Abstract
Western blot experiments showed that sera from mice infected with the mouse hepatitis virus strain A59 (MHV‐A59) contained autoantibodies (autoAb) that bound to a 40‐kDa protein present in liver and kidney extracts. No reaction was observed with extracts of the heart, muscles, spleen, brain and lung. The Ab cross‐reacted with a 40‐kDa protein from human, rat and sheep liver, but not withliver extracts from the silver side fish (Odontesthes bonariensis). No correlation was found between the development of the hypergammaglobulinemia that followed the viral infection and theoccurrence of the autoAb. Reactive immunoglobulins pertained to the IgG1, IgG2a and IgG2b subclasses, recognized cryptic epitopes and were detected from 10 days up to 8 weeks after MHV‐infection. The 40‐kDa protein was purified from mouse liver extracts by ion‐exchange chromatography, gel filtration and SDS‐PAGE. Because the N‐terminal was blocked, we digested the protein in‐gel with trypsin and sequenced various peptides. Results indicated a 100% homology of sequence between the protein recognized by the autoAb and liver fumarylacetoacetate hydrolase (FAH), the enzyme that mediates the last step of tyrosine catabolism. Additionally, a second protein recognized by the autoAb was detected during FAH purification steps and was identified as liver alcohol dehydrogenase.
Keywords: Mouse hepatitis virus, Autoantibody, Fumarylacetoacetate hydrolase, Cryptic epitope
Abbreviations:
- ADH:
Alcohol dehydrogenase
- autoAb:
Auto‐antibody
- FAH:
Fumarylacetoacetate hydrolase
- MHV:
Mouse hepatitis virus
1 Introduction
Mouse hepatitis viruses (MHV) are known to be lymphotropic and, depending on the viral strain and on the mouse genetic background, to induce diverse alterations of immune responses 1–3. The MHV strain A59 (MHV‐A59) is a coronavirus that triggers in susceptible mice various pathologies, including hepatitis and thymus involution, B lymphocyte polyclonalactivation and, after intra‐cerebral inoculation, transient demyelination 3. MHV‐A59 causes IgG2a‐restricted hypergammaglobulinemia, although only a small fraction displays an antibody (Ab) activity against the virus itself 4. Furthermore, anti‐erythrocyte autoantibodies (autoAb) induced by immunization against rat red blood cells increased in MHV‐infected mice, indicating that the virus may enhance a concomitant autoimmune response 4.
Here we report that autoAb occurring in sera from mice infected with MHV‐A59 recognize mainly a 40‐kDa protein present in liver and kidney lysates from mice, rats, sheep and humans. The protein was identified as fumarylacetoacetate hydrolase (FAH), a key enzyme involved in tyrosine catabolism. Furthermore, during FAH purification we found that the autoAb detected another liver protein, which turned to be the enzyme alcohol dehydrogenase (ADH).
2 Results
2.1 Western blot analysis
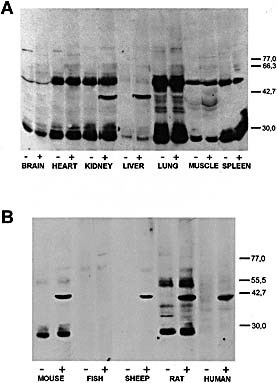
Sera from control and MHV‐infected CBA/Ht, BALB/c or 129/Sv mice were studied individually by immunoblots on brain, heart, kidney, liver, lung, muscle and spleen extracts from the corresponding strain. Fig. 1A shows a typical result obtained in CBA/Ht mice. MHV‐infected BALB/c and 129/Sv mice showed similar results (data not shown). Sera from 8 weeks infected animals recognized a protein of approximately 40 kDa present in kidney and liver lysates, whereas no specific binding was observed with mouse sera obtained before infection (Fig. 1A). The 25‐ and 50‐kDa bands were immunoglobulin fragments detected without any primary antibody (data not shown).
Figure 1.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Reactivity of sera from MHV‐infected CBA/Ht mice with extracts of several tissues from non‐infected CBA/Ht mice (A) or liver lysate from different sources (B). Organ lysates were prepared as indicated in Sect. 4 and run in SDS‐PAGE using 10% gels, transferred onto nitrocellulose sheets and incubated with a 1:100 serum dilution. Bound antibodies were revealed with peroxidase‐labeled IgG anti‐mouse IgG and ECL reagents. Results were obtained with pooled serum from four mice. Control serum (–) means serum obtained before infection and (+) indicates serum corresponding to 8 weeks post‐infection with MHV. The positions of molecular mass markers (kDa) are shown at right.
To investigate the specificity of the autoAb elicited in mice infected with MHV we tested hepatic extracts from human beings and different animals (Fig. 1B). The mouse sera reacted with the 40‐kDa protein in human liver as well as in sheep and rat liver, but not with hepatic extracts from silver side fish.
2.2 Purification and identification of the 40‐kDa autoantigen
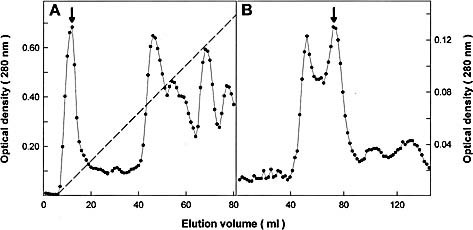
The 40‐kDa protein was first purified from mouse liver extracts by chromatography on DEAE‐cellulose and Sephadex G‐100 (Fig. 2), followed by SDS‐PAGE in 10–12% gradient of monomer concentration (Fig. 3). The 40‐kDa band was excised from the membrane and submitted to amino acid sequence analysis. Since the N terminus was found to be blocked (data not shown), the protein was digested in‐gel 5, 6 and the resulting peptides were separated by reverse‐phase HPLC. Sequence determination of three peptides indicated 100% identity with mouse liver FAH (EC 3.7.1.2) (Fig. 4).
Figure 2.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Purification of the mouse liver 40‐kDa protein recognized by serum from MHV‐infected mice. The arrows indicate the fractions containing the protein. (A) DEAE‐cellulose chromatography of mouse liver 100,000×g supernatant. Elution was done in buffer Tris‐HCl 20 mM, pH 8.0, and a 0–0.3 M NaCl gradient (dotted line). (B) The material isolated from the fraction indicated in (A) was chromatographied in a Sephadex G‐100 column equilibrated with Tris 5 mM, NaCl 150 mM, pH 7.4.
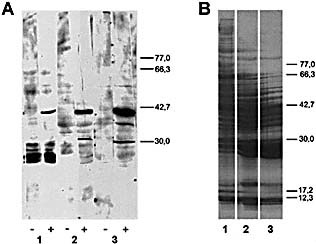
Figure 3.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Western blot analysis (A) and Coomasie blue staining (B) of different mouse liver fractions. According with Sect. 4 and Fig. 2: liver supernatant obtained after 100,000×g centrifugation (lane 1), DEAE‐cellulose (lane 2) or Sephadex G‐100 chromatography (lane 3). SDS‐PAGE was performed in 14×16 cm gels using a gradient of 10‐12.5% of polyacrylamide. Transferred proteins were incubated with serum from non‐infected (‐) or MHV‐infected (+) mice. The positions of molecular mass markers (kDa) are shown at right.
Figure 4.

This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Sequence alignment of mouse (m) and rat (r) liver fumarylacetoacetate hydrolase. Different residues between both proteins are shadowed in the rat sequence. Peptides generated by trypsin‐cleavage and submitted to amino acid sequencing are shown in bold.
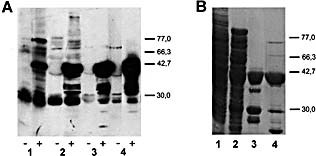
As liver FAH from mouse and rat shared 97% of identity according to Swiss‐Prot databases, we purified the enzyme from rat livers following the method of Hsiang et al. 7. In‐gel digestion and peptide sequencing of the protein isolated from lane 4 (Fig. 5) confirmed that the 40‐kDa protein recognized by sera from MHV‐infected mice was liver FAH (Fig. 4).
Figure 5.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Western blot analysis (A) and Coomasie blue staining (B) of different fractions from rat liver FAH purification. SDS‐PAGE was performed in 5×9 cm gels using 10% (w/v) of polyacrylamide. According with Sect. 4: liver supernatant obtained after 100,000×g centrifugation (lane 1), Fraction I (lane 2), Fraction II (lane 3) and Fraction III (lane 4). Transferred proteins were incubated with serum from non‐infected (–) or MHV‐infected (+) mice. The positions of molecular mass markers (kDa) are shown at right.
It should be noted that Western‐blotting of the purification fractions revealed that other proteins reacted with the Ab elicited by MHV infection, mainly a protein of approximately 38 kDa (see Fig. 3A, lane 3 and Fig. 5A, lane 4). After in‐gel digestion, four peptides from this protein were isolated and sequenced. Results indicated that the 38‐kDa autoantigen was rat alcohol dehydrogenase (ADH, aldehyde reductase, EC 1.1.1.2) (data not shown). Accordingly, Western‐blot of fractions II and III showed two reactive bands of molecular mases lower than that of the 38‐kDa protein (Fig. 5A, lanes 3 and 4), but these proteins were in trace amounts because their presence was not detected in the Coomasie‐stained gel (Fig. 5B, lanes 3 and 4).
2.3 Relationship between anti‐FAH Ab and hypergammaglobulinemia
The time course of appearance of Ab to liver FAH was studied in individual CBA/Ht mice (Fig. 6). Total IgG values were significantly higher than controls at days 10 and 13 after virus infection and autoAb were detected at day 10. At day 56 the level of mouse serum IgG came back to normal although anti‐enzyme Ab were still present (Fig. 6).
Figure 6.
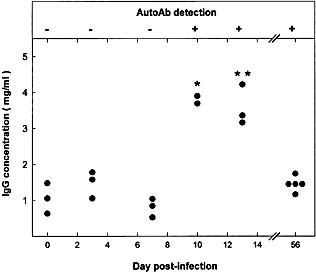
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Time course of Ab to the 40‐kDa liver protein and plasma Ig level in MHV‐infected CBA/Ht mice. Upper panel: Western‐blot reactivity of serum from MHV‐infected CBA/Ht with the 40‐kDa liver protein (see Fig. 1 for details); (+) means that serum from all animals tested reacted with the mouse liver 40‐kDa protein; (–) means absence of reactivity. Lower panel: Total IgG concentration of the corresponding sera was determined as indicated in Sect. 4. Values correspond to individual mice. Results were subject to statistical analysis by using the Mann‐Whitney U‐test. *p<0.1, **p<0.05.
Furthermore, since it has been reported that MHV induced the production of IgG2a 4, we investigated whether this IgG subclass was responsible for the binding to FAH in Western‐blots. Besides IgG2a subclass, IgG1 and IgG2b Ab were found to bind to the enzyme (Table 1).
Table 1.
Immunoglobulins binding to liver FAH in serum from MHV‐infected mice
|
Bound Iga) |
CBA/Ht miceb) |
BALB/c micec) |
|---|---|---|
|
IgG1 |
+ |
+ |
|
IgG2a |
++ |
++ |
|
IgG2b |
– |
+ |
|
IgG3 |
– |
– |
|
IgM |
– |
– |
a) Bound Ig to the liver 40‐kDa protein were detected by Western blot experiments as described in Sect.4.
b) Pooled serum from six CBA/Ht mice after 5–7 weeks of MHV‐infection.
c) Pooled serum from 6 BALB/c mice after 8 weeks of MHV‐infection.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
By the same token, hypergammaglobulinemic sera from LDV‐infected mice were tested for their ability to recognize the purified enzyme. IgG control values were 1.0 ± 0.2 mg/ml whereas after 12 weeks of LDV infection, immunoglobulin levels in sera from BALB/c and CBA/Ht mice raised to 10.0 ± 5.5 and 13.0 ± 3.6 mg/ml, respectively. Results from Western blot experiments carried out as indicated in Fig. 5, lane 3, indicated the absence of anti‐FAH Ab (data not shown), thus discarding a mere augmentation of serum IgG as the source of the autoAb detected in MHV‐infected mice.
2.4 Native versus cryptic epitopes
To check whether the autoAb occurring in MHV‐infected mice were directed to native or to cryptic epitopes, we performed competition Western blots, a procedure previously validated by two different anti‐human growth hormone monoclonal antibodies 8. Representative experiments carried out with serum from MHV‐infected CBA/ Ht mice indicated that simultaneous incubation of Ab with either mouse liver extracts or with the semi‐purified rat liver protein did not affect the Ab binding to the insolubilized antigen (Fig. 7).
Figure 7.
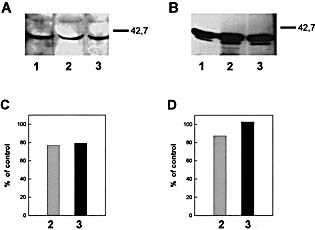
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Western blot competition assay to explore the reactivity of serum from MHV‐infected CBA/Ht mice with liver tissue antigens insolubilized or in solution. (A) Liver lysate (100 μg of protein) was prepared from non‐infected mice and run in SDS‐PAGE using 10% gels, transferred onto nitrocellulose sheets and incubated with 1:100 diluted serum from MHV‐infected CBA/Ht mouse previously incubated overnight at 4°C in the absence (lane 1) or in the presence of liver lysate: 0.5 mg/ml (lane 2) or 1 mg/ml protein (lane 3). (B) 5 μg of the semi‐purified 40‐kDa rat liver protein (Fraction II, see Sect 4 and Fig. 5) was submitted to competition Western blot as indicated in (A) for liver lysate. Lane 1, no competitor, lanes 2 and 3, 10 and 300 μg/ml of the semi‐purified protein (Fraction II), respectively. (C and D) Densitometric analysis of the corresponding 40‐kDa bands revealed by serum from MHV‐infected animals. Control means the band intensity in lane 1 for each assay.
2.5 AutoAb to FAH and metabolic disorders
Hepatorenal tyrosinemia is an autosomal recessive disease caused by deficiency of FAH 9. Symptoms are highly variable and include acute liver failure, cirrhosis, hepatocellular carcinoma, renal Fanconi syndrome, glomerulosclerosis, and crises of peripheral neuropathy 9. Hypertyrosinemia is present in untreated patients.
Mice infected with MHV elicited Ab towards liver FAH but did not exhibit tyrosinemia symptoms. Analysis of plasmatic amino acid concentration indicated that serum from MHV‐infected mice had similar values to those showed by control animals (data not shown).
3 Discussion
Viruses are highly suspected to be involved in the generation of autoimmune disorders 10. It has been proposed that these infectious agents trigger an autoimmune humoral response by diverse mechanisms, including polyclonal B lymphocyte activation, antigenic mimicry, modification of self‐antigen, or enhancement of major histocompatibility complex molecule expression 11, 12. Thus, since MHV‐A59 causes polyclonal activation, IgG2a restriction and modification of the isotypic distribution of Ab raised against an unrelated protein antigen 11, 13, we wondered whether this pathogen could induce an autoimmune response.
We detected autoAb binding to a protein of apparent molecular mass of 40 kDa present in mouse kidney and in the liver of mice, rats, sheep and human beings. The autoAb were found in CBA/Ht, BALB/c or 129/Sv mice 10 days after the MHV infection. The 40‐kDa protein was isolated from mouse liver and partially sequenced. Although only a few peptides representing 10% of the sequence were identified, data indicated 100% of identity with mouse liver FAH, a soluble cytosolic enzyme, also present in kidney, that mediates the hydrolytic formation of fumarate and acetoacetate 9. The enzyme was then purified from rat liver and the protein analysis confirmed the previous results. Actually, the addition of peptides identified for both the mouse and rat proteins account for 30% of the FAH sequence. Since the enzyme is a homodimer 9, our results are compatible with the identification of a subunit of a molecular weight of 46,100.
Western blot experiments showed that sera from MHV‐infected mice were able to recognize the presence of FAH in liver from humans, sheep, rats and mice but not in liver from the fish Odontesthes bonariensis. To the best of our knowledge, the enzyme has been detected and even purified from the liver of some mammals 9, but its presence in fishes has not yet been reported 14.
A second autoantigen was detected in final steps of liver FAH purification. This protein of apparent 38 kDa was identified as rat alcohol dehydrogenase class I (ADH1), a cytosolic, dimeric, NAD‐dependent enzyme with a subunit molecular mass of 36.4 kDa 15.
There was no correlation between the IgG titers and the presence of autoAb to liver FAH in CBA/Ht mice. Furthermore, hypergammaglobulinemic sera from LDV‐infected mice did not recognize the enzyme, suggesting that the autoAb production was not related to the nonspecific polyclonal activation of B lymphocytes produced after viral infection.
As reported in Sect. 4, mice were immunized with extracts of either mouse or human liver. Results indicated that the animals did not develop Ab neither to the 40‐kDa nor to the 38‐kDa liver proteins (data not shown), thus discarding the recognition of a self‐antigen release as source of the autoAb.
Molecular mimicry between MHV proteins, FAH and ADH seems the most probable mechanism. Thus, the search for homologies between the various proteins indicated the existence of a sequence sharedby human, mouse or rat FAH (residues 14–22), rat ADH (residues 110–117) and amino‐acids 103–111 from the MHV spike protein S. The homology between the several peptides is similar to that found between a Semliki Forest virus peptide and a myelin oligodendrocyte glycoprotein 16 and Chlamydia peptides with a heart muscle‐specific protein 17. In the former case a viral peptide is responsible of an autoimmune demyelinating disease in mice 16, whereas heart disease was induced by a Chlamydia‐derived sequence 17, indicating that molecular mimicry could be involved in the induction of autoimmune disease by pathogens.
The absence of pathological effects of the autoAb described in this work could be related to the fact that they are directed to cryptic antigenic determinants, the characteristics shared with Ab from the natural repertoire 18, 19. Accordingly, we have reported that autoAb with a wide variety of specificities were detected in the serum of CBA/Ht mice but not in BALB/c when the animals were infected with LDV. Most autoAb were found to bind to autoantigenic proteins only when the latter were denatured, and no specific tissue lesions were detected 8.
Although MHV, JHM strain, induced a retinal disease after intravitreal and intraaqueal inoculations 20, the autoAb elicited by MHV‐A59 did not show pathological effects. Theobservation that the pathogenicity of autoAb depends on their microenviroment, including the microbiological status of the host 12, 21, 22, opens new ways to explore the relationship between infections and autoimmune disease using the experimental model described in this paper.
4 Materials and methods
4.1 Mice
Female CBA/Ht, BALB/c and 129/Sv mice were bred in isolators at the Ludwig Institute for Cancer Research (Brussels Branch) by Dr. G. Warnier and were used for experiments at the age of 8 to 10weeks. Their microbiological status was described previously 23.
4.2 Viral infection
Mice were inoculated intraperitoneally with either 50 tissue culture infectious doses (TCID50 ) of MHV‐A59, grown in NCTC1469 cells 4, or with 2 × 10750% infectious doses (ID50) of LDV (Riley strain; from the American Type Culture Collection, Rockville, MD) 11, 12. Efficiency of MHV infection was checked by testing antiviral Ab by ELISA 4 whereas LDV infection was assessed by determination of lactate dehydrogenase plasma activity 8.
4.3 Immunoglobulin determinations
For total IgG assay in mouse serum, microplates (Nunc Maxi‐Sorb) were coated with 100 μl of phosphate buffer saline (PBS) containing a 1:500 diluted rabbit antiserum directed against mouseIg. The plates were blocked 2 h at 37°C with 0.01 M Tris, 0.13 M NaCl, pH 7.4 (TMS) containing 5% of non‐fat milk (TMS‐M) and were incubated with serial dilutions of mouse serum in the same medium. After overnight incubation at room temperature and washing with PBS containing 0.125 ml of Tween 20/l (PBS‐Tween), the plates were incubated 2 h at 37°C with peroxidase‐labeled donkey anti‐mouse IgG Ab. These donkey IgG Ab (Jackson Immunoresearch Laboratories, Inc., West Grove, PA) were used at a 1:10,000 dilution in TMS‐M. The amount of IgG in each serum was calculated from paralleled dose response curves obtained from test samples and Protein A‐Sepharose purified mouse IgG of known concentration as a reference.
4.4 Preparation of organ lysates
The spleen, brain, heart, liver, muscle, lungs and kidneys from non‐infected CBA/Ht, BALB/c or 129/Sv mice were removed, soaked in chilled PBS and ground in a Dounce homogenizer at 4°C with 5 vol. of PBS containing 1 mM PMSF. The homogenates were centrifuged for 15 min at 400 × g and the clarified extracts kept at –20°C until used. A sample of each suspension was solubilized by heating for 30 min at 100°C in 1 M NaOH and protein concentration was determined by the method of Lowry et al. 24.
A biopsy from human liver as well as livers from Wistar rats, a 5‐year‐old female Corridale sheep and silver side fish (Odontesthes bonariensis) were homogenized and processed as indicated above for mouse tissues.
4.5 Western blot analysis
Each organ extract (30–100 μg of protein) or semi‐purified proteins (2–10 μg) were subjected to SDS‐PAGE using 10% gels (w/v) for small (5×9 cm) gels and a gradient of 10‐12.5% ofpolyacrylamide was used for the biggest (14×16 cm) 25 and then transferred onto nitrocellulose sheets (Amersham, GB). After reversible staining with Ponceau S to check satisfactory transfer, nonspecific Ab‐binding sites were blocked by incubating the sheets with 10% non‐fat milk in 0.03 M Tris, 0.14 M NaCl, pH 8.0 (TBS) for 1 h at room temperature with shaking. The strips were then incubated for 1 h at 20° C with an Ab dilution in TBS, i.e. either control or MHV‐infected mouse serum. After several washings with TBS containing 0.1% Tween 20, bound Ab were revealed with peroxidase labeled donkey IgG anti‐mouse IgG and ECL reagents (Amersham, GB). The apparent molecular mass (kDa) of the detected bands was always determined using a wide range proteinstandard (BDH Laboratory Supplies Poole BH15 1TD,GB).
To determine the subclass of bound immunoglobulins, the strips were incubated first with mouse Ab and then 1 h at 20° C with goat IgG anti‐mouse IgM, IgA, IgG1, IgG2a, IgG2b and IgG3 (Sigma Chemical Co, St. Louis, MO). Bound Ig were revealed with peroxidase‐labeled rabbit Ig anti‐goat IgG (Organon Tekuika corporation, Cappel Research Products, Durham, Germany) and ECL reagents.
To perform competition experiments the antibodies were incubated overnight at 4° C with different concentrations of liver lysate or the semi‐purified 40‐kDa protein prior to incubation with the antigens insolubilized on nitrocellulose sheets. The intensity of the bands was quantified by densitometric scan of the autoradiograms and the results expressed as percent of control, i.e. band intensity in the absence of competitor.
4.6 Liver membranes and supernatant preparation
Mouse livers were homogenized in 5 vol. (v/w) of chilled 0.3 M sucrose, 5 mM Tris/HCl buffer containing 0.5 mM CaCl2, 1 U/ml of trypsin inhibitor and 1 mM PMSF, pH 7.4. After centrifugation at 10,000×g for 20 min and then at 100,000×g for 1 h, the pellet was resuspended in 25 mM Tris/HCl buffer, pH 7.4. Protein concentration in supernatant and in the pellet suspension was determined by the method of Lowry et al. 24. Each fraction was frozen at –20°C until being used.
4.7 Purification of the mouse liver 40‐kDa protein recognized by the autoAb
Two milliliters of liver 100,000×g supernatant were subject to chromatography in a DEAE‐cellulose column (1×25 cm) previously equilibrated with 20 mM Tris, 1 U/ml of trypsin inhibitor and 1 mM PMSF, pH 8.0. Proteins were eluted at a flow rate of 0.4 ml/min by using a continuous 0‐0.3 M NaCl gradient. Protein concentration in the effluent fractions (1 ml) was determined at 280 nm. Main fractions were concentrated and then submitted to Western blot analysis.
Effluent fractions containing the 40‐kDa protein recognized by serum from MHV‐infected mice were pooled and concentrated and then applied to a Sephadex G‐100 column (1.6×70 cm) equilibrated with 5 mM Tris, 150 mM NaCl, 1 U/ml of trypsin inhibitor and 1 mM PMSF, pH 7.4. Elution was carried out at a flow rate of 16 ml/h with the buffer cited before and the protein concentration in theeffluent fractions (1.8 ml) was determined at 280 nm. Main fractions were concentrated and analyzed by Western blot.
4.8 Rat liver FAH purification
Enzyme purification was carried out from Wistar rats livers following essentially the method reported by Hsiang et al. 7 to isolate bovine liver fumarylacetoacetate hydrolase. The various steps were conducted at 0 – 4° C and all buffers contained 1 U/ml of trypsin inhibitor and 1 mM PMSF.
Rat liver supernatants were prepared as described above for mouse liver and ethyl alcohol was added as to obtain a final concentration of 50% ethanol. This mixture was allowed to stand overnight at 4° C and then centrifuged at 16,300×g for 5 min. The supernatant was mixed with 95% ethyl alcohol to yield a final alcohol concentration of 70%. After overnight at 4° C the precipitatedenzyme was packed by centrifugation at 16,300×g for 5 min.
The pellet was resuspended in 25 mM phosphate buffer pH 7.2 and stirred for 30 min. The supernatant fluid was recovered after centrifugation of the suspension at 16,300×g for 10 min and solid ammonium sulfate was added as to obtain a final salt concentration of 40%. After 1 h at 4° C and centrifugation at 16,300×g for 10 min the pellet was resuspended in 20 mM Tris/HCl buffer.To specify the results obtained in Western experiments this last preparation is named Fraction I.
An aliquot of Fraction I was chromatographied in a MonoQ HR 10/10 column (Pharmacia LKB Biotechnology) equilibrated with 20 mM Tris pH 8.0. Proteins were eluted at a flow rate of 1.0 ml/min byusing a continuous 0 – 0.2 M NaCl gradient. Protein concentration in the effluent fractions (1 ml) was determined at 280 nm (Akta purifier 10, Amersham Pharmacia Biotechnology). Main fractions were concentrated and then submitted to Western blot analysis. The second peak eluted from the column is called Fraction II.
As indicated before for mouse liver, fractions containing the 40‐kDa protein recognized by serum from MHV‐infected mice were pooled and chromatographied in a Sephadex G‐100 column. The main positive fraction was named Fraction III.
4.9 In‐gel digestion of the 40‐kDa liver protein for internal sequence analysis
The positive fractions eluted from the Sephadex column were pooled, concentrated, and then subjected to SDS‐PAGE in a 10 × 15 cm Hoefer SE 600 Dual Cooled Vertical Slab Cell using a 10–12%gradient of monomer concentration. The gel was stained with Coomasie Blue and the specific 40‐kDa band was excised and submitted to in‐gel digestion with trypsin following the procedure of Rosenfeld et al. 5 as modified by Hellman et al. 6.
4.10 Reverse‐phase HPLC peptide purification
Peptides eluted from the polyacrilamide matrices were fractionated on a Brownlee C18 column (220×2.1 mm) equilibrated with 0.1 % trifluoracetic acid (TFA). Elution was performed at a flowrate of 150 μl/min with a 0–90% acetonitrile (80%) and TFA (0.08%) gradient over a period of 75 min. Peaks were manually collected.
Amino acid sequencing was carried out on an Applied Biosystems 477A protein sequencer equipped with an on‐line 120A phenylthiohydantoin analyzer. The amino acid sequence was screened for homologies with known sequences of the Swiss‐Prot using the BLAST Program.
4.11 Amino acid analysis
Mouse plasma amino acid composition was determined using an Applied Biosystem amino acid analyzer 422A that performs derivatization and HPLC of the phenylthio‐hydantoin‐amino acids automatically.
4.12 Immunization of mice with liver extracts
Three‐month‐old BALB/c mice were immunized subcutaneously on days 0, 10 and 20 with either human or mouse liver lysate (300 μg of total protein) emulsifed in an equal volume of complete Freund's adjuvant (Sigma Chemical Co, St. Louis, MO). Animals were bled 10 days after the last inoculation and serum was analyzed for binding to the 40‐kDa liver protein in Western blot experiments.
4.13 Alignment of peptide sequences
LALIGN program was utilized to find multiple matching subsegments in two protein sequences. This program is part of the FASTA package of sequence analysis program that is available from ftp.virginia.edu.
Acknowledgements
The authors are indebted to Drs. Pierre L. Masson (ICP, Brussels, Belgium) and Leonor P. Roguin (IQUIFIB, Buenos Aires, Argentina) for helpful discussions and critical revision of the manuscript and to M.D. Gonzalez for expert technical assistance. Amino acid sequence analysis was performed at the LANAIS‐PRO (National Protein Sequencing Facility, UBA‐CONICET, Buenos Aires, Argentina) and we thank S.B. Linskens and E.V. Dacci for skilful technical work. This work was supported by grants from Fundación Antorchas, CONICET, FONCYT and Universidad deBuenos Aires, Argentina, and Fonds National de la Recherche Scientifique (FNRS), Fonds de la Recherche Scientifique Médicale (FRSM), the State‐Prime Minister's Office – S.S.T.C. (interuniversity attraction poles, grant n°44) and the French Community (concerted actions, grant n° 99/04–239), Belgium. J.‐P. Coutelier is a research director with the FRNS and K.A. Gómez is a "Societé Générale de Belgique" fellow.
Footnotes
WILEY‐VCH
WILEY‐VCH
WILEY‐VCH
WILEY‐VCH
WILEY‐VCH
WILEY‐VCH
WILEY‐VCH
References
- 1. Kraft, L. M., Viral diseases of the digestive system. In Foster, H. L., Small, J. D. and Fox, J. G. (Eds.). The mouse in biomedical research. II Diseases. Academic Press New York1982, pp 159–191. [Google Scholar]
- 2. Godfraind, C., Havaux, N., Holmes, K. V. and Coutelier, J.‐P., Role of virus receptor‐bearing endothelial cells of the blood‐brain barrier in preventing the spread of mouse hepatitis virus‐A59 into the central nervous system. J. NeuroVirology 1997. 3: 428–434. [DOI] [PubMed] [Google Scholar]
- 3. Godfraind, C. and Coutelier, J.‐P., Morphological analysis of mouse hepatitis virus A59‐induced pathology with regard to viral receptor expression. Histol. Histopathol .1998. 13: 181–199. [DOI] [PubMed] [Google Scholar]
- 4. Lardans, V., Godfraind, C., van der Logt, J. T. M., Heessen, F. W. A., Gonzalez, M.‐D. and Coutelier, J.‐P., Polyclonal B lymphocyte activation induced by mouse hepatitis virus A59 infection. J. Gen. Virol .1996. 77: 1005–1009. [DOI] [PubMed] [Google Scholar]
- 5. Rosenfeld, J., Capdevielle, J., Guillemot, J. C. and Ferrara, P., In‐gel digestion of protein for internal sequence analysis after one‐ or two‐dimensional electrophoresis. Anal. Biochem .1992. 203: 173–179. [DOI] [PubMed] [Google Scholar]
- 6. Hellman, U., Wernsted, C., Goñez, J. and Heldin, C. H., Improvementof an in‐gel digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem .1995. 224: 451–455. [DOI] [PubMed] [Google Scholar]
- 7. Hsiang, H. H., Sim, S. S., Mahuran, D. J. and Schmidt, E. Jr., Purification and properties of a diketo acid hydrolase from beef liver. Biochemistry 1972. 11: 2098–2102. [DOI] [PubMed] [Google Scholar]
- 8. Gómez, K. A., Coutelier, J,‐P., Mathieu, P. A., Lustig, L. and Retegui, L. A., Autoantibodies to cryptic epitopes elicited by infection with lactate dehydrogenase‐elevating virus. Scand. J. Immunol .2000. 51: 447–453. [DOI] [PubMed] [Google Scholar]
- 9. Mitchell, G. A., Lambert, M. and Tanguay, R. M., Hypertyrosinemia. In Scriver, C.R. Beaudet, A.L., Sly, W. and Valle, D (Eds.). The Metabolic and Molecular Bases of Inherited Disease. McGraw‐Hill, Inc. NY.1993. pp 1077–1106.
- 10. Cohen, A. D. and Shoenfeld, Y., The viral‐autoimmunity relationship. Viral Immunology 1995. 8: 1–9. [DOI] [PubMed] [Google Scholar]
- 11. Coutelier, J.‐P., Coulie, P. G., Wauters, P., Heremans, H.and van der Logt, J. T. M., In vivo polyclonal B‐lymphocyte activation elicited by murine viruses. J. Virol .1990. 64: 5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rose, N. R., The role of infection in the pathogenesis of autoimmune disease. Semin. Immunol. 1998. 10: 5–13. [DOI] [PubMed] [Google Scholar]
- 13. Coutelier, J.‐P., van der Logt, J. T. M., Heessen, F. W. A., Warnier, G. and Van Snick, J., IgG2a restriction of murine antibodies elicited by viral infections. J. Exp. Med .1987. 165:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hochachka, P. W. and Mommsen, T. P., (Eds.) Biochemistry and molecular biology of fishes. Elsevier, New York1995.
- 15. Duester, G., Farrés, J., Felder, M. R., Holmes, R. S., Höög, J.‐O., Parés, X., Plapp, B. V., Yin, S.‐J. and Jörnvall, H., Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 1999. 58: 389–395. [DOI] [PubMed] [Google Scholar]
- 16. Mokhtarian, F., Zhang, Z., Shi, Y., Gonzales, E. and Sobel, R.A., Molecular mimicry between a viral peptide and a myelin oligodendrocyte glycoprotein peptide induces autoimmune demyelinating disease in mice. J. Neuroimmunol .1999. 95: 43–54. [DOI] [PubMed] [Google Scholar]
- 17. Bachmaier, K., Neu, N., de la Maza, L.M., Pal, S., Hessel, A. and Penninger, J.M., Chlamydia infections and heart disease linked through antigenic mimicry. Science 1999. 283: 1335–1339. [DOI] [PubMed] [Google Scholar]
- 18. Coutinho, A., Kazatchkine, M.D. and Avrameas, S., Natural autoantibodies. Current Opin. Immunol. 1995. 7: 812–818. [DOI] [PubMed] [Google Scholar]
- 19. Dighiero, G. and Rose, N.R., Critical self‐epitopes are key to the understanding of self‐tolerance and autoimmunity. Immunol. Today 1999. 20: 423–428. [DOI] [PubMed] [Google Scholar]
- 20. Hooks, J.J., Percopo, C., Wang, Y., and Detrick, B., Retina and retinal pigment epithelial cell autoantibodies are produced during murine coronavirus retinopathy. J. Immunol .1993. 151: 3381–3389. [PubMed] [Google Scholar]
- 21. Stevenson, F.K. and Natvig, J., Autoantibodies revealed: the role of B cells in autoimmune disease. Immunol. Today 1999. 20: 296–298. [DOI] [PubMed] [Google Scholar]
- 22. Meite, M., Léonard, S., El Azami El Idrissi, M., Izui, S., Masson, P. L. and Coutelier, J‐P., Exacerbation of autoantibody‐mediated hemolytic anemia by viral infection. J. Virol .2000. 74: 6045–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coutelier, J‐P., van der Logt, J.T.M., Heessen, F.W.A., Warnier, G. and Van Snick, J., Rheumatoid factor production in 129/Sv mice: involvement of an intestinal infectious agent. J. Immunol .1986. 137: 337–340 [PubMed] [Google Scholar]
- 24. Lowry, O.H., Rosebrough, N.J., Farr, A.L. and Randall, R.J. Proteinmeasurements with the folin phenol reagent. J. Biol. Chem. 1951. 193: 265–275. [PubMed] [Google Scholar]
- 25. Laemmli, U.K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970. 227: 680–685. [DOI] [PubMed] [Google Scholar]


