Abstract
This nonrandomized clinical trial tested [18F]FluorThanatrace as a method for measuring regional poly–(adenosine diphosphate–ribose) polymerase 1 expression in breast cancer and as a potential functional biomarker for breast cancer poly–(adenosine diphosphate–ribose) polymerase inhibitor response.
Clinical trial data demonstrate poly–(adenosine diphosphate–ribose) polymerase (PARP) inhibitor drug efficacy in individuals with BRCA1/2 pathogenic variants, but not all germline pathogenic variant carriers respond, and some without germline pathogenic variants also derive significant benefit. [18F]FluorThanatrace ([18F]FTT) is a radiolabeled PARP inhibitor that enables noninvasive quantification of PARP-1.1,2 In vitro data demonstrate that the level of PARP-1 correlates positively with cytotoxicity of PARP inhibitors2 and that PARP-1 expression is required for PARP inhibitor efficacy.1,2,3,4 In this prospective nonrandomized clinical trial, we report on PARP-1 positron emission tomography imaging using [18F]FTT in patients with breast cancer across a range of breast cancer phenotypes. These proof-of-concept results provide support for testing [18F]FTT as a method for measuring regional PARP-1 expression in breast cancer and as a potential functional biomarker for breast cancer PARP inhibitor response.
Methods
This trial (NCT03083288) took place at the University of Pennsylvania from May 2017 to October 2018. The institutional review board at the University of Pennsylvania approved the protocol (Supplement), and written informed consent was obtained from 30 women with untreated stage I through IV breast cancer. Uptake in primary breast tumor was quantified as the partial volume corrected, maximum standardized uptake value (SUV) acquired 50 to 55 minutes postinjection of 351 to 434 MBq (9.5-11.7 mCi) of [18F]FTT. [18F]FTT uptake was compared across breast cancer subtypes segregated by receptor and pathogenic variant carrier status. Germline (n = 29) and tumor (n = 24) DNA were extracted by standard laboratory protocols, or clinical genetic testing was performed (n = 1). DNA repair pathogenic variants were identified, and allele-specific loss of heterozygosity (LOH) was determined as previously described.5 Kruskal-Wallis (nonparametric analysis of variance) or t test was used to determine statistical significance of differences in cancer subtype or BRCA 1/2 pathogenic variant carrier status, respectively (2-sided α = .05), using IBM SPSS Statistics, version 25 (IBM Corp). This study followed the Transparent Reporting of Evaluations With Nonrandomized Designs (TREND) reporting guidelines.
Results
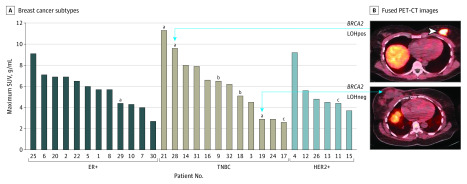
[18F]FTT uptake demonstrated a considerable range (maximum SUV, 2.6-11.3 g/mL); tracer uptake was independent of subtype (estrogen receptor–positive, human epidermal growth factor receptor 2–positive, or triple negative) (Table; P = .73). Maximum SUV among BRCA1/2 pathogenic variant carriers was highly variable (range, 2.9-11.3 g/mL), with levels of uptake overlapping the range of BRCA1/2 pathogenic variant noncarriers (Table; P = .50). Anecdotally, higher uptake was noted in tumors of BRCA2 pathogenic variant carriers with allele-specific LOH (patients 21 and 28 were LOH positive; patients 19 and 29 were LOH negative), as might be expected in tumors with functional homologous repair deficiency (Figure). No somatic pathogenic variants in DNA repair genes were identified.
Table. [18F]FluorThanatrace Uptake, Receptor, and Germline Pathogenic Variant Status .
| Patient identification No. (n = 30) | Maximum SUV, g/mL | Receptor | Germline status |
|---|---|---|---|
| 1 | 5.7 | ER+ | Negative |
| 2 | 6.9 | ER+ | Negative |
| 3 | 4.5 | TNBC | Negative |
| 4 | 9.2 | HER2+ | Negative |
| 5 | 6.0 | ER+ | Negative |
| 6 | 7.1 | ER+ | Negative |
| 7 | 4.0 | ER+ | Negative |
| 8 | 5.7 | ER+ | Negative |
| 9 | 6.5 | TNBC | BRCA1 positive |
| 10 | 4.3 | ER+ | Negative |
| 11 | 4.4 | HER2+ | CHEK2 positive |
| 12 | 5.6 | HER2+ | Negative |
| 13 | 4.5 | HER2+ | Negative |
| 14 | 8.0 | TNBC | Negative |
| 15 | 3.7 | HER2+ | Negative |
| 16 | 6.6 | TNBC | Negative |
| 17 | 2.6 | TNBC | CHEK2 positive |
| 18 | 5.1 | TNBC | BRCA1 positive |
| 19 | 2.9 | TNBC | BRCA2 positive |
| 20 | 6.9 | ER+ | Negative |
| 21 | 11.3 | TNBC | BRCA2 positive |
| 22 | 6.5 | ER+ | Negative |
| 24 | 2.9 | TNBC | Negative |
| 25 | 9.1 | ER+ | Negative |
| 26 | 4.8 | HER2+ | Negative |
| 28 | 9.6 | TNBC | BRCA2 positive |
| 29 | 4.4 | ER+ | BRCA2 positive |
| 30 | 2.7 | ER+ | Negative |
| 31 | 7.9 | TNBC | Negative |
| 32 | 6.2 | TNBC | Negative |
Abbreviations: ER+, estrogen receptor–positive; HER2+, human epidermal growth factor receptor 2–positive; SUV, standardized uptake value; TNBC, triple negative breast cancer.
Figure. [18F]FluorThanatrace Maximum Standardized Uptake Value (SUV) vs Tumor Subtype.
A, Plots by breast cancer subtype show overlapping ranges of tracer uptake. B, Representative fused positron emission tomography–computed tomography (PET-CT) of patient 28 (top) and patient 19 (bottom) illustrate degree of [18F]FluorThanatrace tumor uptake. ER+ indicates estrogen receptor–positive; HER2+, human epidermal growth factor receptor 2–positive; LOHneg, loss of heterozygosity negative; LOHpos, loss of heterozygosity positive; TNBC, triple negative breast cancer.
aBRCA2.
bBRCA1.
cCHEK2.
Discussion
While germline BRCA1/2 pathogenic variant carrier status is currently the accepted biomarker of PARP inhibitor sensitivity, there is variability in response, and overall, more benefit derived by BRCA1 pathogenic variant carriers compared with BRCA2 pathogenic variant carriers. Additionally, multiple ongoing trials examining PARP inhibitor use outside of germline BRCA1/2 indicate a need to find biomarkers for patient selection. Initial results for in vitro biomarkers that include homologous recombination deficiency score, poly–(adenosine diphosphate–ribose) polymers, Rad-51, p53, and microRNAs have been mixed.6 In addition, intratumoral and intertumoral heterogeneity can lead to sampling error, especially in the case of metastatic disease, and pathologic measurements require tissue sampling, which limits the ability for repeat measurements. The study has limitations such as only 30 participants included and the validity of this biomarker to predict response to PARP inhibitors in humans has not been tested, although studies are ongoing and in vitro models have had promising results.1,2
Results of this study demonstrate that in vivo PARP-1 expression is highly variable in breast cancer, and variability appears independent of traditional predictors of increased expression. Although the number of patients is limited, a similar range of [18F]FTT uptake was seen in BRCA pathogenic variant carriers and noncarriers. These early observations support the ability of positron emission tomography to quantitate regional PARP-1 expression. When taken together with preclinical model results,1,2 these findings suggest this approach might help identify patients who will respond to PARP inhibitors. PARP imaging probes might also be used to gain insight into drug pharmacokinetics and pharmacodynamics, including drug delivery. Overall, these early results provide rationale for continued development of [18F]FTT as a companion diagnostic for breast cancer PARP inhibitor therapy.
Trial Protocol
References
- 1.Makvandi M, Pantel A, Schwartz L, et al. A PET imaging agent for evaluating PARP-1 expression in ovarian cancer. J Clin Invest. 2018;128(5):2116-2126. doi: 10.1172/JCI97992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makvandi M, Xu K, Lieberman BP, et al. A radiotracer strategy to quantify parp-1 expression in vivo provides a biomarker that can enable patient selection for PARP inhibitor therapy. Cancer Res. 2016;76(15):4516-4524. doi: 10.1158/0008-5472.CAN-16-0416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins TA, Ainsworth WB, Ellis PA, et al. PARP1 trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol Cancer Res. 2019;17(2):409-419. doi: 10.1158/1541-7786.MCR-18-0138 [DOI] [PubMed] [Google Scholar]
- 4.Pettitt SJ, Rehman FL, Bajrami I, et al. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PLoS One. 2013;8(4):e61520. doi: 10.1371/journal.pone.0061520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maxwell KN, Wubbenhorst B, Wenz BM, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun. 2017;8(1):319. doi: 10.1038/s41467-017-00388-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartron E, Theillet C, Guiu S, Jacot W. Targeting homologous repair deficiency in breast and ovarian cancers: biological pathways, preclinical and clinical data. Crit Rev Oncol Hematol. 2019;133:58-73. doi: 10.1016/j.critrevonc.2018.10.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial Protocol