Abstract
The human (h) zinc transporter ZIP4 is expressed on the plasma membrane and functions to increase cytosolic zinc levels. Mutations in hZIP4 cause the disease acrodermatitis enteropathica. Dysfunction in the regulation of hZIP4 has also been indicated in solid tissue cancers, including pancreatic and prostate cancer. Although structural studies of the extracellular domain and computational modeling of the membrane domain suggest hZIP4 exists as a dimer, the oligomerization status of hZIP4 in the plasma membrane of mammalian cells has not been directly quantified in vivo. Here, the oligomeric state of hZIP4 expressed in HEK293 cells was quantified using fluorescence correlation spectroscopy. hZIP4 was tagged with eGFP, and by comparing brightness values (ε) of monomer and tandem eGFP constructs to that of an hZIP4/eGFP, we show that hZIP4 is a dimer. Determining that hZIP4 is a dimer is an important step toward understanding the function and processing of the protein, which can provide more insight into how diseases affected by hZIP4 occur and can be managed.
Zinc is an essential metal in the body that is involved in enzymatic catalysis, gene regulation, and signaling.1 Intracellular zinc levels are controlled in humans via the zinc- and iron-regulated transport protein (ZIP) and zinc transport (ZnT) protein families.1 These families increase and decrease cytosolic zinc levels, respectively. There are 14 human ZIP proteins that are subdivided into four subfamilies (ZIPI, ZIPII, gufA, and LIV-1).1
ZIP4 (LIV-1 subfamily) has a critical role in the dietary uptake of zinc, cellular signaling, and embryonic development.1 Mutations in the human (h) ZIP4 transporter can lead to the disease acrodermatitis enteropathica (AE).2 AE-causing mutations are found throughout the hZIP4 sequence. These mutations can result in retention of hZIP4 on the endoplasmic reticulum (ER) and/or dysfunction of zinc transport.2,3 Additionally, hZIP4 overexpression has been shown to play a direct role in solid tissue tumors, including pancreatic, liver, and brain cancer.4,5
hZIP4 encodes eight transmembrane (TM) domains with a 327-residue N-terminal extracellular domain (ECD).1 The ECD is cleaved from the holotransporter when cells are deficient in zinc.3 Mutations within the N-terminal ECD have been implicated in protein dysfunction.2 For example, AE-causing mutations that occur near the N-terminal cleavage site prevent processing of ZIP4, resulting in decreased cellular uptake of zinc.3 The roles of AE-causing mutations in the ECD were further elucidated in the crystal structure of the ECD from the black fruit bat Pteropus alecto pZIP4.6 The pZIP4 ECD crystal structure revealed a homodimer with many AE mutations mapping to sites suggested to be important for structural stability, particularly mutations at or near a highly conserved proline-alanine-leucine (PAL) motif. The PAL motif is at the dimerization interface and likely plays a key role in protein stabilization.6 The high degree of conservation of the PAL motif, as well as other regions of the ECD, suggests that, like pZIP4 ECD, other ZIP homologues, including hZIP4, may form dimers via the ECD.6
Recently, the architecture of the membrane domain of ZIP proteins has been investigated using computational modeling and structural approaches.7,8 Inspection of models generated using co-evolution contact data showed a core structure of TMs II, IV, V, and VII with peripheral TMs I, III, VI, and VIII. Alanine substitution of histidine residues within TMs IV and V demonstrated that these residues contribute to transition metal selectivity and transport.7 This hZIP4 TM computational model was confirmed by inspection of the prokaryotic Bordetella bronchiseptica ZIP (BbZIP) crystal structure.8 The BbZIP crystal structure highlighted a binuclear metal center that was proposed to form the metal translocation pathway.8 The crystal structure of the BbZIP transmembrane domain matched well with the hZIP4 computational model, with the notable exception of the oligomeric state. The BbZIP protein crystallized as a monomer, whereas the computational modeling predicted a dimeric hZIP4 structure.7,8 Neither crystallography nor computational modeling is a definitive way to quantify the oligomerization state as the protein is not within the native plasma membrane. Additionally, bacterial ZIP proteins are homologous to the ZIP guf-A subfamily, while hZIP4 is a LIV-1 subfamily protein.9
To quantify the oligomeric state of hZIP4 in vivo, we determined the quaternary structure of hZIP4 within the plasma membrane of mammalian cells using fluorescence correlation spectroscopy (FCS). FCS is a single-molecule detection method that measures changes in fluorescence intensity as fluorescent molecules diffuse through a precise small volume.10‘11 Previous studies have shown that monomers and dimers show a comparative brightness where the brightness of a dimer eGFP2 is double that of a monomer eGFP regardless of the concentration of the plasmid.10 Because brightness values (ε) correspond to the number of fluorescent molecules in a protein complex, ε values can be used to determine the oligomeric state of a protein, using known monomer and dimer construct controls.12,13 Here we use known monomeric (CD86/eGFP) and tandem (CD86/eGFP/eGFP) plasma membrane receptors with C-terminal eGFP.12 Compared to other methods, such as chemical crosslinking or native polyacrylamide gel electrophoresis, each of which is used to determine the oligomeric state of membrane proteins, FCS observes proteins in their native state on the plasma membrane surface of living cells.12–16
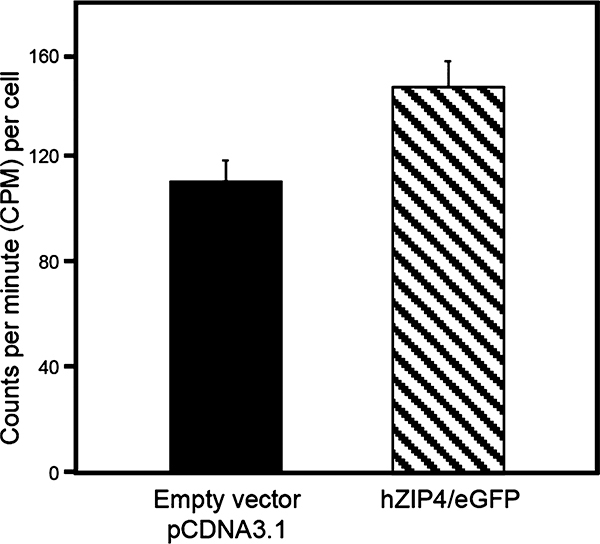
The level of accumulation of zinc in cells transfected with hZIP4/eGFP (147 ± 10.4 cpm per cell) was significantly greater than that for cells transfected with the empty vector [112 ± 8.0 cpm per cell (see the Supporting Information for Methodology)] (Figure 1). These data, which are equivalent to transport function of HA-tagged hZIP4 in HEK293 cells, demonstrate that the hZIP4/eGFP fusion protein is functionally expressed at the plasma membrane of HEK293 cells.17
Figure 1.
hZIP4 is functional with the eGFP tag. HEK293 cells were transfected with either the empty vector (pcDNA3.1) as a negative control or hZIP4/eGFP (black or dashed bar, respectively). After 48 h, cells were lysed and incubated with 5 μM 65Zn for 10 min. The cells were then washed, and the amount of 65Zn that accumulated was measured. This is a representative data set in which n = 8; a total of three independent experiments were completed with similar results. Error bars here represent the standard error of the mean. Data were found to be significant using a t test with p < 0.05.
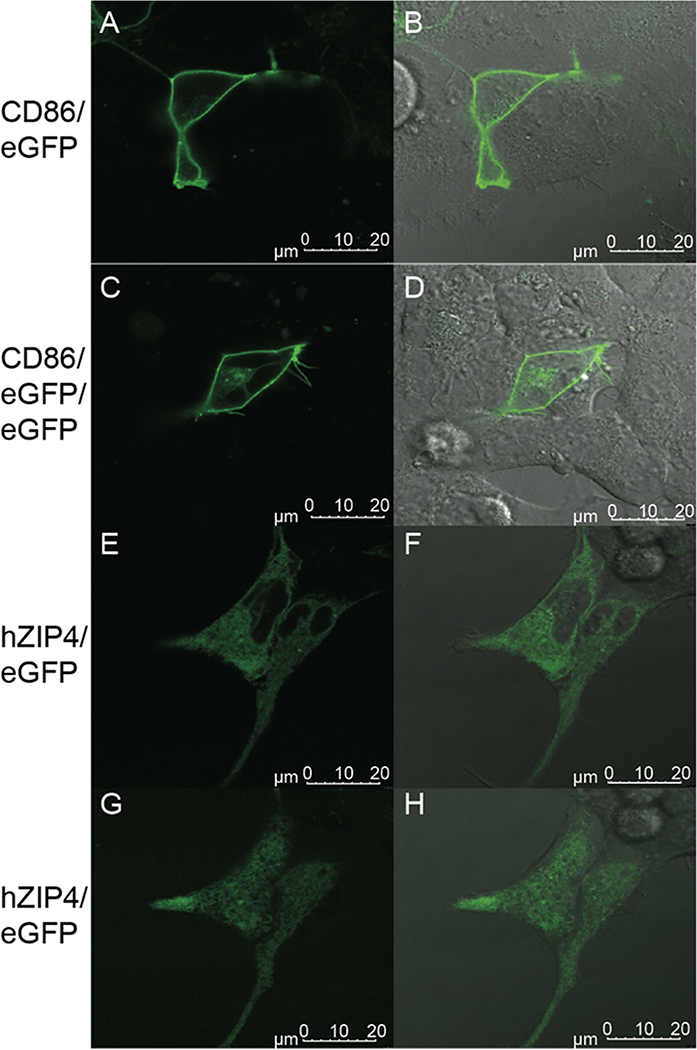
To determine the oligomerization state of hZIP4 in the plasma membrane of HEK293 cells, FCS measurements were recorded on cells expressing hZIP4/eGFP as described previously.12,18 Sample images of cells transfected with CD-86/eGFP, CD-86/eGFP/eGFP, or hZIP4/eGFP constructs were taken (Figure 2). Although a significant amount of hZIP4/eGFP is retained intracellularly, when we focus on the plasma membrane, hZIP4 is surface-expressed (Figure 2G,H). Some hZIP4 is internalized due to the fetal bovine serum in the medium contributing high levels of external zinc, leading to some endocytosis of the hZIP4/eGFP protein. Additionally, variance among membrane proteins can occur in their distribution in the ER, Golgi, endosomes, and plasma membrane due to different rates of trafficking after synthesis. Because of this, the hZIP4/eGFP protein concentration on the membrane is not as pronounced as that of the receptor controls (Figure 2A–D); however, FCS experiments are still possible due to the single-molecule specificity of the method, which requires only small amounts of the molecule in a very small volume.11
Figure 2.
Confocal microscopy showing plasma membrane localization of proteins. HEK293 cells were transfected with either (A and B) monomeric CD-86/eGFP, (C and D) tandem CD-86/eGFP/eGFP, or (E–H) hZIP4/eGFP. After 24 h, cells were imaged using confocal microscopy. Images A–F represent the focus on the center of a cell on the z-axis, while images G and H show the focus on the upper plasma membrane, where FCS measurements were taken. Images G and H also demonstrate the high level of expression of hZIP4/eGFP on the cell membrane. Images B, D, F, and H show an overlay of fluorescence excitation with a DIC image of the cells.
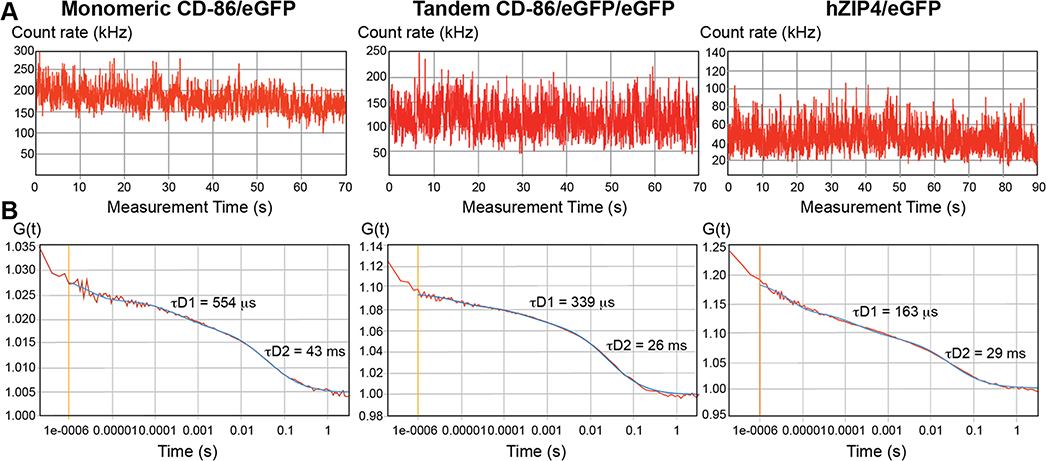
FCS measurements (see the Supporting Information for Methodology) were taken on the upper plasma membrane above the nucleus, as shown in Figure 2G and as has been previously described for quantification of the oligomeric state of membrane proteins.12,13,18 FCS measures fluorescent fluctuations as the membrane molecules diffuse in and out of the path of laser excitation.10,11 Fluorescence-tagged proteins that associate with each other or belong to the same protein complex diffuse through the membrane together. Raw counts of fluorescence intensity (Figure 3A) are plotted as an autocorrelation curve G(τ) (Figure 3B). The amplitude of the autocorrelation curve (y-intercept) is inversely proportional to the number of molecules, N, observed through that measurement.18 As FCS measurements are highly sensitive to the number of molecules in the observation volume, N values for each curve were kept low and were, on average, 5–15 molecules. Additionally, to avoid unusually bright or dim cells, cells with low to medium brightness and count rates (k) between 50 and 250 kHz were chosen.18
Figure 3.
Fluorescence correlation spectroscopy (FCS) autocorrelation curves of eGFP-tagged membrane proteins. FCS recordings were made on the plasma membrane of transiently transfected HEK293 cells expressing the indicated eGFP-tagged proteins. These graphs are representative examples taken from one of three independent studies with multiple samples. (A) Fluorescence intensity fluctuations from which recordings outside the range of 50–250 kHz were excluded. (B) Autocorrelation curve of FCS data (red line) with a model curve fitting a two-dimensional two-component model (blue line). Diffusion times for the fast (τD1) and slow (τD2) components are shown.
The brightness values for cytosolic eGFP and membrane-bound monomeric CD86/eGFP were 3200 ± 300 and 3800 ± 100 counts per second per molecule (CPSM), respectively. These values were expected to be similar, because they are both monomeric, regardless of the orientation in the cell (Table 1). Furthermore, the brightness values for the CD86/eGFP/eGFP tandem and for the hZIP4/eGFP protein are similar (7800 ± 200 and 6700 ± 200 CPSM, respectively) (Table 1). It is also clear that the hZIP/eGFP brightness value is approximately 2 times the amount of the monomeric CD86/eGFP protein. Together, these data demonstrate that, on the surface of HEK293 cells, hZIP4/eGFP is a dimer (Table 1).
Table 1.
FCS Analysis of HEK293 Cells Expressing either Cytosolic, Membrane-Bound Monomeric, or Tandem eGFP Controls or eGFP-Tagged hZIP4 Protein
| sample | brightness (CPSM)a | reduced χ2b | n |
|---|---|---|---|
| cytosolic eGFP | 3200 ± 300 | (5 ± 3) × 10−7 | 11 |
| monomeric CD-86/eGFP | 3800 ± 100 | (9 ± 2) × 10−7 | 19 |
| tandem CD-86/eGFP/eGFP | 7800 ± 200 | (2 ± 3) × 10−7 | 18 |
| hZIP4/eGFP | 6700 ± 200 | (3 ± 1) × 10−7 | 17 |
Brightness values are reported as counts per second per molecule (CPSM), which are determined by dividing the photon count rate by the number of molecules as seen in eq 1 in the Supporting Information.
Data were fit to a two-dimensional two-component model, and resulting χ2 values are listed. χ2 refers to the goodness of fit of the data to the model. Data represent the mean ± the standard error of the mean for the number of cells examined (n).
These results have several implications. The hZIP4/eGFP brightness values are not exactly double the monomer value; this could possibly be due to hZIP4/eGFP forming dimers with native hZIP4 proteins that do not have the eGFP tag, causing an overall decrease in brightness values. Additionally, hZIP4/eGFP may associate with other ZIP proteins on the membrane, as many, including ZIP7 and ZIP13, are indicated as structurally being dimers themselves and ZIP6 and ZIP10 have been shown to form a heteromer.19–21
The establishment of hZIP4 as a homodimer on the plasma membrane has broad implications, particularly in the determination of how zinc is transported across the membrane. Although the structural unit is a dimer, the functional unit, whether monomer or dimer, remains to be elucidated. The structural evidence strongly supports a metal translocation pathway located within a protomer.7,8 However, many membrane protein transporters with translocation pathways contained within individual protomers exhibit functional cooperativity between subunits.22,23 Additionally, oligomerization has been shown to be required for proper processing and plasma membrane targeting for some membrane proteins. Understanding the role of hZIP4 dimerization in function and localization of the protein will yield more insight into how diseases affected by hZIP4 occur and can be managed. Furthermore, elucidating the hZIP4 dimer interface will provide potential drug targets for diseases, such as cancers, where ZIP proteins are upregulated.4,5
Although determination of the oligomeric state of membrane proteins remains a highly debated topic, FCS is a robust real-time, in vivo approach to quantifying the oligomeric states of many integral membrane proteins.12–15,24 Using the molecular brightness, proportional to the number of fluorescent molecules in a protein complex, compared to structurally known control proteins, the oligomerization state for a given protein can be determined. Molecular brightness values of hZIP4/eGFP were similar to those of the tandem (dimeric) control and twice that of the monomeric control, indicating a homodimeric structure within the plasma membrane. These data are not only consistent with previous studies of ZIP4 homologue structures but also similar to data from other studies indicating dimer structures in other ZIP proteins.19–21 This is the first study to directly measure the oligomerization status of any ZIP protein in living cells and to successfully demonstrate that hZIP4 is a homodimer on the plasma membrane of mammalian cells.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Herrick-Davis for CD86/GFP and CD86/GFP/GFP plasmids and the staff at the CCAM Microscopy Facility for help with initial FCS measurements. This study made use of the LSM 780 confocal microscope, at the CCAM Microscopy Facility, at the University of Connecticut Health Center.
Funding
Funding was provided by the Worcester Polytechnic Institute Research Foundation and National Institute of Health Grant R01 GM105964 to R.E.D.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.9b00131.
Transport assay method and FCS analysis method (PDF)
Accession Codes:
hZIP4, entry Q6P5W5
REFERENCES
- (1).Jeong J, and Eide DJ (2013) The SLC39 family of zinc transporters. Mol. Aspects Med. 34, 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Andrews GK (2008) Regulation and function of Zip4, the acrodermatitis enteropathica gene. Biochem. Soc. Trans. 36, 1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kambe T, and Andrews GK (2009) Novel proteolytic processing of the ectodomain of the zinc transporter ZIP4 (SLC39A4) during zinc deficiency is inhibited by acrodermatitis enteropathica mtations. Mol. Cell. Biol. 29, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, and Andrews GK (2010) Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS One 5, e13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lin Y, Chen Y, Wang Y, Yang J, Zhu VF, Liu Y, Cui X, Chen L, Yan W, Jiang T, Hergenroeder GW, Fletcher SA, Levine JM, Kim DH, Tandon N, Zhu J, and Li M (2013) ZIP4 is a novel molecular marker for glioma. Neuro-Oncol. 15, 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhang T, Sui D, and Hu J (2016) Structural insights of ZIP4 extracellular domain critical for optimal zinc transport. Nat. Commun. 7, 11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Antala S, Ovchinnikov S, Kamisetty H, Baker D, and Dempski RE (2015) Computational modeling and functional studies provide a structural scaffold for the zinc transporter hZIP4. J. Biol. Chem. 290, 17796–17805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhang T, Liu J, Fellner M, Zhang C, Sui D, and Hu J (2017) Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 3, No. e1700344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gaither LA, and Eide DJ (2001) Eukaryotic zinc transporters and their regulation. BioMetals 14, 251–270. [DOI] [PubMed] [Google Scholar]
- (10).Jameson DM, Ross JA, and Albanesi JP (2009) Fluorescence fluctuation spectroscopy: ushering in a new age of enlightenment for cellular dynamics. Biophys. Rev. 1, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kim SA, Heinze KG, and Schwille P (2007) Fluorescence correlation spectroscopy in living cells. Nat. Methods 4, 963–973. [DOI] [PubMed] [Google Scholar]
- (12).Herrick-Davis K, Grinde E, Cowan A, and Mazurkiewicz JE (2013) Fluorescence correlation spectroscopy analysis of serotonin, adrenergic, muscarinic, and dopamine receptor dimerization: The oligomer number puzzle. Mol. Pharmacol. 84, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Parmar VK, Grinde E, Mazurkiewicz JE, and Herrick-Davis K (2017) Beta2-adrenergic receptor homodimers: Role of transmembrane domain 1 and helix 8 in dimerization and cell surface expression. Biochim. Biophys. Acta, Biomembr. 1859, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wu B, Chen Y, and Muller JD (2009) Fluorescence fluctuation spectroscopy of mCherry in living cells. Biophys. J. 96, 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen Y, Wei L-N, and Müller JD (2003) Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 100, 15492–15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).García-Marcos A, Sánchez SA, Parada P, Eid J, Jameson DM, Remacha M, Gratton E, and Ballesta JPG (2008) Yeast ribosomal stalk heterogeneity in vivo shown by two-photon FCS and molecular brightness analysis. Biophys. J. 94, 2884–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mao X, Kim B-E, Wang F, Eide DJ, and Petris MJ (2007) A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J. Biol. Chem. 282, 6992–7000. [DOI] [PubMed] [Google Scholar]
- (18).Herrick-Davis K, and Mazurkiewicz JE (2013) Fluorescence correlation spectroscopy and photon-counting histogram analysis of receptor-receptor interactions In Methods in Cell Biology (Conn PM, Ed.) Vol 117, pp 181–196, Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- (19).Bin B-H, Fukada T, Hosaka T, Yamasaki S, Ohashi W, Hojyo S, Miyai T, Nishida K, Yokoyama S, and Hirano T (2011) Biochemical characterization of human ZIP13 protein. J. Biol. Chem. 286, 40255–40265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Taylor KM, Hiscox S, Nicholson RI, Hogstrand C, and Kille P (2012) Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci. Signaling 5, ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Taylor KM, Muraina IA, Brethour D, Schmitt-Ulms G, Nimmanon T, Ziliotto S, Kille P, and Hogstrand C (2016) Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem. J. 473, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Veenhoff LM, Heuberger EHML, and Poolman B (2002) Quaternary structure and function of transport proteins. Trends Biochem. Sci. 27, 242–249. [DOI] [PubMed] [Google Scholar]
- (23).Alguel Y, Cameron AD, Diallinas G, and Byrne B (2016) Transporter oligomerization: form and function. Biochem. Soc. Trans. 44, 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Herrick-Davis K, Grinde E, Lindsley T, Cowan A, and Mazurkiewicz JE (2012) Oligomer size of the serotonin 5-hydroxytryptamine 2C (5-HT 2C) receptor revealed by fluorescence correlation spectroscopy with photon counting histogram analysis. J. Biol. Chem. 287, 23604–23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.