2404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA, Zou J, Xie X, Giraldo MI, Vasconcelos PFC, Weaver SC, Wang T, Rajsbaum R, Shi PY. 2018. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun 9:414. doi: 10.1038/s41467-017-02816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West AP, Shadel GS, Ghosh S. 2011. Mitochondria in innate immune responses. Nat Rev Immunol 11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Paludan SR. 2015. Activation and regulation of DNA-driven immune responses. Microbiol Mol Biol Rev 79:225–241. doi: 10.1128/MMBR.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. 2019. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567:394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu K, Huang J, Jia R, Zhang B, Wang M, Zhu D, Chen S, Liu M, Yin Z, Cheng A. 2015. Identification and molecular characterization of a novel duck Tembusu virus isolate from Southwest China. Arch Virol 160:2781–2790. doi: 10.1007/s00705-015-2513-0. [DOI] [PubMed] [Google Scholar]
- 48.Zhou H, Chen S, Qi Y, Wang M, Jia R, Zhu D, Liu M, Liu F, Chen X, Cheng A. 2015. Development and validation of a SYBR Green real-time PCR assay for rapid and quantitative detection of goose interferons and proinflammatory cytokines. Poult Sci 94:2382–2387. doi: 10.3382/ps/pev241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for huSTING and muSTING are available in GenBank under accession numbers NP_938023 and NP_082537, respectively.