Abstract
The innate immune system initiates immune responses by pattern‐recognition receptors (PRR). Virus‐derived nucleic acids are sensed by the retinoic acid‐inducible gene I (RIG‐I)‐like receptor (RLR) family and the toll‐like receptor (TLR) family as well as the DNA sensor cyclic GMP‐AMP (cGAMP) synthase (cGAS). These receptors activate IRF3/7 and NF‐κB signaling pathways to induce the expression of type I interferons (IFNs) and other cytokines firing antiviral responses within the cell. However, to achieve a favorable outcome for the host, a balanced production of IFNs and activation of antiviral responses is required. Post‐translational modifications (PTMs), such as the covalent linkage of functional groups to amino acid chains, are crucial for this immune homeostasis in antiviral responses. Canonical PTMs including phosphorylation and ubiquitination have been extensively studied and other PTMs such as methylation, acetylation, SUMOylation, ADP‐ribosylation and glutamylation are being increasingly implicated in antiviral innate immunity. Here we summarize our recent understanding of the most important PTMs regulating the antiviral innate immune response, and their role in virus‐related immune pathogenesis.
Keywords: Antiviral immunity, Interferons, Phosphorylation, Post‐translational modifications, PRR, Ubiquitination
Interferon is the central cytokine of antiviral innate immunity. Cells initiate interferon production by a list of pattern recognition receptor (PRR) pathways. In this review, we focus on that how cells tightly control interferon production with post‐translational modifications (PTMs) of PRR signaling molecules.2
Introduction
Infectious diseases, especially virus infections, are still a serious threat to humanity and the host innate immune system represents a critical defense against invading viruses. Host cells can initiate these innate immune responses by detecting viral DNA and RNA with a set of pattern recognition receptors (PRRs) including the toll‐like receptor (TLR) family and the retinoic acid‐inducible gene I (RIG‐I)‐like receptor (RLR) family, as well as cytosolic DNA sensors such as cyclic GMP‐AMP (cGAMP) synthase (cGAS), IFI16 and DDX41 1, 2. After recognition of viral nucleic acids, these PRRs trigger the production of proinflammatory cytokines, chemokines and type I interferons (IFNs), which subsequently induce synthesis of antiviral proteins, death of infected cells and activation of the adaptive immune response 3, 4. These antiviral signals must be spatially and temporally orchestrated to achieve an optimal outcome for the host and much attention has been raised to understanding the signaling pathways and regulatory factors in the antiviral innate immunity.
Toll‐like receptors, including TLR3, TLR7, TLR8 and TLR9, sense endosomal nucleic acids derived from the enclosed microbes and infected apoptotic cells. While TLR9 detects unmethylated CpG DNA species, TLR3 and TLR7/8 recognize double‐stranded RNA (dsRNA) and single‐stranded RNA (ssRNA), respectively 5 (Fig. 1). Following ligand binding, the TLRs form a signaling platform in which distinct Toll/interleukin‐1 receptor (TIR) domain‐containing adaptors are engaged. For instance, TLR3 signals via TIR‐domain‐containing adaptor protein inducing interferon beta (TRIF), and TLR7/8/9 rely on myeloid differentiation factor‐88 (MyD88) 6. For the TRIF‐dependent pathway, the ubiquitin E3 ligase TNF receptor‐associated factor 3(TRAF3) is recruited and hence activates TANK‐binding kinase‐1 (TBK1) and Inhibitor‐κB kinase ε (IKKε) 7. Activated TBK1 and IKKε then phosphorylate IFN regulatory factor (IRF) transcription factors IRF3 and IRF7 to drive the expression of type I IFNs 8. For the MyD88‐dependent pathway, TRAF6 is engaged to the MyD88 signal platform, leading to the activation of the kinase complex composed of IKKα and IKKβ. Activated IKK complex induces NF‐κB activation, which induces the expression of proinflammatory cytokines such as TNF, IL‐6 and IL‐12 6 (Fig. 1). In addition, Myd88 also facilitates IFN‐α production by promoting IKKα‐dependent IRF7 phosphorylation, which is particularly important for the antiviral activities of plasmacytoid dendritic cells 9, 10.
Figure 1.
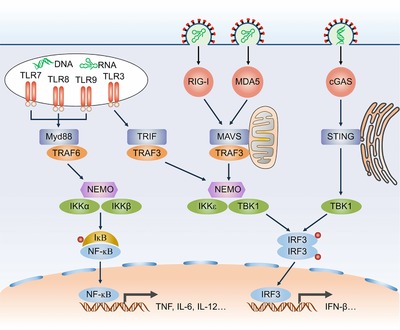
Antiviral signaling pathways. PRRs (red) are activated by endosomal and cytosolic viral RNA and DNA species. TLR7, TLR8 and TLR9 recruit MyD88 and MyD88 in turn activates TRAF6. TLR3 recruits TRIF and subsequently activates TRAF3. TRAF6 and TRAF3 then induce the formation of NEMO‐IKKα/β and NEMO‐IKKε/TBK1 complex respectively. IKKα/β activate the transcription factor NF‐κB and IKKε/TBK1 phosphorylates the transcription factor IRF3. NF‐κB and IRF3 then translocate into the nucleus and drive proinflammatory cytokines and type I IFNs expression. For the RLR pathway, RIG‐I and MDA5 activate TRAF3‐TBK1 axis through the mitochondria‐located adaptor MAVS. For the cGAS pathway, cGAS recognize cytosolic DNA and activate the ER‐located adaptor STING, which then translocates to and activates TBK1. Red circle, phosphorylation.
Aside from endosomal sensing by TLRs, viral RNA in the cytosol is also recognized by RLRs, including Retinoic acid‐inducible gene I (RIG‐I), melanoma differentiation‐associated protein 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) 11. Notably, the RLRs initiate antiviral immune responses in most cell types, which is in contrast to TLR3 and TLR7, which mainly show phagocyte‐restricted expression 12, 13. Upon sensing pathogen‐specific molecular features of viral RNA such as 5′‐triphosphate (5′‐ppp) for RIG‐I and long double‐stranded segments for MDA5, these RLRs translocate to mitochondria and interact with the mitochondrial antiviral‐signaling protein (MAVS) 14, 15 (Fig. 1). This interaction causes the aggregation of MAVS to form a huge prion‐like protein complex for TRAF3 and TRAF6 engaging, which transmit signals to TBK1‐IRF3 and IKKα/β‐ NF‐κB pathways 3, 16 (Fig. 1).
The immunostimulatory effect of foreign DNA was reported more than 50 years ago, but only recently the sensors that link foreign DNA to type I IFNs have been identified. Cytosolic viral DNA is mainly recognized by cyclic GMP‐AMP (cGAMP) synthase (cGAS) that contains a nucleotidyltransferase (NTase) domain. Following DNA binding, cGAS synthesizes a second messenger molecule, cyclic GMP‐AMP (cGAMP), which then activates the stimulator of interferon genes (STING) 17, 18. STING is an ER‐located adaptor protein that plays an essential role upstream of TBK1 in the cytosolic viral DNA sensing pathway 19. Although some other proteins such as IFI16, DDX41 and Mre11 are also reported to be receptors mediating DNA‐induced IFN‐β production in a STING‐dependent manner, only cGAS, which enzymatically generates cGAMP as a second messenger that activates STING, provides a clear molecular mechanism for DNA‐stimulated IFN‐β production 20 (Fig. 1).
Post‐translational modifications (PTMs), involving the covalent linkage of new functional groups to amino acid chains, greatly expand the function of proteins and thus play crucial roles in numerous physiological activities. PTMs control immune responses via regulating protein folding, stability, location and interaction with other molecules 21, 22. The best characterized PTMs, phosphorylation and ubiquitination, as well as other PTMs such as SUMOylation, methylation and acetylation, have been reported to control antiviral signaling via reversible post‐translational modification of virus sensors and downstream signaling molecules 21, 23. Moreover, emerging studies have shown that different PTMs mutually interact, indicating a precise and elaborate PTMs’ regulatory network in the antiviral immunity 24, 25. In this review, we focus on our recent knowledge how individual PTMs regulate antiviral innate immune responses and their functions in virus‐related immune pathogenesis and disorders.
Phosphorylation
In eukaryote, phosphorylation is the kinase‐catalyzed conjugation of a phosphate group to the serine (Ser), threonine (Thr) or tyrosine (Tyr) residues of proteins. In this process, introducing a single phosphoryl group with a ‐2 charge at physiological pH always results in significant protein conformation change, which can influence protein properties and/or create binding motifs for other molecules. Nevertheless, this rigid PTM can be reversed through enzyme‐catalyzed hydrolysis by specific phosphatases 26.
In the TLR pathway, Bruton's tyrosine kinase (BTK), EGFR and Src have been shown to phosphorylate tyrosine residues in the cytoplasmic domain of TLR3 to enhance IFN‐β and proinflammatory cytokine production in macrophages in response to poly (I:C) and LPS stimulation 27, 28, 29. Moreover, in the late phase of infections, the iNOS/Src signal axis retains TLR3 activation through Src‐induced Tyr759 phosphorylation to support sustained IFN‐β transcription 30 (Fig. 2). However, whether and which kinases might phosphorylate TLR7, TLR8 or TLR9 is not known, though tyrosine phosphorylation of the TLR8 cytosolic domain is reported to be essential for downstream signal transduction based on a site‐directed mutation study 31. TRAF3 and TRAF6 possess important functions in antiviral signaling, but only recently have studies revealed that phosphorylation regulates TRAF3 and TRAF6 activation. In particular, the casein kinase CK1ε directly phosphorylates TRAF3 at Ser349 upon VSV and HSV infections in macrophages, which promotes TRAF3 K63‐linked ubiquitination and ultimately enhances IFN‐β production 32 (Fig. 2). However, TRAF6 was found to be suppressed by phosphorylation. Mechanistically, the mammalian STE20‐like protein kinase 4 (MST4) catalyzes TRAF6 Thr463 and Thr486 phosphorylation in THP1 cells, leading to attenuated oligomerization and ubiquitination of TRAF6, as well as reduced production of IL‐6 and TNF‐α 33 (Fig. 2). TBK1 undergoes self‐association and autophosphorylation at Ser172 after being recruited to the TRAF3 complex upon RNA virus infections, a process in which glycogen synthase kinase 3B (GSK3B) functions as an important physical partner 34, 35. By contrast, the phosphatase PPM1B removes TBK1 autophosphorylation to eliminate downstream IRF3 activation and IFN‐β production in HEK293T cells 36 (Fig. 2). In addition, the dual‐specificity tyrosine‐phosphorylation regulated kinase 2 (DYRK2) negatively regulates antiviral responses by inducing TBK1 Ser527 phosphorylation and subsequent K48‐linked ubiquitination and degradation in HEK293 cells 37 (Fig. 2). Activated IKKε and TBK1 directly phosphorylates IRF3, leading to IRF3 dimerization and nuclear translocation (Fig. 1). Multiple phosphorylated Ser and Thr residues on IRF3 were detected in HEK293T cells by in vitro kinase assay and Ser386 phosphorylation is suggested to be crucial for IRF3 activation 8, 38, 39. Recently, a study demonstrated that PTEN, a known tumor suppressive phosphatase, unexpectedly promotes antiviral immunity by reversing IRF3 phosphorylation at Ser97, thereby driving IRF3 nuclear transport 40 (Fig. 2). In addition, the DNA‐dependent protein kinase (DNA‐PK) binds with and phosphorylates IRF3 on Thr135, causing IRF3 to be retained in the nucleus and extending the half‐life of IRF3 transcriptional activity in HEC1B cells 41 (Fig. 2). Other signaling molecules, such as MAPKs, IKKα, IKKβ and IkBα, are also regulated by phosphorylation and we note that several excellent reviews on this subject have been published 42, 43.
Figure 2.
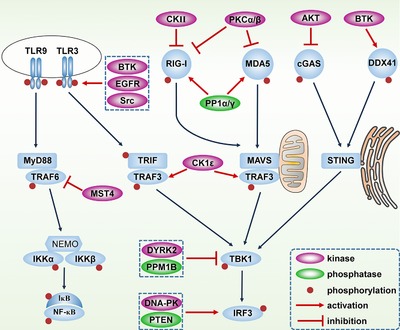
Regulation of antiviral signaling pathways by phosphorylation. For the TLR pathways, TLR3 cytoplasmic domain is phosphorylated and activated by BTK, EGFR and Src. TRAF6 is phosphorylated and inhibited by MST4 while TRAF3 is phosphorylated and activated by CK1ε. TBK1 is inhibited by DYRK2‐mediated phosphorylation and PPM1B‐mediated dephosphorylation. IRF3 is activated by DNA‐PK‐mediated phosphorylation and PTEN‐mediated dephosphorylation. For the RLR pathways, CKII‐ and PKCα/β‐mediated phosphorylation inhibit RIG‐I and MDA5 activation. PP1α/γ dephosphorylate and activate RIG‐I and MDA5. For the cytosolic DNA sensing pathways, BTK phosphorylates and activates DDX41 while AKT inhibits cGAS by phosphorylation.
Multiple phosphorylation‐dependent processes are also critical for type I IFN production in the RLR pathway. Phosphorylation of RIG‐I by protein kinase C α (PKCα) and PKCβ keeps RIG‐I silent in resting cells thus avoiding premature activation of RIG‐I signaling 44 (Fig. 2). The casein kinase II (CK II) constitutively phosphorylates RIG‐I and hence maintains a quiescent state of RIG‐I in HEK293T cells, however, when cells are stimulated with viral RNA, RIG‐I is rapidly dephosphorylated 45. Through phosphatase RNAi screening, PP1α and PP1γ were identified as primary phosphatases responsible for RIG‐I and MDA5 dephosphorylation in HEK293T and NHLF cells 46 (Fig. 2). Furthermore, it is reported that IKKε negatively regulates antiviral signaling by phosphorylating RIG‐I at Ser855 in HEK293T and MEF cells 47. MAVS occupies a central position within the RLR signaling cascade and Tyr9 on N‐terminal is proved as an essential phosphorylation site for the recruitment of TRAF3 and TRAF6, as well as IFN‐β production. However, a definite kinase responsible for Tyr9 phosphorylation has yet to be identified 48.
Among the cytosolic viral DNA signaling, both DDX41 and cGAS are regulated by phosphorylation. Phosphorylation of Tyr414 on DDX41 by BTK was found to be essential for dsDNA recognition and recruitment of STING in BMDMs, HEK293T and MEF cells 49 (Fig. 2). By contrast, Akt phosphorylates cGAS on Ser291 (for mouse) and Ser305 (for human), which inhibits cGAS enzymatic activity in synthesizing cGAMP, leading to a moderate immune responses for the host 50 (Fig. 2).
Notably, a general mechanism of IRF3 activation was recently proposed in which different adaptor proteins (including TRIF for TLR3, MAVS for RIG‐I and STING for cGAS) are phosphorylated by cognate downstream IKKε and TBK1 at conserved motif‐pLxIS (x for any amino acid, S is phosphorylation site) 51. These negatively charged motifs then attract the positive charged surface of IRF3, leading to efficient IRF3 phosphorylation by TBK1. Phosphorylated IRF3 then dissociates from the adaptors and forms a homo‐dimer that enters the nucleus to turn on type I interferon transcription 51. In line with this, protein phosphatase magnesium‐dependent 1A (PPM1A) was reported to dampen the phosphorylation cascade by catalytically dephosphorylating MAVS, STING and TBK1 52, 53. This model suggests a general phosphorylation‐mediated signal transduction in three distinct innate immune pathways, which is impressively similar to the activation mechanism of TGF‐β/Smad and JAK/STAT pathways, indicating an intrinsic unity of signal transduction. A detailed list about discussed phosphorylation of the target proteins, enzymes and signaling functions are included in Table 1.
Table 1.
Phosphorylation of PRR signaling pathways
| Signaling protein | Enzymes | Functions | Ref. |
|---|---|---|---|
| TLR3 | BTK | Phosphorylating TLR3 at Tyr759 and promoting the activation of MAPKs, NF‐κB and IRF3. | 27 |
| TLR3 | EGFR | Promoting the induction of antiviral genes | 28 |
| TLR3 | Src | Inducing and maintaining TLR3 activation | 30 |
| TLR8 | unknown | TLR8 phosphorylation promoting its interaction with PI3K | 31 |
| TRIF | TBK1, IKKε | Phosphorylating TRIF at conserved motif‐pLxIS to recruit IRF3 | 51 |
| TRAF3 | CK1ε | Phosphorylating TRAF3 at Ser349 and promoting its K63‐linked ubiquitination | 32 |
| TRAF6 | MST4 | Phosphorylating TRAF6 to prevent its activation | 33 |
| TBK1 | PPM1B | Remove TBK1 autophosphorylation | 36 |
| TBK1 | DYRK2 | Inducing TBK1 Ser527 phosphorylation and K48‐linked ubiquitination | 37 |
| IRF3 | PTEN | Depleting IRF3 Ser97 phosphorylation and promoting its nuclear translocation | 40 |
| IRF3 | DNA‐PK | Phosphorylating IRF3 on Thr135 and maintaining IRF3 activation | 41 |
| RIG‐I | PKCα, PKCβ | Phosphorylating RIG‐I to keep it silent before viral RNA stimulation | 44 |
| RIG‐I | CK II | Maintaining a quiescent state of RIG‐I before viral RNA binding | 45 |
| RIG‐I | PP1α, PP1γ | Removing RIG‐I phosphorylation and initiating RIG‐I activation | 46 |
| MDA5 | PP1α, PP1γ | Removing MDA5 phosphorylation and initiating MDA5 activation | 46 |
| RIG‐I | IKKε | feedback inhibiting RIG‐I activation | 47 |
| MAVS | unknown | MAVS Tyr9 phosphorylation is essential for TRAF3 and TRAF6 recruitment | 48 |
| MAVS | TBK1, IKKε | Phosphorylating MAVS at conserved motif‐pLxIS to recruit IRF3 | 51 |
| cGAS | AKT | abrogate cGAS activity | 50 |
| DDX41 | BTK | Promoting DDX41 activation | 49 |
| STING | TBK1, IKKε | Phosphorylating STING at conserved motif‐pLxIS to recruit IRF3 | 51 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Ubiquitination
Ubiquitination involves the covalent and reversible addition of a 76 amino acid protein named ubiquitin to lysine or other residues in target proteins. A step‐wise catalyzation by the ubiquitin‐activating enzyme (E1), ubiquitin‐conjugating enzyme (E2), and ubiquitin protein ligase (E3) is needed for this PTM. Ubiquitin can undergo ubiquitination itself at its seven lysine residues (K6/K11/K27/K29/K33/K48/K63) or its amino‐terminal methionine, which generates different types of ubiquitin chains with distinct functions. Ubiquitin chains can be topologically classified into four types of architectures: homogeneous chains, multiple chains in which one substrate is separately modified by distinct chains, mixed chains in which a tandem chain contains two linkage types, and branched chains 54, 55. Different ubiquitin chains have distinct functions, for instance, K48‐linked ubiquitin chain often targets protein for protease degradation while K63‐linked ubiquitin chain mediates protein–protein interaction 54, 55.
In the TLR pathway, several TLRs including TLR3 and TLR9 are K48‐linked ubiquitinated by Triad3A, which induces protease degradation of these receptors 56 (Fig. 3). Whether other E3 ligases could modify TLRs directly, and whether the remaining TLRs are also regulated by ubiquitination, is still unknown. However, in contrast to the limited data available for the direct ubiquitination of TLRs, their downstream signaling proteins are reported to be extensively ubiquitinated. Myd88 is K48‐linked ubiquitinated by the E3 ligase Nrdp1 in the TLR7 pathway, leading to attenuated TNF production by macrophages in response to VSV infection 57. TRIF is targeted for K48‐linked ubiquitination by the E3 ligase WWP2, and the abolition of WWP2 boosts IFN‐β, TNF‐α, and IL‐6 expression after TLR3 agonist poly (I:C) stimulation in 293‐TLR3 and BMDM cells 58 (Fig. 3).
Figure 3.
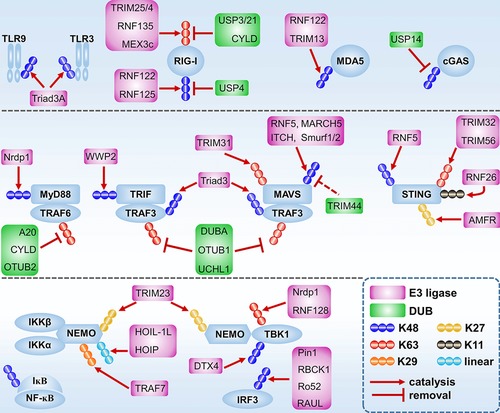
Regulation of antiviral signaling pathways by ubiquitination. TLR9 and TLR3 are K48‐linked ubiquitinated by Triad3A and subsequently degraded by proteasome. RIG‐I is K48‐linked ubiquitinated by RNF122 and RNF125 and then undergoes proteolytic degradation. USP4 remove the K48‐linked ubiquitination of RIG‐I and promotes RIG‐I signaling. TRIM25, TRIM4, RNF135 and MEX3c catalyzed RIG‐I K63‐linked ubiquitin chain and promote RIG‐I activation. USP3, USP21 and CYLD inhibit RIG‐I K63‐linked ubiquitination and activation. MDA5 is K48‐linked ubiquitinated by RNF122 and TRIM13 and then degraded by proteasome. USP14 promotes cGAS signaling by removing K48‐linked ubiquitin chain of cGAS. MyD88 and TRIF are K48‐linked ubiquitinated by Nrdp1 and WWP2 respectively, and then they are sequestered to proteolytic degradation. TRAF6 are inhibited by A20, CYLD and OTUB2 that specially remove TRAF6 K63‐linked ubiquitination. TRAF3 are inhibited by DUBA, OTUB1 and UCHL1 that specilally remove TRAF3 K63‐linked ubiquitination. TRAF3 are also targeted for proteolytic degradation by Triad3‐mediated K48‐linked ubiquitination. MAVS is activated by TRIM31‐medited K63‐linked ubiquitination. RNF5, MARCH5, ITCH and Smurf1/2 catalyze MAVS K48‐linked ubiquitination and induce MAVS degradation by protease. TRIM44 removes ITCH‐induced K48‐linked ubiquitination of MAVS and thus promotes RIG‐I signaling. TRIM32 and TRIM56 promote cGAS signaling by catalyzing K63‐linked ubiquitination of STING. RNF5 catalyzes STING K48‐linked ubiquitination and promotes proteolytic degradation of STING. RNF26 catalyzes STING K11‐linked ubiquitination and inhibit STING degradation. AMFR catalyzes STING K27‐linked ubiquitination and promotes STING activation. TRIM23 promotes NF‐κB and IRF3 activation by adding K27‐linked ubiquitinatin chain to NEMO. HOIL‐1L/HOIP and TRAF7 promote NF‐κB signaling by catalyzing NEMO linear and K29‐linked ubiquitination respectively. Nrdp1 and RNF128 catalyzing TBK1 K63‐linked ubiquitination and thus promoting TBK1 activation. DTX4 induces TBK1 proteolytic degradation by adding K48‐linked ubiquitin chain to it. Pin1, RBCK1, Ro52 and RAUL catalyze IRF3 K48‐linked ubiquitination and promote IRF3 proteolytic degradation.
K63‐linked ubiquitination of TRAF6 and TRAF3 is required for NF‐κB and IRF3 activation. Mechanistically, following activation by TLR signal, TRAF6 not only catalyzes its own K63‐linked ubiquitination, but also generates unanchored K63‐linked ubiquitin chains, both of which serve as scaffolds for TAB2/TAB3‐TAK1 and NEMO‐IKKα/IKKβ complex binding 59. TRAF3 K63‐linked ubiquitination also creates a platform for NEMO‐TBK1/IKKε complex recruitment and activation 60. Notably, a recent study unveiled a novel K63‐K48 branched ubiquitination of TRAF6 contributing to IL‐1β signaling, which led us to consider whether branched or mixed ubiquitination are also relevant for antiviral immunity 61. Several ubiquitin‐modifying enzymes, including A20, CYLD and OTUB2, are shown to remove the K63‐linked ubiquitination of TRAF6, and a set of deubiquitinating enzymes including DUBA, OTUB1 and UCHL1 abolish TRAF3 K63‐ubiquitination, which moderates TRAF6 and TRAF3 activation respectively 62, 63, 64, 65, 66, 67 (Fig. 3).
In addition, NEMO, the cognate binding partner of the IKKα/IKKβ complex and TBK1/IKKε kinase complex, has been shown to undergo K27‐, K29‐ and linear ubiquitination, and these PTMs regulate NF‐κB and IRF3 activation 68, 69, 70 (Fig. 3). TBK1 is targeted for K63‐linked ubiquitination by RNF128 and Nrdp1, which facilitate TBK1 activation 57, 71 (Fig. 3). By contrast, Triad3A and DTX4 catalyze K48‐linked ubiquitination of TRAF3 and TBK1, respectively, leading to target protein degradation and moderate IFN‐β production 72, 73, 74, 75 (Fig. 3). IRF3 is reported to be K48‐linked ubiquitinated by Pin1, RBCK1 and Ro52 as well as RAUL, which also catalyzes K48‐linked ubiquitination of IRF7 76, 77, 78, 79, 80 (Fig. 3).
Ubiquitination is a key regulatory mechanism for RLR pathways as well. For instance, TRIM25, an IFN‐inducible E3 ligase belonging to the tripartite motif (TRIM) protein family, catalyzes RIG‐I K63‐linked ubiquitination at K172, which promotes RIG‐I's interaction with MAVS and ultimately IFN‐β production 81 (Fig. 3). Genetic and structural analyses show that TRIM25‐mediated ubiquitination at K172 is crucial for RIG‐I activation in mammalian cells in response to NDV, Sev, VSV and influenza infections 81, 82, 83. Moreover, RIG‐I is also K63‐linked ubiquitinated by RNF135, TRIM4 and MEX3C, whose abrogation dampens virus‐induced IFN production 84, 85, 86, 87 (Fig. 3). Several reports suggested that it is unanchored K63‐linked ubiquitin chain that binds with RIG‐I and triggers downstream IRF3 phosphorylation 88, 89. An unsolved question is which type of K63‐linked ubiquitin chain, covalent or non‐covalent, is essential for RIG‐I activation in vivo.
As K63‐linked ubiquitination is crucial for RIG‐I activation, it is not surprising that this PTM is tightly controlled by deubiquitinating enzymes. For example, CYLD, USP21 and USP3 are shown to remove K63‐linked ubiquitin chains of RIG‐I, thereby moderating IRF3 phosphorylation and IFN‐β production 90, 91, 92 (Fig. 3). Moreover, the LUBAC complex, which is composed of HOIL‐1L and HOIP, counteracts TRIM25‐mediated RIG‐I activation by sequestering TRIM25 to K48‐linked ubiquitination and degradation 93. MAVS also undergoes K63‐linked ubiquitination at K500, yet the E3 ligase has not been identified 94. Interestingly, a recent study in Raw264.7, HEK293T and BMDM cells suggests that K500 of MAVS is also K48‐linked ubiquitinated by MARCH5, a mitochondria‐resident E3 ligase 95 (Fig. 3). Perhaps a spatial and/or temporal regulation of K500 ubiquitination on MAVS is in place to control this. Besides, TRIM31 is shown to serve as a positive regulator of MAVS aggregation by promoting K63‐linked ubiquitination of MAVS at K10, K311 and K461 in HEK293T and BMDM cells, thus driving IFN‐β production 96 (Fig. 3). The K48‐linked ubiquitination, often leading to protease degradation of target proteins, also plays an important role in the RLR pathway as the abundance of receptors and signaling proteins must be tightly controlled. Indeed, RIG‐I, MDA5 and MAVS are K48‐linked ubiquitinated and thus repressed by RNF125 while ubiquitin‐specific protease 4 (USP4) specifically removes the K48‐linked ubiquitin chain of RIG‐I, thus stabilizing the RIG‐I protein and facilitate RIG‐I activation 97, 98 (Fig. 3). RIG‐I is also reported to be K48‐linked ubiquitinated by RNF122 and CHIP, while another E3 ligase, TRIM13, catalyzes MDA5 K48‐linked ubiquitination and promotes its degradation 99, 100, 101 (Fig. 3). MAVS is targeted for K48‐linked ubiquitination by multiple E3 ligase including Smurf1/2, RNF5 and ITCH, which induces MAVS degradation and RLR signal termination 102, 103, 104 (Fig. 3). Interestingly, TRIM44 dampens ITCH‐induced K48‐linked ubiquitination of MAVS, thus facilitating IFN production 105 (Fig. 3).
It has become evident that the cGAS‐STING pathway is also regulated by ubiquitination. STING is targeted for K63‐linked ubiquitination by TRIM32 and TRIM56 at K150, which potentiates STING dimerization and interaction with TBK1 106 (Fig. 3). Interestingly, the STING K150 residue also undergoes K48‐linked ubiquitination and K11‐linked ubiquitination by RNF5 and RNF26, respectively 107, 108 (Fig. 3). RNF5‐mediated K48‐linked ubiquitination at K150 is dampened by RNF26‐induced K11‐linked ubiquitination. In contrast, RNF26 did not affect the K63‐linked ubiquitination of STING at K150 108. Besides, AMFR, an endoplasmic reticulum (ER) located E3 ligase, catalyzes the K27‐linked ubiquitination at K150 as well, which promotes TBK1 activation and IFN‐β production in MEF and BMDM cells in response to HSV infection 109 (Fig. 3). The discrepancies of ubiquitination at K150 of STING might be caused by different experimental methods and/or a temporal regulation of homogeneous/mixed/branched ubiquitin chain during STING activation. More detailed studies are required to fully understand the dynamic interplay of these ubiquitin‐related processes.
Autophagy is a central cell homeostatic process by which damaged organelles, protein aggregates and invading microbes are sequestered in autophagosomes and delivered to the lysosome for degradation, however, the interplay between antiviral immunity and autophagy is largely unknown. A recent study showed that cGAS is targeted for K48‐linked ubiquitination and subsequent selective autophagic degradation 110. Mechanistically, cGAS is constitutively K48‐linked ubiquitinated and recognized by autophagy receptor p62, leading to selective autophagic degradation 110. Upon viral infection, TRIM14 recruits USP14 to cleave the K48‐linked ubiquitin chain of cGAS, which disrupts the cGAS‐p62 interaction, thereby preventing cGAS autophagic degradation 110 (Fig. 3). An interesting question is whether other signal molecules in antiviral innate immunity are also regulated by ubiquitination‐mediated selective autophagy. A detailed list of ubiquitination in antiviral signaling is included in Table 2.
Table 2.
Ubiquitination of PRR signaling pathways
| Signaling protein | Enzymes | Functions | Ref. |
|---|---|---|---|
| TLR3, TLR9 | Triad3A | Catalyzing K48‐linked ubiquitination and degradation | 56 |
| Myd88 | Nrdp1 | Inducing K48‐linked ubiquitination and degradation | 57 |
| TRIF | WWP2 | Mediating K48‐linked ubiquitination and degradation | 58 |
| TRAF6 | A20, CYLD and OTUB2 | Removing the K63‐linked ubiquitination of TRAF6 to inhibit TRAF6 activation | 62, 63, 64, 65 |
| TRAF3 | DUBA, OTUB1 and UCHL1 | Depleting the K63‐linked ubiquitination of TRAF3 to suppressTRAF3 activation | 65, 66, 67 |
| TRAF3 | Triad3 | Mediating K48‐linked ubiquitination and degradation | 72 |
| NEMO | TRIM23 | Catalyzing K27‐linked ubiquitination and activation of NF‐κB and IRF3 | 68 |
| NEMO | HOIL‐1L, HOIP | Inducing linear ubiquitination and NF‐κB activation | 69 |
| NEMO | TRAF7 | Catalyzing K29‐linked ubiquitination to suppress NF‐κB activation | 70 |
| TBK1 | RNF128 | Mediating K63‐linked ubiquitination and activation | 71 |
| TBK1 | DTX4 | Catalyzing K48‐linked ubiquitination and degradation | 74, 75 |
| IRF3 | Pin1, RBCK1, Ro52 and RAUL | Catalyzing K48‐linked ubiquitination and degradation | 76, 77, 78, 79, 80 |
| IRF7 | RAUL | Inducing K48‐linked ubiquitination and degradation | 80 |
| RIG‐I | TRIM25, RNF135, TRIM4 and MEX3c | Catalyzing RIG‐I K63‐linked ubiquitination and RIG‐I K63‐linked ubiquitination is critical for RIG‐I activation | 81, 82, 83, 84, 85, 86, 87 |
| RIG‐I | CYLD, USP21, USP3, | Removing RIG‐I K63‐linked ubiquitination to inhibit RIG‐I activation | 90, 91, 92 |
| RIG‐I | RNF125, RNF122 | Inducing K48‐linked ubiquitination and degradation of RIG‐I | 97, 99, 100 |
| CHIP | |||
| RIG‐I | USP4 | Removing RIG‐I K48‐linked ubiquitination to promote RIG‐I activation | 98 |
| MDA5 | RNF125 TRIM13 | Inducing K48‐linked ubiquitination and degradation of MDA5 | 97, 101 |
| TRIM25 | HOIL‐1L, HOIP | Catalyzing K48‐linked ubiquitination of TRIM25 to inhibit RIG‐I activation | 93 |
| MAVS | TRIM31 | Inducing K63‐linked ubiquitination of MAVS and activation of MAVS | 96 |
| MAVS | MARCH5, | Catalyzing K48‐linked ubiquitination of MAVS to inhibit MAVS | 95, 102, 103, 104 |
| Smurf1/2, RNF5 and ITCH | |||
| MAVS | TRIM44 | Removing ITCH‐induced K48‐linked ubiquitination and degradation of MAVS | 105 |
| cGAS | USP14 | Removing K48‐linked ubiquitination of cGAS to suppress autophagic degradation of cGAS | 110 |
| STING | TRIM32, TRIM56 | Catalyzing K63‐linked ubiquitination of STING to activate STING | 106 |
| STING | RNF5 | Catalyzing K48‐linked ubiquitination of STING and degradation | 107 |
| STING | RNF26 | Catalyzing K11‐linked ubiquitination and inhibiting K48‐linked ubiquitination of STING | 108 |
| STING | AMFR, INSIG1 | Catalyzing K27‐linked ubiquitination of STING to activate STING | 109 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Unconventional PTMs
In light of the development of mass spectrometry (MS) technology, and in the context of the extensively investigated antiviral signaling pathways, the importance of some unconventional PTMs in the innate response has been revealed. These unconventional PTMs include methylation, acetylation, SUMOylation and ADP‐ribosylation.
Methylation refers to the transfer of a methyl group from the donor S‐adenosylmethionine (SAM) to the lysine or arginine residues of target proteins. The histones are the first and the best described target proteins of methylation, and notably, several studies have emphasized the importance of non‐histone methylation in regulating innate immunity 111. For instance, the NF‐κB subunit, p65, is hyper methylated and has distinct functions when methylated. In particular, the protein lysine methyltransferases (PKMT) SETD6 catalyzes monomethylation of p65 at K310 and dampens p65‐driven inflammatory responses 112, 113. Mechanistically, K310‐methylated p65 recruits the ankryin repeat of the histone methyltransferase GLP to maintain a repressed chromatin state of p65 target genes, thus inhibiting the transcription of proinflammatory cytokines such as TNF and IL‐1α 112, 113. Moreover, the protein kinase C‐ζ (PKC‐ζ) phosphorylates p65 at Ser311, blocking the interaction of p65 with GLP and promoting proinflammatory gene expression 112. In contrast, methylation of p65 at K218 and K221 by NSD1, as well as at K37 by SET9 potentiates transcription of p65‐regulated genes 114, 115. Notably, the NDS1‐mediated methylation of p65 is clearly induced upon double‐stranded RNA stimulation, yet the complete function of p65 methylation in viral infections is still unknown 114. An interesting question is whether other signaling proteins are also methylated in antiviral immunity, since the role of methylation beyond histones is of comprehensive concern.
Acetylation is the inverse introduction of an acetyl group to protein lysine residues. Several studies have highlighted the importance of acetylation in antiviral responses. MAPK phosphatase 1 (MKP1), a dual‐specificity phosphatase that inactivates p38 MAPK, is acetylated by p300, resulting in enhanced MKP1 activity and moderate proinflammatory cytokine expression 116. RIG‐I is shown to be acetylated at K909 in resting Huh7 and HEK293 cells, and HCV or VSV infections induce a quick deacetylation of RIG‐I by histone deacetylase 6 (HDAC6) 117, 118. This deacetylation is crucial for RIG‐I activation and downstream IFN‐β production in vivo, since HDAC6 knockout mice show increased susceptibility to HCV and VSV infection 117, 118 (Fig. 4). Notably, another histone deacetylase, HDAC9, deacetylates TBK1 and enhances its kinase activity 119 (Fig. 4). Moreover, the DNA methyltransferase Dnmt3a contributes to IFN‐β production by maintaining high HDAC9 expression, which uncovers the crosstalk between epigenetics and post‐translational modifications in antiviral immunity 119.
Figure 4.
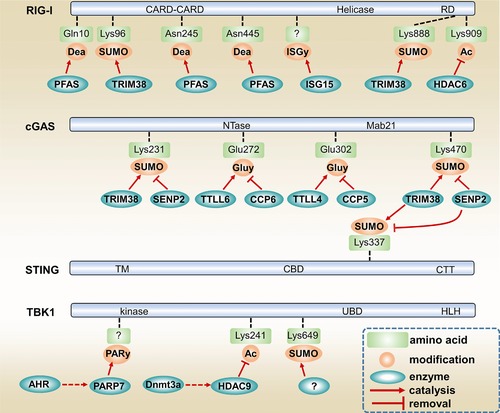
Non‐canonical PTMs of RIG‐I, cGAS, STING and TBK1. RIG‐I is deamidated by PFAS at Q10, N245 and N445. RIG‐I's deamidation is crucial for RIG‐I signaling during γHV68 infection. TRIM38 promotes RIG‐I activation by inducing RIG‐I SUMOylation at K96 and K888. ISG15 limits RIG‐I activation by catalyzing RIG‐I ISGylation. HDAC6 is critical for RIG‐I activation by removing RIG‐I K909 acetylation. TRIM38 also promotes cGAS signaling by catalyzing cGAS K231 and K470 SUMOylation as well as STING K337 SUMOylation. SENP2 inhibits cGAS signaling by counteracting TRIM38‐mediated SUMOylation of cGAS and STING. AHR signaling moderates antiviral response by promoting PARP7‐mediated TBK1 ADP‐ribosylation. Dnmt3a contributes to IFN‐β production by promoting HDAC9 expression. HDAC9 removes TBK1 K241 acetylation to boost TBK1 kinase activity. Dea, deamidation. SUMO, SUMOylation. ISGy, ISGylation. Ac, acetylation. Gluy, glutamylation. PARy, ADP‐ribosylation. CARD, caspase recruitment domain. RD, regulatory domain. Mab21, Mab‐21 domain. TM, transmembrane domain. CBD, c‐di‐GMP binding domain. CTT, c‐terminal tail. UBD, ubiquitin‐binding domain. HLH, helix‐loop‐helix domain.
One of the features of immune homeostasis is the negative feedback regulation of signaling pathways by varying PTMs. Apart from IKKε‐mediated RIG‐I phosphorylation in preventing excessive RIG‐I activation 46, ISG15, an IFN‐stimulated ubiquitin‐like protein, suppresses RIG‐I signaling by conjugating to RIG‐I 120, 121 (Fig. 4). The biological function of this process, termed ISGylation, is still poorly understood. In addition, during murine gamma herpesvirus 68 (γHV68) infection of MEF cells, RIG‐I is also reported to be deamidated by PFAS, which converts glutamine or asparagine residues to glutamate or aspartate respectively 122. Moreover, the viral homologues of PFAS, named vGAT, promote RIG‐I deamidation and subsequent MAVS and IKKβ activation 122 (Fig. 4). A probable question is whether the deamidation of RIG‐I is a general or γHV68 infection specific regulation process.
Interestingly, a recent study suggested that the aryl hydrocarbon receptor (AHR) pathway, known for mediating the toxic activity of many environmental xenobiotics, is unexpectedly linked to antiviral responses via ADP‐ribosylation, a PTM referring the addition of one or more ADP‐ribose moieties to proteins 123. AHR signaling was shown to specifically induce the expression of TIPARP/PARP7 which in turn dampens TBK1 activity via catalyzing TBK1 ADP‐ribosylation 123 (Fig. 4). The study highlighted the function of PTMs as an access point connecting different signaling pathways and physiological processes such as nutritional dysregulation stress and innate resistance against viral infections.
The cGAS pathway has been characterized for a few years and its regulatory mechanism is still not clear. Recently, a published work showed that cGAS activity is controlled by reversible glutamylation, a PTM which involves the addition of glutamate side chains to the γ‐carboxyl groups of glutamic acid residues of the target proteins 124. The tubulin tyrosine ligase–like (TTLL) 4 and 6 were shown to promote IFN‐β production by catalyzing monoglutamylation and polyglutamylation of cGAS, respectively, while the carboxypeptidase CCP5 and CCP6 both remove the glutamate chain of cGAS in macrophages 124 (Fig. 4).
Processed by specific small ubiquitin related modifier (SUMO) enzymes E1, E2, and E3, SUMOylation is a PTM resembling ubiquitination as ubiquitination use ubiquitin to covalently attach to target protein while SUMOylation use SUMO 125. A number of studies have highlighted the importance of SUMOylation in antiviral signaling. For example, SUMOylation of RIG‐I and MDA5 by the E3 ligase PIAS2β and TRIM38 promotes type I IFN production and antiviral activities in the cell 126, 127, 128. cGAS and STING are SUMOylated by the ubiquitin ligase TRIM38, which prevents their K48‐linked ubiquitination and degradation, hence facilitating antiviral innate immune responses 129 (Fig. 4). In agreement, the deSUMOylating enzyme Sentrin/SUMO‐specific protease (SENP) 2 alleviates antiviral responses by deSUMOylating cGAS, STING and IRF3 129 (Fig. 4). Furthermore, TBK1 K694 SUMOylation contributes to its antiviral activities as K694 mutated TBK1 presents enforced interaction with TANK1, a known negative regulator of TBK1 130 (Fig. 4). The E3 ligase responsible for TBK1 SUMOylation has not been identified yet, but at least it is known that the adenoviral protein Gam1 inhibits SUMOylation of TBK1 130. Notably, SUMOylation also represses antiviral responses via targeting the Ifnb1 gene promoter 131. It seems that SUMOylation leads to both enhancing and suppressive functions in antiviral innate immunity, depending on the substrate specificity. Overall, however, an intrinsic question is why the cells extensively utilize the “long peptide chains” and “multi‐enzymes” PTMs, such as ubiquitination, SUMOylation, ISGylation, polyglutamylation and ADP‐ribosylation, to control antiviral signaling.
PTMs in virus related immune disorders and therapy
Due to the critical role of PTMs in antiviral responses, it is not surprising that virus have evolved strategies to actively interfere or hijack the PTMs of host proteins. Influenza A viruses (IAVs) are serious infectious RNA viruses that cause grievous economic and health consequences. The influenza non‐structural protein 1 (NS1) is known to act as a virulence factor inhibiting host immune responses. The NS1 encoded by human, avian, swine and mouse‐adapted influenza viruses can bind with human TRIM25 and inhibit TRIM25‐catalyzed RIG‐I ubiquitination 132. However, the NS1 from mouse‐adapted influenza viruses targets RNF135, but not TRIM25, to inhibit RIG‐I K63‐linked ubiquitination, which indicates a species divergence between human and mice influenza infections and immune responses 132. The papillomaviruses induce expression of the cellular protein ubiquitin carboxyl‐terminal hydrolase L1 (UCHL1) in keratinocytes, which inhibits TRAF3 K63‐linked ubiquitination and subsequent IRF3 phosphorylation, as well as IFN‐β expression 67. UCHL1 also promotes K48‐linked ubiquitination of NEMO, leading to attenuated proinflammatory cytokines production 67. In addition, a large number of viruses, such as severe acute respiratory syndrome (SARS) coronavirus, human coronavirus NL63, mouse hepatitis virus (MHV) A59, herpes simplex virus 1 (HSV‐1), Kaposi's sarcoma‐associated herpesvirus (KSHV), HBV, Epstein‐Barr virus and some arteriviruses, have been shown to block IFN‐β production by encoding DUB motif‐containing proteins 133, 134. Given the distinct functions of ubiquitination in the same or different signaling proteins, further investigation is urgent to characterize how these virus DUB‐containing proteins interact with target proteins and remove the “bad” type of ubiquitin chains on specific residues. Apart from ubiquitination, the Ebola Zaire virus dampens IRF7 activity and IFN‐β transcription via VP35, which increases PIAS1‐mediated SUMOylation of IRF7 and thus impairs the recruitment of IRF7 to type I IFN gene promoters 135.
Conclusion
The idea of PAMPs and PRRs was started two decades ago 136. At that time, protein phosphorylation was already extensively explored and ubiquitination was just around the corner from receiving wide‐spread interest 137. Twenty years of study have not only highlighted the cornerstone role of the PRR pathway in infection immunity, but also reveals the importance of PTMs in regulating numerous cell activities. For the antiviral innate immune responses, several intriguing and crucial aspects still await further investigation.
Though much progress has been made in characterizing PTMs and enzymes, which regulate type I interferon production, the in vivo relevance of these enzymes and modifications to antiviral immunity still need to be addressed in future studies, as a considerable part of these functions and mechanisms are characterized in vitro or in non‐immune cells. More detailed genetic and biochemical evidence is also needed for a full understanding of PTM role in antiviral responses. Moreover, while multiple kinases and E3 ligases contributing to IFN‐β production are identified, not much is known about the phosphatases and deubiquitinating enzymes which functions as IFN‐β production brake. Though insufficient interferon production may cause chronic infections, excessive interferon production often causes autoimmune and/or inflammatory diseases such as Aicardi–Goutières Syndrome, systemic lupus erythematosus, arthritis and influenza pneumonia. In this regard, we must address significant gaps in knowledge of PTMs and enzymes that are moderating antiviral responses, many of which could be new and favorable antiviral and autoimmune therapeutic targets.
While phosphorylation and ubiquitination are shown to regulate antiviral innate immunity, the exact roles of other PTMs, such as acetylation, methylation, ADP‐ribosylation, SUMOylation, ISGylation and palmitoylation, are still poorly addressed. Even for phosphorylation and ubiquitination, a global map of regulation network and the detailed mechanisms of each signaling protein cannot clearly be drawn. Moreover, differences in observations generated from studies using different virus species, host species and host cell lineages make the questions even trickier. Meanwhile, whether additional novel PTMs might exist in antiviral signaling is unknown. In this regard, advanced technological platforms such as mass spectrometry, fluorescence imaging, nuclear magnetic resonance (NMR) and Raman spectroscopy are eagerly required for both in vitro and in vivo characterizing the dynamics and interplay of different PTMs.
Genome sequencing has unveiled the importance of antiviral signaling in various human diseases such as IRF7 in influenza infection, TRAF3 in herpes simplex encephalitis and MDA5 in systemic lupus erythematous 138, 139, 140. Given the pivotal role of PTMs in modulating antiviral innate immune responses, we eagerly await the identification of disease‐relevant mutations in numerous PTM enzymes and the possible diagnostic markers and therapeutic targets for virus‐related immune disorders.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Abbreviations
- AHR
aryl hydrocarbon receptor
- BTK
Bruton's tyrosine kinase
- cGAMP
cyclic GMP‐AMP
- HDAC6
histone deacetylase 6
- HSV
herpes simplex virus
- IFNs
interferons
- IKKε
Inhibitor‐κB kinase ε
- IRF
IFN regulatory factor
- LGP2
laboratory of genetics and physiology 2
- MAVS
mitochondrial antiviral‐signaling
- MDA5
melanoma differentiation‐associated protein 5
- MST4
mammalian STE20‐like protein kinase 4
- MyD88
myeloid differentiation factor‐88
- NS1
non‐structural protein 1
- PKCα
protein kinase C α
- PRR
pattern‐recognition receptors
- PTMs
post‐translational modifications
- RIG‐I
retinoic acid‐inducible gene I
- RLR
RIG‐I‐like receptor
- Sev
Sendai virus
- STING
stimulator of interferon genes
- SUMO
small ubiquitin related modifiers
- TBK1
TANK‐binding kinase‐1
- TLR
toll‐like receptor
- TRAF3
TNF receptor‐associated factor 3
- TRIF
TIR‐domain‐containing adaptor protein inducing interferon beta
- TRIM
tripartite motif
- USP4
ubiquitin‐specific protease 4
- VSV
vesicular stomatitis virus
Acknowledgements
This work was supported by National Natural Science Foundation of China (No.91542111 and 81330069).
References
- 1. Takeuchi, O. and Akira, S. , Pattern recognition receptors and inflammation. Cell 2010. 140: 805–820. [DOI] [PubMed] [Google Scholar]
- 2. Barrat, F. J. , Elkon, K. B. and Fitzgerald, K. A. , Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 2016. 67: 323–336. [DOI] [PubMed] [Google Scholar]
- 3. Ivashkiv, L. B. and Donlin, L. T. , Regulation of type I interferon responses. Nat. Rev. Immunol. 2014. 14: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sin, W. X. , Li, P. , Yeong, J. P. and Chin, K. C. , Activation and regulation of interferon‐beta in immune responses. Immunol Res. 2012. 53: 25–40. [DOI] [PubMed] [Google Scholar]
- 5. Kawai, T. and Akira, S. , The role of pattern‐recognition receptors in innate immunity: update on Toll‐like receptors. Nat. Immunol. 2010. 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 6. Gay, N. J. , Symmons, M. F. , Gangloff, M. and Bryant, C. E. , Assembly and localization of Toll‐like receptor signalling complexes. Nat. Rev. Immunol. 2014. 14: 546–558. [DOI] [PubMed] [Google Scholar]
- 7. Saha, S. K. and Cheng, G. , TRAF3: a new regulator of type I interferons. Cell Cycle 2006. 5: 804–807. [DOI] [PubMed] [Google Scholar]
- 8. Fitzgerald, K. A. , McWhirter, S. M. , Faia, K. L. , Rowe, D. C. , Latz, E. , Golenbock, D. T. , Coyle, A. J. et al, IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003. 4: 491–496. [DOI] [PubMed] [Google Scholar]
- 9. Honda, K. , Ohba, Y. , Yanai, H. , Negishi, H. , Mizutani, T. , Takaoka, A. , Taya, C. et al, Spatiotemporal regulation of MyD88‐IRF‐7 signalling for robust type‐I interferon induction. Nature 2005. 434: 1035–1040. [DOI] [PubMed] [Google Scholar]
- 10. Wang, L. , Zhao, J. , Ren, J. , Hall, K. H. , Moorman, J. P. , Yao, Z. Q. and Ning, S. , Protein phosphatase 1 abrogates IRF7‐mediated type I IFN response in antiviral immunity. Eur. J. Immunol 2016. 46: 2409–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rehwinkel, J. and Reis e Sousa, C. , RIGorous detection: exposing virus through RNA sensing. Science 2010. 327: 284–286. [DOI] [PubMed] [Google Scholar]
- 12. Kato, H. , Sato, S. , Yoneyama, M. , Yamamoto, M. , Uematsu, S. , Matsui, K. , Tsujimura, T. et al, Cell type‐specific involvement of RIG‐I in antiviral response. Immunity 2005. 23: 19–28. [DOI] [PubMed] [Google Scholar]
- 13. Muzio, M. , Bosisio, D. , Polentarutti, N. , D'Amico, G. , Stoppacciaro, A. , Mancinelli, R. , van't Veer, C. et al, Differential expression and regulation of toll‐like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J. Immunol 2000. 164: 5998–6004. [DOI] [PubMed] [Google Scholar]
- 14. Schlee, M. , Master sensors of pathogenic RNA ‐ RIG‐I like receptors. Immunobiology 2013. 218: 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belgnaoui, S. M. , Paz, S. and Hiscott, J. , Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol 2011. 23: 564–572. [DOI] [PubMed] [Google Scholar]
- 16. Kawasaki, T. , Kawai, T. and Akira, S. , Recognition of nucleic acids by pattern‐recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011. 243: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun, L. , Wu, J. , Du, F. , Chen, X. and Chen, Z. J. , Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013. 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu, J. , Sun, L. , Chen, X. , Du, F. , Shi, H. , Chen, C. and Chen, Z. J. , Cyclic GMP‐AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013. 339: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao, T. S. and Fitzgerald, K. A. , The cGAS‐STING pathway for DNA sensing. Mol. Cell 2013. 51: 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gray, E. E. , Winship, D. , Snyder, J. M. , Child, S. J. , Geballe, A. P. and Stetson, D. B. , The AIM2‐like receptors are dispensable for the interferon response to intracellular DNA. Immunity 2016. 45: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu, J. , Qian, C. and Cao, X. , Post‐translational modification control of innate immunity. Immunity 2016. 45: 15–30. [DOI] [PubMed] [Google Scholar]
- 22. Deribe, Y. L. , Pawson, T. and Dikic, I. , Post‐translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010. 17: 666–672. [DOI] [PubMed] [Google Scholar]
- 23. Mowen, K. A. and David, M. , Unconventional post‐translational modifications in immunological signaling. Nat. Immunol 2014. 15: 512–520. [DOI] [PubMed] [Google Scholar]
- 24. Hunter, T. , The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 2007. 28: 730–738. [DOI] [PubMed] [Google Scholar]
- 25. Beltrao, P. , Bork, P. , Krogan, N. J. and van Noort, V. , Evolution and functional cross‐talk of protein post‐translational modifications. Mol. Syst. Biol. 2013. 9: 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hunter, T. , Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2012. 367: 2513–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee, K. G. , Xu, S. , Kang, Z. H. , Huo, J. , Huang, M. , Liu, D. , Takeuchi, O. et al, Bruton's tyrosine kinase phosphorylates Toll‐like receptor 3 to initiate antiviral response. Proc. Natl. Acad. Sci. USA 2012. 109: 5791–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamashita, M. , Chattopadhyay, S. , Fensterl, V. , Saikia, P. , Wetzel, J. L. and Sen, G. C. , Epidermal growth factor receptor is essential for Toll‐like receptor 3 signaling. Sci. Signal 2012. 5: ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarkar, S. N. , Smith, H. L. , Rowe, T. M. and Sen, G. C. , Double‐stranded RNA signaling by Toll‐like receptor 3 requires specific tyrosine residues in its cytoplasmic domain. J. Biol. Chem. 2003. 278: 4393–4396. [DOI] [PubMed] [Google Scholar]
- 30. Hsieh, M. Y. , Chang, M. Y. , Chen, Y. J. , Li, Y. K. , Chuang, T. H. , Yu, G. Y. , Cheung, C. H. et al, The inducible nitric‐oxide synthase (iNOS)/Src axis mediates Toll‐like receptor 3 tyrosine 759 phosphorylation and enhances its signal transduction, leading to interferon‐beta synthesis in macrophages. J. Biol. Chem. 2014. 289: 9208–9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rajagopal, R. , Waller, A. S. , Mendoza, J. D. and Wightman, P. D. , The covalent modification and regulation of TLR8 in HEK‐293 cells stimulated with imidazoquinoline agonists. Biochem. J. 2008. 409: 275–287. [DOI] [PubMed] [Google Scholar]
- 32. Zhou, Y. , He, C. , Yan, D. , Liu, F. , Liu, H. , Chen, J. , Cao, T. et al, The kinase CK1varepsilon controls the antiviral immune response by phosphorylating the signaling adaptor TRAF3. Nat. Immunol. 2016. 17: 397–405. [DOI] [PubMed] [Google Scholar]
- 33. Jiao, S. , Zhang, Z. , Li, C. , Huang, M. , Shi, Z. , Wang, Y. , Song, X. et al, The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat. Immunol 2015. 16: 246–257. [DOI] [PubMed] [Google Scholar]
- 34. Kishore, N. , Huynh, Q. K. , Mathialagan, S. , Hall, T. , Rouw, S. , Creely, D. , Lange, G. et al, IKK‐i and TBK‐1 are enzymatically distinct from the homologous enzyme IKK‐2: comparative analysis of recombinant human IKK‐i, TBK‐1, and IKK‐2. J. Biol. Chem. 2002. 277: 13840–13847. [DOI] [PubMed] [Google Scholar]
- 35. Lei, C. Q. , Zhong, B. , Zhang, Y. , Zhang, J. , Wang, S. and Shu, H. B. , Glycogen synthase kinase 3beta regulates IRF3 transcription factor‐mediated antiviral response via activation of the kinase TBK1. Immunity 2010. 33: 878–889. [DOI] [PubMed] [Google Scholar]
- 36. Zhao, Y. , Liang, L. , Fan, Y. , Sun, S. , An, L. , Shi, Z. , Cheng, J. et al, PPM1B negatively regulates antiviral response via dephosphorylating TBK1. Cell Signal 2012. 24: 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. An, T. , Li, S. , Pan, W. , Tien, P. , Zhong, B. , Shu, H. B. and Wu, S. , DYRK2 negatively regulates type I interferon induction by promoting TBK1 degradation via Ser527 phosphorylation. PLoS Pathog. 2015. 11: e1005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mori, M. , Yoneyama, M. , Ito, T. , Takahashi, K. , Inagaki, F. and Fujita, T. , Identification of Ser‐386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 2004. 279: 9698–9702. [DOI] [PubMed] [Google Scholar]
- 39. Lin, R. , Heylbroeck, C. , Pitha, P. M. and Hiscott, J. , Virus‐dependent phosphorylation of the IRF‐3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome‐mediated degradation. Mol. Cell Biol. 1998. 18: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li, S. , Zhu, M. , Pan, R. , Fang, T. , Cao, Y. Y. , Chen, S. , Zhao, X. et al, The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol 2016. 17: 241–249. [DOI] [PubMed] [Google Scholar]
- 41. Karpova, A. Y. , Trost, M. , Murray, J. M. , Cantley, L. C. and Howley, P. M. , Interferon regulatory factor‐3 is an in vivo target of DNA‐PK. Proc. Natl. Acad. Sci. U S A 2002. 99: 2818–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arthur, J. S. and Ley, S. C. , Mitogen‐activated protein kinases in innate immunity. Nat. Rev. Immunol 2013. 13: 679–692. [DOI] [PubMed] [Google Scholar]
- 43. Sun, S. C. , Non‐canonical NF‐kappaB signaling pathway. Cell Res. 2011. 21: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maharaj, N. P. , Wies, E. , Stoll, A. and Gack, M. U. , Conventional protein kinase C‐alpha (PKC‐alpha) and PKC‐beta negatively regulate RIG‐I antiviral signal transduction. J. Virol. 2012. 86: 1358–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun, Z. , Ren, H. , Liu, Y. , Teeling, J. L. and Gu, J. , Phosphorylation of RIG‐I by casein kinase II inhibits its antiviral response. J. Virol. 2011. 85: 1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wies, E. , Wang, M. K. , Maharaj, N. P. , Chen, K. , Zhou, S. , Finberg, R. W. and Gack, M. U. , Dephosphorylation of the RNA sensors RIG‐I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 2013. 38: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang, X. , Yu, H. , Zhao, J. , Li, X. , Li, J. , He, J. , Xia, Z. et al, IKK negatively regulates RIG‐I via direct phosphorylation. J. Med. Virol 2016. 88: 712–718. [DOI] [PubMed] [Google Scholar]
- 48. Wen, C. , Yan, Z. , Yang, X. , Guan, K. , Xu, C. , Song, T. , Zheng, Z. et al, Identification of tyrosine‐9 of MAVS as critical target for inducible phosphorylation that determines activation. PLoS One 2012. 7: e41687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee, K. G. , Kim, S. S. , Kui, L. , Voon, D. C. , Mauduit, M. , Bist, P. , Bi, X. et al, Bruton's tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep. 2015. 10: 1055–1065. [DOI] [PubMed] [Google Scholar]
- 50. Seo, G. J. , Yang, A. , Tan, B. , Kim, S. , Liang, Q. , Choi, Y. , Yuan, W. et al, Akt kinase‐mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015. 13: 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu, S. , Cai, X. , Wu, J. , Cong, Q. , Chen, X. , Li, T. , Du, F. et al, Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015. 347: aaa2630. [DOI] [PubMed] [Google Scholar]
- 52. Li, Z. , Liu, G. , Sun, L. , Teng, Y. , Guo, X. , Jia, J. , Sha, J. et al, PPM1A regulates antiviral signaling by antagonizing TBK1‐mediated STING phosphorylation and aggregation. PLoS Pathog. 2015. 11: e1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiang, W. , Zhang, Q. , Lin, X. , Wu, S. , Zhou, Y. , Meng, F. , Fan, Y. et al, PPM1A silences cytosolic RNA sensing and antiviral defense through direct dephosphorylation of MAVS and TBK1. Sci. Adv. 2016. 2: e1501889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Komander, D. and Rape, M. , The ubiquitin code. Annu. Rev. Biochem. 2012. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- 55. Yau, R. and Rape, M. , The increasing complexity of the ubiquitin code. Nat. Cell. Biol. 2016. 18: 579–586. [DOI] [PubMed] [Google Scholar]
- 56. Chuang, T. H. and Ulevitch, R. J. , Triad3A, an E3 ubiquitin‐protein ligase regulating Toll‐like receptors. Nat. Immunol 2004. 5: 495–502. [DOI] [PubMed] [Google Scholar]
- 57. Wang, C. , Chen, T. , Zhang, J. , Yang, M. , Li, N. , Xu, X. and Cao, X. , The E3 ubiquitin ligase Nrdp1 'preferentially' promotes TLR‐mediated production of type I interferon. Nat. Immunol 2009. 10: 744–752. [DOI] [PubMed] [Google Scholar]
- 58. Yang, Y. , Liao, B. , Wang, S. , Yan, B. , Jin, Y. , Shu, H. B. and Wang, Y. Y. , E3 ligase WWP2 negatively regulates TLR3‐mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc. Natl. Acad. Sci. U S A 2013. 110: 5115–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen, J. and Chen, Z. J. , Regulation of NF‐kappaB by ubiquitination. Curr. Opin. Immunol 2013. 25: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hacker, H. , Tseng, P. H. and Karin, M. , Expanding TRAF function: TRAF3 as a tri‐faced immune regulator. Nat. Rev. Immunol 2011. 11: 457–468. [DOI] [PubMed] [Google Scholar]
- 61. Ohtake, F. , Saeki, Y. , Ishido, S. , Kanno, J. and Tanaka, K. , The K48‐K63 branched ubiquitin chain regulates NF‐kappaB signaling. Mol Cell 2016. 64: 251–266. [DOI] [PubMed] [Google Scholar]
- 62. Wertz, I. E. , Newton, K. , Seshasayee, D. , Kusam, S. , Lam, C. , Zhang, J. , Popovych, N. , Helgason, E. et al, Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 2015. 528: 370–375. [DOI] [PubMed] [Google Scholar]
- 63. Trompouki, E. , Hatzivassiliou, E. , Tsichritzis, T. , Farmer, H. , Ashworth, A. and Mosialos, G. , CYLD is a deubiquitinating enzyme that negatively regulates NF‐kappaB activation by TNFR family members. Nature 2003. 424: 793–796. [DOI] [PubMed] [Google Scholar]
- 64. Zhang, J. , Stirling, B. , Temmerman, S. T. , Ma, C. A. , Fuss, I. J. , Derry, J. M. and Jain, A. , Impaired regulation of NF‐kappaB and increased susceptibility to colitis‐associated tumorigenesis in CYLD‐deficient mice. J. Clin. Invest. 2006. 116: 3042–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li, S. , Zheng, H. , Mao, A. P. , Zhong, B. , Li, Y. , Liu, Y. , Gao, Y. et al, Regulation of virus‐triggered signaling by OTUB1‐ and OTUB2‐mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 2010. 285: 4291–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kayagaki, N. , Phung, Q. , Chan, S. , Chaudhari, R. , Quan, C. , O'Rourke, K. M. , Eby, M. et al, DUBA: a deubiquitinase that regulates type I interferon production. Science 2007. 318: 1628–1632. [DOI] [PubMed] [Google Scholar]
- 67. Karim, R. , Tummers, B. , Meyers, C. , Biryukov, J. L. , Alam, S. , Backendorf, C. , Jha, V. et al, Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog 2013. 9: e1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arimoto, K. , Funami, K. , Saeki, Y. , Tanaka, K. , Okawa, K. , Takeuchi, O. , Akira, S. et al, Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. U S A 2010. 107: 15856–15861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tokunaga, F. , Sakata, S. , Saeki, Y. , Satomi, Y. , Kirisako, T. , Kamei, K. , Nakagawa, T. et al, Involvement of linear polyubiquitylation of NEMO in NF‐kappaB activation. Nat. Cell Biol. 2009. 11: 123–132. [DOI] [PubMed] [Google Scholar]
- 70. Zotti, T. , Uva, A. , Ferravante, A. , Vessichelli, M. , Scudiero, I. , Ceccarelli, M. , Vito, P. et al, TRAF7 protein promotes Lys‐29‐linked polyubiquitination of IkappaB kinase (IKKgamma)/NF‐kappaB essential modulator (NEMO) and p65/RelA protein and represses NF‐kappaB activation. J. Biol. Chem. 2011. 286: 22924–22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Song, G. , Liu, B. , Li, Z. , Wu, H. , Wang, P. , Zhao, K. , Jiang, G. et al, E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63‐linked ubiquitination of TBK1. Nat. Immunol. 2016. 17: 1342–1351. [DOI] [PubMed] [Google Scholar]
- 72. Nakhaei, P. , Mesplede, T. , Solis, M. , Sun, Q. , Zhao, T. , Yang, L. , Chuang, T. H. et al, The E3 ubiquitin ligase Triad3A negatively regulates the RIG‐I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog 2009. 5: e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao, K. , Zhang, M. , Zhang, L. , Wang, P. , Song, G. , Liu, B. , Wu, H. et al, Intracellular osteopontin stabilizes TRAF3 to positively regulate innate antiviral response. Sci. Rep. 2016. 6: 23771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cui, J. , Li, Y. , Zhu, L. , Liu, D. , Songyang, Z. , Wang, H. Y. and Wang, R. F. , NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol 2012. 13: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lin, M. , Zhao, Z. , Yang, Z. , Meng, Q. , Tan, P. , Xie, W. , Qin, Y. et al, USP38 inhibits Type I interferon signaling by editing TBK1 ubiquitination through NLRP4 signalosome. Mol Cell 2016. 64: 267–281. [DOI] [PubMed] [Google Scholar]
- 76. Saitoh, T. , Tun‐Kyi, A. , Ryo, A. , Yamamoto, M. , Finn, G. , Fujita, T. , Akira, S. et al, Negative regulation of interferon‐regulatory factor 3‐dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol 2006. 7: 598–605. [DOI] [PubMed] [Google Scholar]
- 77. Zhang, M. , Tian, Y. , Wang, R. P. , Gao, D. , Zhang, Y. , Diao, F. C. , Chen, D. Y. et al, Negative feedback regulation of cellular antiviral signaling by RBCK1‐mediated degradation of IRF3. Cell Res. 2008. 18: 1096–1104. [DOI] [PubMed] [Google Scholar]
- 78. Lei, C. Q. , Zhang, Y. , Xia, T. , Jiang, L. Q. , Zhong, B. and Shu, H. B. , FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J. Biol. Chem. 2013. 288: 12596–12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Higgs, R. , Ni Gabhann, J. , Ben Larbi, N. , Breen, E. P. , Fitzgerald, K. A. and Jefferies, C. A. , The E3 ubiquitin ligase Ro52 negatively regulates IFN‐beta production post‐pathogen recognition by polyubiquitin‐mediated degradation of IRF3. J. Immunol. 2008. 181: 1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yu, Y. and Hayward, G. S. , The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 2010. 33: 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gack, M. U. , Shin, Y. C. , Joo, C. H. , Urano, T. , Liang, C. , Sun, L. , Takeuchi, O. et al, TRIM25 RING‐finger E3 ubiquitin ligase is essential for RIG‐I‐mediated antiviral activity. Nature 2007. 446: 916–920. [DOI] [PubMed] [Google Scholar]
- 82. Gack, M. U. , Kirchhofer, A. , Shin, Y. C. , Inn, K. S. , Liang, C. , Cui, S. , Myong, S. et al, Roles of RIG‐I N‐terminal tandem CARD and splice variant in TRIM25‐mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U S A 2008. 105: 16743–16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sanchez, J. G. , Chiang, J. J. , Sparrer, K. M. , Alam, S. L. , Chi, M. , Roganowicz, M. D. , Sankaran, B. et al, Mechanism of TRIM25 catalytic activation in the antiviral RIG‐I pathway. Cell Rep. 2016. 16: 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Oshiumi, H. , Miyashita, M. , Inoue, N. , Okabe, M. , Matsumoto, M. and Seya, T. , The ubiquitin ligase Riplet is essential for RIG‐I‐dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010. 8: 496–509. [DOI] [PubMed] [Google Scholar]
- 85. Oshiumi, H. , Matsumoto, M. , Hatakeyama, S. and Seya, T. , Riplet/RNF135, a RING finger protein, ubiquitinates RIG‐I to promote interferon‐beta induction during the early phase of viral infection. J. Biol. Chem. 2009. 284: 807–817. [DOI] [PubMed] [Google Scholar]
- 86. Yan, J. , Li, Q. , Mao, A. P. , Hu, M. M. and Shu, H. B. , TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG‐I for K63‐linked ubiquitination. J. Mol. Cell Biol. 2014. 6: 154–163. [DOI] [PubMed] [Google Scholar]
- 87. Kuniyoshi, K. , Takeuchi, O. , Pandey, S. , Satoh, T. , Iwasaki, H. , Akira, S. and Kawai, T. , Pivotal role of RNA‐binding E3 ubiquitin ligase MEX3C in RIG‐I‐mediated antiviral innate immunity. Proc. Natl. Acad. Sci. U S A 2014. 111: 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zeng, W. , Sun, L. , Jiang, X. , Chen, X. , Hou, F. , Adhikari, A. , Xu, M. and Chen, Z. J. , Reconstitution of the RIG‐I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 2010. 141: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jiang, X. , Kinch, L. N. , Brautigam, C. A. , Chen, X. , Du, F. , Grishin, N. V. and Chen, Z. J. , Ubiquitin‐induced oligomerization of the RNA sensors RIG‐I and MDA5 activates antiviral innate immune response. Immunity 2012. 36: 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Friedman, C. S. , O'Donnell, M. A. , Legarda‐Addison, D. , Ng, A. , Cardenas, W. B. , Yount, J. S. , Moran, T. M. et al, The tumour suppressor CYLD is a negative regulator of RIG‐I‐mediated antiviral response. EMBO Rep 2008. 9: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fan, Y. , Mao, R. , Yu, Y. , Liu, S. , Shi, Z. , Cheng, J. , Zhang, H. et al, USP21 negatively regulates antiviral response by acting as a RIG‐I deubiquitinase. J. Exp. Med. 2014. 211: 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cui, J. , Song, Y. , Li, Y. , Zhu, Q. , Tan, P. , Qin, Y. , Wang, H. Y. et al, USP3 inhibits type I interferon signaling by deubiquitinating RIG‐I‐like receptors. Cell Res. 2014. 24: 400–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Inn, K. S. , Gack, M. U. , Tokunaga, F. , Shi, M. , Wong, L. Y. , Iwai, K. and Jung, J. U. , Linear ubiquitin assembly complex negatively regulates RIG‐I‐ and TRIM25‐mediated type I interferon induction. Mol Cell 2011. 41: 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Paz, S. , Vilasco, M. , Arguello, M. , Sun, Q. , Lacoste, J. , Nguyen, T. L. , Zhao, T. et al, Ubiquitin‐regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol 2009. 29: 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yoo, Y. S. , Park, Y. Y. , Kim, J. H. , Cho, H. , Kim, S. H. , Lee, H. S. , Kim, T. H. et al, The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat. Commun. 2015. 6: 7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu, B. , Zhang, M. , Chu, H. , Zhang, H. , Wu, H. , Song, G. , Wang, P. et al, The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63‐linked polyubiquitination. Nat Immunol 2017. 18: 214–224. [DOI] [PubMed] [Google Scholar]
- 97. Arimoto, K. , Takahashi, H. , Hishiki, T. , Konishi, H. , Fujita, T. and Shimotohno, K. , Negative regulation of the RIG‐I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U S A 2007. 104: 7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang, L. , Zhao, W. , Zhang, M. , Wang, P. , Zhao, K. , Zhao, X. , Yang, S. et al, USP4 positively regulates RIG‐I‐mediated antiviral response through deubiquitination and stabilization of RIG‐I. J Virol 2013. 87: 4507–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang, W. , Jiang, M. , Liu, S. , Zhang, S. , Liu, W. , Ma, Y. , Zhang, L. et al, RNF122 suppresses antiviral type I interferon production by targeting RIG‐I CARDs to mediate RIG‐I degradation. Proc. Natl. Acad. Sci. U S A 2016. 113: 9581–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhao, K. , Zhang, Q. , Li, X. , Zhao, D. , Liu, Y. , Shen, Q. , Yang, M. et al, Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP‐mediated degradation of RIG‐I. J. Immunol 2016. 196: 1209–1217. [DOI] [PubMed] [Google Scholar]
- 101. Narayan, K. , Waggoner, L. , Pham, S. T. , Hendricks, G. L. , Waggoner, S. N. , Conlon, J. , Wang, J. P. et al, TRIM13 is a negative regulator of MDA5‐mediated type I interferon production. J. Virol 2014. 88: 10748–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang, Y. , Tong, X. and Ye, X. , Ndfip1 negatively regulates RIG‐I‐dependent immune signaling by enhancing E3 ligase Smurf1‐mediated MAVS degradation. J. Immunol. 2012. 189: 5304–5313. [DOI] [PubMed] [Google Scholar]
- 103. Zhong, B. , Zhang, Y. , Tan, B. , Liu, T. T. , Wang, Y. Y. and Shu, H. B. , The E3 ubiquitin ligase RNF5 targets virus‐induced signaling adaptor for ubiquitination and degradation. J. Immunol 2010. 184: 6249–6255. [DOI] [PubMed] [Google Scholar]
- 104. You, F. , Sun, H. , Zhou, X. , Sun, W. , Liang, S. , Zhai, Z. and Jiang, Z. , PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 2009. 10: 1300–1308. [DOI] [PubMed] [Google Scholar]
- 105. Yang, B. , Wang, J. , Wang, Y. , Zhou, H. , Wu, X. , Tian, Z. and Sun, B. , Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J. Immunol. 2013. 190: 3613–3619. [DOI] [PubMed] [Google Scholar]
- 106. Zhang, J. , Hu, M. M. , Wang, Y. Y. and Shu, H. B. , TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63‐linked ubiquitination. J. Biol. Chem. 2012. 287: 28646–28655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhong, B. , Zhang, L. , Lei, C. , Li, Y. , Mao, A. P. , Yang, Y. , Wang, Y. Y. et al, The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 2009. 30: 397–407. [DOI] [PubMed] [Google Scholar]
- 108. Qin, Y. , Zhou, M. T. , Hu, M. M. , Hu, Y. H. , Zhang, J. , Guo, L. , Zhong, B. et al, RNF26 temporally regulates virus‐triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 2014. 10: e1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang, Q. , Liu, X. , Cui, Y. , Tang, Y. , Chen, W. , Li, S. , Yu, H. et al, The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014. 41: 919–933. [DOI] [PubMed] [Google Scholar]
- 110. Chen, M. , Meng, Q. , Qin, Y. , Liang, P. , Tan, P. , He, L. , Zhou, Y. et al, TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell 2016. 64: 105–119. [DOI] [PubMed] [Google Scholar]
- 111. Biggar, K. K. and Li, S. S. , Non‐histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015. 16: 5–17. [DOI] [PubMed] [Google Scholar]
- 112. Levy, D. , Kuo, A. J. , Chang, Y. , Schaefer, U. , Kitson, C. , Cheung, P. , Espejo, A. et al, Lysine methylation of the NF‐kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF‐kappaB signaling. Nat. Immunol. 2011. 12: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mukherjee, N. , Cardenas, E. , Bedolla, R. and Ghosh, R. , SETD6 regulates NF‐kappaB signaling in urothelial cell survival: implications for bladder cancer. Oncotarget 2017. 8: 15114–15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lu, T. , Jackson, M. W. , Wang, B. , Yang, M. , Chance, M. R. , Miyagi, M. , Gudkov, A. V. et al, Regulation of NF‐kappaB by NSD1/FBXL11‐dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. U S A 2010. 107: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ea, C. K. and Baltimore, D. , Regulation of NF‐kappaB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. U S A 2009. 106: 18972–18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cao, W. , Bao, C. , Padalko, E. and Lowenstein, C. J. , Acetylation of mitogen‐activated protein kinase phosphatase‐1 inhibits Toll‐like receptor signaling. J. Exp. Med. 2008. 205: 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Choi, S. J. , Lee, H. C. , Kim, J. H. , Park, S. Y. , Kim, T. H. , Lee, W. K. , Jang, D. J. et al, HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG‐I. EMBO J 2016. 35: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu, H. M. , Jiang, F. , Loo, Y. M. , Hsu, S. , Hsiang, T. Y. , Marcotrigiano, J. and Gale, M. , Jr., Regulation of retinoic acid inducible gene‐I (RIG‐I) activation by the histone deacetylase 6. EBioMedicine 2016. 9: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li, X. , Zhang, Q. , Ding, Y. , Liu, Y. , Zhao, D. , Zhao, K. , Shen, Q. et al, Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 2016. 17: 806–815. [DOI] [PubMed] [Google Scholar]
- 120. Arimoto, K. , Konishi, H. and Shimotohno, K. , UbcH8 regulates ubiquitin and ISG15 conjugation to RIG‐I. Mol Immunol 2008. 45: 1078–1084. [DOI] [PubMed] [Google Scholar]
- 121. Kim, M. J. , Hwang, S. Y. , Imaizumi, T. and Yoo, J. Y. , Negative feedback regulation of RIG‐I‐mediated antiviral signaling by interferon‐induced ISG15 conjugation. J. Virol 2008. 82: 1474–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. He, S. , Zhao, J. , Song, S. , He, X. , Minassian, A. , Zhou, Y. , Zhang, J. et al, Viral pseudo‐enzymes activate RIG‐I via deamidation to evade cytokine production. Mol Cell 2015. 58: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yamada, T. , Horimoto, H. , Kameyama, T. , Hayakawa, S. , Yamato, H. , Dazai, M. , Takada, A. et al, Constitutive aryl hydrocarbon receptor signaling constrains type I interferon‐mediated antiviral innate defense. Nat Immunol 2016. 17: 687–694. [DOI] [PubMed] [Google Scholar]
- 124. Xia, P. , Ye, B. , Wang, S. , Zhu, X. , Du, Y. , Xiong, Z. , Tian, Y. et al, Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol 2016. 17: 369–378. [DOI] [PubMed] [Google Scholar]
- 125. Flotho, A. and Melchior, F. , Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013. 82: 357–385. [DOI] [PubMed] [Google Scholar]
- 126. Fu, J. , Xiong, Y. , Xu, Y. , Cheng, G. and Tang, H. , MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol Immunol 2011. 48: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mi, Z. , Fu, J. , Xiong, Y. and Tang, H. , SUMOylation of RIG‐I positively regulates the type I interferon signaling. Protein Cell 2010. 1: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hu, M. M. , Liao, C. Y. , Yang, Q. , Xie, X. Q. and Shu, H. B. , Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG‐I and MDA5. J. Exp. Med. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hu, M. M. , Yang, Q. , Xie, X. Q. , Liao, C. Y. , Lin, H. , Liu, T. T. , Yin, L. et al, Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016. 45: 555–569. [DOI] [PubMed] [Google Scholar]
- 130. Saul, V. V. , Niedenthal, R. , Pich, A. , Weber, F. and Schmitz, M. L. , SUMO modification of TBK1 at the adaptor‐binding C‐terminal coiled‐coil domain contributes to its antiviral activity. Biochim Biophys Acta 2015. 1853: 136–143. [DOI] [PubMed] [Google Scholar]
- 131. Decque, A. , Joffre, O. , Magalhaes, J. G. , Cossec, J. C. , Blecher‐Gonen, R. , Lapaquette, P. , Silvin, A. et al, Sumoylation coordinates the repression of inflammatory and anti‐viral gene‐expression programs during innate sensing. Nat Immunol 2016. 17: 140–149. [DOI] [PubMed] [Google Scholar]
- 132. Rajsbaum, R. , Albrecht, R. A. , Wang, M. K. , Maharaj, N. P. , Versteeg, G. A. , Nistal‐Villan, E. , Garcia‐Sastre, A. et al, Species‐specific inhibition of RIG‐I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 2012. 8: e1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Heaton, S. M. , Borg, N. A. and Dixit, V. M. , Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016. 213: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Calistri, A. , Munegato, D. , Carli, I. , Parolin, C. and Palu, G. , The ubiquitin‐conjugating system: multiple roles in viral replication and infection. Cells 2014. 3: 386–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chang, T. H. , Kubota, T. , Matsuoka, M. , Jones, S. , Bradfute, S. B. , Bray, M. and Ozato, K. , Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog 2009. 5: e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lemaitre, B. , Nicolas, E. , Michaut, L. , Reichhart, J. M. and Hoffmann, J. A. , The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996. 86: 973–983. [DOI] [PubMed] [Google Scholar]
- 137. Bailly, V. , Lauder, S. , Prakash, S. and Prakash, L. , Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997. 272: 23360–23365. [DOI] [PubMed] [Google Scholar]
- 138. Ciancanelli, M. J. , Huang, S. X. , Luthra, P. , Garner, H. , Itan, Y. , Volpi, S. , Lafaille, F. G. et al, Infectious disease. Life‐threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 2015. 348: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Perez de Diego, R ., Sancho‐Shimizu, V. , Lorenzo, L. , Puel, A. , Plancoulaine, S. , Picard, C. , Herman, M. et al, Human TRAF3 adaptor molecule deficiency leads to impaired Toll‐like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 2010. 33: 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rice, G. I. , del Toro Duany, Y. , Jenkinson, E. M. , Forte, G. M. , Anderson, B. H. , Ariaudo, G. , Bader‐Meunier, B. et al, Gain‐of‐function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 2014. 46: 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]


