SUMMARY
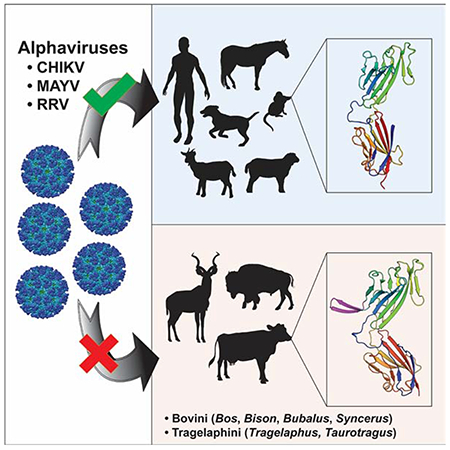
Alphaviruses are emerging, mosquito-transmitted RNA viruses with poorly understood cellular tropism and species selectivity. Mxra8 is an entry receptor for multiple alphaviruses including chikungunya virus (CHIKV). We discovered that while expression of mouse, rat, chimpanzee, dog, horse, goat, sheep, and human Mxra8 enables alphavirus infection in cell culture, cattle Mxra8 does not. Cattle Mxra8 encodes a 15-amino acid insertion in its ectodomain that prevents Mxra8 binding to CHIKV. Identical insertions are present in zebu, yak, and the extinct auroch. As other Bovinae lineages contain related Mxra8 sequences, this insertion likely occurred at least 5 million years ago. Removing the Mxra8 insertion in Bovinae enhances alphavirus binding and infection, while introducing the insertion into mouse Mxra8 blocks CHIKV binding, prevents infection by multiple alphaviruses in cells, and mitigates CHIKV-induced pathogenesis in mice. Our studies showing this insertion provides resistance to CHIKV infection could facilitate countermeasures that disrupt Mxra8 interactions with alphaviruses.
eTOC Blurb
Kim et al. identify a sequence insertion in the Mxra8 receptor of Bovinae species that prevents alphavirus binding. Deletion of the Bovinae Mxra8 insertion restores alphavirus infection, while mice engineered with the Mxra8 insertion exhibit reduced CHIKV infection. Identification of this insertion could facilitate countermeasures preventing Mxra8 engagement of alphaviruses.
Graphical Abstract
INTRODUCTION
Alphaviruses are emerging, mosquito-transmitted positive-sense RNA viruses that cause explosive disease outbreaks in humans and animals. These viruses are classified into groups based on their genetic relatedness and historical boundaries. Old World alphaviruses, including chikungunya (CHIKV), Mayaro (MAYV), O’nyong’nyong (ONNV), and Ross River (RRV), cause acute and chronic musculoskeletal disease affecting millions of people globally. New World alphaviruses, including Eastern (EEEV), Venezuelan (VEEV), and Western (WEEV) equine encephalitis viruses, infect the central nervous system in humans and some animals. Despite the epidemic potential of alphaviruses, there are no licensed therapies or vaccines for any family member.
The alphavirus RNA genome encodes four non-structural and five structural proteins using two open reading frames (Strauss et al., 1994). The non-structural proteins are required for virus translation, replication, and immune evasion, and the structural proteins (capsid [C] and envelope [E3-E2-6K-E1]) form the virion. The E1 glycoprotein participates in pH-dependent fusion in the acidified endosome (Lescar et al., 2001), and the E2 glycoprotein binds to entry factors (Smith et al., 1995; Zhang et al., 2005) and facilitates endocytosis (DeTulleo and Kirchhausen, 1998; Lee et al., 2013). The E3 protein is necessary for the folding of the E2-E1 heterodimer (Carleton et al., 1997; Mulvey and Brown, 1995) but is cleaved during the maturation process (Heidner et al., 1996). Mature enveloped alphaviruses form at the plasma membrane with 240 E2-E1 heterodimers assembled into 80 trimeric icosahedral spikes (Cheng et al., 1995; Kostyuchenko et al., 2011; Paredes et al., 1993; Voss et al., 2010).
The basis for receptor engagement, cellular tropism, and species selectivity of alphaviruses is poorly understood. Attachment factors including heparan sulfates have been shown to enhance infection of some alphaviruses (Gardner et al., 2011; Klimstra et al., 1998; Wang et al., 1992). Natural resistance-associated macrophage protein (NRAMP2) has been described as a receptor for SINV but not for CHIKV or RRV (Rose et al., 2011). We identified Mxra8 as a cellular entry receptor for multiple Old World arthritogenic alphaviruses including CHIKV, MAYV, ONNV, and RRV (Zhang et al., 2018a). Expression of Mxra8 facilitated alphavirus binding to and infection of mouse and human fibroblasts, skeletal muscle cells, and chondrocytes, and was required for virulence in mice (Zhang et al., 2019; Zhang et al., 2018a). Mxra8 binds to alphaviruses by wedging into a cleft formed by adjacent E2-E1 heterodimers in one trimeric spike and engaging a neighboring spike (Basore et al., 2019; Song et al., 2019). Apart from its role as an alphavirus receptor, the physiological function of Mxra8 is uncertain. Mxra8 also has been termed adipocyte specific protein 3 (ASP3), limitrin, and DICAM because of reported functions in mesenchymal cell differentiation, blood-brain barrier homeostasis, osteoclast development, and angiogenesis (Han et al., 2013; Jung et al., 2012; Jung et al., 2004; Jung et al., 2008; Yonezawa et al., 2003). Although Mxra8 has these ascribed functions, genetically deficient mice are viable and fertile (Han et al., 2019; Zhang et al., 2019).
Because alphaviruses in nature infect humans and several other animal hosts, here, we tested the ability of mammalian Mxra8 orthologs to facilitate infection. Whereas many mammalian Mxra8 genes support CHIKV infection in complementation studies in Mxra8-deficient cells, surprisingly, cattle Mxra8 does not. Genetic and structural analysis reveal that the ectodomain of cattle Mxra8 contains a 15-amino acid insertion in the C’-C’’ loop of domain 1 (D1), which sterically blocks alphavirus binding and infection. Deletion of this insertion restores the ability of cattle Mxra8 to bind CHIKV and promote infection by multiple alphaviruses. Reciprocally, introduction of the insert into the corresponding site of mouse Mxra8 abrogates CHIKV binding and infection. Indeed, CRISPR-Cas9 engineered mice containing mouse Mxra8 with the 15-amino acid cattle insertion phenocopy a Mxra8 knock-out (KO) mouse (Zhang et al., 2019) with markedly diminished CHIKV infection and disease, confirming a loss of function allele in vivo. Detailed evolutionary analysis reveals that the insertion was present at the same site in most Bovinae family members, which dates its origin to the Miocene epoch. Overall, our experiments explain how an evolutionarily ancient sequence insertion impacts alphavirus-Mxra8 receptor interactions and species tropism and provides a path for developing countermeasures to limit these globally concerning and emerging pathogens.
RESULTS AND DISCUSSION
Alphavirus infection of Mxra8 orthologs.
Gene editing of Mxra8 results in markedly diminished alphavirus infection, and viral infectivity is restored following complementation with mouse Mxra8 or human MXRA8 (Zhang et al., 2018a). Because many alphaviruses infect other vertebrate hosts in epizootic cycles (Weaver et al., 2012), we tested whether Mxra8 orthologs (Fig 1A) support infection of arthritogenic alphaviruses. We complemented 3T3 fibroblasts lacking Mxra8 expression (ΔMxra8) with Mxra8 from mouse (Mus musculus, positive control), rat (Rattus norvegicus), chimpanzee (Pan troglodytes), dog (Canis lupus familiaris), horse (Equus caballus), cattle (Bos taurus), goat (Capra hircus), and sheep (Ovis aries), which vary by 7 to 25 percent at the nucleotide level and 6 to 24 percent at the amino acid level (Table S1). We also tested whether Mxra8 of three avian species, turkey (Meleagris gallopavo), duck (Anas platyrhynchos), and chicken (Gallus gallus), which vary by ~45% at the amino acid level from mouse Mxra8, promote CHIKV infection. Although birds are not a common amplifying host for arthritogenic alphaviruses (Suhrbier et al., 2012) they act as a reservoir for some encephalitic alphaviruses (Weaver et al., 1999). Surface expression of the different Mxra8 orthologs was confirmed with cross-reactive monoclonal antibodies (mAbs) against Mxra8 (Zhang et al., 2018a) or antibodies against an N-terminal tag placed downstream of the signal peptide (Fig S1A–B and S1E–F). Complemented cells were inoculated with CHIKV (strain 181/25) and evaluated for infection by quantifying intracellular viral E2 protein expression by flow cytometry. Consistent with previous findings (Zhang et al., 2018a), CHIKV E2 antigen was absent in ΔMxra8 cells but present at high levels in ΔMxra8 cells complemented with mouse Mxra8. Complementation of ΔMxra8 cells with rat, chimpanzee, dog, horse, goat, and sheep Mxra8 orthologs also restored CHIKV infectivity (Fig 1B–C). However, Mxra8 from cattle and the three more distantly related avian species failed to restore infectivity despite comparable surface expression (Fig 1B–C and S1C–D). Of note, the N-terminal tag used to detect the avian constructs did not diminish the ability of mouse Mxra8 to support CHIKV infection (Fig S1C). Similar results were observed with other CHIKV strains (AF15561 and LR-2006) and MAYV and RRV (Fig 1D–H). In contrast, expression of the mammalian Mxra8 orthologs did not enhance infection of VEEV, an encephalitic alphavirus (Fig 1I). These data are consistent with previous results showing that mouse Mxra8 expression does not enhance infectivity of bunyaviruses, flaviviruses, or rhabdoviruses (Zhang et al., 2018a).
Figure 1. Alphavirus infection of cells expressing Mxra8 gene orthologs.
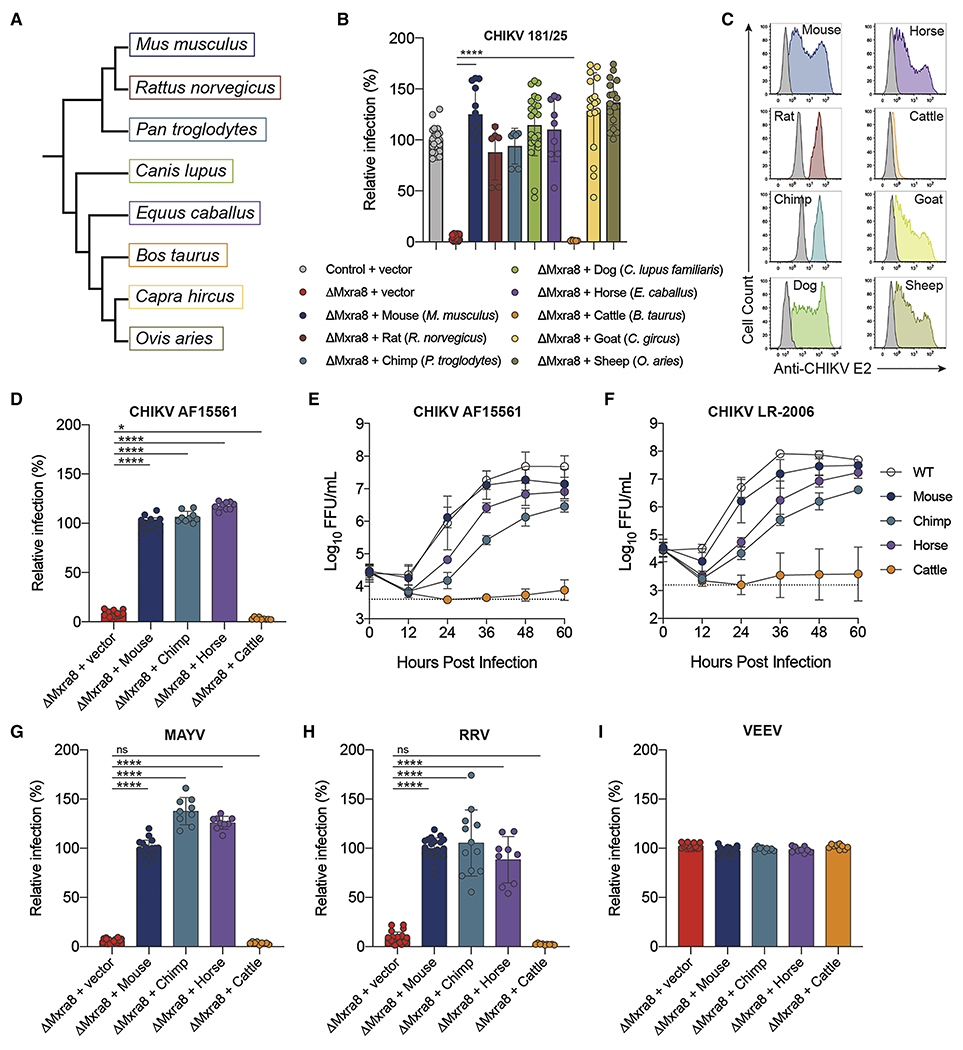
A. Dendrogram of Mxra8 genes in mammals. B-C. Lentivirus complementation of ΔMxra8 3T3 with Mxra8 cDNA from mouse, rat, chimpanzee, dog, horse, cattle, goat, and sheep. Cells were inoculated with CHIKV (181/25) and analyzed for infection by staining with anti-E2 mAbs. Infection data are pooled from three to nine experiments (n = 6 to 24 replicates; one-way ANOVA with Dunnett’s post-test: ****, P < 0.0001) (B) and representative flow cytometry plots of infected cells (C) are shown. Small differences in infection of some species (e.g., chimp and horse) may reflect relative levels of expression of the Mxra8 orthologs (see Fig S1) or inherent differences in affinity of binding. D-I. Lentivirus complementation of ΔMxra8 3T3 with Mxra8 cDNA from mouse, cattle, horse, and chimpanzee. Cells were inoculated with (D-E) CHIKV (AF15561), (F) CHIKV (LR-2006) (G) MAYV (BeH407), (H) RRV (T48), or (I) VEEV (TC-83) and stained with virus-specific anti-E2 mAbs to quantify infection (see STAR Methods). For E-F, a multi-step growth analysis was conducted, and virus was titrated by focus-forming assay. Data are pooled from three experiments performed in triplicate (one-way ANOVA with Dunnett’s post-test: *, P < 0.05; ****, P < 0.0001; n.s., not significant). See Fig S1.
Cattle Mxra8 contains a repeat sequence insertion.
To define the mechanism by which cattle Mxra8 fails to promote alphavirus infectivity, we first aligned its sequence with mouse Mxra8. Cattle Mxra8 contains a 15-amino acid insertion composed of three quasi-identical (GEQRL/V) five-residue repeats that appear to be derived from an immediately adjacent GEQRV gene sequence (Fig S2A). These sequences are encoded by GC-rich tandem repeats and expand a CpG island (Fig S2B) while preserving the protein coding frame. CpG islands often are hypomethylated (Kundaje et al., 2015) and associated with genome instability (Du et al., 2014). Thus, analogous to well-characterized structural variations in human coding exons (Challis et al., 2015; Montgomery et al., 2013), this GC-rich region is prone to form a single-stranded DNA loop (Fig S2C) as a consequence of polymerase slippage during DNA replication (Tian et al., 2011), which may have caused the nucleotide insertion in Mxra8.
Structural analysis of cattle Mxra8.
We expressed cattle Mxra8 protein in E. coli (Fig S3A–B) and obtained a 2.3 Å crystal structure (Fig 2A and Table S2). Cattle Mxra8, like mouse Mxra8, adopts two immunoglobulin (Ig)-like domains (Fig 2A–B) that position in an unusual head-to-head orientation with a disulfide bond linkage between them (Fig S3C and (Basore et al., 2019; Song et al., 2019)). The two domains of cattle Mxra8 are connected by a hinge, with ~ 31° of movement in the position of domain D1 relative to D2 compared with mouse Mxra8 when bound to CHIKV (Basore et al., 2019). This domain rotation is similar to that in unliganded murine Mxra8 (Basore et al., 2019) and unlikely to explain why cattle Mxra8 does not support alphavirus infection. Weak electron density for the 15-amino acid insertion of cattle Mxra8 indicates it can form a β-hairpin loop (herein termed moo’ and moo’’ strands), which projects away from D1 between the C’ and C’’ loops (Fig 2A, 2C, and S3C). This inserted region appears to be stabilized by crystal lattice contacts with an adjacent symmetry mate, suggesting that this might not be the only conformation of the ‘moo’ loop.
Figure 2. Structure of cattle Mxra8.
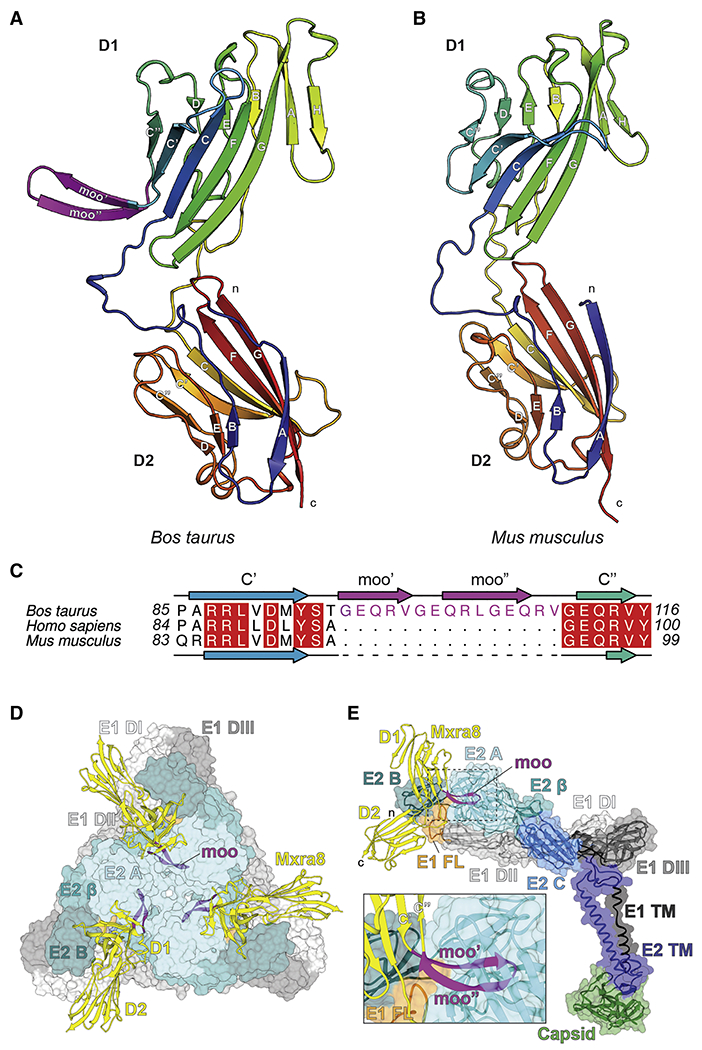
A-B. Ribbon models of the (A) cattle and (B) mouse Mxra8 (PDB 6NK3) structures determined by X-ray crystallography. The two Ig-like domains are colored by Jones’ rainbow scheme with the N-terminus in blue and C-terminus in red. The β-strands of each Ig domain are labeled according to standard convention. The N- and C-termini are labeled in lowercase. C. Structure-based alignment of Bos taurus (cattle), Homo sapiens (human), and Mus musculus (mouse) Mxra8 protein sequences highlighting the 15-amino acid insertion site between the C’-C’’ loop in D1. D-E. Docking of cattle Mxra8 onto a cryo-EM model of mouse Mxra8 bound to CHIKV VLPs, viewed from a trimeric spike (D), an E2-E1 subunit (E), and enlarged to highlight the clash (inset). Cattle Mxra8 is colored yellow and labeled by domain, with the moo insert depicted in magenta. The N- and C-termini are labeled in lowercase. Within the inset, the C’, ‘moo’ insert, and C’’ β-strands are labeled, and the E1 fusion loop is depicted in orange. Structural proteins are colored and labeled by domain. E1: DI, light grey; DII, medium grey; DIII, dark grey; fusion loop, orange; TM region, black. E2: A domain, light cyan; β-linker, medium cyan; domain B, dark cyan; domain C, medium blue; TM region, dark blue. Capsid, green. See Fig S2 and S3.
We docked cattle Mxra8 coordinates onto the cryo-electron microscopy structure of CHIKV virions in complex with mouse Mxra8 (Basore et al., 2019) (Fig 2D–E). These analyses suggest that the modeled 15-amino acid insertion sterically hinders Mxra8 binding to the CHIKV virion, as the insertion physically clashes with residues in domain A of the CHIKV E2 protein on the heterotrimeric spike. Superimposition of cattle Mxra8 into the crystal structure of CHIKV E3-E2-E1 proteins bound to human MXRA8 (Song et al., 2019) also indicates clashes between the insertion and domain A of the CHIKV E2 protein. These docking studies suggest that cattle Mxra8 could not productively engage the CHIKV virion even if a different conformation of the 15-amino acid insertion were adopted, as alternate steric clashes would be generated. In comparison, structure-function analysis of avian Mxra8 orthologs, which also do not promote CHIKV infection, showed no insertions, but instead revealed sequence variation at the virus-receptor interface (Basore et al., 2019; Song et al., 2019) with substitutions in 17 different contact residues (Fig S1G) relative to mouse Mxra8.
The insertion in cattle Mxra8 restricts alphavirus binding and infection.
To determine whether the insertion in cattle Mxra8 disrupts interactions with CHIKV virions, we engineered mouse and cattle Mxra8-Fc fusion proteins with or without the additional 15 residues from cattle (hereafter termed the ‘moo’ insert: mouse Mxra8-Fc, mouse Mxra8 + moo-Fc, cattle Mxra8-Fc, and cattle Mxra8 Δmoo-Fc) (Fig 3A and S3D). We evaluated antigenic integrity of the chimeric proteins by assessing binding to four cross-reactive anti-Mxra8 mAbs (Zhang et al., 2018a) that engage multiple epitopes (Basore et al., 2019). MAbs 3G2.F5 and 9G2.D6 bound to all cattle and mouse Mxra8-Fc variants (Fig 3B and S3E), whereas mAbs 4E7.D10 and 8F7.E1 failed to bind efficiently to cattle Mxra8-Fc but recognized mouse Mxra8-Fc and cattle Mxra8 Δmoo-Fc, suggesting they bind an epitope proximal to the 15-amino acid insertion. Consistent with this idea, 4E7.D10 and 8F7.E1 were mapped by hydrogen-deuterium exchange mass spectrometry to residues 91-107 on Mxra8 (Basore et al., 2019).
Figure 3. Effect of the 15-amino acid insertion in cattle Mxra8 on alphavirus binding and infection.
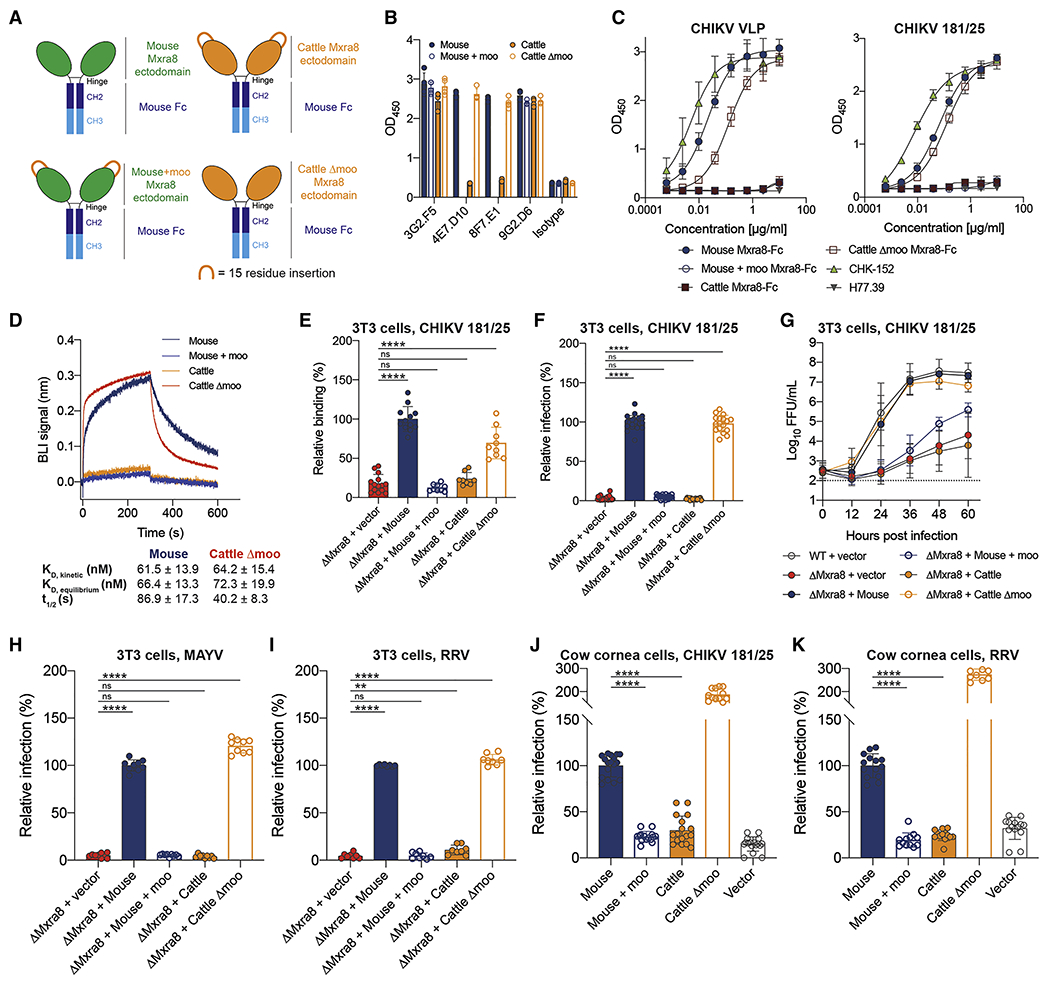
A. Diagram of mouse, cattle, mouse + moo [GEQRVGEQRLGEQRV insert], and cattle Δmoo Mxra8-Fc fusion proteins. B. Binding of anti-Mxra8 mAbs 3G2.F5, 4E7.D10, 8F7.E1, and 9G2.D6 to mouse, cattle, mouse + moo, and cattle Δmoo Mxra8-Fc fusion proteins by ELISA. Data are pooled from four experiments performed in duplicate. C. Binding of increasing concentrations of mouse, cattle, mouse + moo, and cattle Δmoo Mxra8-Fc fusion proteins to antibody-captured CHIKV VLPs and CHIKV 181/25 virions by ELISA. Data are pooled from two to three experiments performed in duplicate. D. Representative kinetic sensograms of bacterially-expressed mouse (blue), mouse + moo (light blue), cattle (orange), and cattle Δmoo (red) Mxra8 binding to CHIKV VLPs as determined by BLI. CHIKV VLPs were captured using an anti-CHIKV mAb (CHK-265) and then incubated with Mxra8 proteins. Binding data represent the average of three experiments. E. Binding of CHIKV 181/25 virions to 3T3 ΔMxra8 cells complemented with mouse, mouse + moo, cattle, or cattle Δmoo Mxra8 cDNA. Virions were incubated with cells at 4°C, and CHIKV antigen staining was analyzed by flow cytometry. Data are pooled from three to four experiments (n = 9 to 14 replicates; one-way ANOVA with Dunnett’s post-test: ****, P < 0.0001). F-I. Lentivirus complementation of 3T3 ΔMxra8 cells with Mxra8 cDNA from mouse, mouse + moo, cattle, and cattle Amoo. Cells were inoculated with (F, G) CHIKV (181/25), (H) MAYV (BeH407), or (I) RRV (T48). For G, a multi-step growth analysis was conducted, and virus was titrated by focus-forming unit assay. Data are pooled from three experiments performed in sextuplicate. For F, H-I, cells were processed at specified time points (see STAR Methods) and stained with virus-specific anti-E2 protein mAbs. J-K. Lentivirus complementation of bovine cornea cells with Mxra8 cDNA of mouse, cattle, mouse + moo, and cattle Amoo. Cells were inoculated with (J) CHIKV (181/25) or (K) RRV (T48) and processed by staining with anti-E2 protein mAbs. The relative increase in CHIKV and RRV infection in bovine cornea cells expressing cattle Δmoo Mxra8 may reflect the higher levels of surface expression (see Fig S5C). For F, H-K, data are pooled from three to nine experiments (n = 6 to 26 replicates; one-way ANOVA with Dunnett’s post-test: **, P < 0.01; ****, p < 0.0001). See Fig S3 and S4.
We tested the Mxra8-Fc fusion proteins for their capacity to bind CHIKV virus-like particles (VLPs) and virions by ELISA. Mouse Mxra8-Fc and cattle Mxra8 Δmoo-Fc bound avidly to CHIKV VLPs and virions, whereas mouse Mxra8 + moo-Fc and cattle Mxra8-Fc did not (Fig 3C). In a complementary approach, we assessed the monovalent binding of purified wild-type and chimeric Mxra8 proteins to recombinant CHIKV VLPs using biolayer interferometry (BLI). Mouse Mxra8 (KD = 66.4 ± 13.3 nM) and cattle Mxra8 Δmoo (KD = 72.3 ± 19.9 nM) but not mouse Mxra8 + moo and cattle Mxra8 bound to CHIKV VLPs (Fig 3D, S3F and Table S3). Consistent with these results, CHIKV virions bound poorly to 3T3 ΔMxra8 cells expressing mouse Mxra8 + moo or cattle Mxra8 compared to mouse or cattle Δmoo Mxra8 (Fig 3E). These data suggest that the 15-amino acid insertion in cattle Mxra8 inhibits binding to CHIKV and support the structure-based hypothesis that cattle Mxra8 cannot engage the CHIKV spike due to the presence of a protruding loop that sterically blocks binding (Fig 2D–E).
To test whether the insertion blocks alphavirus infection, we transduced ΔMxra8 3T3 cells with mouse Mxra8, mouse Mxra8 + moo, cattle Mxra8, or cattle Mxra8 Δmoo. All wild-type and chimeric Mxra8 variants were detected on the cell surface (Fig S4A–B). The addition of the 15-amino acid insertion to mouse Mxra8 abolished infectivity by CHIKV, MAYV, and RRV, whereas its deletion from cattle Mxra8 restored infection to levels observed following transduction of mouse Mxra8 (Fig 3F–I). To confirm these results in a more species-relevant cell type, we transduced bovine endothelial cells with mouse Mxra8, mouse Mxra8 + moo, cattle Mxra8, or cattle Mxra8 Δmoo, followed by inoculation of CHIKV or RRV. These cells lack endogenous surface expression of Mxra8 and at baseline did not support infection of either CHIKV or RRV (Fig S4C–E). Bovine endothelial cells transduced with mouse Mxra8 or cattle Mxra8 Δmoo but not mouse Mxra8 + moo or cattle Mxra8 were susceptible to CHIKV and RRV infection (Fig 3J–K and S4D–E).
The 15-amino acid insertion attenuates CHIKV infection and pathogenesis in vivo in CRISPR-Cas9 engineered mice.
We engineered C57BL/6J knock-in (KI) mice with a Mxra8 allele containing the ‘moo’ insertion (Mxra8moo) to test its function in alphavirus virulence in vivo (Fig 4A). Founder Mxra8moo/moo or WT mice were bred to Mxra8-deficient (Mxra8KO/KO) (Zhang et al., 2019) or each other to establish Mxra8 ‘moo’ KI (Mxra8moo/KO and MXra8moo/moo) mice and corresponding control animals. All mice containing the Mxra8 ‘moo’ allele developed normally. To confirm expression of Mxra8moo in the KI mice, we probed primary mouse embryonic fibroblasts (MEFs) by performing immunoblotting of lysates and flow cytometry of cells. Whereas Mxra8WT/WT and Mxra8WT/KO MEFs had Mxra8 bands at ~50 kDa, no band migrating at this mobility was detected in lysates from Mxra8KO/KO MEFs. Immunoblots of Mxra8moo/KO MEFs, however, displayed a band of ~52 kDa, consistent with the 15-amino-acid ‘moo’ insertion (Fig 4B). We detected cell surface expression of Mxra8 on Mxra8WT/WT, Mxra8WT/KO, and Mxra8moo/KO MEFs but not Mxra8KO/KO MEFs (Fig 4C). In multi-step growth analysis, both Mxra8WT/WT and Mxra8WT/KO MEFs supported robust CHIKV infection whereas Mxra8KO/KO and Mxra8moo/KO MEFs did not (Fig 4D). We then examined disease by inoculating mice with virulent CHIKV (strain AF15561). Markedly reduced ankle joint swelling was observed throughout the acute phase (days 2 to 10) in Mxra8moo/KO and Mxra8KO/KO compared to Mxra8WT/KO and Mxra8WT/WT mice (Fig 4E). Moreover, at day 3 post-inoculation, the Mxra8moo/KO and Mxra8KO/KO mice displayed diminished viral loads in the serum, ipsilateral and contralateral calf muscles, and contralateral ankle relative to Mxra8WT/WT and Mxra8WT/KO mice (Fig 4G–4J). Similar results were observed after CHIKV infection of homozygous Mxra8moo/moo mice with reduced ankle swelling and viral burden compared to Mxra8 mice (Fig 4F, 4K–4N). However, low levels of CHIKV were detected in Mxra8moo/KO, Mxra8moo/moo, and Mxra8KO/KO mice. These results agree with previous cell culture and in vivo experiments (Zhang et al., 2019; Zhang et al., 2018a), and suggest that a Mxra8-independent entry pathway exists for CHIKV, at least in mice, although it is not sufficient to promote clinical disease. Nonetheless, as CHIKV virulence is diminished in, Mxra8moo/KO, Mxra8moo/moo, and Mxra8KO/KO mice relative to Mxra8WT/WT and Mxra8WT/KO mice, the ‘moo’ insertion produces a loss-of-function allele for CHIKV infection in vivo.
Figure 4. In vivo assessment of the Mxra8 ‘moo’ insertion.
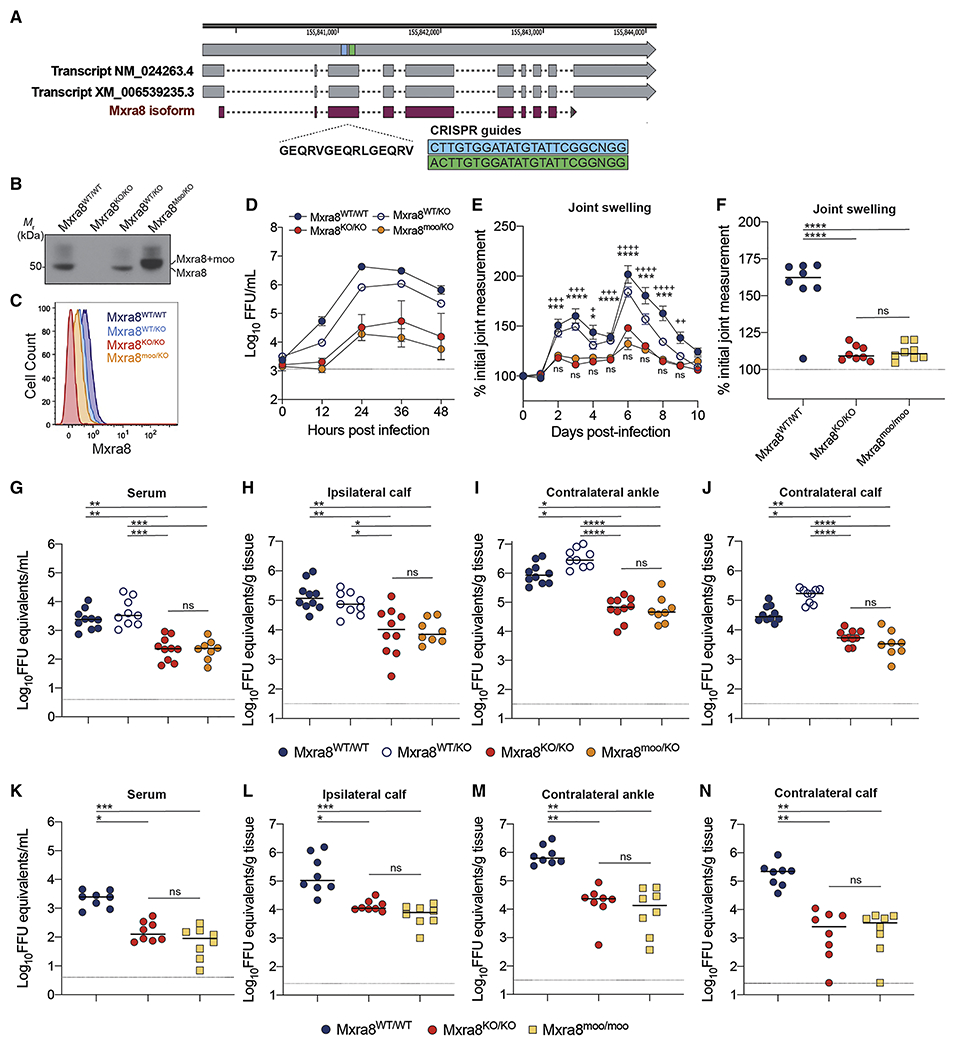
A. Generation of Mxra8-moo knock-in C57BL/6J mice using CRISPR-Cas9 gene editing. The ‘moo’ insertion (GEQRVGEQRLGEQRV) was introduced into exon 3 of the Mxra8 gene using two single guide RNAs (sgRNAs) as indicated by the blue and green boxes. Annotated transcripts are shown in gray and the encoded protein in purple. B. Immunoblotting of cell lysates from primary Mxra8WT/WT, Mxra8KO/KO, Mxra8WT/KO, and Mxra8moo/KO MEFs using anti-Mxra8 mAbs 3G2.F5 and 9G2.D6 (Mr: migration rate). Data are representative of three experiments. C. Surface expression of Mxra8 from primary MEFs (Mxra8WT/WT (dark blue), Mxra8KO/KO (red), Mxra8WT/KO (light blue), and Mxra8moo/KO (orange)) after staining with a pool of anti-Mxra8 mAbs as determined by flow cytometry. Data are representative of two experiments. D. Multi-step growth analysis of CHIKV AF15561 in primary MEFs (Mxra8WT/WT, Mxra8KO/KO, Mxra8WT/KO, and Mxra8moo/KO). Cells were infected at an MOI of 0.01, and virus in supernatants was harvested at the time points shown and titrated by focus-forming assay. Data are representative of three experiments performed in sextuplicate. E-N. Mxra8WT/WT, Mxra8KO/KO, Mxra8WT/KO, and Mxra8moo/KO, and Mxra8moo/moo mice were inoculated in the footpad with 103 focus-forming units (FFU) of CHIKV AF15561. Joint swelling was monitored over 10 days (E) or at day 3 post-infection (F). Viral RNA levels in serum (G, K), ipsilateral calf muscle (H, L), contralateral ankle (I, M), and contralateral calf muscle (J, N) were measured at 3 days post-infection. For E, data are from three experiments (n = 11 to 15; two-way ANOVA with Dunnett’s post-test: * or +, P < 0.05; ** or ++, P < 0.01; *** or +++, P < 0.001; **** or ++++, P < 0.0001; ns, not significant. “*” indicates comparison between Mxra8WT/WT and Mxra8moo/KO “+” indicates comparison between Mxra8WT/KO and Mxra8moo/KO “ns” data are a comparison between Mxra8KO/KO and Mxra8moo/KO For F-N, data are from two experiments (n = 8 to 10; one-way ANOVA with Kruskal-Wallis post-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
Mxra8 insertions are present in most Bovinae species and inhibit alphavirus infection.
We determined whether the 15-amino acid insertion was present in species related to cattle. We evaluated calibrated phylogenetic trees that were developed using autosomal and mitochondrial DNA sequences (MacEachern et al., 2009; Zurano et al., 2019) to identify Bovinae subfamily and Bovidae family members with shared ancestry (Fig 5, left panel). This tree included tribes within the Bovinae subfamily (Bovini, Tragelaphini, and Boselaphini), closely related Bovidae (Ovis, Pseudois, and Capra), and more distantly related Cervidae (Cervus, Muntiacus, and Odocoileus) and Moschidae (Moschus) species. We obtained sequences of Mxra8 gene orthologs (Table S4, S5, and STAR Methods) by: (a) downloading annotated Mxra8 gene sequences from GenBank, (b) assembling deposited unannotated whole genome or exome sequences, and (c) isolating mRNA from tissues obtained at necropsy from animals at the Saint Louis Zoo, samples acquired from commercial vendors, and validated primary fibroblasts of different Bovinae species (Modi et al., 2004).
Figure 5. Phylogenetic relationship of Mxra8 orthologs.
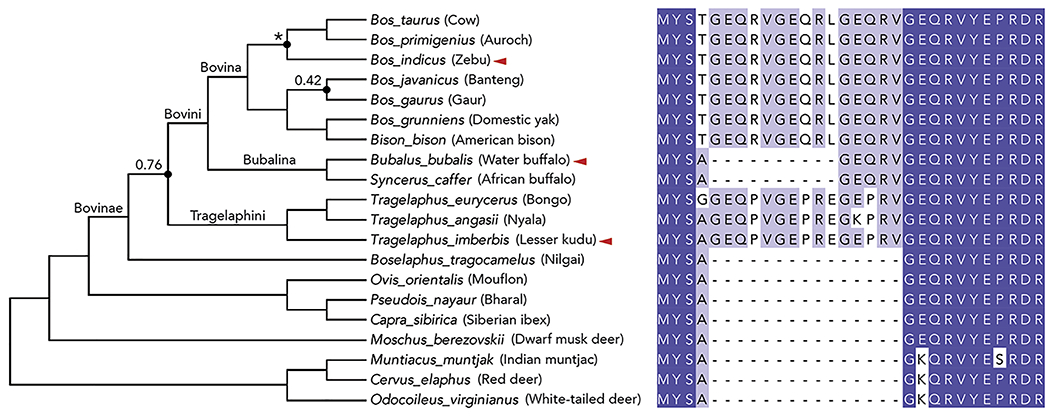
The phylogeny summarizes the relationships for Bovinae, Bovidae, Moschidae, and Cervidae as estimated by (Zurano et al., 2019) but was modified to include the relationships for Bos taurus and Bos indicus (reported by (Wang et al., 2018)) as indicated by asterisks. All nodes have high support with posterior probabilities >95% unless indicated otherwise by values. Species indicated with red arrows are investigated in subsequent functional experiments. The phylogenetic tree and sequence alignment were prepared using FigTree and Jalview (Waterhouse et al., 2009), respectively. Protein sequences of Mxra8 gene orthologs from Bovidae members in the region corresponding to the 15-residue Mxra8 insertion in D1 were aligned using MUSCLE and visualized using Jalview. Amino acid residues are colored according to sequence identity among Bovidae family members: dark blue boxes (100%), light blue boxes (50-90%), and white boxes (<50%). Gaps in the protein sequences are indicated by a dash.
All Bovinae subfamily members except one had insertions in Mxra8 at the same site as observed in cattle. The most distantly related Bovinae member (Boselaphus tragocamelus) lacked the insertion as did representatives of the Bovidae, Cervidae, or Moschidae (Ovis orientalis, Pseudois nayaur, and Capra sibirica) (Fig 5, right panel and S5A). Members of the Bovina subtribe (Bos and Bison) share identical 15-amino acid insertions whereas the Bubalina subtribe (Bubalus and Syncerus) had 5-amino acid insertions (one GEQRV repeat) at the same site as in cattle. The common ancestor of Bubalus bubalis and Syncerus caffer may have lost 30 nucleotides of the ancestral insertion. Alternatively, an independent introduction of the GEQRV insertion could have occurred in the ancestor of Bubalus and Syncerus at the same site in Mxra8. Members of the more distantly related Tragelaphus subtribe of spiral-horned antelopes (e.g., nyala [Tragelaphus angasii], bongo [Tragelaphus eurycerus], and lesser kudu [Tragelaphus imberbis]) all had similar 45-nucleotide (89% identity) and 15-amino acid (73% identity) insertions (GEQPVGEPREGEPRV; with three notable proline substitutions) at the same site in Mxra8.
To determine the significance of the insertions of other Bovinae species in Mxra8, we engineered a panel of Mxra8 Fc-fusion proteins including water buffalo (Bubalus)-Fc, water buffalo-Δ5-Fc, kudu-Fc, and kudu-Δ15-Fc (Fig S5B). We designed three additional variants to define the effect of size and sequence of the insertion on CHIKV binding: mouse-Mxra8+8-Fc and mouse-Mxra8+10-Fc add 8 (QRVGEQRL) and 10 (GEQRVGEQRL) of the 15 amino acids, respectively, from the cattle insertion; and mouse-Mxra8+[GGS]5-Fc adds 15 non-homologous amino acids (GGSGGSGGSGGSGGS) that are predicted to form a flexible loop (Fig S5C). These Mxra8-Fc fusion proteins were recognized by anti-Mxra8 mAbs (Fig S5D). We tested these Mxra8-Fc proteins for their capacity to bind CHIKV VLPs by ELISA. No binding was detected with kudu-Mxra8-Fc, markedly diminished binding was observed with mouse-Mxra8+10-Fc and mouse-Mxra8+[GGS]5-Fc, intermediate binding was detected with water buffalo Mxra8-Fc and mouse-Mxra8+8-Fc, and avid binding was observed with water buffalo-Δ5-Mxra8-Fc, kudu-Δ15-Mxra8-Fc, and mouse Mxra8-Fc (Fig 6A–B). To corroborate these results, we assessed the monovalent binding of purified mouse Mxra8 insertion variants to CHIKV VLPs using BLI. All Mxra8 insertion variants were recognized by an anti-Mxra8 mAb (Fig S5E), suggesting proper folding. Mouse Mxra8 bound strongly to CHIKV VLPs, whereas mouse Mxra8+5, mouse Mxra8+8, mouse Mxra8+9, mouse Mxra8+10, mouse Mxra8 + moo, or mouse Mxra8 +[GGS]5 displayed little or no binding (Fig 6C). These data suggest that insertions of as few as five residues at the ‘moo’ site inhibit interactions of Mxra8 with CHIKV, although receptor blockade does not require a specific sequence.
Figure 6. Functional and evolutionary relationships of Mxra8.
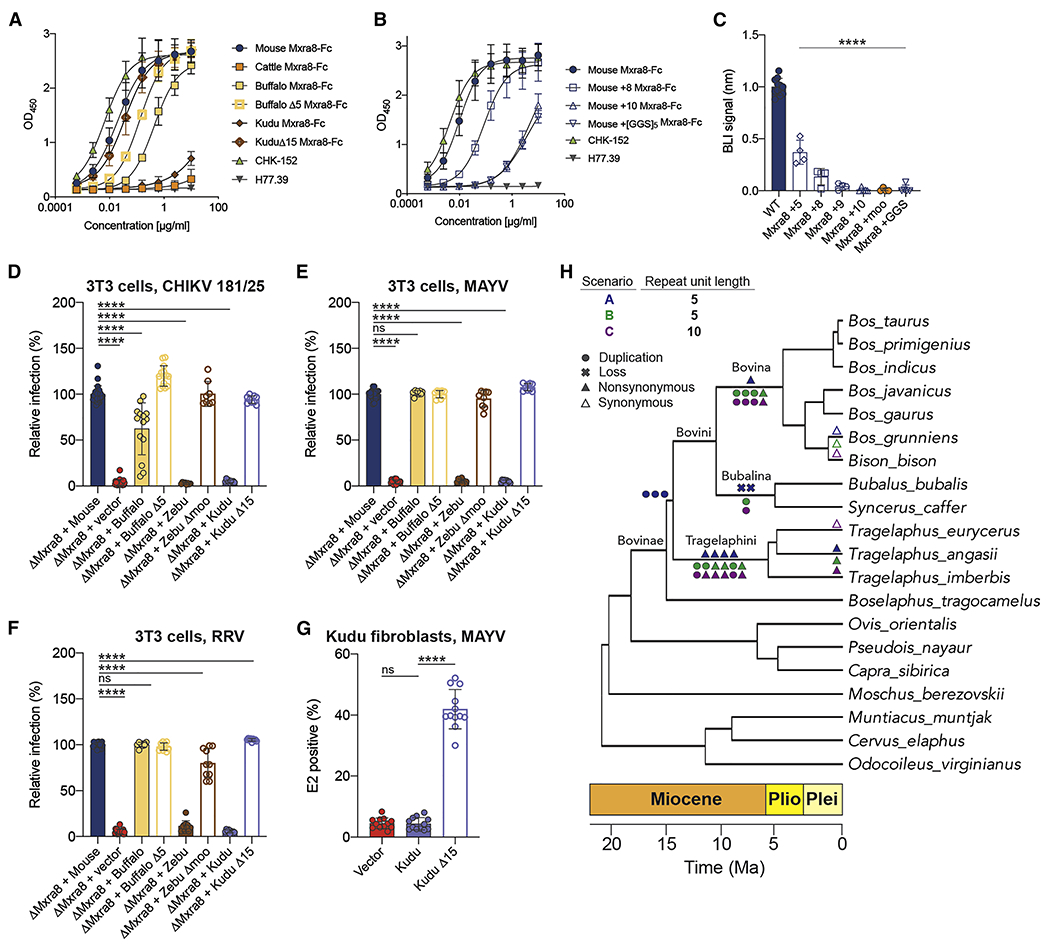
A. Binding of mouse-Fc, cattle-Fc, water buffalo-Fc, water buffalo-Δ5-Fc, kudu-Fc, and kudu-Δ15-Fc Mxra8 fusion proteins to antibody-captured CHIKV VLPs by ELISA. B. Binding of mouse-Fc, mouse-Mxra8+8-Fc, mouse-Mxra8+10-Fc, and mouse-Mxra8+[GGS]5-Fc fusion proteins to antibody-captured CHIKV by ELISA. MAbs CHK-152 (anti-CHIKV) and H77.39 (anti-HCV) were included as positive and negative controls, respectively. Data are pooled from three experiments performed in duplicate. C. Binding response of the bacterially-expressed mouse Mxra8 and insertion variants Mxra8+5, Mxra8+8, and Mxra8+9, Mxra8+10, Mxra8 +moo, and Mxra8 +[GGS]5 to antibody-captured CHIKV VLPs at 1 μM concentration by BLI. Data are the mean and standard deviation of four to five experiments (one-way ANOVA with Dunnett’s post-test: ****, P < 0.0001). D-F. Lentivirus complementation of ΔMxra8 3T3 with Mxra8 cDNA from water buffalo, water buffalo Δ5, zebu, zebu Δmoo, kudu, and kudu Δ15. Cells were inoculated with (D) CHIKV (181/25), (E) MAYV (BeH407), or (F) RRV (T48), harvested, stained with anti-E2 mAbs, and processed by flow cytometry. For D-F, data are pooled from three to eight experiments (n = 8 to 23 replicates; one-way ANOVA with Dunnett’s post-test: ****, P < 0.0001). G. MAYV infection of primary kudu fibroblasts transduced with vector control, kudu Mxra8, or kudu Δ15 Mxra8. Data are from four experiments (n = 12; one-way ANOVA with Dunnett’s post-test: ****, P < 0.0001, ns, not significant). H. Evolutionary scenarios for the Bovinae Mxra8 gene insertions. The three scenarios are based on the length of the repeat unit and whether losses or duplications of the repeat unit are more common. Duplication events are indicated as circles, losses are indicated by an “x”, nonsynonymous substitutions are indicated as filled triangles, and synonymous substitutions are indicated as open triangles. Events are shown in their reconstructed order, from oldest (left) to youngest (right), and in their youngest possible positions, though the age of any event is not precisely known. All scenarios involve one synonymous substitution occurring within Bos grunniens, and one nonsynonymous substitution occurring within Tragelaphus angasii, but differ elsewhere. In scenario A (blue symbols), where loss of the 5-amino acid repeat unit is relatively common, the repeat unit is duplicated three times in the common ancestor of Bovini and Tragelaphini. A nonsynonymous substitution occurs in the ancestor of Bovini followed by two loss events in the ancestor of Bubalina. Four more nonsynonymous substitutions occur in the ancestor of Tragelaphini. In scenario B (green symbols), where duplication of the 5-amino acid repeat unit is more common, three duplication events and a nonsynonymous substitution occur in the ancestor of Bovina. A single duplication event occurs in the Bubalina ancestor. In the ancestor of Tragelaphini, two duplication events are followed by two nonsynonymous substitutions, a duplication event, and a nonsynonymous substitution. Scenario C (purple symbols), where duplication of the 10-amino acid repeat unit is more common, is similar to scenario B with an exception in the Tragelaphini ancestor. Here, a single duplication event is followed by three nonsynonymous substitutions, a duplication event, and a nonsynonymous substitution. A geological time scale indicating the Miocene (orange), Pliocene (yellow), and Pleistocene (light yellow) epochs is shown below. See Fig S7 and Table S6.
To test whether the 15-residue insertion in other Mxra8 orthologs similarly disrupts alphavirus infection, we complemented ΔMxra8 3T3 cells with water buffalo Mxra8, water buffalo Δ5 Mxra8, zebu Mxra8, zebu Δ15 Mxra8, kudu Mxra8, or kudu Δ15 Mxra8, followed by inoculation with CHIKV, MAYV, or RRV. All wild-type and sequence-deleted Mxra8 variants were detected on the cell surface (Fig S6A). Whereas zebu and kudu Mxra8 did not promote infection, expression of the respective 15-amino acid deleted forms enabled infection of the viruses tested (Fig 6D–F and S6B–D). Water buffalo Mxra8 had an intermediate phenotype; while it supported MAYV and RRV infection, the level of CHIKV infection was diminished. However, CHIKV infection was increased in cells expressing water buffalo Δ5 Mxra8. In additional experiments, we transduced primary kudu fibroblasts with kudu Mxra8 or kudu Δ15 Mxra8 (Fig S6E). Whereas kudu Mxra8 did not promote infection of MAYV, kudu Δ15 Mxra8 did (Fig 6G and S6F).
Ancient origin of the Bovine Mxra8 insertion.
The Mxra8 insertion either originated once in a common ancestor of the Bos, Bubalus, and Tragelalphus lineages, or up to three times independently in each of these lineages (Fig 6H and Fig S7). Our phylogenetic reconstruction places at least 80% of the amino acid substitutions within the Mxra8 insertion along internal branches of the Bovinae tree (Fig 6H, Fig S7A–C, and Table S6). This indicates that the Mxra8 insertion, and the conserved amino acid substitutions within it, probably originated during the Miocene or earlier (> 5.3 million years ago [5.3 Ma]) (Zurano et al., 2019), before Tragelaphus diversified (Table S7). Recent, widespread introgression of the insertion appears unlikely, as the Bovinae species tree and the Mxra8 gene tree are highly congruent (Fig S7D–E) and the chromosome counts of Bovinae species differ substantially within and between genera (see Fig S7 and Table S8) (O’Brien, 2005).
At present, we have no evidence suggesting that alphavirus disease resistance selected for the Mxra8 insertion. Although age estimates based on nucleotide sequence divergence suggest alphaviruses evolved recently from a common ancestor (Weaver et al., 1993), RNA viruses may be considerably older than hypothesized (Zhang et al., 2018b). The insertion also could have evolved to enhance an endogenous function of Mxra8 in Bovinae physiology. As Mxra8 reportedly interacts with αVβ3 integrin and possibly, other matrix proteins, the insertion might modulate its attributed roles in cell-cell adhesion, angiogenesis, and/or mesenchymal cell differentiation (Han et al., 2018; Jung et al., 2012; Jung et al., 2008).
The insertion in Mxra8 likely limits alphavirus infection in some Bovinae species. The mechanism of evasion is reminiscent of those postulated for mouse hepatitis virus and some arenaviruses, where alleles of the Ceacam1a or transferrin (TFR1) receptors, respectively, with sequence variations yielding lower binding affinity were selected in subsets of rodents (Demogines et al., 2013; Peng et al., 2017). Analogously, some humans are homozygous for a 32-nucleotide deletion in CCR5, which confers resistance to HIV entry (Martinson et al., 1997). Arthritogenic alphaviruses (e.g., CHIKV and RRV) failed to infect cattle and kudu cells efficiently except when an engineered variant (cattle Δmoo or kudu Δ15) but not wild-type Mxra8 was expressed. Moreover, introducing the ‘moo’ insert into mouse Mxra8 produces a loss-of-function allele for CHIKV infection in vivo, and phenocopies an absence of Mxra8. Consistent with this idea, four Bos taurus calves inoculated with CHIKV failed to develop viremia (Bosco-Lauth et al., 2016), and in an area of Central Africa with epidemic transmission of CHIKV, only 1 of 183 zebus had CHIKV antibodies (Guilherme et al., 1996). Similarly, in a region of New Zealand with active RRV transmission, 0 of 207 cattle were seropositive for RRV (McFadden et al., 2009), and in Australia, RRV has been isolated from horses and goats, but not cattle (Gard et al., 1988). Detailed seroepidemiological studies of Bovinae and related Bovidae species in areas of epidemic alphavirus transmission could clarify the relationship between exposure, infection, and resistance to disease.
Our in vitro and in vivo data suggest that animals retaining the Mxra8 insertion or those modified with an introduced insertion are resistant to infection and disease by Mxra8-dependent contemporary alphaviruses. By combining established resistance patterns of specific animals to viral infection with the sequences and structures of evolutionarily related receptor orthologs, molecular and functional analysis can elucidate how and which sequence variation alters virus-receptor interactions. Moreover, these studies may foster the development of countermeasures, as they provide a strategy for genetically modifying animals or identifying molecules that bind to specific regions of Mxra8 to promote resistance to infection by multiple alphaviruses. Indeed, small molecules that differentially bind cattle and mouse/human Mxra8 in the region proximal to the C’-C’’ loop insertion could block alphavirus attachment and infection.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact author Michael S. Diamond (diamond@wusm.wustl.edu). All plasmids, antibodies, cells, viruses, and mouse lines developed for this study are available under Material Transfer Agreements from Washington University School of Medicine.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells and viruses.
NIH-3T3, HEK-293, and Vero cells were obtained from ATCC and were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum (Hyclone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES. Bos taurus corneal endothelial cells (CRL-2048) were obtained from the ATCC and were cultured at 37°C in DMEM supplemented with 20% fetal bovine serum (Hyclone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES. Expi293 cells were obtained from Thermo Fisher and cultured shaking at 37°C and 8% CO2 in Expi293 Expression Medium. Primary MEFs were generated from embryos obtained on E13. The head and internal organs were dissected from the embryos, washed, minced in the presence of 0.05% trypsin-EDTA, and cultured until confluency. Primary kudu (Tragelaphus imberbis) fibroblasts (generously provided by T. Raudsepp and J. Womack, Texas A&M University) were maintained in DMEM supplemented with 20% fetal bovine serum (Hyclone). The following alphaviruses were propagated in Vero cells and titered by focus forming assay as described previously (Fox et al., 2015): CHIKV 181/25, CHIKV AF15561, MAYV (BeH407), RRV (T48), ONNV (MP30), and VEEV-GFP (TC-83).
Generation of Mxra8 knock-in mice.
All mouse studies were performed after approval by the Institutional Animal Care and Use Committee at the Washington University School of Medicine and the Saint Louis Zoo (Assurance number A3381-01). Gene-edited Mxra8 mice were generated with support from the Genome Engineering and iPSC center and Department of Pathology Micro-Injection Core (Washington University School of Medicine). The 45-nucleotide “moo” insertion sequence (GGCGAGCAGCGCGTGGGCGAGCAGCGCTTGGGCGAGCAGCGCGTG) from cattle Mxra8 was inserted into exon 3 of mouse Mxra8. Two sgRNAs were selected based on a low off-target profile and distance to target site: sgRNA-1: 5’-CTTGTGGATATGTATTCGGCNGG-3’ and sgRNA-2: 5’ACTTGTGGATATGTATTCGGNGG-3’. The two gRNAs were synthesized in vitro (HiScribe T7 In Vitro Transcription Kit, New England BioLabs) and purified (MEGAclear Transcription Clean-Up Kit, Thermo Fisher). Guide RNAs, Cas9 protein, and the oligo donor sequence modified with silent blocks (gtccactgggacctcagcgggggcccgggcagccaacggcgccgacttgtggatatgtatAGTgcgGGCGAGCAGCGC GTGGGCGAGCAGCGCTTGGGCGAGCAGCGCGTGggtgaacagcgcgtgtacgagccgcgcgatcgcga ccgcctcctgctgtcgccttctgct) were complexed and electroporated into C57BL/6 zygotes. After microinjection, founder lines were confirmed by genotyping and next-generation sequencing.
Mouse experiments.
Four-week-old congenic Mxra8WT/WT, Mxra8WT/KO, Mxra8KO/KO, Mxra8moo/KO, Mxra8moo/moo male or female C57BL/6J mice were inoculated subcutaneously in the footpad with 103 FFU of CHIKV AF15561 strain. The Mxra8KO/KO (out of frame 8 nucleotide deletion) were generated by CRISPR-Cas9 editing and characterized previously (Zhang et al., 2019). At 3 days post infection, mice were euthanized, perfused extensively with PBS, and serum and tissues were collected. Foot swelling (width x height) was monitored using digital calipers as previously described. RNA was extracted from serum and tissues using the MagMax-96 Viral RNA Isolation Kit (Thermo Fisher). Viral RNA levels were quantified by qRT-PCR using a TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher), compared to a CHIKV RNA standard curve, and expressed on a log110 scale as viral focus-forming unit (FFU) equivalents per gram of tissue or milliliter of serum. Primers and probes used are as follows: CHIKV-AF FOR: 5’-TCGACGCGCCATCTTTAA-3’; CHIKV-AF REV: 5’-ATCGAATGCACCGCACACT-3’; CHIKV-AF Probe: 5’-/56-FAM/ACCAGCCTG/ZEN/CACCCACTCCTCAGAC/3IABkFQ/-3’.
METHOD DETAILS
Plasmid construction for trans-complementation studies.
Mxra8 cDNA fragments containing a C-terminal FLAG tag were codon-optimized, synthesized, and inserted into the lentivirus vector pLV-EF1a using an In-Fusion HD Cloning Kit (Takara) for the following species: Mus musculus (Genbank accession no. NM_024263), Bos taurus (NM_001075830), Bos indicus (XM_019976191), Bubalus bubalis (XM_006066948.2), Tragelaphus angasii (primary sequence, see Table S4), Capra hircus (XM_018060531), Canis lupus familiaris (XM_546712), Rattus norvegicus (NM_001007002), Pan troglodytes (NM_001280245), Equus caballus (XM_023636045), Ovis aries (XM_027975805), Meleagris gallopavo (XP_010721105.1), Anas platyrhynchos (XM_027443263), and Gallus gallus (NP_989967). The mouse Mxra8 + moo was generated by inserting residues “GEQRVGEQRLGEQRV” after amino acid residue 98. The cattle Δmoo, zebu Δmoo, and kudu Δ15 Mxra8 were generated by removing residues 96-110. The Bubalus Δ5 was generated by removing residues 96-100. All sequences were codon-optimized. Plasmids were transformed into One Shot Stbl3 Chemically Competent E. coli (Thermo Fisher) and bacteria were grown at 30°C on LB Agar plates with carbenicillin (100 μg/ml). Colonies were picked and grown overnight at 30°C in LB supplemented with carbenicillin (100 μg/ml). Plasmids were extracted (Qiagen) and sequenced using the following primers: GCACTTGATGTAATTCTCCTTGGAATTTGC, CTCAAGCCTCAGACAGTGGTTCAAAGT and GGTGGAAAATAACATATAGACAAACGCAC. The following primers were used to sequence the following species: Bos indicus: GTCTATGAGCCTAGGGACCGA and GTCTATGAGCCTAGGGACCGA; Bubalus bubalis: CGCGTATACGAGCCTCGA and ATAGAGTCGCAGTTGAGGCAG; Tragelaphus angasii: TGGGGAAAGACGGGCCT; and Equus caballus: CGATCGAGGTCGACTGCTTC and ACAGAGTTGCCGTAGCTGTAG.
Trans-complementation and infection experiments.
Mxra8 lentiviruses were packaged with psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) vectors in HEK-293 cells using FugeneHD (Promega). ΔMxra8 3T3 cells (Zhang et al., 2018a) were transduced with lentiviruses and selected with blasticidin for 7 days. Surface expression of Mxra8 was assessed by flow cytometry after staining with a pool of seven hamster anti-mouse Mxra8 mAbs (Zhang et al., 2018a) (1 μg/ml for all species except for dog Mxra8 [10 μg/ml]), and Alexa Fluor 647 conjugated goat anti-Armenian hamster IgG (1 μg/ml) at 4°C. Surface expression of avian Mxra8 orthologs was assessed using a FLAG tag located at the N-terminus located downstream of the signal peptide sequence. Mouse Mxra8 expression levels were compared to ΔMxra8 3T3 (negative) and wild-type 3T3 (positive) cells. Mxra8 surface expression levels of all other species were compared to ΔMxra8 3T3 (negative) and ΔMxra8 3T3 cells transduced with mouse Mxra8 (positive). Trans-complemented ΔMxra8 3T3 cells with Mxra8 surface expression levels of less than 90% after blasticidin selection were further enriched by fluorescence activated cell sorting. Cells (2.5 x 105) were incubated with a pool of anti-Mxra8 mAbs (1G11.E6, 1H1.F5, 3G2.F5, 4E7.D10, 7F1.D8, 8F7.E1, and 9G2.D6) (1 μg/ml) in 1% BSA/PBS for 30 min at 4°C. After 30 min, cells were washed and incubated with Alexa Fluor 647 conjugated goat anti-hamster IgG (1 μg/ml). After a 30-min incubation, cells were washed, resuspended in PBS supplemented with 2% FBS and 1 mM EDTA, and sorted using a BD FACSAria II. Cells positive for Mxra8 expression subsequently were expanded in culture.
Trans-complemented ΔMxra8 3T3 cells were inoculated with CHIKV 181/25 (MOI 3, 9.5 h), CHIKV AF15561 (MOI 3, 9.5 h), MAYV (MOI 3, 24 h), RRV (MOI 3, 32 h), or VEEV-GFP (MOI 3, 12 h) in DMEM supplemented with 2% FBS. At indicated time points, cells were harvested, fixed and permeabilized using Foxp3 Transcription Factor Staining Buffer Set (Thermo Fisher), and stained for viral antigen after incubation with the following antibodies: CHIKV (mouse mAb CHK-11 (Pal et al., 2013)), MAYV (mouse mAb CHK-48 (Fox et al., 2015)), RRV (human mAb 1I9 (Smith et al., 2015)), or VEEV-GFP (mouse 3B4C-4 (Hunt et al., 2006)). Cells were washed, incubated with Alexa Fluor 647 conjugated goat anti-mouse IgG or goat anti-human IgG (Thermo Fisher), and analyzed by flow cytometry using a MACSQuant Analyzer 10 (Miltenyi Biotec). For multi-step growth curves, trans-complemented ΔMxra8 3T3 cells or primary MEFs were inoculated (MOI 0.01) with CHIKV 181/25, CHIKV AF15561, or CHIKV LR-2006 for 1 h, washed, and maintained in 2% FBS growth medium. Viral supernatants were harvested at indicated timepoints, titered on Vero cells, fixed, permeabilized, and stained with CHK-11 and horseradish peroxidase-conjugated goat anti-mouse IgG. Infected foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macroanalyzer (Cellular Technologies).
Bovine corneal endothelial cells were transduced with lentiviruses and selected with blasticidin for 7 days. Mxra8 surface expression was confirmed by flow cytometry as described above. To generate a clonal line that expressed high levels of Mxra8, bovine corneal endothelial cells were subjected to fluorescence-activated cell sorting as described above. Bovine corneal endothelial cells were inoculated with CHIKV 181/25 (MOI 3, 12 h) or RRV (MOI 3, 12 h) in 5% FBS growth medium. After infection, cells were harvested, fixed, permeabilized, stained with virus-specific antibodies, and analyzed by flow cytometry.
Primary kudu fibroblasts were transduced with lentiviruses encoding kudu or kudu Δ15 Mxra8. Cell surface expression of Mxra8 was confirmed using a pool of anti-Mxra8 mAbs. Cells with >90% Mxra8 surface expression were inoculated with MAYV (MOI 3, 26 h). After infection, cells were harvested, fixed, permeabilized, stained with biotinylated anti-MAYV antibodies (MAYV-115 and MAYV-134) (Earnest et al., 2019) that contain an N297Q mutation in the Fc region, and analyzed by flow cytometry.
Expression and purification of Mxra8 proteins.
A cDNA fragment encoding residues 24-309 of the Bos taurus Mxra8 extracellular domain was codon-optimized, synthesized, and inserted into the pET21a vector using the NdeI/NotI sites. After sequence confirmation, the plasmid construct was transformed into BL21(DE3) chemically competent cells (Thermo Fisher). Cells were grown at 37°C to an optimal density (600 nm) of 0.8 and induced with 0.1 mM IPTG for 4 h. Cells were harvested and resuspended in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 25% sucrose (TENDS) buffer, and lysed in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 200 mM sodium chloride, 1% sodium deoxycholate and 1% Triton X-100. Inclusion bodies were isolated from the cellular lysate after centrifugation at 6,000 x g for 20 min and washed in TENDS buffer supplemented with 100 mM NaCl and 0.5% Triton X-100. A final wash was performed in the same buffer without 0.5% Triton X-100. Inclusion bodies were denatured in in 100 mM Tris-HCl, 6 M guanidinium chloride and 20 mM β-mercaptoethanol for 1 h. Denatured protein was oxidatively refolded overnight at 4°C in 400 mM L-arginine, 100 mM Tris-HCl, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 10 mM EDTA and 200 mM phenylmethylsulphonyl fluoride. Refolded protein was concentrated using a 10,000-molecular weight cut-off stirred cell concentrator (EMD Millipore). Concentrated protein was further purified by HiLoad 16/600 Superdex 75 size exclusion chromatography (GE Healthcare) and HiTrap Q HP anion exchange chromatography (GE Healthcare). Purity and oligomeric state were confirmed by SDS-PAGE analysis and size exclusion chromatography coupled with multi-angle light scattering.
The mouse Mxra8-Fc fusion protein was generated as previously described where a cDNA fragment encoding Mxra8 (Genbank accession no. NM_024263) and the mouse IgG2b Fc region was synthesized (Integrated DNA Technologies) and inserted into the pCDNA3.4 vector. The mouse +moo Mxra8-Fc, mouse-Mxra8+8-Fc, mouse-Mxra8+10-Fc, and mouse +(GGS)5 proteins were generated as described above with the addition of the amino acids “GEQRVGEQRLGEQRV”, “QRVGEQRL”, “GEQRVGEQRL”, or “(GGS)5” after amino acid residue 98, respectively. Cattle (Genbank accession no. NM_001075830, residues 24-358), cattle Δmoo (Genbank accession no. NM_001075830, residues 24-95 and 111-358), Bubalus (residues 24-348), Bubalus Δmoo (residues 24-95, 101-348), kudu (residues 24-358), and kudu Δ15 (residues 24-95, 111-358) Mxra8-Fc proteins were generated as described above. Mxra8-Fc plasmids were diluted in Opti-MEM, incubated with HYPE-5 reagent (OZ Biosciences), and the complex was transfected into Expi-293 cells (Thermo Fisher, 106 cells/ml). Cells were supplemented daily with Expi293 medium and 2% (w/v) Hyclone Cell Boost. Four days post transfection, the supernatant was harvested by centrifuging at 3,000 x g for 15 min and purified using Protein A Sepharose 4B (Thermo Fisher). After elution, Mxra8-Fc proteins were dialyzed into 20 mM HEPES, 150 mM NaCl, pH 7.5 and stored at −80°C. Purity was confirmed by SDS-PAGE analysis.
Protein crystallization and X-ray structure determination.
Purified, recombinant Bos taurus Mxra8 protein (residues 24-309) produced in E. coli was crystallized by hanging drop vapor diffusion at 15 mg/ml in 0.1 M HEPES, pH 8.0, 6% (w/v) PEG 6000, and 1.0 M LiCl2. Crystals were cryo-protected in the mother liquor supplemented with 25% ethylene glycol and flash-cooled in liquid nitrogen. Diffraction data was collected at the Advanced Light Source MBC Beamline 4.2.2 (100K; 1.0000 Å wavelength) and processed with XDS. Four datasets were collected by translating along a single crystal. Datasets were processed and merged using XDS. Initial phases were obtained through molecular replacement with Phaser using murine Mxra8 coordinates (PDB 6NK3) as a search model. Model building was carried out in COOT (Emsley et al., 2010) and refinement was performed with Phenix (Adams et al., 2010). The overall electron density of the structure was good except in regions corresponding to amino acid residues 79-85 and 95-112, where weak electron density was noted and building of well-restrained geometric models was challenging. For these two regions, refinement was carried out with occupancy values set to 0.5. The final Bos taurus Mxra8 model contains mature residues 33-309 and 117 water molecules with an Rwork of 21.8% and Rfree of 24.2%. Data collection and refinement statistics are reported in Table S2. Structural validation was assessed using Molprobity (Chen et al., 2010), and structural figures were generated using PyMOL (Schrodinger, Version 2.1.0).
Mxra8 ELISA binding assays.
Maxisorp ELISA plates were coated with anti-CHIKV mAb 4N12 (Smith et al., 2015) (2 μg/ml) overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed four times with PBS and blocked with 4% BSA for 1 h at 25°C. CHIKV VLPs were diluted to 1 μg/ml in 2% BSA and added for 1 h at 25°C. Mxra8-Fc proteins were diluted in 2% BSA and incubated for 1 h at 25°C. Plates were washed with PBS and incubated with horseradish peroxide conjugated goat anti-mouse IgG (H + L) (1:2000 dilution, Jackson ImmunoResearch) for 1 h at 25°C. After washing, plates were developed with 3,3’-5,5’ tetramethylbenzidine substrate (Thermo Fisher) and 2N H2SO4. Plates were read at 450 nM using a TriStar Microplate Reader (Berthold). Anti-Mxra8 mAb ELISAs were performed by coating Maxisorp ELISA plates with Mxra8-Fc proteins (2 μg/ml) overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed four times with PBS and 0.05% Tween-20 and blocked with 4% BSA for 1 h at 25°C. Anti-Mxra8 mAbs were diluted in 2% BSA and added for 1 h at 25°C. Plates were washed with PBS and 0.05% Tween-20 and incubated with horseradish peroxidase conjugated goat anti-Armenian hamster IgG (H + L) (1:2000 dilution, Jackson ImmunoResearch) for 1 h at 25°C. After washing, plates were developed and read as described above.
Mxra8 BLI binding assays.
BLI experiments were performed in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% P20 surfactant with 1% BSA at 25°C using an Octet-Red96 device (Pall ForteBio). CHK-265 was biotinylated as previously described (Fox et al., 2015), loaded onto streptavidin biosensors (ForteBio) until saturation, then incubated with CHIKV VLP for 5 min. To measure the affinity of mouse and cattle Δmoo Mxra8 to CHIKV VLP, the loaded streptavidin biosensors were dipped into increasing concentrations of Mxra8 protein (10 nM to 1 μM) for 5 min, followed by a 5 min dissociation. Recombinant cattle and mouse + moo Mxra8 proteins (5 μM) were allowed to associate and dissociate for 5 min at each step. Real-time data was analyzed using BIAevaluation 3.1 (GE Healthcare) and kinetic curves and steady-state equilibrium were fitted using a global 1:1 binding algorithm with drifting baseline.
Virus-cell binding assays.
ΔMxra8 3T3 cells or complemented cells (3 × 104 cells) were incubated with CHIKV 181/25 (MOI 200) for 1 h at 4°C. Cells were washed extensively with cold PBS, and viral antigen staining was performed using a cocktail of CHIKV anti-E1 and anti-E2 antibodies (mAbs CHK-11, CHK-84, CHK-124, and CHK-166 (Pal et al., 2013)). Cells were washed, incubated with Alexa Fluor 647 conjugated goat anti-mouse IgG (Thermo Fisher), and analyzed by flow cytometry.
Sample collection and sequencing.
All experiments were performed after approval by the Institutional Animal Care and Use Committee at the Washington University School of Medicine and the Saint Louis Zoo (Assurance number A3381-01). Muscle and liver samples were obtained from banked samples collected at necropsy. All sample sources are listed in Table S4. Total RNA was extracted using TRIzol and a Direct-zol RNA kit (Zymo Research). First strand cDNA was generated using a Superscript III First-Strand Synthesis System (Thermo Fisher). Species- and gene-specific primers were designed from deposited sequences and assembled whole genome sequences (Table S4 and S5). PCR master mixes were prepared in a nucleic acid-free PCR workstation. Bovinae Mxra8 genes were amplified using nested PCR with 1X Q5 Reaction Buffer, 200 μM dNTPs, 200 nM Forward primer, 200 nM Reverse primer, 2 μl cDNA, 1X Q5 High GC Enhancer, and 1 μl of Q5 High-Fidelity DNA Polymerase (NEB). The following amplification protocol was used for both first- and second-round amplifications: 1) 98°C for 30 sec; 2) 98°C for 20 sec; 3) 60-68°C for 30 sec; 4) 72°C for 60 sec; 5) 72°C for 2 min; with steps 2-4 repeated for 35 cycles. All primer sequences and annealing temperatures are listed in Table S5. PCR products were separated on a 1% agarose gel, and the band of the predicted size was purified by gel-extraction (Qiagen). Purified PCR products were sequenced using amplification primers and primers that target a conserved region in the Mxra8 gene (TGCCACCTGCACCACCACTAC and GTAGTGGTGGTGCAGGTGGCA).
Structural docking analysis.
The cattle Mxra8 crystal structure was superimposed onto the Mxra8 chains in the 1) Mxra8-bound CHIKV-VLP cryo-EM model (PDB 6NK6 and (Basore et al., 2019)) using the MatchMaker tool in USCF Chimera and 2) MXRA8-bound CHIKV p62-E1 glycoprotein crystal structure (PDB 6ORT) using the ‘super’ command in PyMOL (Version 2.1.0, Schrodinger).
Whole genome sequence assembly and alignments.
Whole genome sequences of the species listed in Table S4 were aligned to the Bos taurus genome (Elsik et al., 2016) using BWA-MEM from the Burrows-Wheeler Aligner (Li and Durbin, 2010) to look for reads mapping to the Mxra8 gene and the “GEQRVGEQRLGEQRV” insertion sequence. For non-Bovinae species, reads were mapped to genomes with either a 15-residue insertion (Bos taurus), partial insertion (Bubalus bubalis; AWWX01000000), no insertion (Ovis aries; CM008472) to identify reads overlapping the insert junction. Mxra8 nucleotide sequences were aligned with MUSCLE. Quality for alignments was refined by manual assessment and adjustment of misaligned sites.
Immunoblotting.
Primary MEFs (106 cells) were harvested and lysed in RIPA buffer (Thermo Fisher) containing a protease inhibitor cocktail (Roche). Cell lysates were mixed with LDS buffer (Thermo Fisher), incubated at 70°C for 10 min, and electrophoresed using 10% Bis-Tris gels (Thermo Fisher) in MOPS running buffer. Separated proteins were transferred to a PVDF membrane using an iBlot2 Dry Blotting System (Thermo Fisher). The PVDF membrane was blocked with 5% non-fat milk, probed with hamster anti-Mxra8 mAbs (3G2.F5 or 9G2.D6, 1 μg/ml) and horseradish peroxidase conjugated goat anti-Armenian hamster IgG, and developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher).
Evolutionary analyses.
Phylogenetic analyses were executed in RevBayes (Hohna et al., 2016). The unrooted Mxra8 gene tree topology was estimated under the HKY+Gamma substitution model (Hasegawa et al., 1985; Yang, 1994). (see Fig S7 and Table S6).
Statistical analyses.
Statistical significance was assigned using Prism Version 8 (GraphPad) when P < 0.05. Statistical analysis of viral infection levels was determined by one-way ANOVA with Dunnett’s post-test. Statistical analysis of in vivo experiments was determined by either one-way or two-way ANOVA with a Kruskal-Wallis or Dunnett’s post-test depending on the data distribution and the number of comparison groups. The statistical tests, number of independent experiments, and number of experimental replicates are indicated in the Figure legends.
Data and Code Availability.
The authors declare that all data supporting the findings of this study are available within the paper. Analysis scripts and the sequence alignment are available at: https://github.com/mlandis/mxra8_bovinae. The X-ray crystal structure of cattle Mxra8 has been deposited as PDB 6ORT.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mxra8 mAb 1G11.E6 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 1H1.F5 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 3G2.F5 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 4E7.D10 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 7F1.D8 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 8F7.E1 | Zhang et al., 2018 | N/A |
| Anti-Mxra8 mAb 9G2.D6 | Zhang et al., 2018 | N/A |
| CHK-11 | Pal et al., 2013 | N/A |
| CHK-48 | Pal et al., 2013 | N/A |
| CHK-84 | Pal et al., 2013 | N/A |
| CHK-124 | Pal et al., 2013 | N/A |
| CHK-152 | Pal et al., 2013 | N/A |
| CHK-166 | Pal et al., 2013 | N/A |
| CHK-265 | Pal et al., 2013 | N/A |
| 1I9 | Smith et al., 2015 | N/A |
| 3B4C-4 | Hunt et al., 2015 | N/A |
| MAYV-115 N297Q | Earnest et al., 2019 | N/A |
| MAYV-134 N297Q | Earnest et al., 2019 | N/A |
| Alexa Fluor 488 conjugated goat anti-mouse IgG | Thermo Fisher | A28175; RRID: AB_2536161 |
| Alexa Fluor 647 conjugated goat anti-Armenian hamster IgG | Abcam | ab173004; RRID: AB_2732023 |
| Alexa Fluor 647 conjugated goat anti-mouse IgG | Thermo Fisher | A21235; RRID: AB_2535804 |
| Alexa Fluor 647 conjugated goat anti-human IgG | Thermo Fisher | A21445; RRID: AB_2535862 |
| Peroxidase conjugated goat anti-mouse IgG (H + L) | Jackson ImmunoResearch | 115-035-062; RRID: AB_2338504 |
| Peroxidase conjugated goat anti-Armenian hamster IgG (H + L) | Jackson ImmunoResearch | 127-035-160; RRID: AB_2338976 |
| Bacterial and Virus Strains | ||
| Chikungunya virus (strain 181/25) | Levitt et al., 1986 | AF192908 |
| Chikungunya virus (strain AF15561) | Hawman et al., 2016 | EF452493 |
| Chikungunya virus (strain LR-2006) | Tsetsarkin et al., 2016 | KY575571 |
| Mayaro virus (strain BeH407) | World Reference Center for Emerging Viruses and Arboviruses | AAY45742 |
| Ross River virus (strain T48) | World Reference Center for Emerging Viruses and Arboviruses | ACV67002 |
| O’nyong’nyong virus (strain MP30) | World Reference Center for Emerging Viruses and Arboviruses | AAC97207 |
| Venezuelan equine encephalitis virus (strain TC-83) | World Reference Center for Emerging Viruses and Arboviruses | L01443 |
| Biological Samples | ||
| Muscle tissue, Bos taurus | Whole Foods Market | N/A |
| Liver tissue, Bos javanicus | Saint Louis Zoo | ISIS #108161 |
| Muscle tissue, Bos grunniens | The Yak Boys | YB105 |
| Muscle tissue, Bison bison | Whole Foods Market | N/A |
| Muscle tissue, Bubalus bubalis | Nicky Farms | 4695 |
| Liver tissue, Tragelaphus eurycerus | Saint Louis Zoo | ISIS #103656 |
| Liver tissue, Tragelaphus angasii | Saint Louis Zoo | ISIS #118494 |
| Muscle and liver tissue, Tragelaphus imberbis | Saint Louis Zoo | ISIS #107129 |
| Liver tissue, Muntiacus muntjac | Saint Louis Zoo | ISIS #103117 |
| Muscle tissue, Odocoileus virginianus | Case Farms | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Chikungunya virus-like particles (strain 37997) | Akahata et al., 2010 | N/A |
| Mouse Mxra8 ectodomain23-296 | Basore et al., 2019 | N/A |
| Mouse +moo Mxra8 ectodomain | This study | N/A |
| Mouse +5 Mxra8 ectodomain | This study | N/A |
| Mouse +8 Mxra8 ectodomain | This study | N/A |
| Mouse +9 Mxra8 ectodomain | This study | N/A |
| Mouse +10 Mxra8 ectodomain | This study | N/A |
| Mouse +[GGS]5 Mxra8 ectodomain | This study | N/A |
| Cattle Mxra8 ectodomain24-309 | This study | N/A |
| Cattle Δmoo Mxra8 ectodomain | This study | N/A |
| Mouse Mxra8 fused to mouse IgG2b Fc region | Zhang et al., 2018 | N/A |
| Mouse +moo Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Mouse +8 Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Mouse +10 Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Mouse +[GGS]5 Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Cattle Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Cattle Δmoo Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Buffalo Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Buffalo Δ5 Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Kudu Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Kudu Δ15 Mxra8 fused to mouse IgG2b Fc region | This study | N/A |
| Critical Commercial Assays | ||
| TaqMan RNA-to-Ct 1-Step Kit | Thermo Fisher | 4392939 |
| In-Fusion HD Cloning Plus | Takara | 638910 |
| HiScribe T7 In Vitro Transcription Kit | New England BioLabs | E2040S |
| MEGAclear Transcription Clean-Up Kit | Thermo Fisher | AM1908 |
| Deposited Data | ||
| X-ray crystal structure of murine Mxra8 | Basore et al., 2019 | PDB 6NK3 |
| Electron Cryo-Microscopy of Chikungunya VLP in complex with mouse Mxra8 receptor | Basore et al., 2019 | PDB 6NK6; EMD-9394 |
| Crystal Structure of Bos taurus Mxra8 Ectodomain | This study | PDB 6ORT |
| Bos primigenius isolate:CPC98 Raw sequence reads | Park et al., 2015 | PRJNA294709; SRR2465682 |
| Bos indicus strain:Nelore RefSeq Genome sequencing | Canavez et al., 2012 | PRJNA360096; XM_019976191 |
| Bos javanicus WGS data | Kalbfleisch et al., 2013 | PRJNA325061; SRR4035276 |
| Bos grunniens WGS data | N/A | PRJNA359997; SRR5140177 |
| Bubalus bubalis assembly | N/A | PRJNA207334; AWWX01000000 |
| Syncerus caffer paired end sequencing | N/A | PRJNA341313; SRR4104498 |
| Tragelaphus angasii RNA-seq data | N/A | PRJNA388863; SRR5647659 |
| Ovis vignei paired end sequencing | N/A | PRJEB5463; ERR454948 |
| Pseudois nayaur genome sequencing and assembly | N/A | PRJNA361448; SRR5439716 |
| Capra ibex paired end sequencing | N/A | PRJNA361447; SRR5260693 |
| Moschus berezovskii raw sequence reads | N/A | PRJNA289641; SRR2098995 |
| Cervus elaphus genome sequencing and assembly | Bana et al., 2018 | PRJNA324173; SRR4013902 |
| Experimental Models: Cell Lines | ||
| NIH/3T3 | ATCC | CRL-1658; RRID: CVCL_0594 |
| HEK-293 | ATCC | CRL-1573; RRID: CVCL_0045 |
| Vero | ATCC | CCL-81; RRID: CVCL_0059 |
| Bos taurus corneal endothelial cells | ATCC | CRL-2048; RRID: CVCL_2865 |
| Expi293F | Thermo Fisher | A14527 |
| C57BL/6J primary MEF | This study | N/A |
| C57BL/6J Mxra8WT/KO primary MEF | This study | N/A |
| C57BL/6J Mxra8KO/KO primary MEF | Zhang et al., 2019 | N/A |
| C57BL/6J Mxra8moo/KO primary MEF | This study | N/A |
| C57BL/6J Mxra8moo/moo primary MEF | This study | N/A |
| Bos gaurus primary fibroblasts | Modi et al., 2004 | N/A |
| Syncerus caffer primary fibroblasts | Modi et al., 2004 | N/A |
| Boselaphus tragelaphus primary fibroblasts | Modi et al., 2004 | N/A |
| Tragelaphus imberbis primary fibroblasts | Modi et al., 2004 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | 000664; RRID: IMSR_JAX:000664 |
| Mouse: C57BL/6J Mxra8WT/KO | This study | N/A |
| Mouse: C57BL/6J Mxra8KO/KO | This study | N/A |
| Mouse: C57BL/6J Mxra8moo/KO | This study | N/A |
| Mouse: C57BL/6J Mxra8moo/moo | This study | N/A |
| Oligonucleotides | ||
| Mxra8 sgRNA-1: 5’-CTTGTGGATATGTATTCGGCNGG-3’ | Genome Engineering and iPSC Center, Washington University School of Medicine | N/A |
| Mxra8 sgRNA-2: 5’ACTTGTGGATATGTATTCGGNGG-3’ | Genome Engineering and iPSC Center, Washington University School of Medicine | N/A |
| CHIKV-AF FOR: 5’-TCGACGCGCCATCTTTAA-3’ | Zhang et al., 2019 | N/A |
| CHIKV-AF REV: 5’-ATCGAATGCACCGCACACT-3’ | Zhang et al., 2019 | N/A |
| CHIKV-AF Probe: 5’-/56-FAM/ACCAGCCTG/ZEN/CACCCACTCCTCAGAC/3IABkFQ/-3’ | Zhang et al., 2019 | N/A |
| See Table S5 for primer sequences and annealing temperatures used to amplify Mxra8 from primary tissue samples of different animals | ||
| Recombinant DNA | ||
| pLV-EF1a vector | Hayer et al., 2016 | Addgene 85132; RRID: Addgene_85132 |
| Codon-optimized mouse Mxra8 cloned into pLV-EF1a vector | Zhang et al., 2018 | NM_024263 |
| Codon-optimized mouse +moo Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized cattle (Bos taurus) Mxra8 cloned into pLV-EF1a vector | This study | NM_001075830 |
| Codon-optimized cattle Δmoo Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized zebu (Bos indicus) Mxra8 cloned into pLV-EF1a vector | This study | XM_019976191 |
| Codon-optimized zebu Δmoo Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized water buffalo (Bubalus bubalis) Mxra8 cloned into pLV-EF1a vector | This study | XM_006066948.2 |
| Codon-optimized water buffalo Δ5 Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized lesser kudu (Tragelaphus angasii) Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized lesser kudu Δ15 Mxra8 cloned into pLV-EF1a vector | This study | N/A |
| Codon-optimized goat (Capra hircus) Mxra8 cloned into pLV-EF1a vector | This study | XM_018060531 |
| Codon-optimized dog (Canis lupus familiaris) Mxra8 cloned into pLV-EF1a vector | This study | XM_546712 |
| Codon-optimized rat (Rattus norvegicus) Mxra8 cloned into pLV-EF1a vector | This study | NM_001007002 |
| Codon-optimized chimp (Pan troglodytes) Mxra8 cloned into pLV-EF1a vector | This study | NM_001280245 |
| Codon-optimized horse (Equus caballus) Mxra8 cloned into pLV-EF1a vector | This study | XM_023636045 |
| Codon-optimized sheep (Ovis aries) Mxra8 cloned into pLV-EF1a vector | This study | XM_027975805 |
| Codon-optimized turkey (Meleagris gallopavo) Mxra8 cloned into pLV-EF1a vector | This study | XP_010721105.1 |
| Codon-optimized duck (Anas platyrhynchos) Mxra8 cloned into pLV-EF1a vector | This study | XM_027443263 |
| Codon-optimized chicken (Gallus gallus) Mxra8 cloned into pLV-EF1a vector | This study | NP_989967 |
| psPAX2 | Didier Trono | Addgene 12260; RRID: Addgene_12260 |
| pMD2.G | Didier Trono | Addgene 12259; RRID: Addgene_12259 |
| Codon-optimized mouse Mxra8 ectodomain23-296 cloned into pET21a vector | Basore et al., 2019 | N/A |
| Codon-optimized mouse Mxra8 +5 ectodomain cloned into pET21a vector | This study | N/A |
| Codon-optimized mouse Mxra8 +8 ectodomain cloned into pET21a vector | This study | N/A |
| Codon-optimized mouse Mxra8 +9 ectodomain cloned into pET21a vector | This study | N/A |
| Codon-optimized mouse Mxra8 +10 ectodomain cloned into pET21a vector | This study | N/A |
| Codon-optimized mouse Mxra8 +[GGS]5 ectodomain cloned into pET21a vector | This study | N/A |
| Codon-optimized cattle Mxra8 ectodomain24-309 cloned into pET21a vector | This study | NM_001075830 |
| Codon-optimized cattle Δmoo ectodomain Mxra8 cloned into pET21a vector | This study | N/A |
| Codon optimized mouse Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized mouse +moo Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized mouse +8 Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized mouse +10 Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized mouse +[GGS]5 ectodomain Mxra8 and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized cattle Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized cattle Δmoo Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized buffalo Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized buffalo Δ5 Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized kudu Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Codon optimized kudu Δ15 Mxra8 ectodomain and mouse IgG2b Fc region cloned into pCDNA3.4 vector | This study | N/A |
| Software and Algorithms | ||
| Burrows-Wheeler Aligner | Li et al., 2009 | http://bio-bwa.sourceforge.net/ |
| MUSCLE | Edgar et al., 2004 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| RevBayes | Höhna et al., 2016 | https://revbayes.github.io/ |
| Mesquite | Maddison et al., 2018 | https://www.mesquiteproject.org/ |
| PAML | Yang et al., 2007 | http://abacus.gene.ucl.ac.uk/software/paml.html |
| FigTree | Andrew Rambaut | http://tree.bio.ed.ac.uk/software/figtree/; Version 1.4.4 |
| FlowJo | FlowJo, LLC | Versions 9 and 10 |
| PyMOL | Schrodinger | Version 2.1.0 |
| UCSF Chimera | RVBI | https://www.cgl.ucsf.edu/chimera/; Version 1.13 |
| GraphPad Prism | GraphPad | Version 8.2.1 |
| Other | ||
| Evolution analysis scripts and sequence alignments | This study | https://github.com/mlandis/mxra8_bovinae |
HIGHLIGHTS.
An insertion in Bovinae Mxra8 sterically blocks alphavirus binding and infection
The sequence insertion evolved in the Miocene epoch at least 5 million years ago
Loss of the insertion in Mxra8 in several Bovinae restores alphavirus infection
Introduction of the insertion into Mxra8 of mice prevents alphavirus pathogenesis
ACKNOWLEDGEMENTS
This study was supported by NIH grants R01AI114816, R01AI123348, R01AI095436, T32AI007172, and Center for Structural Genomics of Infectious Diseases contract number HHSN272201700060C. We thank Jay Nix at ALS Beamline 4.2.2 for assistance with data collection and processing, James Womack and Terje Raudsepp for providing the primary Bovine fibroblasts, James Crowe for providing anti-alphavirus mAbs, Mary Duncan for arranging access to the non-domestic bovine tissues, the Genome Engineering and iPSC Center and Department of Pathology Microinjection Facility at Washington University for gRNA design and mouse generation, Larissa Thackray for design of the Mxra8 knock-in mouse, and the Living Earth Collaborative, Jonathan Losos, Joseph Bielawski, and Terence Dermody for editorial suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
M.S.D. is a consultant for Inbios and Emergent BioSolutions and on the Scientific Advisory Board of Moderna. D.H.F. is a founder of Courier Therapeutics.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basore K, Kim AS, Nelson CA, Zhang R, Smith BK, Uranga C, Vang L, Cheng M, Gross ML, Smith J, et al. (2019). Cryo-EM structure of Chikungunya virus in complex with the Mxra8 receptor. Cell 177, 1725–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Lauth AM, Nemeth NM, Kohler DJ, and Bowen RA (2016). Viremia in North American Mammals and Birds After Experimental Infection with Chikungunya Viruses. Am J Trop Med Hyg 94, 504–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Lee H, Mulvey M, and Brown DT (1997). Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J Virol 71, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, Antunes L, Garrison E, Banks E, Evani US, Muzny D, Poplin R, Gibbs RA, Marth G, and Yu F (2015). The distribution and mutagenesis of short coding INDELs from 1,128 whole exomes. BMC Genomics 16, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, and Baker TS (1995). Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, Abraham J, Choe H, Farzan M, and Sawyer SL (2013). Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11, e1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTulleo L, and Kirchhausen T (1998). The clathrin endocytic pathway in viral infection. Embo j 17, 4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Gertz EM, Wojtowicz D, Zhabinskaya D, Levens D, Benham CJ, Schaffer AA, and Przytycka TM (2014). Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation. Nucleic Acids Res 42, 12367–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest JT, Basore K, Roy V, Bailey AL, Wang D, Alter G, Fremont DH, and Diamond MS (2019). Neutralizing antibodies against Mayaro virus require Fc effector functions for protective activity. J Exp Med, 216, 2282–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Unni DR, Diesh CM, Tayal A, Emery ML, Nguyen HN, and Hagen DE (2016). Bovine Genome Database: new tools for gleaning function from the Bos taurus genome. Nucleic Acids Res 44, D834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MK, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. (2015). Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard GP, Shorthose JE, Weir RP, Walsh SJ, and Melville LF (1988). Arboviruses recovered from sentinel livestock in northern Australia. Veterinary microbiology 18, 109–118. [DOI] [PubMed] [Google Scholar]
- Gardner CL, Ebel GD, Ryman KD, and Klimstra WB (2011). Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc Natl Acad Sci U S A 108, 16026–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilherme JM, Gonella-Legall C, Legall F, Nakoume E, and Vincent J (1996). Seroprevalence of five arboviruses in Zebu cattle in the Central African Republic. Transactions of the Royal Society of Tropical Medicine and Hygiene 90, 31–33. [DOI] [PubMed] [Google Scholar]
- Han S, Park HR, Lee EJ, Jang JA, Han MS, Kim GW, Jeong JH, Choi JY, Frank B, and Jung YK (2018). Dicam promotes proliferation and maturation of chondrocyte through Indian hedgehog signaling in primary cilia. Osteoarthritis and cartilage 26, 945–953. [DOI] [PubMed] [Google Scholar]
- Han SW, Jung YK, Lee EJ, Park HR, Kim GW, Jeong JH, Han MS, and Choi JY (2013). DICAM inhibits angiogenesis via suppression of AKT and p38 MAP kinase signalling. Cardiovascular research 98, 73–82. [DOI] [PubMed] [Google Scholar]
- Han SW, Kim JM, Lho Y, Cho HJ, Jung YK, Kim JA, Lee H, Lee YJ, and Kim ES (2019). DICAM Attenuates Experimental Colitis via Stabilizing Junctional Complex in Mucosal Barrier. Inflammatory bowel diseases 25, 853–861. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, and Yano T (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22, 160–174. [DOI] [PubMed] [Google Scholar]
- Heidner HW, Knott TA, and Johnston RE (1996). Differential processing of sindbis virus glycoprotein PE2 in cultured vertebrate and arthropod cells. J Virol 70, 2069–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohna S, Landis MJ, Heath TA, Boussau B, Lartillot N, Moore BR, Huelsenbeck JP, and Ronquist F (2016). RevBayes: Bayesian Phylogenetic Inference Using Graphical Models and an Interactive Model-Specification Language. Systematic Biology 65, 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AR, Frederickson S, Hinkel C, Bowdish KS, and Roehrig JT (2006). A humanized murine monoclonal antibody protects mice either before or after challenge with virulent Venezuelan equine encephalomyelitis virus. J Gen Virol 87, 2467–2476. [DOI] [PubMed] [Google Scholar]
- Jung YK, Han SW, Kim GW, Jeong JH, Kim HJ, and Choi JY (2012). DICAM inhibits osteoclast differentiation through attenuation of the integrin alphaVbeta3 pathway. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 27, 2024–2034. [DOI] [PubMed] [Google Scholar]
- Jung YK, Jeong JH, Ryoo HM, Kim HN, Kim YJ, Park EK, Si HJ, Kim SY, Takigawa M, Lee BH, et al. (2004). Gene expression profile of human chondrocyte HCS-2/8 cell line by EST sequencing analysis. Gene 330, 85–92. [DOI] [PubMed] [Google Scholar]
- Jung YK, Jin JS, Jeong JH, Kim HN, Park NR, and Choi JY (2008). DICAM, a novel dual immunoglobulin domain containing cell adhesion molecule interacts with alphavbeta3 integrin. Journal of Cellular Physiology 216, 603–614. [DOI] [PubMed] [Google Scholar]
- Klimstra WB, Ryman KD, and Johnston RE (1998). Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 72, 7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, and Lok SM (2011). The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J Virol 85, 9327–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hapuarachchi HC, Chen KC, Hussain KM, Chen H, Low SL, Ng LC, Lin R, Ng MM, and Chu JJ (2013). Mosquito cellular factors and functions in mediating the infectious entry of chikungunya virus. PLoS Neglected Tropical Diseases 7, e2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, and Rey FA (2001). The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEachern S, McEwan J, and Goddard M (2009). Phylogenetic reconstruction and the identification of ancient polymorphism in the Bovini tribe (Bovidae, Bovinae). BMC Genomics 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson JJ, Chapman NH, Rees DC, Liu YT, and Clegg JB (1997). Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet 16, 100–103. [DOI] [PubMed] [Google Scholar]
- McFadden AM, McFadden BD, Mackereth GF, Clough RR, Hueston L, Gradwell B, and Dymond M (2009). A serological survey of cattle in the Thames - Coromandel district of New Zealand for antibodies to Ross River virus. New Zealand Veterinary Journal 57, 116–120. [DOI] [PubMed] [Google Scholar]
- Modi WS, Ivanov S, and Gallagher DS (2004). Concerted evolution and higher-order repeat structure of the 1.709 (satellite IV) family in bovids. J Mol Evol 58, 460–465. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Goode DL, Kvikstad E, Albers CA, Zhang ZD, Mu XJ, Ananda G, Howie B, Karczewski KJ, Smith KS, et al. (2013). The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Research 23, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, and Brown DT (1995). Involvement of the molecular chaperone BiP in maturation of Sindbis virus envelope glycoproteins. J Virol 69, 1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SJ (2005). Atlas of Mammalian Chromosomes (John Wiley & Sons, Inc.). [Google Scholar]
- Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MKS, et al. (2013). Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus PLoS Pathog 9, e1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes AM, Brown DT, Rothnagel R, Chiu W, Schoepp RJ, Johnston RE, and Prasad BV (1993). Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA 90, 9095–9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Yang Y, Pasquarella JR, Xu L, Qian Z, Holmes KV, and Li F (2017). Structural and Molecular Evidence Suggesting Coronavirus-driven Evolution of Mouse Receptor. J Biol Chem 292, 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, and Cherry S (2011). Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, Khomadiak S, Ashbrook AW, Kahle KM, Fong RH, et al. (2015). Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host Microbe 18, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Cheng RH, Olson NH, Peterson P, Chase E, Kuhn RJ, and Baker TS (1995). Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc Natl Acad Sci USA 92, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Zhao Z, Chai Y, Jin X, Li C, Yuan F, Liu S, Gao Z, Wang H, Song J, et al. (2019). Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein. Cell 177, 1714–1724.e1712. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Wang KS, Schmaljohn AL, Kuhn RJ, and Strauss EG (1994). Host-cell receptors for Sindbis virus. Arch Virol Suppl 9, 473–484. [DOI] [PubMed] [Google Scholar]
- Suhrbier A, Jaffar-Bandjee MC, and Gasque P (2012). Arthritogenic alphaviruses--an overview. Nat Rev Rheumatol 8, 420–429. [DOI] [PubMed] [Google Scholar]
- Tian X, Strassmann JE, and Queller DC (2011). Genome nucleotide composition shapes variation in simple sequence repeats. Molecular Biology and Evolution 28, 899–909. [DOI] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, and Rey FA (2010). Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712. [DOI] [PubMed] [Google Scholar]
- Wang K, Lenstra JA, Liu L, Hu Q, Ma T, Qiu Q, and Liu J (2018). Incomplete lineage sorting rather than hybridization explains the inconsistent phylogeny of the wisent. Communications Biology 1, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Kuhn RJ, Strauss EG, Ou S, and Strauss JH (1992). High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol 66, 4992–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Hagenbaugh A, Bellew LA, Netesov SV, Volchkov VE, Chang GJ, Clarke DK, Gousset L, Scott TW, Trent DW, et al. (1993). A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197, 375–390. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Powers AM, Brault AC, and Barrett AD (1999). Molecular epidemiological studies of veterinary arboviral encephalitides. Veterinary Journal (London, England : 1997) 157, 123–138. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Winegar R, Manger ID, and Forrester NL (2012). Alphaviruses: population genetics and determinants of emergence. Antiviral Res 94, 242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (1994). Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39, 306–314. [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Ohtsuka A, Yoshitaka T, Hirano S, Nomoto H, Yamamoto K, and Ninomiya Y (2003). Limitrin, a novel immunoglobulin superfamily protein localized to glia limitans formed by astrocyte endfeet. Glia 44, 190–204. [DOI] [PubMed] [Google Scholar]
- Zhang R, Earnest JT, Kim A,S, Winkler EA, Desai P, Adams LJ, Hu G, Bullock C, Gold B, Cherry S, et al. (2019). Expression of the Mxra8 receptor promotes alphavirus infection and pathogenesis in mice and Drosophila. Cell Reports 28, 2647–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, Rimkunas R, Fong RH, Lin H, Poddar S, et al. (2018a). Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Heil M, Kuhn RJ, and Baker TS (2005). Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology 332, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Wu WC, Shi M, and Holmes EC (2018b). The diversity, evolution and origins of vertebrate RNA viruses. Curr Opin Virol 31, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurano JP, Magalhaes FM, Asato AE, Silva G, Bidau CJ, Mesquita DO, and Costa GC (2019). Cetartiodactyla: Updating a time-calibrated molecular phylogeny. Molecular Phylogenetics and Evolution 133, 256–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper. Analysis scripts and the sequence alignment are available at: https://github.com/mlandis/mxra8_bovinae. The X-ray crystal structure of cattle Mxra8 has been deposited as PDB 6ORT.


