Abstract
T regulatory cells (Tregs) are a cell subset that can suppress immune responses to maintain homeostasis and self-tolerance. In some scenarios, the immunosuppressive nature could be associated to other pathological developments such as autoimmune diseases and cancers. Due to the importance of Tregs in disease pathogenesis, we developed and validated an 11-color flow cytometry panel for phenotypic and functional detection of Treg markers using healthy human donor peripheral blood mononuclear cells (PBMCs). Our panel contains 4 Treg surface proteins and 2 functional cytokines as well as T-lymphocyte lineage markers CD3, CD4, and CD8. Our data shows an increase in expression of markers CD25, FoxP3, CTLA4, GITR and intracellular cytokines IL4 and TGFβ when comparing unstimulated samples to CD3/CD28 bead stimulated samples. This 11-color panel can be used to functionally evaluate immunosuppressive Tregs in human PBMC samples.
Keywords: Tregs, flow cytometry, antibody panel, peripheral blood mononuclear cells
INTRODUCTION
T regulatory cells (Tregs) are a subset of lymphocytes that directly or indirectly dampen futile immune responses to self and non-self antigens, thereby allowing the immune system to establish and maintain homeostasis [1]. These immune-suppressive cells can promote tolerance in a cell-to-cell contact-dependent manner, or by producing inhibitory cytokines, such TGF-β [2,3]. When Treg function is disturbed by mutation or quantitative imbalance, numerous complications may arise, such as autoimmune diseases.
Tregs are characterized as CD3+CD4+CD25+CD127neg/low FoxP3+. When Foxp3, a transcription factor essential to the development and function of Tregs, is rendered nonfunctional by mutations, afflicted individuals develop immunodysregulation polyendocrinopathy, enteropathy X-linked syndrome (IPEX). This rare and fatal syndrome functions in the development of autoimmune diseases, such as type 1 diabetes and dermatitis [2,4]. Treg cells are not only important in the context of autoimmune disease; the number of Treg cells is notably higher in patients with hematologic malignancies or solid tumors [5]. This abundance is indicative of responders to cancer immunotherapy: when patients with hematologic malignancies were treated with the anti-CTLA-4 therapy, those with a stable or improving disease state had marked decreases in Treg cell number and significant increases in their number of T effector cells [6].
Prior studies have suggested that Tregs can be characterized as CD4+ T cells that express the transcription factor Foxp3 and high levels of the IL-2 receptor alpha chain, CD25. However, concerns about the validity of using Foxp3 and CD25 as definitive Treg markers remain [7]. T effector populations briefly express high levels of CD25 upon activation, and previous research has shown that Foxp3 is transiently expressed by T cells without suppressive capacity [3,5,8-10]. Epigenetic analyses have revealed the existence of a Treg-specific demethylated region (TSDR) in the Foxp3 promoter and other studies have further expanded Treg characterization to include cells with negative to low levels of CD127 expression [2,11-17]. Studies have also highlighted the importance of GITR, CTLA-4, TGF-β and IL-4. GITR and CTLA-4 are functional markers intertwined with Treg expansion and mechanisms of immune tolerance [18,19]. TGF-β inhibits T effector differentiation, proliferation, and activation [20], while IL-4 supports Treg survival and suppressive capacity [21].
Because Treg dysfunction is central to the pathogenesis of autoimmunity, infectious diseases, transplant-associated diseases, and cancer, complete phenotypic identification of Tregs is of extreme clinical and biological significance. To ensure complete evaluation of Tregs, we created a 11-color flow cytometry panel that includes viability dye and an array of lineage markers (CD3, CD8 and CD4), functional markers (CD25, CD127, CTLA-4, GITR and Foxp3), and cytokines (IL-4 and TGF-β). This panel successfully characterizes Tregs and provides more insight into their functional status and immunosuppressive capacity than previously published panels [2,5,6,11,16]. Antibodies used in this protocol are listed in Table 1. We titrated each antibody using a standardized flow cytometry staining protocol, which is described in Methods section.
Table 1.
Antibody panel for phenotyping using flow cytometry surface and intracellular fluorophore-conjugated antibodies.
| Antibody | Fluorophore | Clone | Cat. # | Vendor | Concentration (μg/100 μl) | Marker/cytokine function |
|---|---|---|---|---|---|---|
| CD3* | PerCP 5.5 | OKT3 | 45-0037-42 | eBioscience | 0.002 | Lineage |
| CD4* | AF700 | RPA-T4 | 56-0049-42 | eBioscience | 0.003 | Lineage |
| CD8* | BV786 | RPA-T8 | 563823 | BD | 0.004 | Lineage |
| CD25 | BV421 | BC96 | 302630 | Biolegend | 0.004 | Lineage/activation |
| CD127* | BV510 | A019D5 | 351332 | Biolegend | 0.0025 | Lymphocyte development (negative gate) |
| GITR | BV650 | V27-580 | 747663 | BD | 0.005 | Checkpoint |
| CTLA-4 | PE | L3D10 | 349906 | Biolegend | 0.004 | Checkpoint |
| FoxP3 | FITC | PCH101 | 11-4776-42 | eBioscience | 0.0025 | Lineage |
| IL-4 | PeCy7 | 8D4-8 | 25-7049-82 | eBioscience | 0.005 | Helper T differentiation/activation (negative gate) |
| TGF-β | APC | TW4-6H10 | 349706 | Biolegend | 0.003 | Developmental regulator of Th17, Treg, and Th9 cells, suppressive cytokine |
| CD56 | APC-Cy7 | HCD56 | 318332 | Biolegend | 0.002 | Dump channel |
| CD19 | APC-Cy7 | SJ25c1 | 348794 | eBioscience | 0.002 | Dump channel |
*Markers were used in a separate FMO panel for gating controls.
METHODS
Collection and isolation of peripheral blood mononuclear cells
Blood collection and peripheral blood mononuclear cell (PBMC) isolation follows the protocol as described by Patel et al., in 2017 [22]. Briefly, six healthy donor apheresis leukoreduction collars containing Anticoagulant Citrate Dextrose Solution USP (ACD) Solution A were obtained from Brigham and Women’s Hospital (Boston, MA, USA). Blood was collected per the blood collection protocol approved by the Institutional Review Board of Brigham and Women’s Hospital and every participant gave written informed consent prior to donation. PBMCs were isolated via density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare Biosciences, Uppsala, Sweden). Each collar, containing 10–25 ml of solution, was diluted to a total volume of 80 ml with phosphate buffered saline (PBS). Ten milliliter of this diluted solution was pipetted over 12 ml Ficoll-Pacque PLUS in a 50 ml conical tube, centrifuged at 845 g for 20 min. The PBMC pellet was resuspended and washed with PBS, centrifuged at 527 g for five minutes, and cells were counted in AO/PI staining solution with a Nexcelom Cellometer (Nexcelom Bioscience, Lawrence, Massachusetts, USA). PBMCs were frozen at –80°C in fetal bovine serum (FBS; ThermoFisher, Waltham, MA, USA) + 15% dimethylsulfoxide (DMSO; ThermoFisher, Waltham, MA, USA). PBMCs were kept in CoolCell (Corning Incorporated, Tewksbury, MA, USA) overnight and transferred to liquid nitrogen for long term storage the next day.
T cell activation
Ninety-six hours prior to staining, samples were removed from liquid nitrogen, fully thawed in a 37°C water bath, and immediately transferred into warm RPMI supplemented with 10% FBS and 10% of 100× Antibiotic-Antimycotic containing penicillin, streptomycin, and amphotericin B (ThermoFisher, Waltham, MA, USA). Cells were counted in AO/PI staining solution and centrifuged at 527 g for five minutes. After centrifugation, cell pellet was resuspended in 12 ml of supplemented RPMI. Cell suspension was plated in a nonpyrogenic polystyrene cell culture flask and Dynabeads Human T activator CD3/CD28 (ThermoFisher, Waltham, MA, USA) were added to the flask at a ratio of 1 bead per 10 cells. Cells were incubated at 37°C, 5% CO2, for 96 h to stimulate T cell activation, expansion, and phenotypic differentiation [23].
Flow cytometry staining and acquisition
Flow cytometry staining, acquisition, and analysis methods were adapted from previously published Dana-Farber Immune Assessment Lab protocols [22,24]. All centrifugation occurred at 758 g at 4°C and cells were kept on ice in the dark throughout all incubations. Unstimulated frozen PBMCs were removed from liquid nitrogen, thawed, and resuspended in supplemented RPMI. Stimulated PBMCs were removed from incubation and the Dynabeads were removed via magnet. Both stimulated and unstimulated cells were counted, resuspended in PBS at a concentration of 100 million cells/ml, and plated into a 96-well plate at a concentration of 10 million cells per well. FMO wells received a one to one mixture of stimulated and unstimulated cells for a final concentration of 100 million cells/ml. After plating, cells were washed with 150 μl of PBS and incubated for 18 min with Zombie NIR Fixable Viability Dye (1:2500 Biolegend, San Diego, CA, USA, Cat. # 423105) on ice in the dark. After this incubation, cells were washed with 150 μl of PBS and incubated in 100 μl FcR blocking reagent for 18 min (1:625 in FACS buffer Miltenyi Biotec, Somerville, MA, USA, Cat. # 130-059-901) on ice in the dark. Next, cells were washed with 150 μl of FACS buffer, and stained with the extracellular antibody panel: CD3, CD4, CD8, CD25, CD127, GITR, CTLA4, CD56, and CD19. After a 45 min incubation, the cells were washed twice with FACs and fixed with 100 μl of BD Cytofix/Cytoperm (BD Biosciences, San Jose, CA, USA, Cat. # 554714), and incubated again for 20 min on ice in the dark. The cells were then resuspended in Perm Wash 1× (BD Biosciences, San Jose, CA, USA, Cat. # 554714), stained with the intracellular antibody panel, FoxP3, IL4, and TGF-β, and incubated for 30 min on ice in the dark. Details of the full antibody panel used can be found in Table 1. The cells were washed once with 150 μl of Perm Wash 1×, fixed in 2% paraformaldehyde (EMS, Hatfield, PA, USA, Cat. # 15712) and transferred to 5 ml round-bottom polystyrene tubes (Corning Inc, Corning, NY, USA, Cat. # 352058). The samples were stored overnight at 4°C.
Cells were resuspended in PBS and acquired on a 4-laser LSRFortessa X-20 cell analyzer using FACS Diva software (BD Biosciences, San Jose, CA, USA). Cells were collected according to the methods described by Cunningham, et al. [24]. To avoid spillover between channels, voltages (Table S1) were set so that the negative population was on-scale and the positive population was at or above 104 median fluorescence intensity (MFIs). Compensation controls (Table 2) were set on single color controls using healthy donor cells.
Table 2.
Averaged compensation from the three acquisitions applied to each fluorophore.
| Alexa Fluor 700-A | BV 786-A | BV 421-A | BV 510-A | BV 650-A | PE-A | FITC-A | PE-Cy7-A | APC-A | PerCP-Cy5-5-A | Zombie NIR-A | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alexa Fluor 700-A | 1.00 | 0.03 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.08 | 0.04 | 0.04 | 0.12 |
| BV 786-A | 0.01 | 1.00 | 0.03 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.04 |
| BV 421-A | 0 | 0 | 1.00 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BV 510-A | 0 | 0.03 | 0.22 | 1.00 | 0.14 | 0 | 0 | 0 | 0 | 0 | 0 |
| BV 650-A | 0.1 | 0.14 | 0.07 | 0.02 | 1.00 | 0 | 0 | 0.05 | 0.21 | 0.03 | 0.02 |
| PE-A | 0 | 0 | 0 | 0 | 0 | 1.00 | 0 | 0.01 | 0 | 0.05 | 0 |
| FITC-A | 0.02 | 0 | 0.02 | 0.05 | 0 | 0.01 | 1.00 | 0 | 0.05 | 0.06 | 0 |
| PE-Cy7-A | 0.02 | 0.03 | 0 | 0 | 0 | 0.02 | 0 | 1.00 | 0.01 | 0 | 0.01 |
| APC-A | 0.36 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0.05 | 1.00 | 0.01 | 0.04 |
| PerCP-Cy5-5-A | 0.29 | 0.10 | 0 | 0 | 0.13 | 0 | 0 | 0.17 | 0.13 | 1.00 | 0.04 |
| Zombie NIR-A | 0.25 | 0.14 | 0 | 0 | 0 | 0 | 0 | 0.55 | 0.06 | 0.01 | 1.00 |
Flow cytometry gating
Populations were manually gated using FlowJo 10.5.3 (BD, Franklin Lakes, NJ, USA). Light scatter parameters, FSC-A and SSC-A, designating size and granularity respectively, were used to identify lymphocytes. Because CD3/CD28 stimulation causes increases in lymphocyte size, these gates differ between time points [25,26]. Cellular debris and doublets were eliminated from this population by gating on FSC-H and FSC-A. Within this single cell population, live, CD19- and CD56- cells were gated based on negative staining for Zombie NIR. Cells were then gated for specific Treg markers using FMO and unstained controls, as well as by comparing stimulated and unstimulated plots; live, single cells were gated on CD3+ cells, then biaxial CD4+CD25+. Next, this double positive population was gated on FoxP3+CD127neg/low cells. These positive events were then gated on GITR+CTL4+ double positive population. Lastly, this double positive population was gated on IL4+TGFβ+ for the double positive population. We included separate FMO controls for each donor. As such, gates based on fluorophore conjugated antibodies differ between donors, but remain the same across conditions for each individual donor. Both dot plots and contour plots were used to establish gates (Fig. 1). FMOs were used to aid in gate establishment by examining checkpoint markers and cytokines on a CD4+CD25- parent population.
Figure 1.
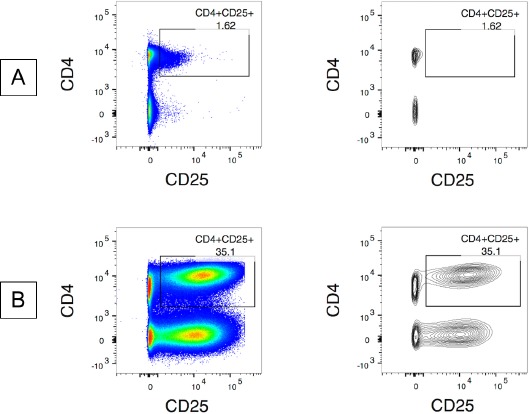
Examples of pseudocolor plots and contour plots for gate establishment. Samples were previously gated on single cells/lymphocytes/live/CD3+. A. Sample in pseudocolor and contour plots. B. 96 h stimulated sample in dot and contour plot.
RESULTS
Expression of Treg markers CD25, CD127, FoxP3, GITR, CTLA-4, IL-4, and TGFβ increased upon CD3/CD28 stimulation (Fig. 2). While the increases in magnitude varied amongst donors, this trend remained constant for all six samples. The most noted increase of expression was in GITR+CTLA4+ cells. For unstimulated samples, there was little to no expression of GITR+CTLA4+ double positive cells, with expression ranging from 0.15% to 3.51% in the 6 donors. In contrast, for 96 h bead-stimulated samples, the expression of GITR+CTLA4+ cells ranged from 5.41% to 40.8% in the 6 donors. Similar increases in expression were seen for IL4 and TGFβ, with increases ranging from 31.7% to 91.3% for the 6 donors. The positive percent of parent population from each run was averaged (Table 3). The table lists classic Treg marker expression comparing unstimulated samples and 96 h stimulated samples. Each column is used to further characterize Tregs. The last two columns characterize the functionality of Tregs, specifically the immunosuppressive capabilities. Expression averages did vary among the 6 donors. The overall percent of parent fold change for the 6 donors was higher in stimulated samples (Fig. 3).
Figure 2.
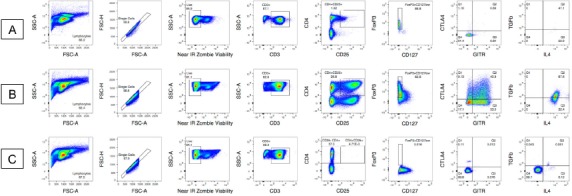
Pseudo color plots for donor A exhibiting increase in expression in unstimulated sample (A) and 96 h stimulated sample (B) with the FMO for reference (C). Gates are conserved for individual donor samples. A. Full staining and gating strategy for Treg specific markers on unstimulated donor A sample. There is low expression of CD3+CD4+CD25+, as well as low FoxP3 and CD127neg/low expression. B. Full staining and gating strategy for Treg specific markers on 96 h stimulated donor A sample. C. The FMO gating strategy for donor (A). CD4+CD25- is the parent population for all subsequent FMO gates. The FMO, unstimulated sample, and 96 h stimulated sample gating strategy for each donor can be found in the supplemental section (Fig. S1).
Table 3.
Averaged positive percent of parent population for Treg-specific phenotypes in unstimulated (no stim) and 96 h stimulated (stim) samples from all three runs*.
| CD3+ | CD3+/CD4+CD25+ | CD3+/CD4+CD25+ /FoxP3+CD127neg/low |
CD3+/CD4+CD25+/FoxP3+ CD127neg/low/GITR+CTLA4+ |
CD3+/CD4+CD25+/FoxP3+CD127neg/low/ GITR+CTLA4+/IL4+TGFβ+ |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Donor | No stim | Stim | No stim | Stim | No stim | Stim | No stim | Stim | Stim |
| A | 86.60 | 95.70 | 2.29 | 23.76 | 50.10 | 48.43 | 0.49 | 27.20 | 73.60 |
| B | 89.10 | 97.76 | 2.68 | 13.26 | 41.25 | 40.61 | 0.37 | 6.31 | 61.40 |
| C | 81.90 | 94.20 | 4.31 | 12.70 | 23.77 | 36.57 | 1.65 | 13.52 | 79.50 |
| D | 85.00 | 94.00 | 5.27 | 28.40 | 30.99 | 38.03 | 1.86 | 22.33 | 80.56 |
| E | 67.53 | 83.70 | 7.21 | 20.27 | 41.03 | 57.53 | 2.17 | 14.79 | 42.71 |
| F | 69.67 | 83.10 | 3.67 | 18.10 | 24.09 | 30.90 | 1.68 | 15.12 | 45.47 |
*Each positive population was previously gated on single cells/lymphocytes/live. IL4+TGFβ+ double positive percentages for unstimulated samples could not be determined due to too low number of events, less than 500 events of parent population, for confident analysis.
Figure 3.
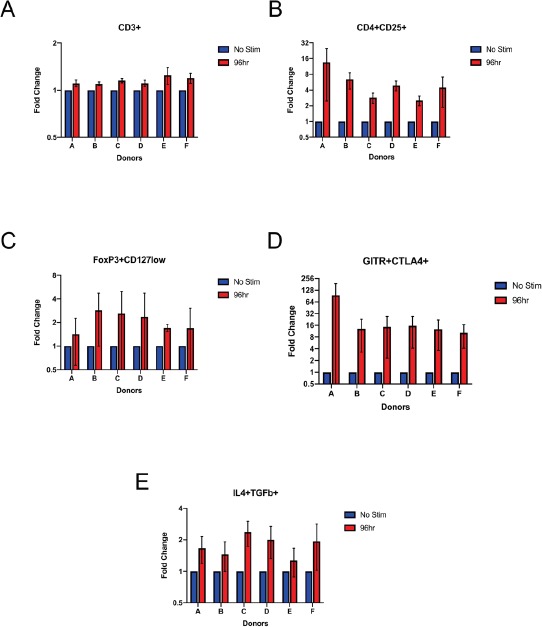
Comparative changes in Treg marker expression as a fold change in percent of parent population. All cells were gated on single cells/lymphocytes/live. Graphs represent gating on CD3+ cells (A), CD3+/CD4+CD25+ (B), CD3+/CD4+CD25+/FoxP3+CD127neg/low (C), CD3+/CD4+CD25+/FoxP3+CD127neg/low/CTLA4+GITR+ (D), and CD3+/CD4+CD25+/FoxP3+CD127neg/low/CTLA4+GITR+/IL4+TGFβ+ (E) populations. Error bars represent standard error of the mean for the averaged fold change from the three acquisitions (n = 6, N = 3).
DISCUSSION
Due to heterogeneity in phenotype and activity of cells expressing CD25 and Foxp3, robust multiparametric analyses are necessary to adequately define the Treg population [3,5,8-10].
Comprehensive flow cytometry panels for identifying Tregs should include markers associated with immunosuppressive function. We developed, optimized, and validated an 11-color antibody panel for the standardized detection of Tregs from cryopreserved human PBMCs. As expected, this panel detects an increase in 4 extracellular and 2 intracellular functional markers in Treg lymphocytes within six T-cell stimulated healthy donor PBMCs.
While our methods detail reproducible experiments, inter-donor variability before and after T cell stimulation highlights the importance of using appropriate controls within experiments and statistical analyses [22,24].
This panel improves upon past studies; in addition to Treg lineage markers, it includes checkpoint markers GITR and CTLA-4, and cytokines IL-4 and TGFβ [2,5,6,11,16]. Upregulation of these markers in CD4+CD25+FoxP3+CD127neg/low cells within six T-cell stimulated healthy donor PBMCs demonstrates the ability to assess immunosuppressive capacity of identified Treg populations. Thus, this panel provides a method for in-depth detection of functional Tregs that is ideal for a clinical context.
Supplementary Material
Table S1. Voltages set on BD LSR Fortessa X-20 using single color controls.
Figure S1. Gating strategy for each of the 6 donors, donors A–F, is shown in the figure with the letter caption corresponding to the donor.
Supplementary information of this article can be found online at http://www.jbmethods.org/jbm/rt/suppFiles/325.
Acknowledgments
The Sharon Crowley Martin Memorial Fund for Melanoma Research (Hodi FS) and the Malcolm and Emily Mac Naught Fund for Melanoma Research (Hodi FS) at Dana-Farber Cancer Institute. We would like to thank Suzan Lazo and John Daley of the Dana-Farber Cancer Institute Flow Cytometry Core for their assistance with protocol development. We would like to thank Joanna Baginska, Ryan Brennick, Alyssa Masciarelli, Emma Hathaway, Matthew Nazzaro, and Carter Smith of the Dana-Farber Cancer Institute Center for Immuno-Oncology for their assistance with flow cytometry, and all of the above for help editing and proofreading the manuscript.
References
- 1.Chatenoud L, Bach J. (2006) Adaptive human regulatory T cells: myth or reality? J Clin Invest 116: 2325-2327. doi: 10.1172/JCI29748. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Amarnath S, Chen W. (2006) Requirement of CD28 signaling in homeostasis/survival of TGF-beta converted CD4+CD25+ Tregs from thymic CD4+CD25- single positive T cells. Transplantation 82: 953-964. doi: 10.1097/01.tp.0000232330.46688.37. PMID: [DOI] [PubMed] [Google Scholar]
- 3.Bluestone JA, Abbas AK. (2003) Natural versus adaptive regulatory T cells. Nat Rev Immunol 3: 253-257. doi: 10.1038/nri1032. PMID: [DOI] [PubMed] [Google Scholar]
- 4.Bin Dhuban K, d’Hennezel E, Nashi E, Bar-Or A, Rieder S, et al. (2015) Coexpression of TIGIT and FCRL3 identifies Helios+ human memory regulatory T cells. J Immunol 194: 3687-3696. doi: 10.4049/jimmunol.1401803. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer M, Classen S, Endl E, Kochanek M, Weihrauch MR, et al. (2011) Comparative approach to define increased regulatory T cells in different cancer subtypes by combined assessment of CD127 and FOXP3. Clin Dev Immunol 2011: 734036-12. doi: 10.1155/2011/734036. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davids MS, Kim HT, Bachireddy P, Costello C, Liguori R, et al. (2016) Ipilimumab for Patients with Relapse after Allogeneic Transplantation. N Engl J Med 375: 143-153. doi: 10.1056/NEJMoa1601202. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. (2004) Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J Exp Med 199: 971-979. doi: 10.1084/jem.20031579. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran DQ, Ramsey H, Shevach EM. (2007) Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood 110: 2983-2990. doi: 10.1182/blood-2007-06-094656. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.d’Hennezel E, Piccirillo CA. (2011) Analysis of human FOXP3+ Treg cells phenotype and function. Methods Mol Biol 707: 199-218. doi: 10.1007/978-1-61737-979-6_13. PMID: [DOI] [PubMed] [Google Scholar]
- 10.Ronchetti S, Ricci E, Petrillo MG, Cari L, Migliorati G, et al. (2015) Glucocorticoid-induced tumour necrosis factor receptor-related protein: a key marker of functional regulatory T cells. J Immunol Res 2015: 171520. doi: 10.1155/2015/171520. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nettenstrom L, Alderson K, Raschke EE, Evans MD, Sondel PM, et al. (2012) An optimized multi-parameter flow cytometry protocol for human T regulatory cell analysis on fresh and viably frozen cells, correlation with epigenetic analysis, and comparison of cord and adult blood. J Immunol Methods 387: 81-88. doi: 10.1016/j.jim.2012.09.014. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron U, Floess S, Wieczorek G, Baumann K, Grützkau A, et al. (2007) DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 37: 2378-2389. doi: 10.1002/eji.200737594. PMID: [DOI] [PubMed] [Google Scholar]
- 13.Floess S, Freyer J, Siewert C, Baron U, Olek S, et al. (2007) Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 5: doi: 10.1371/journal.pbio.0050038. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, et al. (2008) DNA methylation controls Foxp3 gene expression. Eur J Immunol 38: 1654-1663. doi: 10.1002/eji.200838105. PMID: [DOI] [PubMed] [Google Scholar]
- 15.Miyara M, Yoshioka Y, Kitoh A, Shima T, Wing K, et al. (2009) Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30: 899-911. doi: 10.1016/j.immuni.2009.03.019. PMID: [DOI] [PubMed] [Google Scholar]
- 16.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, et al. (2011) Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol 186: 3918-3926. doi: 10.4049/jimmunol.1003099. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H, Longhi MS, Wang P, Vergani D, Ma Y. (2012) Human CD4+CD25(high)CD127 (low/neg) regulatory T cells. Methods Mol Biol 806: 287-299. doi: 10.1007/978-1-61779-367-7_20. PMID: [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Amarnath S, Chen W. (2006) Requirement of CD28 signaling in homeostasis/survival of TGF-beta converted CD4+CD25+ Tregs from thymic CD4+CD25- single positive T cells. Transplantation 82: 953-964. doi: 10.1097/01.tp.0000232330.46688.37. PMID: [DOI] [PubMed] [Google Scholar]
- 19.Ephrem A, Epstein AL, Stephens GL, Thornton AM, Glass D, et al. (2013) Modulation of Treg cells/T effector function by GITR signaling is context-dependent. Eur J Immunol 43: 2421-2429. doi: 10.1002/eji.201343451. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahmani A, Delisle J. (2018) TGF-β in T Cell Biology: Implications for Cancer Immunotherapy. Cancers (Basel) 10: doi: 10.3390/cancers10060194. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Hwang Y, Chen Y, Liu C, Shen C, et al. (2017) Interleukin-4 Supports the Suppressive Immune Responses Elicited by Regulatory T Cells. Front Immunol 8: 1508. doi: 10.3389/fimmu.2017.01508. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel T, Cunningham A, Holland M, Daley J, Lazo S, et al. (2017) Development of an 8-color antibody panel for functional phenotyping of human CD8+ cytotoxic T cells from peripheral blood mononuclear cells. Cytotechnology 70: 1-11. doi: 10.1007/s10616-017-0106-3. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canavan JB, Afzali B, Scottà C, Fazekasova H, Edozie FC, et al. (2012) A rapid diagnostic test for human regulatory T-cell function to enable regulatory T-cell therapy. Blood 119: doi: 10.1182/blood-2011-09-380048. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham RA, Holland M, McWilliams E, Hodi FS, Severgnini M. (2019) Detection of clinically relevant immune checkpoint markers by multicolor flow cytometry. J Biol Methods 6: doi: 10.14440/jbm.2019.283. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heylmann D, Badura J, Becker H, Fahrer J, Kaina B. (2018) Sensitivity of CD3/CD28-stimulated versus non-stimulated lymphocytes to ionizing radiation and genotoxic anticancer drugs: key role of ATM in the differential radiation response. Cell Death Dis 9: 1053. doi: 10.1038/s41419-018-1095-7. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Kurlander RJ. (2010) Comparison of anti-CD3 and anti-CD28-coated beads with soluble anti-CD3 for expanding human T cells: differing impact on CD8 T cell phenotype and responsiveness to restimulation. J Transl Med 8: 104. doi: 10.1186/1479-5876-8-104. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Voltages set on BD LSR Fortessa X-20 using single color controls.
Figure S1. Gating strategy for each of the 6 donors, donors A–F, is shown in the figure with the letter caption corresponding to the donor.
Supplementary information of this article can be found online at http://www.jbmethods.org/jbm/rt/suppFiles/325.


