Abstract
High-dimensional, single-cell cell technologies revolutionized the way to study biological systems, and polychromatic flow cytometry (FC) and mass cytometry (MC) are two of the drivers of this revolution. As up to 30–50 dimensions respectively can be measured per single-cell, they allow deep phenotyping combined with cellular functions studies, like cytokine production or protein phosphorylation. In parallel, the bioinformatics field develops algorithms that are able to process incoming data and extract the most useful and meaningful biological information. However, the success of automated analysis tools depends on the generation of high-quality data. In this review we present the most recent FC and MC computational approaches that are used to prepare, process and interpret high-content cytometry data. We also underscore proper experimental design as a key step for obtaining good quality data.
Keywords: Flow cytometry, Mass cytometry, Bioinformatics, Computational tools, Single-cell proteomics
1. Introduction
High-throughput single-cell technologies are becoming common approaches in daily research. The impressive progression in the number of different molecules that can be measured in a single cell changed the way experiments are done and analyzed. Flow and mass cytometry (FC and MC respectively) are great examples of these changes. Starting from the first flow experiments that measured 2–4 markers which were manually gated, the multiplexing capabilities are currently increasing to 30 [1] and 45 [2] parameters in FC and MC respectively, and strong bioinformatics skills are needed to extract meaningful information.
The general concepts of both technologies are similar; antibodies or probes labeled with fluorochromes (FC) or high atomic mass elements (MC) are used to target desired antigens or biomolecules to characterize certain cell properties like cell phenotype, cell cycle [3] or response to stimulation agents via cytokine production, protein phosphorylation [4] or RNA expression [5], among others.
Following the staining, cells are introduced in single-cell suspension via capillary tubes into the flow cytometer for FC or alternatively into a Cytometry by Time-Of-Flight (CyTOF, Helios) device for MC. The biological information with single cell resolution is obtained via photons or time-of-flight ion’s mass-to-charge ratios for FC and MC respectively, converted into digital values and stored using the same file format called flow cytometry standard (.FCS). Although both technologies are commonly used to measure cell properties, the definition of event is different. In FC every event that emits light and reach the user-defined threshold will be stored in the FCS file. Both light scatters: FSC (forward-scatter), correlated with cell size, and SSC (side-scattered), correlated with cell granularity, together with fluorescence are used to differentiate single cells from noise [6]. In MC the ion cloud that lasts for more than 10 and less than 150 pushes (spectrum scans) and exceeds the lower convolution threshold is recorded in FCS file as an event. MC lacks the power of light scatter, thus cell events are defined using the metals associated with them in form of antibodies or probes [7]. Nucleic acid intercalators like Iridium (Ir) or rhodium (Ro) are used to define nucleated cells and for non-nucleated cells antigen-specific markers must be used. In FC the light can excite some cell components like flavins, folic acid, retinol, which emit the so called autofluorescence, especially in the green spectrum [8]. This does not use to be a problem in MC, since the high atomic mass metals detected are not frequently found within the cells. However, tissue metal contaminations due to environmental exposures, medical procedures or experimental protocols were reported [9], [10], [17] and should also be considered when deciding the most suitable technology, FC or MC.
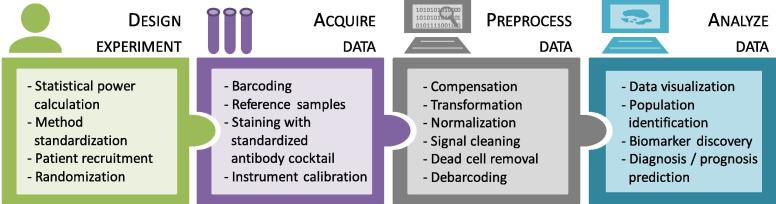
Both techniques benefit from the development of new probes that increase the number of measured parameters. The high dimensionality of the data changed the way to visualize the results; from manually building two-dimensional gating hierarchies to applying automated clustering or dimensionality reduction methods. The automation process requires properly preprocessed high-quality input data free of artifacts. Artifacts may be introduced during sample collection, processing and acquisition, and should be detected and removed. Although these alterations come from different sources in FC and MC, they have similar impact on data quality. In this review we present the most common artifacts and their sources during data preparation and acquisition and show how to manage them (Table 1). Additionally, we summarize the algorithms and workflows that can improve data quality prior to feature extraction, as an update of previously published reviews [11], [12], [13]. Furthermore, we introduce the state-of-the-art clustering and visualization tools that can be applied to the data and point out their strengths and limitations. Finally, we present the main approaches to analyze the extracted features in the context of biomarker discovery and trajectory interference studies. An overview of the computational methods discussed is presented in Table 2. In Fig. 1 we show a typical workflow for the preparation and analysis of multi-parametric FC and MC data.
Table 1.
Artifacts and their prevention in high-dimensional flow and mass cytometry data preparation and analysis.
| Source | Effect/Artifact | Prevention | |
|---|---|---|---|
| Experimental | Change in reagent’s batches e.g. a) Change in fixation reagent;b) Change in antibody lot |
General change in protocol performance that can introduce batch effect. Not predicted contaminations (important in MC study);a) Affect the staining and cell recovery;b) Different fluorochrome or metal conjugation efficiency results in different antibody background and staining intensity |
Order enough quantity needed for the whole experiment if product stability allows it; Re-test new lot and confirm similar/good performance |
| Change in the protocol e.g. a) use of fixed vs fresh blood; b) Change in centrifugation steps;c) Change in staining temperature or time |
a) Change in sample stability; Change in antibody performance; Change in sample background b) Different cell recovery;c) Different background of antibody due to inefficient or different washing step; Instability of fluorochrome-conjugated tandems at RT in light |
Decide staining approach before sample collection; Optimize the protocol, prepare SOP; Use exactly the same protocol as was used for antibody, barcoding optimization, cocktail preparation, including cell preparation, antibody staining (RT vs 4°C, dark vs light) |
|
| Change in cocktail preparation: e.g. lack of one antibody, wrong fluorochromes, different clone | Different staining intensity; Different staining pattern; Problems with population definition if one of the markers is missing |
Prepare one big cocktail of antibody, aliquot and store frozen (in MC), lyophilized or desiccated (MC and FC) | |
| Pipetting errors | Variation in staining intensity, especially problematic when MI will be compared; variation in the number of collected events | Barcode the samples and process them in the batches Create as few batches as possible Include reference sample to track variation in each batch |
|
| Improper antibody titration |
Too little: problems with population definition; no split between positive and negative values Too much: Unspecific binding of the antibodies; Values out of or at the edge of dynamic range |
Perform titration experiments Use tools like AOF or calculate staining index to ensure correct titration |
|
| Unspecific staining | Unknown co-expression of markers, High, “weird” signal in most of the channels |
Block FC receptors, Block hydrophobic biding using heparin Use life/dead staining to exclude dead cells that have high antibody binding properties |
|
| Incorrect panel design | High spreading error that can mask dim populations Inability to define needed population due to the lack of proper cell definition Significant signal spillover especially in MC |
Use tools like Guided Panel Solution or Maxpar panel designer to design your panel; Use databases like [20] to carefully select appropriate markers for the needed cell populations; Use published panels |
|
| *Metal contamination (*Only in the case of MC) | Unspecific signal in .FCS file registered as events; Crosstalk of the contaminant with different channels; Stickiness of the contaminant to the capilar causing clogging; Shorter detector lifespan; Cones contamination and loss of CyTOF sensitivity |
Troubleshooting will depends on the source of contamination: Reagent contamination: check all reagents by running them in the solution mode and at proper concentration as stated in [134]; avoid using autoclaved glass as it can contain barium contaminants; use always filter tips; use previously tested references in MC studies Sample contamination: Due to medical procedure [10], [135], environmental exposure [9] or experimental protocol [17] Be aware of possible contamination; decide if MC can be used; if contamination is possible screen small aliquots of samples as shown in [139]; If contamination was discovered during acquisition dilution of the sample can be considered |
|
| Acquisition | a) Improper cytometer calibration;b) Device decalibration upon acquisition |
a) Loss in antigen resolution; Intensity changes across the runs; Run-to-run variation;b) Decrease in the signal intensity |
Check your machine performance and get to know its resolution; Calibrate device; Control for time changes in the machine calibration; Use calibration beads to correct for signal drop and changes; Include reference samples in every batch |
| Clogging of the device | Signal instability affecting median expression of the markers | Clean device when necessary to remove the clog | |
| High speed of acquisition or changing in acquisition speed | Change in doublets to singlets ratio; Increase in coefficients of variation (CVs) together with the sample speed |
Keep the speed constant, and adequate to obtain good singlets/doublets ratio | |
| Different sample or panel labeling | Errors when analyzing the files or inability to read-in the files | Define labeling strategy, create a template and keep it constant across all the project | |
| Analysis | Not enough statistical power | Lack of significance | Calculate statistical power Consult statistician Include more samples |
| Improper transformation | Inability to distinguish positive and negative events; Improper clustering |
Verify transformation method by visualizing markers e.g. using flowJo Optimize transformation |
|
| Batch effect | Improper data interpretation; Incorrect conclusions |
Visualize batch effect e.g. using dimensionality reduction tools like PCA or t-SNE; Properly design the experiment e.g. include reference sample Correct for batch effect when building the model |
|
| Improper normalization | Changes in marker expression distribution; Improper assignment to the clusters |
Carefully verify the performance of normalization tools by visualizing the markers | |
| Uncleaned changes in signal intensities | Improper assignment to the clusters | Spot problematic files by applying tools like AOF, normalize or discard them from the analysis | |
| Uncleaned bad quality events | Improper assignment to the cluster | Verify the signal stability and clean if necessary using tools like flowAI, flowClean, flowCut or manual gating Remove doublets, debris, and dead cells |
|
| Presence of doublets/high doublets to singlets ratio | Co-expression of the markers biologically incorrect; Improper assignment to the clusters |
Gate out all the doublets | |
| Improper Clustering or dimensional reduction performance | Unstable clusters or cell position, different results with every run of cluster | Verify clustering settings by gating few example files and calculating F1 score Check for run-to-run stability Check for sensitivity to subsampling |
|
Table 2.
Overview of bioinformatics tools for high-dimensional flow and mass cytometry data analysis.
| Application | Source | ||
|---|---|---|---|
| Panel design | Guided Panel Solution[39] | Panel design in Flow Cytometry | BD Biosciences |
| Maxpar Panel Design [40] | Panel design in Mass Cytometry | Fluidigm | |
| Quality control/preprocessing | Average overlap frequency (AOF) [45] | Antibody performance evaluation | R package/Bioconductor package; Astrolab |
| CATALYST [51] | MC data preprocessing (bead-based normalization, debrcoding; compensation: FlowSOM clustering) | R package/Bioconductor package; interactive Shiny-based web application |
|
| flowAI [61] | Signal Cleaning; Flow rate cleaning; Outliers cleaning |
R package/Bioconductor package; GUI; Plugin FlowJo Exchange |
|
| flowClean [63] | Signal Cleaning | R package/Bioconductor package; Plugin FlowJo Exchange |
|
| flowCore [65] | Basic structures for flow cytometry data | R package/Bioconductor package; | |
| flowCut [62] | Signal cleaning | R package (github repository) | |
| flowTrans [58] | Data transformation | R package/Bioconductor package | |
| flowVS [59] | Data transformation | R package/Bioconductor package | |
| flowWorkspace [70] | Representation and interaction with gated and ungated data in R | R package/Bioconductor package | |
| Single-cell deconvolution algorithm [64] | Debarcoding | CATALYST; Matlab; Fludigm stand-alone; Updated Single-cell Debarcoder [140]; R package PREMESSA(gihub repository) | |
| Optimal Sample Assignment Tool (OSAT) [22] | Sample to batch allocation | R package/Bioconductor package | |
| Normalization and batch effect correction | gaussNorm [76] | Normalization | R package/Bioconductor package flowStat |
| fdaNorm [76], [77] | Normalization | R package/Bioconductor package flowStat | |
| CytoNorm [79] | Normalization using reference sample | R package/Bioconductor package; Plugin FlowJo Exchange | |
| CytofBatchAdjust [80] | Normalization using reference sample | R package/Bioconductor package | |
| BatchEffectRemoval [78] | Normalization using reference sample | Python | |
| Dimensionality reduction | Diffusion Maps [91], [101], [103] | Non-linear dimensionality reduction/Trajectory inference | R package/Bioconductor package; |
| Isomap [89], [90] | Non-linear dimensionality reduction/Trajectory inference | R package/CRAN package vegan | |
| PCA [83] | Linear dimensionality reduction | R package stats | |
| t-SNE [84] | Non-linear dimensionality reduction | R package/CRAN package; Plugin FlowJo Exchange; Cytobank; Matlab; Python |
|
| BH-SNE (viSNE) [92], [97] | Non-linear dimensionality reduction | Python; Cytobank; Matlab; R package/CRAN package Rtsne | |
| UMAP [88] | Non-linear dimensionality reduction | Python, R package/CRAN package uwot; Plugin FlowJo Exchange; | |
| One-SENSE [105] | Non-linear dimensionality reduction | R package/Bioconductor package | |
| HSNE [85] | Non-linear dimensionality reduction | Cytosplore | |
| FIt-SNE [86] | Non-linear dimensionality reduction | R package; Matlab; Python; Plugin FlowJo Exchange | |
| EmbedSOM [112] | Non-linear dimensionality reduction | R package/CRAN package ; Plugin FlowJo Exchange | |
| opt-tSNE [87] | Non-linear dimensionality reduction | Python; Cloud opt-SNE | |
| Jensen-Shannon (JS) divergence [92] | Dimensionality reduction comparison | R package/Bioconductor package cytutils | |
| Data clustering and automated gating | CytoCompare [110] | Clustering comparison | R package (github repository) |
| flowClust [136] | Unsupervised Clustering | R package/Bioconductor package; GenePattern Platform [137] |
|
| flowDensity [73] | Supervised clustering | R package/Bioconductor package | |
| flowLearn [71] | Semi supervised Clustering | R package (github repository) | |
| FlowSOM [81] | Unsupervised Clustering | R package/Bioconductor package, Cytofkit [128]; Plugin FlowJo Exchange | |
| flowType [123] | Unsupervised Clustering | R package/Bioconductor package | |
| PhenoGraph [114] | Unsupervised Clustering | Matlab; Python; R package Rphenograph (github repository); Cytofkit [126]; Plugin FlowJo Exchange | |
| SPADE [44] | Unsupervised Clustering | R package/Bioconductor package; Cytobank, Matlab | |
| X-shift [115] | Unsupervised Clustering | Standalone application (VorteX); Plugin FlowJo Exchange | |
| DensVM [90] | Unsupervised Clustering | R package/Bioconductor package cytofkit | |
| ACCSENSE [117] | Unsupervised Clustering | Standalone ACCENSE application | |
| Useful pipelines and approaches | CellCNN [126] | Representation learning approach to detect rare cell subsets associated with disease | Python |
| Citrus [125] | Unsupervised clustering with regularized regression model | R package with GUI; Cytobank | |
| cydar [119] | Unsupervised assignment to hyperspheres, control of the spatial false discovery rate, changes in abundance visualization | R package/Bioconductor package, GUI with Shiny application |
|
| Cytofast [127] | Visual and quantitative analysis of cytometry data to discover immune signatures and correlations | R package/Bioconductor package | |
| Cytosplore [129] | Interactive visual analysis system contain t-SNE, HSNE, SPADE | Interactive tool | |
| Cytofkit [128] | Preprocessing; cell subset detection (DensVM, FlowSOM or Phenograph, ClusterX); data visualization (PCA, t-SNE, Isomap) | R package/Bioconductor package, GUI with Shiny application |
|
| diffcyt [56] | Unsupervised clustering with FlowSOM, empirical Bayes moderated tests for statistical analysis | R package/Bioconductor package | |
| flowType/RchyOptimyx [122] | Unsupervised clustering Construction of cell hierarchy for maximization of an external variable |
R package/Bioconductor package | |
| FloREMI [121] | Preprocessing; feature extraction; feature selection; survival time prediction |
R scripts (github repository) | |
| CyTOF workflow [54] | Unsupervised clustering with FlowSOM, generalized linear mixed models or linear mixed models | R package/Bioconductor package | |
| DAMACY [96] | Multivariate method based on PCA and multivariate regression based on Partial Least Squares (PLS) | Matlab | |
| OpenCyto [138] | Facilitate the automated gating methods | Bioconductor package/GUI with shinyCyto application | |
| Trajectory detection | pCreode [133] | Trajectory inference with multiple branching | Python |
| Wanderlust [131] | Trajectory inference without branching | Matlab based interactive tool cyt | |
| Wishbone [132] | Trajectory inference with two branches | Python/Matlab based interactive tool cyt | |
Fig. 1.
The flow and mass cytometry experimental and data analysis computational workflow. For more information about which tools to use and how to design well each step to avoid artifacts refer to Table 1, Table 2.
2. Obtaining reproducible and high-quality data
To obtain statistical power for both experimental studies and evaluation cohorts sample size estimation is a key step in the design of a cytometry project. This calculation prevents changes in reagent batches, including antibody cocktail, and should be planned upfront, avoiding the introduction of additional variability. A Standard Operating Procedure (SOP) for sample collection and processing is highly recommended, as it significantly improves data reproducibility [14], [15], [16]. For MC the selection of reagents and their storage is critical to avoid metal contamination events (see Table 1 for possible contamination sources) [17]. It is essential to consider if cells should be stained immediately upon collection or preserved until recruitment is completed. If all the samples are obtained at once, they can be stained and acquired immediately. However, in longitudinal studies, or if the cytometry unit is far from the recruitment center, the sample preservation before [19] or after staining [18] should be considered. The goal is to process, stain and acquire as many samples as possible with the same protocol, antibody cocktail, and instrument settings. Each preservation protocol will affect the sample composition and antigen expression [18], [20], [21]; hence benefits and drawbacks will depend on the biological question and should be carefully considered before performing the experiments.
Often, hundreds of samples are included in cytometry studies and are split into multiple experimental groups. This can introduce “batch effects” defined as non-biological differences between them . To minimize this effect, a careful experimental design should ensure the even distribution of biological groups and confounding factors across batches [22]. Packages like OSAT (Optimal Sample Assignment Tool) [22] can be used to optimally distribute the samples into batches. The antibody labeling and sample staining should be consistent across all the groups, as discrepancies can introduce technical differences in mean intensity (MI) values that can be hard to distinguish from biologically meaningful information. This is why strict control of intra- and inter-group variations should be introduced in the experimental design. To limit intra-batch variation, barcoding (labeling of individual cell samples with unique combinatorial barcodes) and sample pooling before antibody staining is used particularly in MC [23], [24], [25], [64], and less often in FC [26], [27]. To minimize inter-batch variation, an experiment-required stability master-mix of the staining cocktail is recommended to be used along the project. Both lyophilized and desiccated antibody cocktails were reported [20], [28], [29] and freezing of the MC cocktail aliquots was also shown to be successful [30]. Unfortunately, even well prepared SOP minimize, but do not resolve the problems with day-to-day reproducibility. Thus, measures allowing estimation and correction of batch effects are needed. The practice of including a reference sample in each barcoded batch is becoming a standard in MC [31] and was reported in FC experiments as well [32]. The reference sample is an aliquot of a bigger volume obtained from one donor at a particular time, aliquoted, and preserved. It carries the information of the technical variability introduced during sample preparation, staining and acquisition, and therefore allows to measure run-to-run variation [31].
In FC and MC the panel optimization is the most critical and difficult step. Both technologies require proper assignment of dim and bright markers depending on the channel sensitivity and its performance in the context of staining index and spillover [1], [33], [34]. The success of automated methods to resolve cell populations depends more on well-selected markers than on the frequency of the cells, thus the probes should be selected carefully [35]. To identify the markers of interest, a recently published antibody staining database could be useful, as it contains staining patterns for 350 antibodies used in fresh and fixed peripheral blood mononuclear cells (PBMC) [20]. Additionally, antibody titration, done at the same conditions as the final experiment, is essential to ensure proper signal intensity allowing population definition. It should be stressed that if a population cannot be defined by manual inspection due to a sub-optimal amount of added antibodies, it will not be detected by most clustering algorithms [35].
In both techniques, signal spill from one channel to another is observed. In FC it is caused by the overlapping emission spectra of different fluorochromes. In MC it can be due to metal impurities from the metal tags; metal oxidation affecting mainly light lanthanides and causing signal spillover to the heavier spectrum of masses; or metal over-abundance when high antibody concentration is used inappropriately and the signal of this particular mass cannot be resolved [36]. In FC the signal crosstalk can be severe and cannot be avoided in multicolor experiments. In MC, maximum spillover does no exceed a few percent and proper panel design can minimize these issues. Inadequate panel design or lack of proper compensation controls, especially in FC, can create false positive events [37]. Additionally, it can introduce spreading error, an artifact produced by the error in photon counting [6], which can mask low or dim fluorescence positive cells. As a higher number of markers requires more sophisticated panel designing skills, tools like Guided Panel Solution offered by BD [39], or Maxpar Panel Designer by Fluidigm [40] can be helpful, but not sufficient, especially in FC where spreading error information is not provided. Spreading errors depend on laser configuration, dye brightness and quality of PMT (photomultiplier tubes). Thus, careful selection of probes and deep understanding of cytometer configuration and its performance are critical in FC [41], [42]. For MC it is also important to be familiar with the instrument performance, as variation in the sensitivity and resolution was observed between different CyTOF devices [43]. During the preparation of the SOP a pilot study including a few samples is strongly recommended, as it can help to fix the protocol limitations [28], [43].
Evaluation of antibody staining, titration, and signal spillover is an important but time-consuming process, especially in high dimensional approaches. Fortunately, a recent study shows that clustering algorithms like SPADE (Spanning Tree Progression with Density Normalized Tree) [44] can be used to evaluate the titration of a panel and track the spillover artifacts. Additionally, metrics like Average Overlap Frequency (AOF) can be applied to verify antibody performance by calculating staining distances between the positive and negative populations, reducing substantially time for calculation and plotting of staining indices [45]. This shows that even at the moment of panel optimization, computational approaches can significantly accelerate bench work and improve data quality. For more details about panel preparation and standardization, readers are directed to the following literature [46], [47], [48], [49].
The capillary introduction system in both FC and MC suffers from cell clogging, altering the flow rate and signal quality over time of acquisition. Sample clogs can be caused by specific biological materials starting from “easily” acquired cell lines or PBMC to whole blood or the most prone to clogging, the disaggregated tissue. In both technologies the disturbances in the acquisition rate affects signal quality. The higher the speed the more coincidence events known as doublets are collected, and the more spread of the signal is seen [6]. The maximum recommended acquisition speed for FC is 25 000 cells/s, while for MC is up to 1000 cell/s [50]. It should be noted that the maximum speed depends on the type or cells that are acquired and on the experimental target. If rare cells that constitute 0.01% frequency are of interest flow rate should be lower and well optimized [6].
For more information about frequent errors and solutions in the experimental part of the workflow, readers are directed to Table 1.
3. Prior to feature analysis: data preprocessing and quality controls
3.1. Data compensation and transformation
As stated before, both FC and MC suffer from signal crosstalk across detection channels. To obtain correct data, a compensation matrix needs to be calculated using appropriate controls [1], [51]. While proper MC panel design can minimize spillover issues [37], it is almost inevitable in standard polychromatic FC above 15 colors. However, as pointed out by Leipold [52], minimal spillover is not equal to zero spillover, so MC data might also require correction. As mentioned before, the reason for signal crosstalk is different for FC and MC, however in both technologies the spills can be defined as a linear function of signal intensity, and can therefore be corrected using spillover coefficients for each channel [51], [53]. Although this method is working for standard FC, in MC this correction introduces negative values, which are normally almost absent in MC data. As an alternative, non-negative least-squares (NNLS) approach used in spectral cytometry, was applied to MC data [51]. If proper, single stained controls and unstained samples are provided, compensation can be automatically calculated using platforms like Diva or FlowJo for FC and CATALYST package for MC [51].
FC and MC raw data are often characterized by skewed distribution with varying ranges of expression. In consequence it can be difficult to distinguish positive and negative populations [54]. As visualization and clustering performance depends on the scale and distribution, it is important to bring the expression peaks as close to a normal distribution as possible [55]. To do so, the expression values are usually transformed using an inverse hyperbolic sine (arcsinh) transformation with the cofactor 5 or 150 for MC and FC, respectively [56]. The arcsinh conversion behaves similarly to a log transformation at high values, but is approximately linear near zero, and a cofactor controls the width of the linear region. FC data contain more negative values due to the correction of background noise, autofluorescence, and compensation; conversely, MC data contains zero values when no ions are detected and few negative values are introduced due to background subtraction and randomization [56], [57]. The type of transformation can be sample and marker-specific, especially in FC data, as shown in [58], [59], and the choice of parameters can be automatically optimized by tools like flowTrans and flowVS. It should be noted that some of the visualization and clustering tools require transformation to be done upfront, while others perform it as a default. It is important to always check the transformation requirements, as this might affect the downstream analysis.
3.2. Signal quality check and cleaning
As mentioned in Section 2, the capillary tubes used for sample introduction in FC and MC can clog resulting in sudden changes in the signal. Other issues such as unstable data acquisition can cause signal shifts and change the mean intensity [60]. These signal disturbances affecting downstream analysis should be identified and removed from the data. Currently, three algorithms can be used to do this: flowAI, [61] that uses change point analysis and allows automatic or interactive analysis; flowCut, [62] that creates summed density measures using mean, median, percentiles, variation, skewness, and removes events based on density curve analysis; and flowClean, [63] that tracks the changes in the frequency of artificially created populations, taking advantage of compositional and change point analysis, flagging outliers with unusual ratio of cell populations. The first two methods are fully automated while flowClean represents a semi-automated approach. In all methods, the signal check is performed for every channel across the time of acquisition. The data are divided into equally sized bins of cell events. For each bin, the models corresponding to each method are calculated and every bin that differs from the rest is flagged in flowClean, or alternatively flagged and removed in flowAI and flowCut. Additionally flowAI can remove outliers from the flow rate and dynamic range [61].
Due to their different implementations, the level of stringency differs across methods. Thus the optimal performance will depend on the data and on the parameter settings [60]. It should be noted that all of the methods mentioned above were designed for FC studies and to our knowledge were not applied to MC data. Due to differences in the FC and MC data, as different time resolution (events in FC are acquired faster and at higher concentrations than in MC), negative values in FC versus “0” values in MC, parameter settings can be different, but up to now no data exists to support this statement. This is an unexplored niche open for further studies.
3.3. Data debarcoding and dead cells/debris gating
In order to obtain de-barcoded data, deconvolution of the raw events needs to be performed. The most common way to debarcode MC data is to use a single-cell deconvolution algorithm [64]. For debarcoding user-friendly programs and R-based functions that can be used are listed in Table 2. For FC data automated deconvolution methods include rectangleGate from the flowCore package [65] and flowClust clustering methods, when the number of wanted clusters is equal to the number of barcoded samples [32].
Doublets, debris, and dead cells introduce noise into the data and should be removed prior to data analysis as these affect clustering results. As mentioned before, the definition of event is quite different for FC and MC and hence the gating strategy will differ. In FC, usually FSC parameters height (H) and area (A) are plotted against each other and used to eliminate doublets. The events that are out of the diagonal are defined as doublets, as they are characterized by the same height but different area of the signal curve [6]. However, debris and dead cells can overlap with the cell populations of interest, and scatter parameters can change depending on the sample processing protocol [66]. Therefore, it is recommended to stain live cells and populations of interest with specific fluorescent probes. For MC, as data are usually acquired with calibration beads [67], they need to be identified using bead specific channels and removed manually, or automatically using e.g. the CATALYST package [51]. The nucleated, intact cells are defined by balanced intensity for Ir, which distinguishes them from Irlow debris and Irhi doublets. If red blood cells, or other non-nucleated particles, need to be defined, the use of specific probes is required. Doublets are a real challenge in MC as FCS and SSC parameters cannot be used. Instead, users define them based on balanced Ir staining and event length [11] or Gaussian parameters, such as residual, offset, center, and width [68], [69]. It is worth noting that barcoding staining with 3 different isotopes per sample helps to identify and remove doublets [64], thus increasing sample quality.
Among other platforms FlowJo and Cytobank can be used for manual gating, or alternatively data can be imported in an R environment using e.g. flowWorkspace package [70]. If gating is provided for some of the files, semi-supervised gating methods like flowLearn could be used to reproduce the gating strategy for the remaining data [71]. This algorithm employs the gating thresholds provided as input and transfers them to the rest of the samples using derivative-based density alignments. Packages like flowStats [72], flowDensity [73] or OpenCyto [138] (a framework for constructing automated gating hierarchy) can be useful to build user-defined gating strategies. Although manual inspection is always advised, the automated approach should be considered for projects generating a high number of files.
3.4. Staining irregularities, data normalization and removal of batch effects
Inspection of marker expression levels across all files and batches is an important step of sample quality control. Staining irregularities, such as a loss of separation between positive and negative values for a given marker, or significant changes in the signal intensity, must be identified and removed, as they can affect event classification into specific clusters [45]. Recently the AOF algorithm, that uses cell frequencies to calculate the average of overlapping cells per channel, was applied to more than 2000 files in MC [20]. Based on calculated sample scores and user-defined thresholds, AOF identified problematic marker expression and affected files were discarded prior to analysis. This algorithm might be a good expansion of the quality control pipeline, however, it should be used with caution, since the signal changes could be due to biological or technical variation. Barcoding and reference samples can help to distinguish between these two possibilities, and the introduction of normalization and batch effect correction can help in saving files instead of discarding them. The technical variability can come from day-to-day differences in experimental and instrumental performances. Instrument variation that cannot be controlled by the users (e.g. differences in daily instrument calibration), are identified and removed by normalization. The variations in the experimental procedure (e.g. slight differences in staining) are identified and removed via a batch effect correction [74]. Both will be discussed below.
The acquisition time in FC and MC differs, from few minutes in FC, up to a whole day in MC for barcoded samples, and therefore requires different approaches for normalizing the data. In FC, the use of single-stained capture beads and rainbow beads, just before sample acquisition was reported [28], [75] to optimize PMT voltages, resulting in similar MIs for the markers. As FC experiments are shorter in acquisition time, it is assumed that the MIs will be equivalent for the samples acquired within the same day. On the other hand, in MC a signal drop caused by progressing CyTOF decalibration is frequently observed, especially when long, barcoded samples are run. In order to correct for it, bead-based normalization was introduced in [67] and modified by Fluidigm. The algorithm uses commercially available calibration beads, spiked and acquired together with the sample. Hence, changes in the signal can be tracked through the acquisition time. Next, the beads are identified and the median intensities of the beads are calculated in defined time intervals across all files. Based on the obtained values, the global mean for each bead is calculated and used as a target value. To obtain the transformation factor, a linear model using the global means and interval-specific intensities is calculated. This factor is then applied to all cell events and interpolated to all markers in the corresponding intervals and files. Although run-to-run machine variation can be optimized for both MC and FC, the technical differences introduced upon sample preparation will remain. Therefore the normalization and batch effect correction play important roles in downstream analysis.
fdaNorm and gaussNorm algorithms were developed to correct the files across the experiments [76]. They both perform density-based normalization per single channel using ungated .FCS files. The algorithms assumes that each marker has its characteristic number of density peaks called landmarks, which are shared by all samples and can be identified even with some changes in MI. During normalization these density peaks are shifted to align the samples. Although algorithms differ in their implementation, they perform similarly in the context of resolution in binary markers like CD3, CD4 or CD8. When using gaussNorm, the number of density peaks needs to be known upfront for each marker, while fdaNorm estimates peaks automatically. The remarks and a extended version of the fdaNorm algorithm can be found in [77]. In this version the reference file provides information about marker distributions together with gating template, and additionally normalization is performed during the gating. The reason for these changes is that the marker densities can differ across distinct populations, affecting the normalization process, and the use of a reference sample with gating upon normalization improves the automation process. These methods perform well for automated gating, as the density peaks alignment facilitates implementation of reproducible gating hierarchy, however it requires previous knowledge of the analyzed cells. This can be useful in clinical studies when known populations are quantified in a relatively short time or for the extraction of cell frequencies identified using binary markers. However, as the intensity of the peaks are shifted, comparison of the MI cannot be performed, and part of the biological information is lost.
As mentioned before the inclusion of reference samples becomes a useful tool to track batch effects introduced during sample preparation. Recently three methods that take advantage of it became available to researchers, and will be discussed. Shaham et al. [78] introduced a deep learning approach called BatchEffectRemoval. This approach is based on Maximum Mean Discrepancy (MMD) and Residual Nets, and corrects the distribution of one sample to its corresponding pair, collected at a different time point. Although it can be a good solution when time point experiments are performed, its performance in MI-sensitive markers is still questionable. CytoNorm [79] and CytofBatchAdjust [80] are two alternatives that use reference samples aliquoted across the batches to obtain batch-specific transformation factors. CytoNorm starts with FlowSOM [81] clustering for each reference file. At the cluster level, quantiles for each marker are computed and the mean quantile distribution is calculated using values from all the reference files. This information is used to learn the appropriate transformations for each batch and to correct for it. One of the CytoNorm assumptions is that the batch effects are small enough to do not impact FlowSOM clustering results. In other words, although samples differ at the cluster level, the metaclustering that defines cell populations should be the same across all reference samples. If not, some artifacts can be introduced to the data [80], and therefore a careful and detailed investigation should be performed before normalizing collected batches. On the other hand, CytofBatchAdjust performs the normalization on ungated files, where batches can be scaled to a user-defined percentile, mean, medium or quantile normalization.
Both algorithms give the advantage of preserving the biological information contained in MI. However, it is important to ensure that the reference sample is prepared using the same protocol as for the studied samples. Therefore upfront assumption of sample composition should be taken into consideration.
4. Data analysis
4.1. Data visualization – dimensionality reduction methods
Manual gating not only aims at extracting the important features, but also gives a good insight into data quality, variability, structure or differences between groups of individuals. In high-dimensional data, the same inspection should be performed using dimensionality reduction or clustering-based approaches.
The goal of the dimensionality reduction methods is to preserve the structure of high-dimensional data in the lower, easier to interpret, 2 or 3 dimensional map. These methods can be divided into linear and non-linear tools. Linear methods represented by PCA (Principal Component Analysis) [82], [83] focus on keeping the maximum variance of the points in the lower space, thus keeping the dissimilar points far from each other [84]. On the other hand non-linear algorithms like t-SNE (t-Stochastic Neighbor Embedding) [84] and its derivatives [85], [86], [87], [92], [97] keep the similar cells close to each other, therefore focusing on local relationship preservation [84]. Some of the tools like t-SNE and UMAP (Uniform Manifold Approximation and Projection) [88] separate well known populations, giving a nice overview of existing cells. Other methods like Isomap (isometric feature mapping) [89], [90] or Diffusion Maps [91] visualize differentiation trajectories, as they are able to preserve both local and global distances between cells.
PCA is designed to preserve the features with the highest variability in the principal components (PC). It assumes that the most prominent variation will be explained by the first two to three PC, making them easily interpretable. As shown by [92], [93], due to the linear assumption, PCA cannot separate well populations in the first two PC, as immune panels are usually designed in the way that each marker brings new and independent information. Nevertheless PCA as an easily scalable and not-stochastic technique, remains a powerful tool and is widely used in biological and clinical cytometry studies, as shown in [94], [95], [96].
t-SNE is a state-of-the-art visualization method that projects high-dimensional information into easily interpretable 2D maps [84]. t-SNE calculates two similarity matrices based on the distance in the high- and low-dimensional space using pairwise comparison across all the points. Next, in a iterative way the algorithm minimizes the difference between two matrices, which results in the optimized position of each cell in the 2D space [55]. t-SNE pairwise comparison has its pros and cons, it is a robust and accurate algorithm, and on the other hand the more cells are analyzed, the more pairs need to be computed and the highest the computational cost. This limits the use of t-SNE in FC/MC studies where thousands or even millions of events are acquired. To overcome this issue random downsampling (generation of a smaller subset of cells), is often used, taking the risk of losing rare populations. Therefore, new implementations were developed, aiming at limiting the computational power required to obtain high-resolution data. Among them BH-SNE (Barnes-Hut-SNE) [97] reduces the number of pair comparisons by constructing a tree-like structure. This implementation is used in viSNE and published by Amir et al. [92]. HSNE (Hierarchical Stochastic Neighbor Embedding) [85] is a combination of A-tSNE (t-SNE approximation) where, instead of computing precise distances, approximated k-nearest neighborhood graph is computed and embedded using BH-SNE. FIt-SNE (Fast Interpolation-based t-SNE) [86] uses Fourier interpolation to speed up the convolution step and opt-SNE [87] allows fine-tuning of t-SNE parameters, like the number of iterations, to obtain high resolution maps in a shorter time. It should be noted that t-SNE is stochastic, which means that every new run will give slightly different visualization. Consequently, researchers should perform multiple runs in order to obtain good data representation. Comparison of multiple maps can be only done if the samples were run simultaneously with the same settings. Jensen-Shannon divergence, a statistical method that measures two probability distributions, can be useful to compare the projection from the same data set as shown in [92], [98].
Recently a new visualization tool called UMAP gained attention in the cytometry field. This tool also preserves global distances between cell types, while t-SNE conserves only close neighborhoods [88], [99]. For this reason UMAP was used to recapitulate human hematopoiesis, and is useful for cell continuity visualization [99]. Additionally both UMAP and FIt-SNE can analyze more cells than t-SNE in a shorter time [99]. Isomap [89] and Diffusion maps [91] also preserve global relatedness and continuity between cells instead of calculating the pairwise Euclidian distance. Isomap uses non-linear geodesic distance [89]. Diffusion map introduced by [91], and adapted to the single cell study by [101], constructs diffusion matrices based on random walk probabilities between cells and generate diffusion components DC (known as eigenvectors), that similarly to the PC correspond to the largest coefficients of the data [102], [103].
Even though some improvements were made on t-SNE implementation and faster algorithms like UMAP were built, the scalability problem remains. Most of the embedding techniques were first used on transcriptomic data where, in contrast to cytometry, a relatively small amount of cells are described by a much larger amount of markers. Although other dimensionality reduction and topology inference algorithms can be used, the lack of good implementations that enable handling of millions of cells prevents researchers to apply them to big files as noted in [104].
Although non-linear dimensionality reduction methods are powerful in projecting phenotypically similar cells, the understanding of the marker contribution to cell segregation can be difficult, as it requires plotting multiple markers in individual maps. In such case, studying marker co-expression is even more challenging as was pointed out in [96], [105]. One-SENSE (One-Dimensional Soli-Expression by Non-Linear Stochastic Embedding) answers to this limitation and propose 2D assignment of the markers to categories that can be then visualized using a combination of t-SNE map and heatmaps [105]. This method was successfully applied to study T cell and dendritic cell heterogeneity [100], [105], [106].
4.2. Data visualization – clustering methods
Clustering-based algorithms group similar cells and use visualization tools to represent them in a lower dimensional space [13]. When choosing the best clustering method several requirements should be considered, such as the need for downsampling, reproducibility, rare cell detection, and running time. These variables were measured by Weber et al., where several of the currently used cytometry clustering algorithms were compared, identifying FlowSOM as a good trade-off between quality and time [107].
Since its publication, FlowSOM [81] became a widely used clustering algorithm in the field of cytometry [54], [108], [109]. This algorithm uses a two-step clustering process: a SOM (Self-Organizing Map) and consensus hierarchical clustering. SOM, a type of artificial neural network, contains a grid of nodes where each node represents a point in a multidimensional space. SOM reproduces the data topology by assigning the most similar cells to the same node or its closest neighbors. Increasing the grid size increases the possibility of finding rare populations. However, as shown by Weber et al., the reproducibility of the data can be compromised and additional splitting of the largest populations can be seen. In the second step, node centers are grouped into metaclusters using a consensus hierarchical clustering, and final cluster labels are obtained. The data can be visualized using a minimal spanning tree, like in SPADE [44], or in a heatmap [54]. Although similar results can be obtained with both methods, the two-step clustering in FlowSOM accelerates analysis and evades downsampling, making it a better choice. Unfortunately, the stochasticity problem remains, and unless the seed (starting analysis point) is pre-defined, the comparison between different runs cannot be done. When comparing clustering performance, the F1 score measuring tests’ accuracy using precision and recall could be applied [107]. Alternatively the algorithm CytoCompare which computes the distance between the clusters using marker distribution [110], or the Jaccard coefficient [111] can also be applied.
Multiple tools and workflows implementing FlowSOM have been recently published: EmbedSOM improves data visualization [112]; diffcyt, a new computational framework for differential discovery analyses [56] will be discussed below; Ek’Balam, a hierarchy-based clustering in the Astrolab Cytometry Platform [20]. All these applications emphasize the broad utility of FlowSOM. However as noticed in [113], one of the major drawbacks of this algorithm is the user-defined number of clusters, which limits the understanding of population diversity and introduces researcher supervision. Other popular clustering approaches could be used instead, like Phenograph, which uses k-nearest-neighborhoods (KNN) to represent phenotypically similar cells as highly interconnected nodes [114] or X-shift, that also applies KNN with density estimation [115]. Both tools ranked high in benchmark studies, especially for rare population detection [107], [116]. They have the ability to predict the number of clusters in a given sample, although they perform poorly in scalability requiring downsampling. Additionally the fusion of both dimensionality reduction methods using t-SNE and density based clustering was also reported and successfully applied in the immune diversity study of lymphoid compartment using ACCENSE (Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding), [117] and of the myeloid compartment using DensVM (Density-based clustering aided by Support Vector Machine), which combines density based algorithm with machine learning techniques [90].
4.3. Looking for the meaning: analysis of cytometry data – biomarker discovery
FC and MC are commonly used as biomarker discovery platforms to improve diagnosis or allow the prediction of response to therapies. Typically the cell abundances and their median marker expressions are extracted using clustering or dimensionality reduction. Then statistical tests are run to associate cell differential abundance (DA) and states (DS) with specific phenotypes, while correcting for different covariates [54]. This approach is presented in various analysis pipelines [54], [56], [118], [119] and the main ones will be briefly discussed.
Nowicka et al. [54] analysis pipeline called CyTOF workflow uses FlowSOM to cluster the data, followed by differential expression analysis to identify cell populations responding to the stimulation. Two different models are applied depending on the type of data: the General Linear Mixed Model (GLMM) and the Linear Mixed Model (LMM) for DA and DS respectively. In both cases the mixed model with random intercept is used to account for random effects caused by variations across the individuals. The general model is used for DA analysis to account for non-normal distribution in cell proportions when samples with lower cell-counts are present. cydar [119] detects diversely abundant cells by assigning them to overlapping ‘hyperspheres’ in the high dimensional space of markers. Cell counts and median marker expression are calculated within each hypersphere for each sample. Finally, the negative binominal generalized linear model from edgeR is used to test the differences between two groups. This model, similarly to GLMM, improves the estimation of dispersion parameters. Both pipelines provide flexibility in the adjustment of experimental covariates like batch, age or sex. However, only CyTOF workflow distinguishes between phenotypical and functional (e.g. phosphorylation state) markers making the analysis easier to interpret in the context of biological knowledge.
Besides regression models, different machine learning approaches were successfully used to identify biomarkers. In the “ Flow Cytometry: Critical Assessment of Population Identification Methods” (FlowCAP) challenge IV [120], two pipelines, FloReMi.1 [121] and flowType-RchyOptimyx [122], provided statistically significant predictive values in the context of patient progression from HIV+ to AIDS. Both methods use flowType [123] to detect cell populations and apply random survival forest, using the ensemble of decision tree in FloReMi or dynamic programming together with graph theory in flowType-RchyOptimyx [124] to find the best gating hierarchy correlated with the clinical outcome. Citrus combines the cell classification obtained by hierarchical clustering with the automated selection of features based on a regularized classification model to associate the obtained features (cell percentages and median marker expressions) with the endpoint of interest. This algorithm was successfully used to identify cell subsets associated with AIDS-free survival [125]. However, as commented by Arvaniti et al. [126], a high amount of irrelevant events used as clustering input can result in either model overfitting or alternatively prevent rare cell detection. To address this issue, the authors developed CellCNN, an algorithm that applies convolutional neural networks in representation learning, making use of the sample classes during population identification. CellCNN is designed to detect rare populations with a frequency lower than 0.01% and was able to identify minor survival-associated cell populations in HIV-infected patients or spiked-in rare leukemic blast populations of two AML subclasses [126].
The use of regression or machine learning approaches are not mutually exclusive and their combination was presented by Krieg et al. [108]. They used FlowSOM together with GLMM to find relevant cell populations that distinguish responders to anti-PD1 therapy in metastatic melanoma patients. These features were further characterized using CellCNN algorithm. The comparison between different regression based methods was integrated in the package diffcyt and compared to other machine learning algorithms [56]. According to this report diffcyt outperforms other tools in rare population and differential state detection between two conditions. However, it is crucial to ensure the proper selection of clustering setting and regression method.
None of the presented approaches is perfect, and their performance will depend on the biological question, type and volume of the data. It is important to consider what type of analysis is required [13]. Different approaches should be used when rare cell populations or activation of a particular known cell population are targeted. Results should be verified with at least two different algorithms, incorporating also traditional methods when verifying the outcome. Various ready-to-go R or python-coded analytical pipelines or user friendly interfaces are nowadays available, with no need for strong programming skills [54], [127], [128], [129]. Benchmarking that incorporates the newest algorithms and both FC and MC data should be organized in order to guide FC and MC users through different analytical approaches, pipelines, and algorithms.
4.4. Looking for the meaning: analysis of cytometry data – trajectory interference
Besides being a biomarker discovery platform, FC and MC are commonly used in the modeling of cell developmental stages with trajectory inference (TI) methods. These methods estimate for each cell a numeric value, called pseudotime, which orders the cells within the dynamic process of interest. This allows to define and study different transition stages [130]. The typical TI workflow comprises a dimensional reduction followed by a trajectory modeling using the tools described below. Most of the earlier algorithms were designed to model fixed topologies, such as one dimensional path, while currently bifurcating points, or tree-like structures can also be detected.
Wanderlust was applied to reconstruct human B cell lymphopoiesis [131]. It is an example of one-path trajectory modeling, and was the first algorithm designed to study developmental stages using MC data. It is a graph-based method where each cell is represented as a node connected to its closest neighborhoods by the edges. To eliminate noise and possibility of introducing short circuits, multiple graphs and trajectories are built using random waypoint cells and l-out-of-k-nearest neighbors (l-k-NNG). The final position of the cell is the average over all graph trajectories. Two main assumptions are taken when using this tool: all cells that represent the non-branching differentiation pathway are present, and the changes in the marker expression are gradual. Therefore a proper marker selection is crucial.
Wishbone [132] was used to track the development of T cells in the mouse thymus. It is an algorithm designed to detect bifurcating developmental trajectories. Similar to Wanderlust, Wishbone is based on k-NNG, where the shortest path between two cells is used as a distance metrics. However, as the bifurcating points are prone to build short circuits due to insufficient marker differences, instead of using subsampling subset of edges like in Wanderlust, Wishbone takes advantage of diffusion maps. Because of this, the major structure is kept in the first diffusion components, leaving out the trend to short circuit noise. The embedded space is used to construct k-NNG. In the case of Wishbone the waypoints have a double role: first, they allow to robustly order the cells along the trajectory, and secondly, together with spectral clustering, they provide information about the placement of waypoints on the same or on a different branch, thus giving the branching point information.
The robustness of both Wanderlust and Wishbone depends on a user-defined starting cell, whereas p-Creode [133] constructs tree-like structure in an unsupervised manner. This algorithm introduces density pre-analysis downsampling, and is also based on graph theory using k-NNG construction with a density-based modification. After the construction of multiple trajectories a new metric called p-Creode scores is used to select the most representative trajectory.
All the above mentioned methods were recently benchmarked using single cell RNA sequencing data [104]. This study provides useful guidelines for choosing proper TI methods. However, due to the different nature of cytometry and sequencing data, the outcome can be different. Therefore, it would be helpful to provide similar comparison using MC/FC data.
5. Conclusions
FC and MC are powerful high-dimensional technologies in single-cell biology. They are becoming important tools in biomarker discovery research, disease monitoring, and medical diagnostics. The rapid increase in dimensionality gives an opportunity to understand cell diversity in detail, narrow the research to fine cell populations, and by doing so, enable precision in the development of new therapies and biomarkers. However, dimensionality reduction and automated analysis require high-quality data, analytical skills, and powerful algorithms to meaningfully process the multidimensional space. As previously discussed, the design and execution of a good cytometry-based study is not a trivial process. Small details like changes in stocks, pipetting errors, shifts in machine performance, and improper data preprocessing can significantly contribute to data variation. Controlling for batch effects, although well adopted in transcriptomic data, is still inefficient and not often applied in MC and FC due to different data structures. It should be noted that inclusion of covariants like “batch effect” in the statistical model does not eliminate the bias introduced upon the clustering, and therefore batch effects should be corrected before data analysis, and ideally prevented when preparing the SOP. Many dimensionality reduction and clustering methods are available and they should be combined to verify and confirm results. To make an analysis accessible to non-programming researchers many packages bringing together various preprocessing and analysis tools that can be used in user-friendly interfaces Table 2. Hence, high-dimensional analysis can be available to both biologist and bioinformaticians. Since the single-cell high-dimensional era is just starting, it is important to take care when interpreting the data. Careful validation with multiple methods and standard approaches like traditional manual gating should be implemented in the analysis pipelines.
Author statement
PR wrote the original draft.
MAR provided critical review of the manuscript.
CM designed the work, supervised PR and revised the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
PR acknowledges support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n° [115565], resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution (MEAR as PI) and in particular the in-cash support from Sanofi/Genzyme to PR. CM was supported by Instituto de Salud Carlos III (Miguel Servet II program, CPII16/00028). The authors also acknowledge support from Instituto de Salud Carlos III (PI18/00082) partly supported by European FEDER funds. Special acknowledge for Yvan Saeys laboratory, especially for Sofie Van Gassen and Katrien Quintelier for the feedback and Robrecht Cannoodt for useful discussions. Also we express our gratitude to Guillermo Barturen for his input.
References
- 1.Mair F., Prlic M. OMIP-044: 28-color immunophenotyping of the human dendritic cell compartment. Cytometry Part A. 2018;93:402–405. doi: 10.1002/cyto.a.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartmann F.J., Bernard-Valnet R., Quériault C., Mrdjen D., Weber L.M., Galli E. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J Exp Med. 2016;213:2621–2633. doi: 10.1084/jem.20160897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behbehani G.K., Bendall S.C., Clutter M.R., Fantl W.J., Nolan G.P. Single cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Gorman W.E., Hsieh E.W.Y., Savig E.S., Gherardini P.F., Hernandez J.D., Hansmann L. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 2015;136:1326–1336. doi: 10.1016/j.jaci.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frei A.P., Bava F.A., Zunder E.R., Hsieh E.W., Chen S.-Y., Nolan G.P. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossarizza A., Chang H.-D., Radbruch A., Akdis M., Andrä I., Annunziato F. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immunol. 2017;47:1584–1797. doi: 10.1002/eji.201646632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stern A.D., Rahman A.H., Birtwistle M.R. Cell size assays for mass cytometry. Cytometry A. 2017;91:14–24. doi: 10.1002/cyto.a.23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun Y.W., Kim H.R., Reo Y.J., Dai M., Ahn K.H. Addressing the autofluorescence issue in deep tissue imaging by two-photon microscopy: the significance of far-red emitting dyes. Chem Sci. 2017;8:7696–7704. doi: 10.1039/C7SC03362A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman A.H., Lavin Y., Kobayashi S., Leader A., Merad M. High-dimensional single cell mapping of cerium distribution in the lung immune microenvironment of an active smoker. Cytometry B Clin Cytom. 2018;94:941–945. doi: 10.1002/cyto.b.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller B.C., Presti R.M., Byers D.E., Atkinson J.J. Significant interference in mass cytometry from medicinal iodine in human lung. Am J Respir Cell Mol Biol. 2016;55:150–151. doi: 10.1165/rcmb.2015-0403LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen L.R., Leipold M.D., Pedersen C.B., Maecker H.T. The anatomy of single cell mass cytometry data. Cytometry Part A. 2019;95:156–172. doi: 10.1002/cyto.a.23621. [DOI] [PubMed] [Google Scholar]
- 12.Newell E.W., Cheng Y. Mass cytometry: blessed with the curse of dimensionality. Nat Immunol. 2016 doi: 10.1038/ni.3485. [DOI] [PubMed] [Google Scholar]
- 13.Saeys Y., Gassen S.V., Lambrecht B.N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 14.Duffy D., Rouilly V., Libri V., Hasan M., Beitz B., David M. Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–450. doi: 10.1016/j.immuni.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Duffy D., Rouilly V., Braudeau C., Corbière V., Djebali R., Ungeheuer M.-N. Standardized whole blood stimulation improves immunomonitoring of induced immune responses in multi-center study. Clin Immunol. 2017;183:325–335. doi: 10.1016/j.clim.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Mikes J., Olin A., Lakshmikanth T., Chen Y., Brodin P. Automated cell processing for mass cytometry experiments. Methods Mol Biol. 2019;1989:111–123. doi: 10.1007/978-1-4939-9454-0_8. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y., Ahmed S., Harari F., Vahter M. Impact of Ficoll density gradient centrifugation on major and trace element concentrations in erythrocytes and blood plasma. J Trace Elem Med Biol. 2015;29:249–254. doi: 10.1016/j.jtemb.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Sumatoh H.R., Teng K.W.W., Cheng Y., Newell E.W. Optimization of mass cytometry sample cryopreservation after staining. Cytometry Part A. 2017;91:48–61. doi: 10.1002/cyto.a.23014. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez R., Maecker H. Cytokine-stimulated phosphoflow of whole blood using CyTOF mass cytometry. Bio Protoc. 2015;5 [PMC free article] [PubMed] [Google Scholar]
- 20.Amir E.D., Lee B., Badoual P., Gordon M., Guo X.V., Merad M. Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front Immunol. 2019;10. doi: 10.3389/fimmu.2019.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakkestad S.T., Skavland J., Hanevik K. Whole blood preservation methods alter chemokine receptor detection in mass cytometry experiments. J Immunol Methods. 2019;112673 doi: 10.1016/j.jim.2019.112673. [DOI] [PubMed] [Google Scholar]
- 22.Yan L., Ma C., Wang D., Hu Q., Qin M., Conroy J.M. OSAT: a tool for sample-to-batch allocations in genomics experiments. BMC Genomics. 2012;13:689. doi: 10.1186/1471-2164-13-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai L., Ong R., Li J., Albani S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytometry Part A. 2015;87:369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann F.J., Simonds E.F., Bendall S.C. A universal live cell barcoding-platform for multiplexed human single cell analysis. Sci Rep. 2018;8 doi: 10.1038/s41598-018-28791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz A.R., Mei H.E. Surface barcoding of live PBMC for multiplexed mass cytometry. Methods Mol Biol. 2019;1989:93–108. doi: 10.1007/978-1-4939-9454-0_7. [DOI] [PubMed] [Google Scholar]
- 26.Krutzik P.O., Nolan G.P. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 27.Giudice V., Feng X., Kajigaya S., Biancotto A., Young N.S. Fluorescent cell barcoding as new flow cytometric technique for multiplexed phenotyping and signaling profiling in hematologic patients. Blood. 2016;128 5033 5033. [Google Scholar]
- 28.Jamin C., Le Lann L., Alvarez-Errico D., Barbarroja N., Cantaert T., Ducreux J. Multi-center harmonization of flow cytometers in the context of the European “PRECISESADS” project. Autoimmun Rev. 2016;15:1038–1045. doi: 10.1016/j.autrev.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Pitoiset F., Cassard L., Soufi K.E., Boselli L., Grivel J., Roux A. Deep phenotyping of immune cell populations by optimized and standardized flow cytometry analyses. Cytometry Part A. 2018;93:793–802. doi: 10.1002/cyto.a.23570. [DOI] [PubMed] [Google Scholar]
- 30.Schulz A.R., Baumgart S., Schulze J., Urbicht M., Grützkau A., Mei H.E. Stabilizing antibody cocktails for mass cytometry. Cytometry Part A. 2019;95:910–916. doi: 10.1002/cyto.a.23781. [DOI] [PubMed] [Google Scholar]
- 31.Kleinsteuber K., Corleis B., Rashidi N., Nchinda N., Lisanti A., Cho J.L. Standardization and quality control for high-dimensional mass cytometry studies of human samples. Cytometry A. 2016;89:903–913. doi: 10.1002/cyto.a.22935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giudice V., Fantoni G., Biancotto A. Fluorescent cell barcoding for immunophenotyping. Methods Mol Biol. 2019;2032:53–68. doi: 10.1007/978-1-4939-9650-6_3. [DOI] [PubMed] [Google Scholar]
- 33.Ryherd M., Plassmeyer M., Alexander C., Eugenio I., Kleschenko Y., Badger A. Improved panels for clinical immune phenotyping: utilization of the violet laser. Cytometry, Part B. 2018;94:827–835. doi: 10.1002/cyto.b.21532. [DOI] [PubMed] [Google Scholar]
- 34.Leipold M.D., Newell E.W., Maecker H.T. Multiparameter phenotyping of human PBMCs using mass cytometry. Methods Mol Biol. 2015;1343:81–95. doi: 10.1007/978-1-4939-2963-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finak G., Langweiler M., Jaimes M., Malek M., Taghiyar J., Korin Y. Standardizing flow cytometry immunophenotyping analysis from the Human ImmunoPhenotyping Consortium. Sci Rep. 2016;6 doi: 10.1038/srep20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han G., Spitzer M.H., Bendall S.C., Fantl W.J., Nolan G.P. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat Protoc. 2018;13:2121–2148. doi: 10.1038/s41596-018-0016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi C., Au-Yeung A., Fuh F., Ramirez-Montagut T., Bolen C., Mathews W. Mass cytometry panel optimization through the designed distribution of signal interference. Cytometry Part A. 2017;91:39–47. doi: 10.1002/cyto.a.22977. [DOI] [PubMed] [Google Scholar]
- 39.BD Biosciences. BD HorizonTM Guided Panel Solution (GPS) tool. https://www.bdbiosciences.com/en-us/applications/research-applications/multicolor-flow-cytometry/product-selection-tools/horizon-gps-tool.
- 40.Maxpar Panel Fluidigm.Designer. https://www.fluidigm.com/binaries/content/documents/fluidigm/search/hippo%3Aresultset/maxpar-panel-designer/fluidigm%3Afile
- 41.Perfetto S.P., Roederer M. Increased immunofluorescence sensitivity using 532 nm laser excitation. Cytometry A. 2007;71:73–79. doi: 10.1002/cyto.a.20358. [DOI] [PubMed] [Google Scholar]
- 42.Giesecke C., Feher K., von Volkmann K., Kirsch J., Radbruch A., Kaiser T. Determination of background, signal-to-noise, and dynamic range of a flow cytometer: a novel practical method for instrument characterization and standardization. Cytometry Part A. 2017;91:1104–1114. doi: 10.1002/cyto.a.23250. [DOI] [PubMed] [Google Scholar]
- 43.Leipold M.D., Obermoser G., Fenwick C., Kleinstuber K., Rashidi N., McNevin J.P. Comparison of CyTOF assays across sites: results of a six-center pilot study. J Immunol Methods. 2018;453:37–43. doi: 10.1016/j.jim.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu P., Simonds E.F., Bendall S.C., Gibbs K.D., Bruggner R.V., Linderman M.D. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amir E.D., Guo X.V., Mayovska O., Rahman A.H. Average Overlap Frequency: a simple metric to evaluate staining quality and community identification in high dimensional mass cytometry experiments. J Immunol Methods. 2018;453:20–29. doi: 10.1016/j.jim.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Mair F., Tyznik A.J. High-dimensional immunophenotyping with fluorescence-based cytometry: a practical guidebook. Methods Mol Biol. 2019;2032:1–29. doi: 10.1007/978-1-4939-9650-6_1. [DOI] [PubMed] [Google Scholar]
- 47.Brodie T.M., Tosevski V., Medová M. OMIP-045: characterizing human head and neck tumors and cancer cell lines with mass cytometry. Cytometry Part A. 2018;93:406–410. doi: 10.1002/cyto.a.23336. [DOI] [PubMed] [Google Scholar]
- 48.Brummelman J., Haftmann C., Núñez N.G., Alvisi G., Mazza E.M.C., Becher B. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat Protoc. 2019;14:1946–1969. doi: 10.1038/s41596-019-0166-2. [DOI] [PubMed] [Google Scholar]
- 49.Kalina T. Reproducibility of flow cytometry through standardization: opportunities and challenges. Cytometry. 2020;97:137–147. doi: 10.1002/cyto.a.23901. [DOI] [PubMed] [Google Scholar]
- 50.Bendall S.C., Nolan G.P., Roederer M., Chattopadhyay P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chevrier S., Crowell H.L., Zanotelli V.R.T., Engler S., Robinson M.D., Bodenmiller B. Compensation of signal spillover in suspension and imaging mass cytometry. Cels. 2018;6(612–620) doi: 10.1016/j.cels.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leipold M.D. Another step on the path to mass cytometry standardization. Cytometry Part A. 2015;87:380–382. doi: 10.1002/cyto.a.22661. [DOI] [PubMed] [Google Scholar]
- 53.Szalóki G., Goda K. Compensation in multicolor flow cytometry. Cytometry Part A. 2015;87:982–985. doi: 10.1002/cyto.a.22736. [DOI] [PubMed] [Google Scholar]
- 54.Nowicka M., Krieg C., Crowell H.L., Weber L.M., Hartmann F.J., Guglietta S. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 2019;6:748. doi: 10.12688/f1000research.11622.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mair F., Hartmann F.J., Mrdjen D., Tosevski V., Krieg C., Becher B. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol. 2016;46:34–43. doi: 10.1002/eji.201545774. [DOI] [PubMed] [Google Scholar]
- 56.Weber L.M., Nowicka M., Soneson C., Robinson M.D. diffcyt: differential discovery in high-dimensional cytometry via high-resolution clustering. Commun Biol. 2019;2:1–11. doi: 10.1038/s42003-019-0415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bendall S.C., Simonds E.F., Qiu P., Amir E.D., Krutzik P.O., Finck R. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finak G., Perez J.-M., Weng A., Gottardo R. Optimizing transformations for automated, high throughput analysis of flow cytometry data. BMC Bioinf. 2010;11:546. doi: 10.1186/1471-2105-11-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Azad A., Rajwa B., Pothen A. flowVS: channel-specific variance stabilization in flow cytometry. BMC Bioinf. 2016;17:291. doi: 10.1186/s12859-016-1083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S., Brinkman R.R. Data-driven flow cytometry analysis. Methods Mol Biol. 2019;1989:245–265. doi: 10.1007/978-1-4939-9454-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monaco G., Chen H., Poidinger M., Chen J., de Magalhães J.P., Larbi A. flowAI: automatic and interactive anomaly discerning tools for flow cytometry data. Bioinformatics. 2016;32:2473–2480. doi: 10.1093/bioinformatics/btw191. [DOI] [PubMed] [Google Scholar]
- 62.Meskas J, Wang S. Precise and accurate automated removal of outlier events and flagging of files based on time versus fluorescence analysis. Github Repository: https://github.com/jmeskas/flowCut. [DOI] [PMC free article] [PubMed]
- 63.Fletez-Brant K., Špidlen J., Brinkman R.R., Roederer M., Chattopadhyay P.K. flowClean: automated identification and removal of fluorescence anomalies in flow cytometry data. Cytometry A. 2016;89:461–471. doi: 10.1002/cyto.a.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zunder E.R., Finck R., Behbehani G.K., Amir E.-A.D., Krishnaswamy S., Gonzalez V.D. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellis B, Haal P, Hahne F, Meur NL, Gopalakrishnan N, Spidlen J, et al. flowCore: flowCore: Basic structures for flow cytometry data. Bioconductor version: Release (3.9); 2019. https://doi.org/10.18129/B9.bioc.flowCore.
- 66.Chen X., Zhang H., Mou W., Qi Z., Ren X., Wang G. Flow cytometric analyses of the viability, surface marker expression and function of lymphocytes from children following cryopreservation. Mol Med Rep. 2016;14:4301–4308. doi: 10.3892/mmr.2016.5780. [DOI] [PubMed] [Google Scholar]
- 67.Finck R., Simonds E.F., Jager A., Krishnaswamy S., Sachs K., Fantl W. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee B.H., Rahman A.H. Acquisition, processing, and quality control of mass cytometry data. In: McGuire H.M., Ashhurst T.M., editors. Mass cytometry: methods and protocols. Springer New York; New York, NY: 2019. pp. 13–31. [DOI] [PubMed] [Google Scholar]
- 69.Bagwell C.B., Inokuma M., Hunsberger B., Herbert D., Bray C., Hill B. Automated data cleanup for mass cytometry. Cytometry A. 2019 doi: 10.1002/cyto.a.23926. [DOI] [PubMed] [Google Scholar]
- 70.Finak G, Jiang M. flowWorkspace: Infrastructure for representing and interacting with gated and ungated cytometry data sets. Bioconductor version: Release (3.9); 2019. https://doi.org/10.18129/B9.bioc.flowWorkspace.
- 71.Lux M., Brinkman R.R., Chauve C., Laing A., Lorenc A., Abeler-Dörner L. flowLearn: fast and precise identification and quality checking of cell populations in flow cytometry. Bioinformatics. 2018;34:2245–2253. doi: 10.1093/bioinformatics/bty082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hahne F, Gopalakrishnan N, Khodabakhshi AH, Wong C-J, Lee K. flowStats: statistical methods for the analysis of flow cytometry data. Bioconductor version: Release (3.9); 2019. https://doi.org/10.18129/B9.bioc.flowStats.
- 73.Malek M., Taghiyar M.J., Chong L., Finak G., Gottardo R., Brinkman R.R. flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics. 2015;31:606–607. doi: 10.1093/bioinformatics/btu677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mertens B.J.A. Transformation, normalization, and batch effect in the analysis of mass spectrometry data for omics studies. In: Datta S., Mertens B.J.A., editors. Statistical analysis of proteomics, metabolomics, and lipidomics data using mass spectrometry. Springer International Publishing; Cham: 2017. pp. 1–21. https://doi.org/10.1007/978-3-319-45809-0_1. [Google Scholar]
- 75.Kalina T., Flores-Montero J., van der Velden V.H.J., Martin-Ayuso M., Böttcher S., Ritgen M. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia. 2012;26:1986–2010. doi: 10.1038/leu.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hahne F., Khodabakhshi A.H., Bashashati A., Wong C.-J., Gascoyne R.D., Weng A.P. Per-channel basis normalization methods for flow cytometry data. Cytometry Part A. 2010;77A:121–131. doi: 10.1002/cyto.a.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finak G., Jiang W., Krouse K., Wei C., Sanz I., Phippard D. High throughput flow cytometry data normalization for clinical trials. Cytometry A. 2014;85:277–286. doi: 10.1002/cyto.a.22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaham U., Stanton K.P., Zhao J., Li H., Raddassi K., Montgomery R. Removal of batch effects using distribution-matching residual networks. Bioinformatics. 2017;33:2539–2546. doi: 10.1093/bioinformatics/btx196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Gassen S, Gaudilliere B, Angst M, Saeys Y, Aghaeepour N. CytoNorm: A normalization algorithm for cytometry data. Github Repository: Https://GithubCom/Saeyslab/CytoNorm/ 2019. [DOI] [PMC free article] [PubMed]
- 80.Schuyler R.P., Jackson C., Garcia-Perez J.E., Baxter R.M., Ogolla S.O., Rochford R. Minimizing batch effects in mass cytometry data. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 82.Lever J., Krzywinski M., Altman N. Principal component analysis. Nat Methods. 2017;14:641–642. doi: 10.1038/nmeth.4346. [DOI] [Google Scholar]
- 83.Hotelling H. Analysis of a complex of statistical variables into principal components. J Educ Psychol. 1933;24:417–441. doi: 10.1037/h0071325. [DOI] [Google Scholar]
- 84.van der Maaten L., Hinton G. Visualizing data using t-SNE. J Machine Learn Res. 2008;9:2579–2605. [Google Scholar]
- 85.van Unen V., Höllt T., Pezzotti N., Li N., Reinders M.J.T., Eisemann E. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun. 2017;8:1–10. doi: 10.1038/s41467-017-01689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linderman G.C., Rachh M., Hoskins J.G., Steinerberger S., Kluger Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods. 2019;16:243–245. doi: 10.1038/s41592-018-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Belkina A.C., Ciccolella C.O., Anno R., Halpert R., Spidlen J., Snyder-Cappione J.E. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-13055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J Open Source Software. 2018 https://doi.org/10.21105/joss.00861. [Google Scholar]
- 89.Tenenbaum J.B. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 90.Becher B., Schlitzer A., Chen J., Mair F., Sumatoh H.R., Teng K.W.W. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 91.Coifman R.R., Lafon S. Diffusion maps. Appl Comput Harmon Anal. 2006;21:5–30. doi: 10.1016/j.acha.2006.04.006. [DOI] [Google Scholar]
- 92.Amir E.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Konstorum A, Jekel N, Vidal E, Laubenbacher R. Comparative analysis of linear and nonlinear dimension reduction techniques on mass cytometry data. BioRxiv 2018:273862. https://doi.org/10.1101/273862.
- 94.Costa E.S., Pedreira C.E., Barrena S., Lecrevisse Q., Flores J., Quijano S. Automated pattern-guided principal component analysis vs expert-based immunophenotypic classification of B-cell chronic lymphoproliferative disorders: a step forward in the standardization of clinical immunophenotyping. Leukemia. 2010;24:1927–1933. doi: 10.1038/leu.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jansen J.J., Hilvering B., van den Doel A., Pickkers P., Koenderman L., Buydens L.M.C. FLOOD: flow cytometric orthogonal orientation for diagnosis. Chemometrics Intelligent Laboratory Systems. 2016;151:126–135. doi: 10.1016/j.chemolab.2015.12.001. [DOI] [Google Scholar]
- 96.Tinnevelt G.H., Kokla M., Hilvering B., van Staveren S., Folcarelli R., Xue L. Novel data analysis method for multicolour flow cytometry links variability of multiple markers on single cells to a clinical phenotype. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-05714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
- 98.Olin A., Henckel E., Chen Y., Lakshmikanth T., Pou C., Mikes J. Stereotypic immune system development in newborn children. Cell. 2018;174(1277–1292) doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 100.Wong M.T., Ong D.E.H., Lim F.S.H., Teng K.W.W., McGovern N., Narayanan S. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity. 2016;45:442–456. doi: 10.1016/j.immuni.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 101.Haghverdi L., Buettner F., Theis F.J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 102.Chen Y., Lakshmikanth T., Olin A., Mikes J., Remberger M., Brodin P. Continuous immune cell differentiation inferred from single-cell measurements following allogeneic stem cell transplantation. Front Mol Biosci. 2018;5 doi: 10.3389/fmolb.2018.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Angerer P., Haghverdi L., Büttner M., Theis F.J., Marr C., Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 104.Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 105.Cheng Y., Wong M.T., van der Maaten L., Newell E.W. Categorical analysis of human T cell heterogeneity with one-dimensional soli-expression by nonlinear stochastic embedding. J Immunol. 2016;196:924–932. doi: 10.4049/jimmunol.1501928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guilliams M., Dutertre C.-A., Scott C.L., McGovern N., Sichien D., Chakarov S. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weber L.M., Robinson M.D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A. 2016;89:1084–1096. doi: 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
- 108.Krieg C., Nowicka M., Guglietta S., Schindler S., Hartmann F.J., Weber L.M. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 109.Emmaneel A., Bogaert D.J., Van Gassen S., Tavernier S.J., Dullaers M., Haerynck F. A computational pipeline for the diagnosis of CVID patients. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Platon L., Pejoski D., Gautreau G., Targat B., Le Grand R., Beignon A.-S. A computational approach for phenotypic comparisons of cell populations in high-dimensional cytometry data. Methods. 2018;132:66–75. doi: 10.1016/j.ymeth.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Melchiotti R., Gracio F., Kordasti S., Todd A.K., de Rinaldis E. Cluster stability in the analysis of mass cytometry data. Cytometry A. 2017;91:73–84. doi: 10.1002/cyto.a.23001. [DOI] [PubMed] [Google Scholar]
- 112.Kratochvíl M, Koladiya A, Balounova J, Novosadova V, Fišer K, Sedlacek R, et al. Rapid single-cell cytometry data visualization with EmbedSOM. BioRxiv 2018:496869. https://doi.org/10.1101/496869.
- 113.Kimball A.K., Oko L.M., Bullock B.L., Nemenoff R.A., van Dyk L.F., Clambey E.T. A beginner’s guide to analyzing and visualizing mass cytometry data. J Immunol. 2018;200:3–22. doi: 10.4049/jimmunol.1701494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.D., Tadmor M.D. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Samusik N., Good Z., Spitzer M.H., Davis K.L., Nolan G.P. Automated mapping of phenotype space with single-cell data. Nat Methods. 2016;13:493–496. doi: 10.1038/nmeth.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X., Song W., Wong B.Y., Zhang T., Yu S., Lin G.N. A comparison framework and guideline of clustering methods for mass cytometry data. Genome Biol. 2019;20:297. doi: 10.1186/s13059-019-1917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shekhar K., Brodin P., Davis M.M., Chakraborty A.K. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE) Proc Natl Acad Sci USA. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fonseka C.Y., Rao D.A., Teslovich N.C., Korsunsky I., Hannes S.K., Slowikowski K. Mixed-effects association of single cells identifies an expanded effector CD4 + T cell subset in rheumatoid arthritis. Sci Transl Med. 2018;10:eaaq0305. doi: 10.1126/scitranslmed.aaq0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lun A.T.L., Richard A.C., Marioni J.C. Testing for differential abundance in mass cytometry data. Nat Methods. 2017;14:707–709. doi: 10.1038/nmeth.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aghaeepour N., Chattopadhyay P., Chikina M., Dhaene T., Van Gassen S., Kursa M. A benchmark for evaluation of algorithms for identification of cellular correlates of clinical outcomes. Cytometry A. 2016;89:16–21. doi: 10.1002/cyto.a.22732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gassen S.V., Vens C., Dhaene T., Lambrecht B.N., Saeys Y. FloReMi: flow density survival regression using minimal feature redundancy. Cytometry Part A. 2016;89:22–29. doi: 10.1002/cyto.a.22734. [DOI] [PubMed] [Google Scholar]
- 122.O’Neill K., Jalali A., Aghaeepour N., Hoos H., Brinkman R.R. Enhanced flowType/RchyOptimyx: a BioConductor pipeline for discovery in high-dimensional cytometry data. Bioinformatics. 2014;30:1329–1330. doi: 10.1093/bioinformatics/btt770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aghaeepour N., Chattopadhyay P.K., Ganesan A., O’Neill K., Zare H., Jalali A. Early immunologic correlates of HIV protection can be identified from computational analysis of complex multivariate T-cell flow cytometry assays. Bioinformatics. 2012;28:1009–1016. doi: 10.1093/bioinformatics/bts082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aghaeepour N., Jalali A., O’Neill K., Chattopadhyay P.K., Roederer M., Hoos H.H. RchyOptimyx: cellular hierarchy optimization for flow cytometry. Cytometry A. 2012;81:1022–1030. doi: 10.1002/cyto.a.22209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bruggner R.V., Bodenmiller B., Dill D.L., Tibshirani R.J., Nolan G.P. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Arvaniti E., Claassen M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nat Commun. 2017;8:1–10. doi: 10.1038/ncomms14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beyrend G., Stam K., Höllt T., Ossendorp F., Arens R. Cytofast: A workflow for visual and quantitative analysis of flow and mass cytometry data to discover immune signatures and correlations. Comput Struct Biotechnol J. 2018;16:435–442. doi: 10.1016/j.csbj.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen H., Lau M.C., Wong M.T., Newell E.W., Poidinger M., Chen J. Cytofkit: A bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Höllt T., Pezzotti N., van Unen V., Koning F., Eisemann E., Lelieveldt B. Cytosplore: interactive immune cell phenotyping for large single-cell datasets. Comput Graphics Forum. 2016;35:171–180. doi: 10.1111/cgf.12893. [DOI] [Google Scholar]
- 130.Cannoodt R., Saelens W., Saeys Y. Computational methods for trajectory inference from single-cell transcriptomics. Eur J Immunol. 2016;46:2496–2506. doi: 10.1002/eji.201646347. [DOI] [PubMed] [Google Scholar]
- 131.Bendall S.C., Davis K.L., Amir E.D., Tadmor M.D., Simonds E.F., Chen T.J. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Herring C.A., Banerjee A., McKinley E.T., Simmons A.J., Ping J., Roland J.T. Unsupervised trajectory analysis of single-cell RNA-Seq and imaging data reveals alternative tuft cell origins in the gut. Cell Systems. 2018;6(37–51) doi: 10.1016/j.cels.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mitchell A.J., Ivask A., Ju Y. Quantitative measurement of cell-nanoparticle interactions using mass cytometry. Methods Mol Biol. 2019;1989:227–241. doi: 10.1007/978-1-4939-9454-0_15. [DOI] [PubMed] [Google Scholar]
- 135.Fricker S.P. Metal based drugs: from serendipity to design. Dalton Trans. 2007:4903–4917. doi: 10.1039/b705551j. [DOI] [PubMed] [Google Scholar]
- 136.Lo K., Hahne F., Brinkman R.R., Gottardo R. flowClust: a bioconductor package for automated gating of flow cytometry data. BMC Bioinf. 2009;10:145. doi: 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.GenePattern. https://software.broadinstitute.org/cancer/software/genepattern/
- 138.Finak G., Frelinger J., Jiang W., Newell E.W., Ramey J., Davis M.M. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chevrier S., Levine J., Tomaso Zanotelli V., Silina K., Schulz D., Bacac M. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–749. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fread K.I., Strickland W.D., Nolan G.P., Zunder E.R. An updated debarcoding tool for mass cytometry with cell type-specific and cell sample-specific stringency adjustment. Pacific Symposium on Biocomputing. 2017;22:588–598. doi: 10.1142/9789813207813_0054. [DOI] [PubMed] [Google Scholar]