Abstract
Dupilumab is a fully human antibody to interleukin-4 receptor α that improves the signs and symptoms of moderate to severe atopic dermatitis (AD). To determine the effects of dupilumab on Staphylococcus aureus colonization and microbial diversity on the skin, bacterial DNA was analyzed from swabs collected from lesional and nonlesional skin in a double-blind, placebo-controlled study of 54 patients with moderate to severe AD randomized (1:1) and treated with either dupilumab (200 mg weekly) or placebo for 16 weeks. Microbial diversity and relative abundance of Staphylococcus were assessed by DNA sequencing of 16S ribosomal RNA, and absolute S. aureus abundance was measured by quantitative PCR. Before treatment, lesional skin had lower microbial diversity and higher overall abundance of S. aureus than nonlesional skin. During dupilumab treatment, microbial diversity increased and the abundance of S. aureus decreased. Pronounced changes were seen in nonlesional and lesional skin. Decreased S. aureus abundance during dupilumab treatment correlated with clinical improvement of AD and biomarkers of type 2 immunity. We conclude that clinical improvement of AD that is mediated by interleukin-4 receptor α inhibition and the subsequent suppression of type 2 inflammation is correlated with increased microbial diversity and reduced abundance of S. aureus.
INTRODUCTION
Atopic dermatitis (AD) is a complex disease characterized by chronic inflammation, barrier dysfunction, and microbial dysbiosis in the skin (Brunner et al., 2018; Brunner et al., 2017a; Werfel et al., 2016; Eichenfield et al., 2014; Cork et al., 2009; Bieber, 2008). Inflammation is driven predominantly by the type 2–T helper type 2 cytokine pathway (Brandt and Sivaprasad, 2011), in which the cytokines IL-4 and IL-13 play a major role (Brunner et al., 2017a). Type 2–T helper type 2 cytokines can inhibit key components of skin barrier function, such as filaggrin and loricrin (Leung and Guttman-Yassky, 2014; Howell et al., 2009; Kim et al., 2008), alter skin lipid metabolism (Berdyshev et al., 2018), and inhibit antimicrobial peptide synthesis (Guttman-Yassky et al., 2008; Nomura et al., 2003; Kisich et al., 2007; Ong et al., 2002). These conditions facilitate binding and colonization by S. aureus (Cho et al., 2001), which is associated with increased inflammation and disease severity (Kobayashi et al., 2015; Bjerre et al., 2017; Hepburn et al., 2017; Gong et al., 2006; Leyden et al., 1974).
Dupilumab is a fully human VelocImmune-derived mAb (MacDonald et al., 2014; Murphy et al., 2014) that blocks the shared receptor subunit for IL-4 and IL-13, thus inhibiting signaling of IL-4 and IL-13 (Gandhi et al., 2015). It is approved for the treatment of adults with inadequately controlled moderate to severe AD and for asthma. In early phase and phase 3 clinical trials, dupilumab administered as monotherapy for up to 16 weeks, or with concomitant topical corticosteroids for 4 or 52 weeks, significantly improved signs and symptoms of AD in adults with moderate to severe AD, with an acceptable safety profile (Beck et al., 2014; Thaçi et al., 2016; Simpson et al., 2016; Blauvelt et al., 2017). Additional studies further characterized the impact of dupilumab-mediated IL-4Rα blockade on the molecular and cellular features of lesional and nonlesional skin in patients with moderate to severe AD (Hamilton et al., 2014; Guttman-Yassky et al., 2019). In a randomized 16-week, phase 2 trial (AD-1307 EXPLORE, registered with ClinicalTrials.gov NCT01979016 on November 1, 2013), dupilumab treatment significantly improved epidermal differentiation in lesional skin (Guttman-Yassky et al., 2019), suggesting normalization of skin barrier function. Here, we report additional findings from AD-1307 EXPLORE regarding microbial diversity, S. aureus abundance, and the impact of dupilumab treatment on these factors. The relation between S. aureus abundance and clinical and molecular markers of AD severity was also assessed.
RESULTS
Patients
A total of 54 patients were randomized to dupilumab 200 mg weekly (n = 27) or placebo (n = 27). Most patients completed the treatment phase: 26 of 27 (96.3%) in the dupilumab group and 25 of 27 (92.6%) in the placebo group. The number of patients completing both the treatment and follow-up phases (up to week 32) was 17 of 27 (63.0%) in the dupilumab group and 13 of 27 (48.1%) in the placebo group (additional information is available from Guttman-Yassky et al., 2019). Skin swabs were available from all 26 patients in the dupilumab group and from 27 patients in the placebo group at prespecified times.
Pretreatment characterization of lesional and nonlesional skin
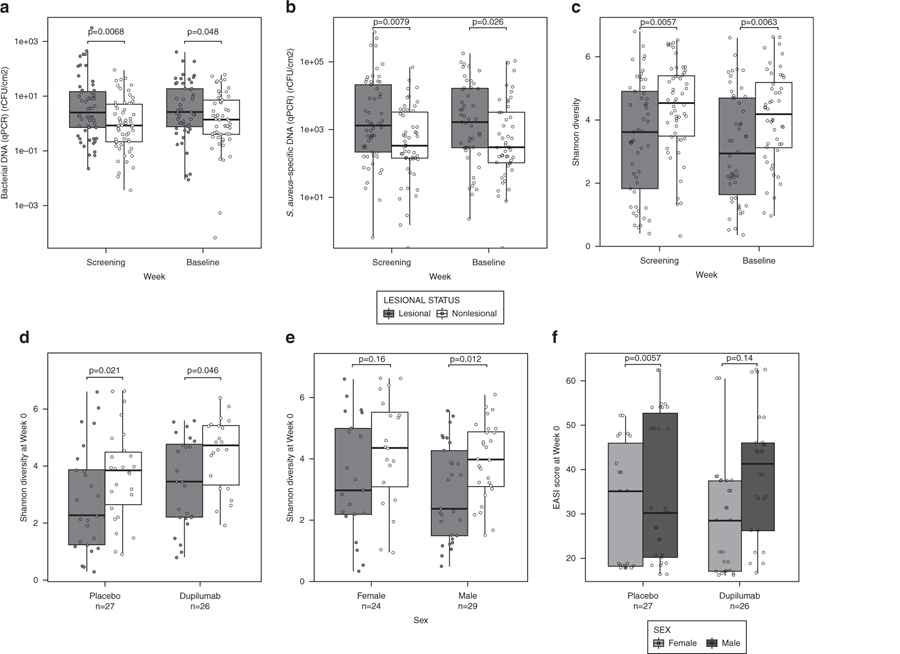
At screening and baseline, lesional skin contained significantly higher quantities of bacteria than did nonlesional skin, based on absolute quantification of 16S ribosomal RNA (rRNA) using quantitative PCR (qPCR) (P < 0.05) (Figure 1a). Lesional skin also contained significantly higher quantities of S. aureus compared with nonlesional skin (P < 0.05) (Figure 1b). Shannon diversity results from 16S rRNA sequencing indicated that lesional skin had significantly lower microbial diversity than did nonlesional skin (Figure 1c). Alpha diversity scores for lesional and nonlesional skin were consistent in the two treatment groups, confirming that both groups were similar in terms of bacterial diversity at baseline (Figure 1d) and were consistent in male and female patients at baseline (Figure 1e). Figure 1f shows baseline Eczema Area and Severity Index (EASI) scores for male and female patients in each treatment group.
Figure 1. Pretreatment status of lesional and nonlesional skin.
(a) Absolute quantification of 16S rRNA using qPCR of skin samples at screening (week −2) and baseline (week 0). (b) Absolute quantification of Staphylococcus aureus using qPCR of skin samples at screening and baseline. (c) Shannon diversity results from 16S rRNA sequencing of skin samples at screening and baseline. (d) Shannon diversity results from 16S rRNA sequencing of skin samples at baseline (week 0) from the placebo and dupilumab-treated groups. (e) Shannon diversity results from 16S rRNA sequencing of skin samples at baseline of male and female patients. (f) EASI score at baseline for the placebo group and the dupilumab-treated group for male and female patients. Statistical differences between lesional and nonlesional skin, between treatment groups, and between males and females were assessed using the nonparametric Mann–Whitney U test, with Bonferroni correction for multiple comparison. EASI, Eczema Area and Severity Index; qPCR, quantitative PCR; rCFU, relative colony-forming units; rRNA, ribosomal RNA.
Relative microbial abundance
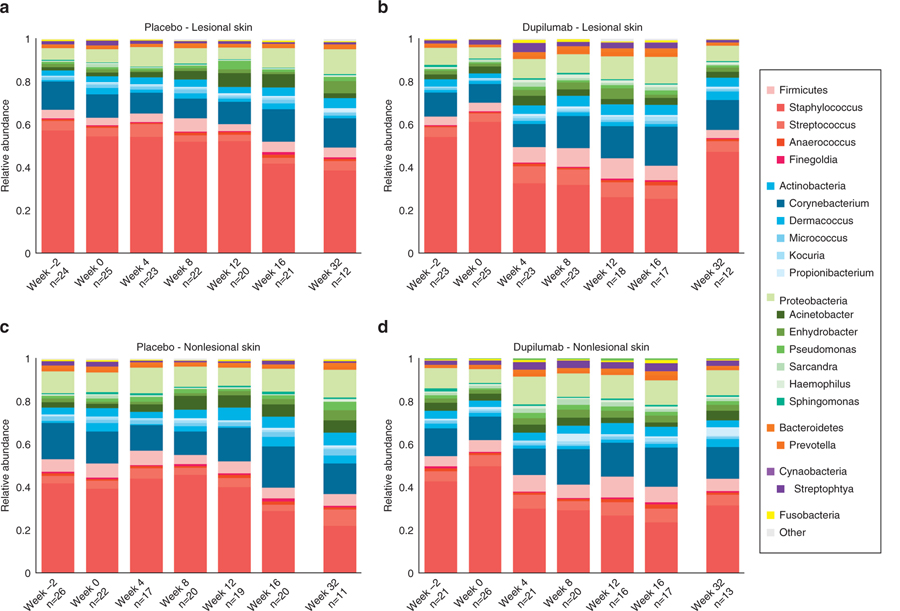
Figure 2 shows relative microbial abundance in lesional and nonlesional skin according to treatment group. The most abundant bacterial phyla were Firmicutes (mostly Staphylococcus), Actinobacteria (mostly Corynebacterium), and Proteobacteria (mostly Acinetobacter). The relative abundance of Staphylococcus decreased during dupilumab treatment in both lesional (P < 0.001) and nonlesional (P < 0.01) skin (Supplementary Figure S1), but the most pronounced reduction was seen in lesional skin samples (Supplementary Figure S2). This decrease in Staphylococcus abundance was observed as early as week 4 and was sustained until week 16. A relative abundance of bacterial taxa in lesional skin for individual dupilumab-treated patients over time is shown in Supplementary Figure S3.
Figure 2. Relative abundance of the most dominant phyla and genera over time in lesional and nonlesional skin according to treatment group.
The most abundant operational taxonomic units are presented. (a) Microbial variability over time of placebo group in lesional skin. (b) Microbial variability over time of dupilumab-treated group in lesional skin. (c) Microbial variability over time of dupilumab-treated group in nonlesional skin. (d) Microbial variability over time of placebo group in nonlesional skin. During treatment (weeks 4–16), an overall decrease in relative abundance of Staphylococcus is noted mainly in the dupilumab-treated patients in lesional skin.
Microbial diversity
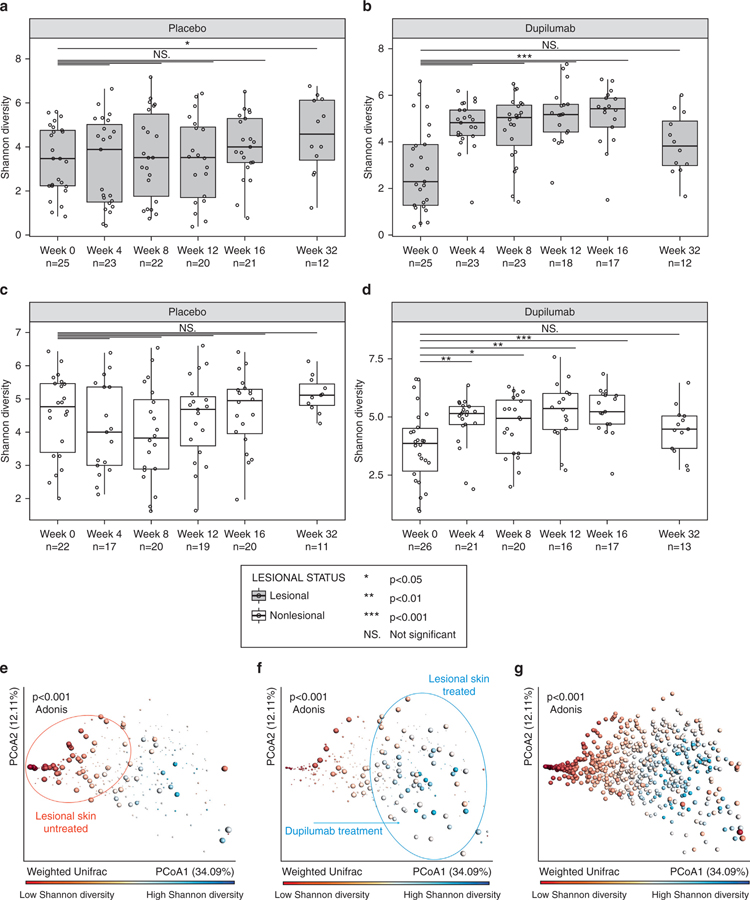
Shannon microbial α-diversity over time is shown in Figures 3a–d. In the placebo group, microbial diversity was relatively consistent throughout the study for both lesional (Figure 3a) and nonlesional skin (Figure 3c); the only time point at which α-diversity was significantly different from baseline was at week 32 for lesional skin (16 weeks after treatment). In contrast, in the dupilumab-treated group, α-diversity in lesional skin increased significantly from week 0 (includes screening and baseline) to week 4 and at all time points through week 16 (during treatment; P < 0.001). The effect was lost at week 32 (i.e., 18 weeks after the last injection) (Figure 3b); similarly, in nonlesional skin, there was a significant increase (P < 0.05) in α-diversity from baseline at weeks 4, 8, 12, and 16 (Figure 3d). Higher microbial α-diversity is associated with a lower relative abundance of Staphylococcus (P < 0.001, Spearman correlation; results not shown).
Figure 3. Shannon microbial α-diversity and β-diversity.
(a–d) α-diversity: (a) Lesional skin, placebo group; (b) Lesional skin, dupilumab-treated group; (c) Nonlesional skin, placebo group; (d) Nonlesional skin, dupilumab-treated group. (e–g) β-diversity: PCoA plots showing Shannon diversity of skin samples; red indicates low diversity and blue indicates high diversity. (e) Large dots depict lesional skin samples from the dupilumab-treated group in periods without treatment (weeks −2, 0, and 32) and small dots represent other samples (nonlesional skin during periods of treatment), showing that without dupilumab treatment, samples are mainly located on the left side with a low Shannon diversity (red circle). (f) Large dots depict lesional skin samples from the dupilumab-treated group taken during the treatment period (weeks 4, 8, 12, and 16); small dots represent other samples (nonlesional skin, no treatment) showing that with dupilumab treatment, samples shift to the right with higher Shannon diversity (blue circle). (g) All samples (placebo and dupilumab, lesional and nonlesional skin) at all time points, indicating a clear separation between lesional skin (left, low Shannon diversity [red]) and nonlesional skin (right, high Shannon diversity [blue]) and significant separation between low and high Shannon diversity (P < 0.001, Adonis test). Each dot represents an individual sample; the color of the dot reflects the bacterial Shannon diversity index on a continuous scale (red indicates low diversity and blue indicates high diversity). *P < 0.05, **P < 0.01, ***P < 0.001; Mann–Whitney U test, with Bonferroni correction for multiple comparison. PCoA, Principal coordinates analysis.
Figures 3e–g summarizes weighted UniFrac microbial β-diversity. In the dupilumab-treated group, samples taken from lesional skin during periods when no treatment was given (weeks 0 and 32) cluster to the left of the plot and are highlighted in red, indicating low Shannon diversity and a return to pretreatment conditions (Figure 3e). During treatment, however, lesional skin samples from the dupilumab-treated group are shifted to the right and are highlighted in blue, indicating high Shannon diversity (Figure 3f). This shift in Shannon diversity can also be observed in plots color-coded according to treatment week; lesional skin samples from the dupilumab-treated group are shifted to the right after 4–16 weeks of dupilumab treatment compared with samples that received no dupilumab treatment (weeks −2, 0, or 32) which are clustered to the left (Supplementary Figure S4a). This distinction is not observed for nonlesional skin samples or placebo-treated skin samples (Supplementary Figures S4b–d). The principal coordinates analysis plot for all samples (placebo and dupilumab, lesional and nonlesional) at all time points showed a separation between lesional and nonlesional skin samples with significant separation between low and high Shannon diversity (P < 0.001) (Figure 3g).
Absolute bacterial abundance over time
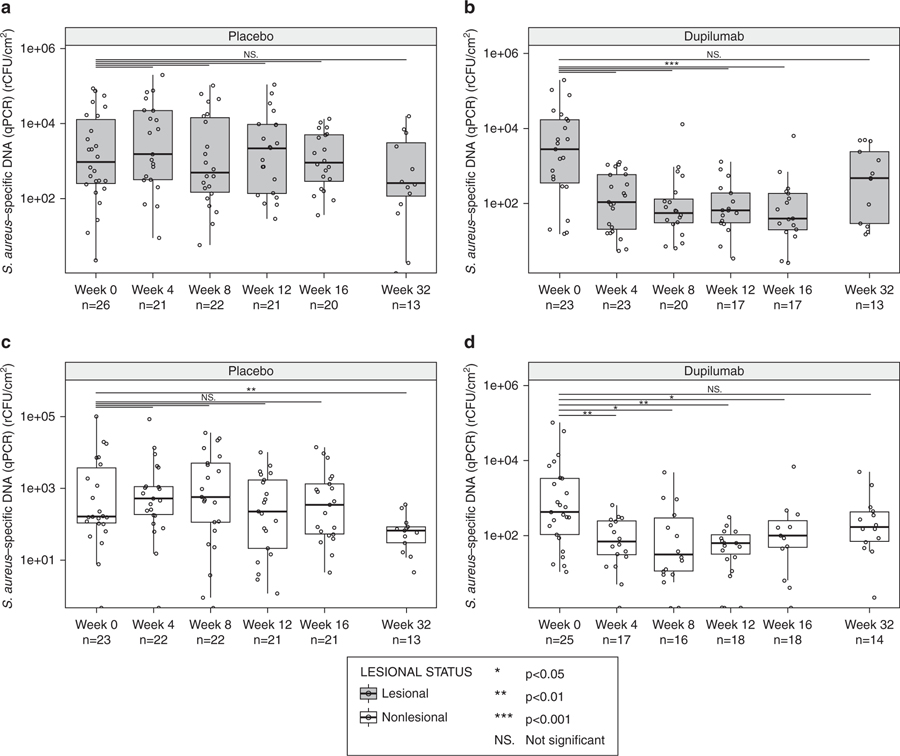
Absolute abundance of total bacteria and S. aureus in lesional skin samples is shown in Figure 4 and Supplementary Figure S1. Absolute abundance of total bacteria as measured by qPCR of 16S rRNA decreased during treatment with dupilumab (P < 0.01) (Supplementary Figure S1a) but not with placebo (Supplementary Figure S1b). The decreased abundance of total bacteria during dupilumab treatment was observed as early as week 4 and was sustained through week 32. Absolute abundance of S. aureus measured by qPCR also decreased during treatment with dupilumab (P < 0.001) (Figure 4a). This decrease was observed as early as week 4 (the first post-treatment time point tested) and continued through the end of treatment (week 16). By week 32 (16 weeks after treatment), S. aureus abundance had returned to baseline levels. In the placebo group, absolute abundance of S. aureus did not change consistently or significantly during dupilumab treatment (Figure 4b). Similar results were observed in nonlesional skin (Figures 4c–d); however, the only time point at which there was a significant decrease in absolute abundance of S. aureus in the placebo-treated group was at week 32, 16 weeks after treatment. Upon comparing the absolute abundance of S. aureus in lesional skin between the dupilumab and placebo treatment groups over time, the differences were statistically significant at weeks 4, 8, 12, and 16 (Supplementary Figure S5). Relative abundance of Staphylococcus also decreased with dupilumab treatment compared with placebo (Supplementary Figures S1c–f). There was a strong positive and significant correlation between relative abundance of Staphylococcus assessed using 16S rRNA gene sequencing and absolute S. aureus abundance determined using qPCR (P < 0.001) (Supplementary Figure S6), which demonstrated that although it is semi-quantitative, relative abundance of Staphylococcus is at least predictive for the abundance of S. aureus.
Figure 4. Absolute abundance Staphylococcus aureus over time.
(a) Lesional skin samples, placebo group. (b) Lesional skin, dupilumab-treated group. (c) Nonlesional skin samples, placebo group. (d) Nonlesional skin samples, dupilumab-treated group. *P < 0.05, **P < 0.01, ***P < 0.001; Mann–Whitney U test, with Bonferroni correction for multiple comparison. qPCR, quantitative PCR; rCFU, relative colony-forming units.
Staphylococcus aureus and clinical score
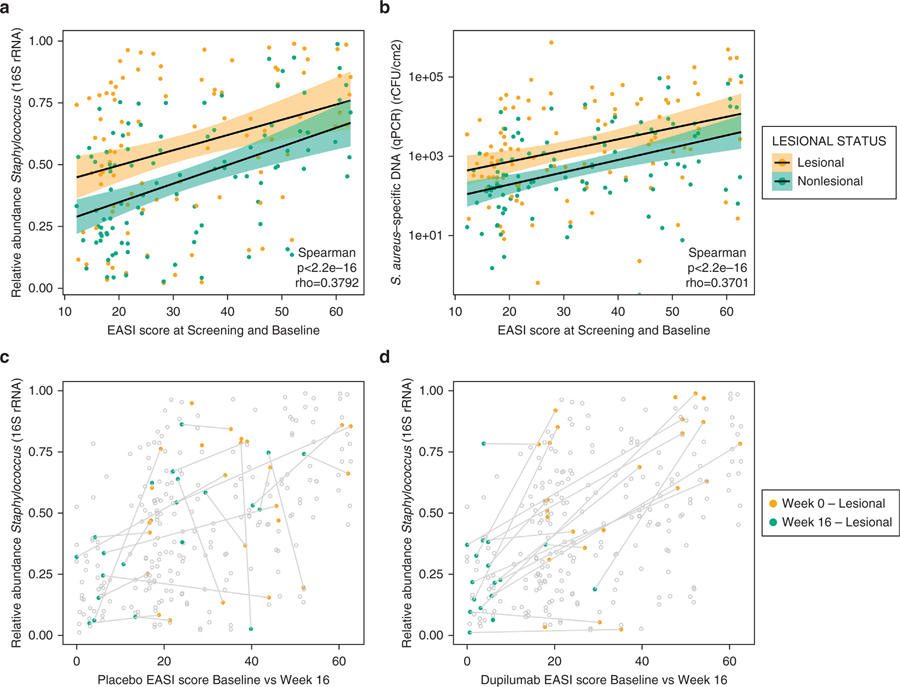
There was a significant correlation between both EASI and SCORAD scores and the relative abundance of Staphylococcus (P < 0.001, Spearman correlation) (Figure 5a and Supplementary Figure S7a) and absolute abundance of S. aureus (P < 0.001, Spearman correlation) (Figure 5b and Supplementary Figure S7b) in both lesional and nonlesional samples before treatment (weeks −2 and 0). The correlation between total bacterial abundance and SCORAD scores was significant (P < 0.001) in both lesional and nonlesional samples (Supplementary Figure S7c). In the dupilumab-treated group, samples clustered in the lower left corner of the plot at week 16, suggesting a reduction in both EASI scores and S. aureus abundance (Figure 5c), whereas samples from the placebo-treated group did not show this trend (Figure 5d). Furthermore, in lesional skin samples of the dupilumab-treated group, absolute abundance of S. aureus and EASI scores demonstrated similar dupilumab-mediated reductions over time in both females and males (Supplementary Figure S8a–d), although the overall absolute abundance was higher in males than females.
Figure 5. Correlation of EASI score and relative and absolute abundance of Staphylococcus aureus.
At screening (week −2) and baseline (week 0): correlation of EASI score and (a) relative and (b) absolute abundance of S. aureus. Shaded areas around the regression line indicate confidence intervals. (c) Correlation of EASI scores and relative abundance of S. aureus at baseline and week 16 for both treatment groups and both lesional and nonlesional samples, with values from the placebo group at week 0 in yellow and week 16 in green. (d) Same as (c), with values from the dupilumab-treated group at week 0 in yellow and week 16 in green. Values for the same patient from weeks 0 and 16 are connected with a line. EASI, Eczema Area and Severity Index; qPCR, quantitative PCR; rCFU, relative colony-forming units; rRNA, ribosomal RNA.
Staphylococcus aureus and serum biomarkers
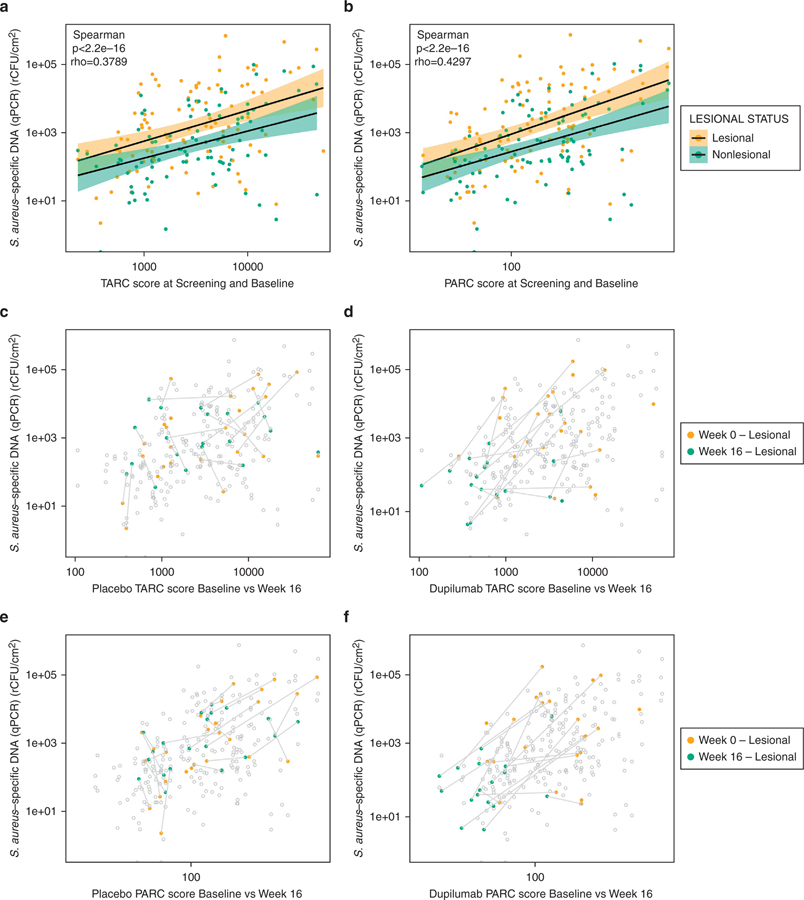
Pulmonary and activation-regulated chemokine (PARC/CCL18) and serum thymus and activation-regulated chemokine (TARC/CCL17), markers of type 2 inflammation, were shown to be associated with AD (Thijs et al., 2015; Pivarcsi et al., 2004). There was a significant correlation between absolute abundance of S. aureus in both lesional and nonlesional samples before treatment (weeks −2 and 0) and serum concentrations of TARC (Figure 6a) and PARC (Figure 6b) in both lesional and nonlesional samples (P < 0.001, Spearman correlation). In the dupilumab-treated group, samples clustered in the lower left corner of the TARC plot at week 16, suggesting a reduction in both TARC score and S. aureus abundance with dupilumab treatment (Figure 6c), whereas samples from the placebo-treated group did not show this trend (Figure 6d). A similar pattern was observed for PARC (Figure 6e–f).
Figure 6. Correlation of absolute abundance of Staphylococcus aureus with biomarkers TARC and PARC.
(a) Correlation of TARC score and absolute abundance of S. aureus at screening (week −2) and baseline (week 0). (b) Correlation of PARC score and absolute abundance of S. aureus at screening (week −2) and baseline (week 0). Shaded areas around the regression line indicate confidence intervals. Correlation of TARC score and absolute abundance of S. aureus at baseline (yellow) and week 16 (green) in the (c) placebo group and the (d) dupilumab-treated group. Correlation of PARC score and absolute abundance of S. aureus at baseline (yellow) and week 16 (green) in the (e) placebo group and (f) dupilumab-treated group. Values for the same patient from weeks 0 and 16 are connected with a line. PARC, pulmonary and activation-regulated chemokine; qPCR, quantitative PCR; rCFU, relative colony-forming units; TARC, thymus and activation-regulated chemokine.
DISCUSSION
Previous findings from this AD-1307 EXPLORE study of the impact of dupilumab treatment on skin in patients with moderate to severe AD indicated that dupilumab treatment reduces clinical signs and symptoms of AD and inflammatory cell infiltrates in lesional skin, inhibits type 2/T helper type 2 polarizing chemokines including PARC and TARC, and induces a progressive shift from a lesional to nonlesional molecular phenotype (Hamilton et al., 2014; Guttman-Yassky et al., 2019). This analysis of skin surface microbial DNA measurements confirmed that before treatment, the skin microbiome of AD is characterized by low microbial diversity and an increased abundance of S. aureus. Treatment with dupilumab for 16 weeks significantly changed the composition of the microbiome and increased microbial diversity while reducing the abundance of S. aureus. Increased diversity was observed as early as week 4 (the earliest time point tested); although this was more pronounced in lesional skin, clinically normal appearing (nonlesional) skin also showed a reduced amount of S. aureus. In contrast, no significant change in microbial diversity or S. aureus abundance was seen in the placebo-treated group. These findings suggest that targeting IL-4/IL-13 signaling via IL-4Rα blockade potentially enables the reduction of S. aureus colonization, a factor known to be associated with more severe AD in humans (Williams and Gallo, 2015) and with an AD-like phenotype in mice (Kobayashi et al., 2015; Nakatsuji et al., 2016).
Our characterization of the baseline skin microbiome in patients with AD is consistent with findings from other studies. A recent systematic review of studies assessing the skin microbiome found that AD is associated with frequent S. aureus colonization and low microbial diversity (Bjerre et al., 2017). Conventional culture-based studies showed that 70% of lesional and 30–40% of nonlesional skin sites are predominantly colonized by S. aureus (Totté et al., 2016; Gong et al., 2006), which is associated with increased numbers of inflammatory cells in the skin (Boguniewicz and Leung, 2011) and more severe forms of AD (Bjerre et al., 2017). Previous studies also indicated that absolute levels of S. aureus correlate with EASI score and other markers of disease severity, including PARC and TARC (Thijs et al., 2015; Pivarcsi et al., 2004). Finally, Kong et al. (Kong et al., 2012) showed that AD flares are associated with increased S. aureus and decreased microbial diversity. Therefore, microbial diversity may be a useful biomarker in AD. However, AD remains a multifactorial disease, and the role of the skin microbiome in AD pathogenesis is still being studied (Bjerre et al., 2017; Williams and Gallo, 2015). Studies in animal models of AD investigating the response to topical S. aureus (Kobayashi et al., 2015; Nakatsuji et al., 2016) and observations that S. aureus colonization or microbiome dysbiosis can precede AD in early childhood (Lebon et al., 2009; Meylan et al., 2017) support a role for the microbiome in exacerbation underlying inflammation that drives early AD pathogenesis.
Most currently available treatment options for AD do not address changes in the skin microbiome (Williams and Gallo, 2015; Hepburn et al., 2017). Topical antibiotic therapy may not reduce bacteria counts (Francis et al., 2017) and theoretically would not protect the normal skin microbiome (Williams and Gallo, 2015). Furthermore, frequent use of pharmaceutical antibiotics may promote antibiotic resistance (Hepburn et al., 2017; Salah and Faergemann, 2015). Several novel strategies are in development that target the skin microbiome, including the use of probiotics and transplantation of microbiota from healthy volunteers; further investigation of these strategies is needed (Nakatsuji et al., 2017; Myles et al., 2016). A promising treatment approach for AD will prevent dysbiosis by promoting or reestablishing a normal skin microbial flora (Williams and Gallo, 2015; Hepburn et al., 2017).
Limitations of this exploratory analysis were the relatively small numbers of patients in both the dupilumab- and placebo-treated groups. However, efforts were made to minimize potential bias: skin swabs were collected from all patients participating in the clinical study (rather than being optional) and all samples were processed and evaluated in a blinded fashion. The use of a global EASI score compared with a target lesion score of the area of microbiome sampling is a potential methodological limitation of this study; however, the lesional area selected for skin swabbing was representative of overall disease severity as quantified by EASI score. Finally, analysis of DNA from skin surface microbial swabs does not assess bacterial survival or whether the microbes are metabolically active. Indeed, comparisons of S. aureus abundance measurements from lesional atopic to normal skin have shown that DNA assessments overestimate the capacity to culture S. aureus from healthy normal skin (Nakatsuji et al., 2017). These observations suggest that healthy skin can kill S. aureus more effectively than can skin from AD. Indeed, type 2 cytokines have been shown to suppress antimicrobial peptide production from keratinocytes (Ong et al., 2002). Future studies should examine the metabolic activity of bacteria and the host antimicrobial response of AD patients treated with dupilumab to assess the mechanism responsible for the improvement in the microbiome with treatment.
16S rRNA analysis of skin in adults with moderate to severe AD demonstrates decreased microbial α-diversity and a relative increase in the proportion of S. aureus. These changes are more pronounced in lesional than nonlesional areas. Dupilumab treatment reversed the AD-associated microbial (dysbiotic) signature for as long as patients were receiving treatment. Normalization of the microbial signature correlated with decreased concentration of serum biomarkers (TARC/PARC) and improvement in clinical signs and symptoms of AD. These results reveal strong links between the skin microbial community, type 2 inflammation, and AD severity. To our knowledge, this characterization of the effect of a targeted treatment on the skin microbiome and type 2 inflammation has not been previously reported. An improvement of host defense against S. aureus colonization after inhibition of type 2 inflammation is consistent with the effects of type 2 cytokines to inhibit host antimicrobial peptide expression and impair barrier function (Guttman-Yassky et al., 2008; Nomura et al., 2003; Kisich et al., 2007; Ong et al., 2002). Future studies, including whole metagenomic sequencing, will better define the role of S. aureus colonization in the initiation of AD pathogenesis and determine whether prolonged suppression of excessive type 2 immunity facilitates the reestablishment of a functional normobiotic shield in patients with AD.
MATERIALS AND METHODS
Trial design
AD-LIBERTY EXPLORE was a randomized, placebo-controlled, double-blind phase 2 trial (NCT01979016) conducted at five medical centers in the United States and Canada (Guttman-Yassky et al., 2019). Eligible patients were aged ≥18 years and had moderate to severe AD, defined as EASI ≥16; Investigator’s Global Assessment ≥3 (on the 0–4 Investigator’s Global Assessment scale); body surface area ≥10%; AD present for ≥3 years; and inadequate response to topical medications within 6 months of screening. More detailed information on study patients is provided in Guttman-Yassky et al. (2019).
Fifty-four patients were randomized (1:1) to weekly subcutaneous injections of dupilumab or placebo. Patients received a loading dose of 400 mg (or placebo) followed by 200 mg (or placebo) every week for 16 weeks. Thereafter, patients entered a 16-week safety follow-up until week 32. The primary end point was percent change in EASI score from baseline to week 16 (continuous variable). Exploratory analyses included correlations between clinical severity (AD phenotype) and the host AD transcriptome, defined by lesional versus nonlesional skin gene expression differences, as well as changes from baseline to week 16 in S. aureus abundance and skin microbial diversity. This study was conducted in accordance with the Declaration of Helsinki International Conference on Harmonization, Good Clinical Practice guidelines, and applicable regulatory requirements. All study documents and procedures were approved by the appropriate institutional review boards and ethics committees at each study site; all participants provided written informed consent under institutional review board–approved protocols (Guttman-Yassky et al., 2019).
Pretreatment characterization of lesional and nonlesional skin
Abundance and diversity of bacterial colonization were characterized at weeks 0, 2, 4, 8, 12, 16, and 32. Bacteria were collected from premeasured areas (10 × 10 cm) of lesional skin on the antecubital fossa and nonlesional skin of the upper arm at least 2 cm separated from the lesional site. Bacterial collection was performed by rubbing 50 strokes with a Catch-All nylon swab (Illumina, San Diego, CA) premoistened with Tris-EDTA buffer containing 0.1% TritonX-100 and 0.05% Tween-20 (w/v) (Millepore-Sigma, St. Louis, MO). The swab was stored at −80°C until DNA extraction. Bacterial cells in the swab were lysed and total genomic DNA was purified. To characterize lesional and nonlesional skin before treatment, absolute quantification of bacterial abundance was performed using qPCR for skin samples collected at screening (week −2) and baseline (week 0). The abundance of S. aureus was performed using qPCR of the femA gene. Overall bacterial abundance was quantified using qPCR of the 16S rRNA. Shannon diversity results were also assessed based on 16S rRNA sequencing of skin samples collected at screening and baseline; diversity results were compared according to sex and treatment group.
DNA extraction and 16S rRNA amplicon sequencing
16S rRNA sequencing was performed following the Earth Microbiome Project protocols (Gilbert et al., 2014; Caporaso et al., 2012). Briefly, DNA was extracted using the MO BIO PowerMag Soil DNA Isolation Kit (Qiagen, Carlsbad, CA) and the V4 region of the 16S rRNA gene was amplified using barcoded primers (Walters et al., 2016). The primers were shown to identify Staphylococcus genus sufficiently in mock communities (Gohl et al., 2016). PCR was performed in triplicate for each sample, and V4 paired-end sequencing was performed using the Illumina MiSeq platform (San Diego, CA). Raw sequence reads were demultiplexed and quality controlled using the defaults, as provided by QIIME (San Diego, CA) (version 1.9.1) (Caporaso et al., 2010). The primary OTU table was generated using Qiita (San Diego, CA) (qiita.ucsd.edu), using Sort-MeRNA (Villeneuve d’Ascq, France) (Kopylova et al., 2012) and closed-reference OTU picking method against the GreenGenes database (San Francisco, CA version 13.8) (McDonald et al., 2012). OTU tables can be found in Qiita (qiita.ucsd.edu) as study ID 10403. Resulting OTU tables were then rarefied to 10,000 sequences/sample for downstream analyses.
Assessment of bacterial diversity
α- and β-diversity were assessed on the basis of 16S rRNA V4 amplicon DNA sequencing results from lesional and nonlesional skin samples collected at weeks −2, 0, 4, 8, 12, 16, and 32. Microbial α-diversity was assessed using the Shannon index. Change in diversity from baseline was measured using the nonparametric Mann–Whitney U test. Microbial β-diversity was measured using the weighted UniFrac distance metric and results are presented using multivariate analysis (principal coordinates analysis). Statistical significance was tested using nonparametric permutational multivariate analysis of variance using the Adonis function in R.
Quantitative PCR
Bacterial DNA was extracted from a 10 × 10-cm skin area by swabbing and eluted in 50 μl elution buffer. The abundance of total bacterial DNA in 1 μl elution was estimated by qPCR using a universal 16S rRNA primer set (1048-F: GTGSTGCAYGGYTGTCGTCA and 1175-R: ACGTCRTCCMCACCTTCCTC) with Power SYBR green master mix (Applied Biosystems, Foster City, CA). The universal primer set covers 79% of Proteobacteria, 85% of Actinobacteria, 72% of Firmicutes, and 61% of Bacteroidetes (Horz et al., 2005). Because of a lack of standard bacteria that can be used as an external control of absolute quantification for universal 16S rRNA gene abundance, the abundance of 16S rRNA gene was calculated with a ∆Ct method and then normalized to the swabbed area. The abundance of S. aureus DNA was quantified using a species-specific primer set targeting femA (S. aureus femA-qPCR-2F: AACTGTTGGCCACTATGAGT; S. aureus femA-qPCR-2R: CCAG-CATTACCTGTAATCTCG) (Paule et al., 2004). The specificity of all primer pairs was confirmed by melting curve analysis and comparison with standard curves. To determine relative colony-forming unit of S. aureusespecific DNA, a standard curve was generated with genomic DNA extracted from standards derived from known amounts of colony-forming units of S. aureus (ATCC35556), and then the quantity was normalized to the swabbed area.
Assessment of microbial abundance
Relative microbial abundance was assessed based on DNA sequencing of 16S rRNA isolated from lesional and nonlesional skin at weeks −2, 0, 4, 8, 12, 16, and 32; assessment of relative S. aureus abundance using 16S rRNA region V4 sequencing was semi-quantitative and is therefore described according to the genus level Staphylococcus. Change in the relative abundance of all bacteria was calculated. Absolute bacterial abundance over time was assessed using qPCR and primers for all bacteria. Absolute abundance of S. aureus was assessed as relative colony-forming units per square centimeter using qPCR and primers specific to S. aureus as described earlier. Statistical significance compared with baseline absolute abundance or between treatment groups was assessed using the nonparametric Mann–Whitney U test. No corrections on statistical tests were performed.
To assess bacterial abundance and microbial diversity, some samples were excluded due to rarefaction (i.e., sequence count was too low, not enough DNA was amplified). Also, in later weeks, not all patients provided a sample, with a decrease from week 4 to week 32.
Staphylococcus aureus and markers of disease severity
Correlation of EASI score with relative and absolute abundance of S. aureus was assessed. Relative abundance was measured using 16S rRNA amplicon sequencing, and absolute abundance was measured using qPCR and a primer specific to S. aureus. Correlation was also assessed between the absolute abundance of S. aureus and biomarkers of disease severity TARC and PARC (Ungar et al., 2017; Brunner et al., 2017b; Thijs et al., 2015; Pivarcsi et al., 2004). Serum TARC and PARC were measured using Quantikine ELISA kits (R&D Systems, Minneapolis, MN). Statistical significance was assessed using nonparametric Spearman rank correlation coefficient.
Data availability statement
Qualified researchers may request access to study documents (including the clinical study report, study protocol with any amendments, blank case report form, and statistical analysis plan) that support the methods and findings reported in this article. Individual anonymized participant data will be considered for sharing once the indication has been approved by a regulatory body, if there is legal authority to share the data and there is not a reasonable likelihood of participant re-identification. Submit requests to https://errs.regeneron.com/external. Datasets related to this article can be found at https://www.ebi.ac.uk/ena/data/view/PRJEB32646, hosted at EBI.
Supplementary Material
ACKNOWLEDGMENTS
Research was sponsored by Sanofi and Regeneron Pharmaceuticals, Inc., ClinicalTrials.gov Identifier: NCT01979016. Medical writing/editorial assistance was provided by Lola MacRae, PhD, of Excerpta Medica, funded by Sanofi Genzyme and Regeneron Pharmaceuticals, Inc. The authors thank Amnon Amir and Usman Chaudhry for their contributions to this study.
Abbreviations:
- AD
atopic dermatitis
- EASI
Eczema Area and Severity Index
- PARC
pulmonary and activation-regulated chemokine
- qPCR
quantitative PCR
- rRNA
ribosomal RNA
- TARC
thymus and activation-regulated chemokine
Footnotes
CONFLICT OF INTEREST
Outside of the submitted work, NT was an investigator for and has received research grants from Regeneron Pharmaceuticals, Inc.; RK was an scientific advisory board member and consultant for Janssen R&D LLC, Commense, Inc., and Prometheus Lab Inc.; MA, NMHG, and JH were employees and shareholders of Regeneron Pharmaceuticals, Inc.; TH and GP were employees of Sanofi and may hold stock and/or stock options in the company; EG was an investigator for AbbVie, Celgene, Eli Lilly and Company, GlaxoSmithKline, Regeneron Pharmaceuticals, Inc., Sanofi, Leo Pharma, Pfizer, Galderma, and Glenmark; served as a consultant for AbbVie, Anacor, Eli Lilly and Company, Galderma, GlaxoSmithKline, Leo Pharma, Menlo Therapeutics, Kiniksa, Pfizer, Realm, Regeneron Pharmaceuticals, Inc., Sanofi, Asana Biosciences, DBV, Kyowa, Daiichi Sankyo, Glenmark, Novartis, Dermira; and received research grants from Regeneron Pharmaceuticals, Inc., Sanofi, Pfizer, Galderma, Janssen, Celgene, Novartis, Dermira, AbbVie, Innovaderm, and Leo Pharma. RB was an investigator, consultant, advisory board member, speaker, and/or received honoraria from Antibiotx, Aquinox Pharma, Asana, Astellas, Brickell Biotech, Dermavant, Dermira, Dignity Sciences, Galderma, Glenmark, GSK-Stiefel, Hoffman-LaRoche Ltd., Kiniksa, Leo Pharma, Neokera, Pfizer, Regeneron Pharmaceuticals, Inc., Sienna Biopharmaceuticals, and Vitae Pharmaceuticals; and was a shareholder of Innovaderm Research; JIS was an investigator for AbbVie, Celgene, Chugai, Eli Lilly and Company, GlaxoSmithKline, Regeneron Pharmaceuticals, Inc., and Sanofi; a consultant for AbbVie, Anacor, Eli Lilly and Company, Galderma, GlaxoSmithKline, Incyte, Leo Pharma, Menlo, Kiniksa, Pfizer, Realm, Regeneron Pharmaceuticals, Inc., and Sanofi; and a speaker for Regeneron Pharmaceuticals, Inc., and Sanofi; JK was a consultant and recipient of research grants from Amgen, Bristol-Myers Squibb, Boehringer Ingelheim, Dermira, Innovaderm, Janssen, Kadmon, Kyowa, Eli Lilly and Company, Merck, Novartis, Parexel, and Pfizer; was consultant and received personal fees from AbbVie, Baxter, Biogen Idec, Delenex, Kineta, Sanofi, Serono, and XenoPort; and was an investigator for Regeneron Pharmaceuticals, Inc.; AM served on the advisory board and was a consultant, investigator, and speaker and received grants and honoraria from AbbVie, Amgen, Janssen Biotech, Inc., and Leo Pharma; served on the advisory board and was a consultant and investigator and received grants and honoraria from Allergan; served on the advisory board and was an investigator and recipient of grants and honoraria from Boehringer Ingelheim; served as a consultant and investigator and was a recipient of grants and honoraria from Novartis, Pfizer, and XenoPort; was investigator and recipient of grants from Anacor, Celgene, Dermira, Merck, Neothetics, Regeneron Pharmaceuticals, Inc., and Symbio/Maruho; served on the advisory board, was an consultant and investigator, and received honoraria from Eli Lilly and Company; and was consultant and received honoraria from Galderma and Vitae Pharmaceuticals; RG was an investigator and recipient of research grants from Regeneron Pharmaceuticals, Inc.; had equity interest in and was co-founder and SAB of MatriSys Bioscience; had equity interest in and was SAB of Sente, Inc.; was a consultant for Roche; received a research grant from Novan; and received a research grant from Colgate Palmolive. The other authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2019.05.024.
REFERENCES
- Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 2014;371:130–9. [DOI] [PubMed] [Google Scholar]
- Berdyshev E, Goleva E, Bronova I, Dyjack N, Rios C, Jung J, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed]
- Bieber T Atopic dermatitis. N Engl J Med 2008;358:1483–94. [DOI] [PubMed] [Google Scholar]
- Bjerre RD, Bandier J, Skov L, Engstrand L, Johansen JD. The role of the skin microbiome in atopic dermatitis: a systematic review. Br J Dermatol 2017;177:1272–8. [DOI] [PubMed] [Google Scholar]
- Blauvelt A, de Bruin-Weller M, Gooderham M, Cather JC, Weisman J, Pariser D, et al. Long-term management of moderate-to-severe atopic dermatitis with dupilumab and concomitant topical corticosteroids (LIBERTY AD CHRONOS): a 1-year, randomised, double-blinded, placebo-controlled, phase 3 trial. Lancet 2017;389:2287–303. [DOI] [PubMed] [Google Scholar]
- Boguniewicz M, Leung DYM. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev 2011;24:233–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt EB, Sivaprasad U. Th2 cytokines and atopic dermatitis. J Clin Cell Immunol 2011;2. [DOI] [PMC free article] [PubMed]
- Brunner PM, Guttman-Yassky E, Leung DYM. The immunology of AD and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol 2017a;139(Suppl. 4):S65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Leung DYM, Guttman-Yassky E. Immunologic, microbial, and epithelial interactions in atopic dermatitis. Ann Allergy Asthma Immunol 2018;120:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner PM, Suárez-Fariñas M, He H, Malik K, Wen HC, Gonzalez J, et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci Rep 2017b;7:8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Strickland I, Tomkinson A, Fehringer AP, Gelfand EW, Leung DY. Preferential binding of Staphylococcus aureus to skin sites of Th2-mediated inflammation in a murine model. J Invest Dermatol 2001;116:658–63. [DOI] [PubMed] [Google Scholar]
- Cork MJ, Danby SG, Vasilopoulos Y, Hadgraft J, Lane ME, Moustafa M, et al. Epidermal barrier dysfunction in atopic dermatitis. J Invest Dermatol 2009;129:1892–908. [DOI] [PubMed] [Google Scholar]
- Eichenfield LF, Tom WL, Chamlin SL, Feldman SR, Hanifin JM, Simpson EL, et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. J Am Acad Dermatol 2014;70:338–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NA, Ridd MJ, Thomas-Jones E, Butler CC, Hood K, Shepherd V, et al. Oral and topical antibiotics for clinically infected eczema in children: a pragmatic randomized controlled trial in ambulatory care. Ann Fam Med 2017;15:124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi NA, Bennett BL, Graham NMH, Pirozzi G, Stahl N, Yancopoulos GD. Targeting key proximal drivers in type 2 inflammation in disease. Nat Rev Drug Discov 2015;15:35–50. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Jansson JK, Knight R. The Earth Microbiome Project: successes and aspirations. BMC Biol 2014;12:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 2016;34:942–9. [DOI] [PubMed] [Google Scholar]
- Gong JQ, Lin L, Lin T, Hao F, Zeng FQ, Bi ZG, et al. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br J Dermatol 2006;155:680–7. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Bissonnette R, Ungar B, Suárez-Fariñas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in atopic dermatitis patients. J Allergy Clin Immunol 2019;143:155–72. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Zaba LC, Cardinale I, Nograles KE, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol 2008;181:7420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JD, Suárez-Fariñas M, Dhingra N, Cardinale I, Li X, Kostic A, et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 2014;134: 1293–300. [DOI] [PubMed] [Google Scholar]
- Hepburn L, Hijnen DJ, Sellman BR, Mustelin T, Sleeman MA, May RD, et al. The complex biology and contribution of Staphylococcus aureus in atopic dermatitis, current and future therapies. Br J Dermatol 2017;177:63–71. [DOI] [PubMed] [Google Scholar]
- Horz HP, Vianna ME, Gomes BP, Conrads G. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J Clin Microbiol 2005;43:5332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, DeBenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol 2009;124(3 suppl 2):R7–12. [DOI] [PubMed] [Google Scholar]
- Kim BE, Leung DYM, Boguniewicz M, Howell MD. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol 2008;126:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisich KO, Howell MD, Boguniewicz M, Heizer HR, Watson NU, Leung DY. The constitutive capacity of human keratinocytes to kill Staphylococcus aureus is dependent on beta-defensin 3. J Invest Dermatol 2007;127: 2368–80. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 2015;42:756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012;22:850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012;28: 3211–7. [DOI] [PubMed] [Google Scholar]
- Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel WJ, et al. Role of Staphylococcus aureus nasal colonization in atopic dermatitis in infants: the Generation R Study. Arch Pediatr Adolesc Med 2009;163: 745–9. [DOI] [PubMed] [Google Scholar]
- Leung DYM, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol 2014;134:769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol 1974;90:525–30. [DOI] [PubMed] [Google Scholar]
- MacDonald LE, Karow M, Stevens S, Auerbach W, Poueymirou WT, Yasenchak J, et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc Natl Acad Sci U S A 2014;111:5147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol 2017;137:2497–504. [DOI] [PubMed] [Google Scholar]
- Murphy AJ, Macdonald LE, Stevens S, Karow M, Dore AT, Pobursky K, et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci U S A 2014;111:5153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles IA, Williams KW, Reckhow JD, Jammeh ML, Pincus NB, Sastalla I, et al. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight 2016;1:e86955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed]
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol 2016;136: 2192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol 2003;171:3262–9. [DOI] [PubMed] [Google Scholar]
- Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med 2002;347:1151e60. [DOI] [PubMed] [Google Scholar]
- Paule SM, Pasquariello AC, Hacek DM, Fisher AG, Thomson RB Jr, Kaul KL, et al. Direct detection of Staphylococcus aureus from adult and neonate nasal swab specimens using real-time polymerase chain reaction. J Mol Diagn 2004;6:191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivarcsi A, Gombert M, Dieu-Nosjean MC, Lauerma A, Kubitza R, Meller S, et al. CC chemokine ligand 18, a atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. J Immunol 2004;173:5810–7. [DOI] [PubMed] [Google Scholar]
- Salah LA, Faergemann J. A retrospective analysis of skin bacterial colonisation, susceptibility and resistance in atopic dermatitis and impetigo patients. Acta Derm Venereol 2015;95:532–5. [DOI] [PubMed] [Google Scholar]
- Simpson EL, Bieber T, Guttman-Yassky E, Beck LA, Blauvelt A, Cork MJ, et al. Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med 2016;375:2335–48. [DOI] [PubMed] [Google Scholar]
- Thaçi D, Simpson EL, Beck LA, Bieber T, Blauvelt A, Papp K, et al. Efficacy and safety of dupilumab in adults with moderate-to-severe atopic dermatitis inadequately controlled by topical treatments: a randomised, placebo-controlled, dose-ranging phase 2b trial. Lancet 2016;387: 40–52. [DOI] [PubMed] [Google Scholar]
- Thijs J, Krastev T, Weidinger S, Buckens CF, de Bruin-Weller M, Bruijnzeel-Koomen C, et al. Biomarkers for atopic dermatitis: a systematic review and meta-analysis. Curr Opin Allergy Clin Immunol 2015;15:453–60. [DOI] [PubMed] [Google Scholar]
- Totté JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol 2016;175:687–95. [DOI] [PubMed] [Google Scholar]
- Ungar B, Garcet S, Gonzalez J, Dhingra N, Correa da Rosa J, Shemer A, et al. An integrated model of atopic dermatitis biomarkers highlights the systemic nature of the disease. J Invest Dermatol 2017;137:603–13. [DOI] [PubMed] [Google Scholar]
- Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved bacterial 16S rRNA gene (V4 and V4–5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 2016;1:e00009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werfel T, Allam JP, Biedermann T, Eyerich K, Gilles S, Guttman-Yassky E, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol 2016;138:336–49. [DOI] [PubMed] [Google Scholar]
- Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep 2015;15:65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Qualified researchers may request access to study documents (including the clinical study report, study protocol with any amendments, blank case report form, and statistical analysis plan) that support the methods and findings reported in this article. Individual anonymized participant data will be considered for sharing once the indication has been approved by a regulatory body, if there is legal authority to share the data and there is not a reasonable likelihood of participant re-identification. Submit requests to https://errs.regeneron.com/external. Datasets related to this article can be found at https://www.ebi.ac.uk/ena/data/view/PRJEB32646, hosted at EBI.