Abstract
Dendritic cell‐specific intracellular adhesion molecule‐3‐grabbing nonintegrin (DC‐SIGN) is a member of the C‐type lectin family selectively expressed on immune‐related cells. In the present study, we performed a systematic interaction analysis of DC‐SIGN and its related receptors, DC‐SIGN‐related protein (DC‐SIGNR) and liver and lymph node sinusoidal endothelial cell C‐type lectin (LSECtin) using frontal affinity chromatography (FAC). Carbohydrate‐recognition domains of the lectins, expressed as Fc–fusion chimeras, were immobilized to Protein A–Sepharose and subjected to quantitative FAC analysis using 157 pyridylaminated glycans. Both DC‐SIGN–Fc and DC‐SIGNR–Fc showed similar specificities for glycans containing terminal mannose and fucose, but great difference in affinity under the given experimental conditions. By contrast, LSECtin–Fc showed no affinity to these glycans. As a common feature, the DC‐SIGN‐related lectin–Fc chimeras, including LSECtin, exhibited binding affinity to mono‐ and/or bi‐antennary agalactosylated N‐glycans. The detailed FAC analysis further implied that the presence of terminal GlcNAc at the N‐acetylglucosaminyltransferase I position is a key determinant for the binding of these lectins to agalactosylated N‐glycans. By contrast, none of the lectins showed significant affinity to highly branched agalactosylated N‐glycans. All of the lectins expressed on the cells were able to mediate cellular adhesion to agalactosylated cells and endocytosis of a model glycoprotein, agalactosylated α1‐acid glycoprotein. In this context, we also identified three agalactosylated serum glycoproteins recognized by DC‐SIGN‐Fc (i.e. α‐2‐macroglobulin, serotransferrin and IgG heavy chain), by lectin blotting and MS analysis. Hence, we propose that ‘agalactosylated N‐glycans’ are candidate ligands common to these lectins.
Keywords: agalactosylated N‐glycan, C‐type lectin, DC‐SIGN, DC‐SIGNR, LSECtin
Abbreviations
- αAGP
α1‐acid glycoprotein
- Bt
effective ligand content
- CHO
Chinese hamster ovary
- CRD
carbohydrate‐recognition domain
- DC
dendritic cell
- DC‐SIGN
dendritic cell‐specific intracellular adhesion molecule‐3 (ICAM‐3)‐grabbing nonintegrin
- DC‐SIGNR
DC‐SIGN‐related protein
- dTHP‐1 cells
differentiated THP‐1 cells
- FAC
frontal affinity chromatography
- FITC
fluorescein isothiocyanate
- Fuc
fucose
- Gal
galactose
- GlcNAc
N‐acetylglucosamine
- GnT
N‐acetylglucosaminyltransferase
- ICAM
intracellular adhesion molecule
- LPS
lipopolysaccharide
- LSECtin
liver and lymph node sinusoidal endothelial cell C‐type lectin
- Man
mannose
- MFI
mean fluorescence intensity
- PA
pyridylaminated
- PE
phycoerythrin
- PVL
GlcNAc‐binding from Psathyrella velutina lectin
- TF
transferrin
Introduction
Dendritic cell‐specific intracellular adhesion molecule‐3 (ICAM‐3)‐grabbing nonintegrin (DC‐SIGN, CD209) is a member of the C‐type lectin family, which is mainly expressed on dendritic cells (DCs) [1, 2]. DC‐SIGN consists of an N‐terminal cytoplasmic tail, a transmembrane domain, an extracellular C‐terminal neck region and a C‐type carbohydrate‐recognition domain (CRD) [3]. Characteristic of C‐type lectins with the CRD containing an EPN (Glu‐Pro‐Asn) motif, the receptor recognizes glycans containing terminal nonreducing mannose (Man), N‐acetylglucosamine (GlcNAc) and fucose (Fuc) in a Ca2+‐dependent manner [4, 5, 6]. There are lines of evidence which indicate that, through this basic specificity, DC‐SIGN recognizes endogenous self, exogenous nonself or tumor‐specific ligands, and mediates various functions in the immune system. In the first line of evidence, DC‐SIGN was found to bind to immune cells in a carbohydrate‐dependent manner. In fact, DC‐SIGN was reported to recognize naïve T cells through ICAM‐3 in a LewisX‐dependent manner, resulting in the initiation of an adaptive immune response [2, 7]. DC‐SIGN also mediates interactions between DCs and neutrophils through binding to LewisX of Mac‐1 expressed on neutrophils, and hence regulates DC maturation [8]. Second, DC‐SIGN recognizes invading pathogens via pathogen‐specific glycan structures, and acts as a scavenging receptor for them. These pathogens include viruses (HIV, Ebola and dengue), bacteria (Mycobacterium, Neisseria), fungi (Candida, Aspergillus) and parasitic protozoa (Leishmania, Schistosoma) [9, 10, 11, 12, 13, 14, 15, 16, 17, 18]. As a contrasting feature, DC‐SIGN has also been reported to function as a target for HIV entry, thus facilitating its infection [9]. Third, DC‐SIGN recognizes tumor‐specific glycans. DC‐SIGN has been reported to interact with carcinoembryonic antigen via Lewis structures expressed on colorectal cancer cells, and attenuates DC maturation [19, 20].
Based on the genomic analysis of chromosome 19p13.3, DC‐SIGN‐related protein (DC‐SIGNR, also known as L‐SIGN and CD209L) has been cloned from human placenta (77% amino‐acid sequence identity to DC‐SIGN) [21]. Unlike the broad expression pattern of DC‐SIGN, DC‐SIGNR is exclusively expressed on endothelial cells in lymph‐node sinuses and on liver sinusoidal endothelial cells, but not on myeloid cells [22], whereas it showed a similar binding feature to DC‐SIGN (i.e. Man‐ and Fuc‐specificity) [4, 23]. DC‐SIGNR binds to and takes up exogenous ligands, including viruses (e.g. HIV and Ebola) and parasites (e.g. Schistosoma), and mediates HIV dissemination [10, 22, 23]. Similarly to DC‐SIGN, DC‐SIGNR also recognizes endogenous ligands, such as ICAM molecules [24], although their glycan epitopes have not been fully characterized.
As a novel member of the DC‐SIGN‐related lectin subfamily, liver and lymph node sinusoidal endothelial cell C‐type lectin (LSECtin) has been found in the DC‐SIGN gene cluster of chromosome 19p13.3 [25]. The receptor is specifically expressed on sinusoidal endothelial cells of human liver and lymph node, showing a distribution similar to that of DC‐SIGNR. Recently, however, LSECtin was found to be expressed in macrophages, DCs and Kupffer cells, where the lectin was reported to function as an endocytic receptor [26, 27]. LSECtin also functions as an attachment factor for viruses, such as Ebola virus, Marburgvirus and severe acute respiratory syndrome coronavirus (SARS CoV), but not for HIV and hepatitis C virus (HCV) [26, 28, 29]. In a more recent paper by Powlesland et al., [30] LSECtin was reported to bind to an Ebola virus surface glycoprotein through GlcNAcβ1‐2Man structures. Undoubtedly, the DC‐SIGN‐related lectins mediate diverse functions in extensive immunobiological phenomena via the C‐type CRDs. However, there has been no report on the quantitative analysis of sugar–protein interactions, in terms of affinity constants (K d or K a), of DC‐SIGN, DC‐SIGNR and LSECtin.
Previously, we developed an automated frontal affinity chromatography (FAC) system, which allows high‐throughput determination of affinity constants of immobilized lectins to a panel of oligosaccharides [31, 32]. In the present study we utilized this automated system to provide a detailed quantitative analysis of the binding specificities of DC‐SIGN and its related receptors, DC‐SIGNR and LSECtin to 157 pyridylaminated (PA) glycans, including high‐mannose‐type and agalactosylated complex‐type N‐glycans, and blood‐antigen‐type glycans. The DC‐SIGN‐related lectins were found to exhibit a common specificity to agalactosylated complex‐type N‐glycans, but with different affinity (K d). Further analysis by glycoconjugate arrays and cell‐based biological assays using flow cytometry confirmed the observed preferences of the lectins for agalactosylated N‐glycans. The specificity to agalactosylated N‐glycans should help our understanding of the previously unknown mechanism of the functions of the DC‐SIGN‐related lectins.
Results
Quantitative analysis of glycan‐binding specificities of DC‐SIGN‐related lectins by FAC
To elucidate the mechanism of cellular functions mediated by the DC‐SIGN‐related lectins, it is fundamental to understand the basic aspects of their glycan‐binding specificities. Glycan‐microarray analyses of the DC‐SIGN‐related lectins have been reported [4, 30], but no quantitative data are available on the binding specificities in terms of K d (or K a). Therefore, we analyzed the oligosaccharide‐binding specificities of the DC‐SIGN‐related lectins using the automated FAC system [31, 32] and 157 PA glycans (Fig. S1). It should be noted, however, that in this study we adopted substantially different conditions of lectin columns in terms of effective ligand content (Bt) (see below). Under such conditions with very different lectin densities, direct comparison of K d/K a values among the three lectins may be inappropriate. Therefore, as a compromise, we used the term ‘apparent’ affinity, or app K a/app K d (meaning it is restrictive to the given conditions) in relevant contexts throughout this paper.
The C‐type CRDs of DC‐SIGN, DC‐SIGNR and LSECtin were expressed as Fc–protein fusions and were immobilized on N‐hydroxysuccinimide‐activated Sepharose 4FF using amine‐coupling chemistry, according to the standard protocol [31]. However, with this immobilization strategy, no substantial binding was observed, even when the Fc–protein fusions were used at a high concentration (8 mg·mL−1). We then immobilized the Fc–protein fusions on Protein A–Sepharose via the Fc region, and could finally observe binding activity of the Fc‐fusion proteins to positive oligosaccharides. To identify the effective ligand contents (Bt values) of the DC‐SIGN‐, DC‐SIGNR‐ and LSECtin–Fc‐immobilized columns, concentration‐dependence analyses were performed using the following oligosaccharide derivatives: Man9GlcNAc2‐methotrexate for DC‐SIGN, Manα1‐3Man‐PA for DC‐SIGNR and NGA2‐Fmoc for LSECtin (Fig. S2). As shown in Fig. 1, the Bt and app K d values were 1.72 nmol and 49.4 μm for DC‐SIGN, 4.25 nmol and 136.4 μm for DC‐SIGNR, and 0.39 nmol and 8 μm for LSECtin, respectively.
Figure 1.

Woolf–Hofstee‐type plots for DC‐SIGN–Fc‐, DC‐SIGNR–Fc‐ and LSECtin–Fc‐immobilized columns. The Bt and app K d values were determined for immobilized DC‐SIGN–Fc (Man9GlcNAc2‐methotrexate), DC‐SIGNR–Fc (Manα1‐3Man‐PA) and LSECtin–Fc (NGA2‐Fmoc) by concentration‐dependence analysis, and then Woolf–Hofstee‐type plots were generated for each lectin column. The glycan structures used for the analysis are shown in Fig. S2.
The overall binding features of the DC‐SIGN‐related lectin–Fc chimeras are summarized in Fig. 2 and Table S1. Apparently, their glycan‐binding properties are different in terms of both apparent affinity and specificity, but they were found to share a common preference for agalactosylated complex‐type N‐glycans (described below). From a global viewpoint, DC‐SIGNR–Fc showed the lowest affinity among the three C‐type lectins, while LSECtin–Fc showed the highest affinity under the present experimental conditions. In terms of specificity, DC‐SIGN–Fc and DC‐SIGNR–Fc apparently exhibited similar profiles for high‐mannose‐type N‐glycans (004‐016, 913‐915, where Arabic numbers correspond to glycan structures in Fig. 2.) and Fuc‐containing glycans represented by blood‐type antigens (723, 726, 727, 730, 731, 740, 910, 932). Furthermore, both recognized a certain group of agalactosylated complex‐type N‐glycans. By contrast, LSECtin–Fc showed remarkable selectivity towards agalactosylated complex‐type N‐glycans.
Figure 2.

Quantitative analysis of DC‐SIGN‐related lectin–Fc chimeras by FAC. Bar graph representation of app K a values of DC‐SIGN–Fc, DC‐SIGNR–Fc and LSECtin–Fc for 157 PA oligosaccharides. Arabic numbers at the bottom of the graphs correspond to the sugar numbers indicated in Fig. S1.
Recognition mechanism of agalactosylated complex‐type N‐glycans by DC‐SIGN‐related lectins
The detailed specificity to agalactosylated complex‐type N‐glycans were analyzed with the aid of the GRYP code representation described previously (Fig. 3) [33]. In this system, branch positions of each complex‐type N‐glycan are numbered from I to VI according to the corresponding mammalian N‐acetylglucosaminyltransferases (GnTs), whereas nonreducing end sugars are shown in different colors: Man in white, GlcNAc in blue, galactose (Gal) in yellow and α1‐6Fuc in red. A concise survey sheet presenting a comparison of app K d values between DC‐SIGN‐related lectins and representative high‐mannose‐type and agalactosylated N‐glycans is shown in Table 1.
Figure 3.

Detailed specificities of DC‐SIGN‐related lectin–Fc chimeras to agalactosylated complex‐type N‐glycans analyzed using the GRYP code representation. (A) Definition of the GRYP code to represent nonreducing end residues and branch positions. Nonreducing end sugars and core Fuc are indicated in different colors, as shown in the left panel. Each branch is numbered from I to VI, corresponding to the GnTs shown in the right panel. (B) Bar graph representation of K a values of the DC‐SIGN‐related lectins to agalactosylated complex‐type N‐glycans. A corresponding GRYP code for each glycan is shown under the bar graph.
Table 1.
Comparison of app K d values, in μm, of DC‐SIGN‐related lectins to representative N‐glycans. The values shown in parentheses are the relative affinities compared with 103 (denoted in bold).

While DC‐SIGN did not bind to the trimannosyl core structure (003), strong binding was observed for agalactosylated complex‐type N‐glycans up to bi‐antenna (102‐104, 202, 203, 304, 403; app K d > 55 μm), indicating that DC‐SIGN preferentially recognizes agalactosylated complex‐type N‐glycans. However, no binding was observed to highly branched N‐glycans (tri‐, tetra‐, and penta‐antenna, 105‐108, 204, 205) or to chitin‐related oligosaccharides (906, 907). In fact, DC‐SIGN gave the highest affinity to 102 (app K d, 55 μm), where the GlcNAc residue is attached at the GnT‐I position of the trimannosyl core structure. The binding affinity to 102 was similar to that to 014 (55 μm), which showed the highest affinity among the high‐mannose‐type N‐glycans tested. By contrast, no detectable binding was observed for its positioning isomer, 101, containing the GlcNAc residue at the GnT‐II position. Other oligosaccharide structures containing the terminal GlcNAc residue at the GnT‐I position (202, 68 μm; 103, 104 μm; 304, 140 μm; 403, 156 μm; 203, 322 μm; 104, 492 μm) were also high‐affinity ligands for DC‐SIGN. Binding was abolished by galactosylation of the GlcNAc residue at the GnT‐I position (302), indicating that the terminal GlcNAc residue at the GnT‐I position is important for DC‐SIGN binding. Addition of the GlcNAc residue at the GnT‐II position (e.g. 102 versus 103) resulted in only a moderate inhibitory effect, while the addition of the bisecting GlcNAc at the GnT‐III position greatly reduced the binding to approx. 20% (104). Addition of the GlcNAc residue at the GnT‐IV position (105) abolished the binding of DC‐SIGN, indicating that highly branched N‐glycans are not ligands for DC‐SIGN. No significant effect was observed for core fucosylation on DC‐SIGN binding (e.g. 103 versus 202). These results indicate that the presence of the terminal GlcNAc residue at the GnT‐I position is essential for DC‐SIGN binding to agalactosylated complex‐type N‐glycans.
Among agalactosylated complex‐type N‐glycans, DC‐SIGNR–Fc binding was detected for only two structures: bi‐antennary, agalactosylated complex‐type N‐glycans with the GlcNAc residues at both GnT‐I and GnT‐II positions, with (202, 944 μm) or without (103, 964 μm) core‐fucosylation. Under the experimental conditions of this study, no binding was detected for 102, the best ligand for DC‐SIGN. No detectable binding was observed for other mono‐antennary, agalactosylated complex‐type N‐glycans (101, 201), highly branched agalactosylated complex‐type N‐glycans (105‐108, 204, 205) or chito‐oligosaccharides (906, 907). The binding affinities for agalactosylated complex‐type N‐glycans were significantly lower than those for high‐mannose‐type N‐glycans (005‐017, > 292 μm), unlike the case of DC‐SIGN. Addition of Gal on either GlcNAc residue of 202 or 103 abolished the binding affinity (304, 306, 307, 403‐405). Addition of the bisecting GlcNAc transferred by GnT‐III (104, 203) also abolished the affinity. These results demonstrate that DC‐SIGNR has broadly similar, but different, specificity from DC‐SIGN towards agalactosylated complex‐type N‐glycans.
LSECtin gave selective affinity for agalactosylated complex‐type N‐glycans, while no binding was observed for high‐mannose‐type N‐glycans. Among agalactosylated complex‐type N‐glycans, LSECtin exhibited binding affinities to mono‐ and bi‐antennary structures (103, 23 μm; 202, 28 μm; 304, 38 μm; 403, 48 μm; 102, 49 μm), but not to tri‐, tetra‐ and penta‐antennary forms (105‐108, 204, 205), consistent with the results of DC‐SIGN and DC‐SIGNR. Also, the presence of the terminal GlcNAc at the GnT‐I position was essential for LSECtin binding, and addition of the GlcNAc residue transferred by GnT‐IV abolished the binding affinity (105, 204). However, addition of the bisecting GlcNAc (104) abolished the binding in the case of LSECtin. The specificity of the DC‐SIGN‐related lectins to agalactosylated complex‐type N‐glycans is summarized as follows: (a) the presence of a terminal GlcNAc at the GnT‐I position is essential, (b) the presence of a GlcNAc residue at the GnT‐IV position abrogates binding (and therefore highly branched agalactosylated complex‐type N‐glycans are not recognized), (c) there is little or no effect of core fucosylation and (d) there is a significant inhibitory effect of the addition of bisecting GlcNAc.
Binding of DC‐SIGN‐related lectins to agalactosylated glycoproteins
In order to investigate the binding of DC‐SIGN‐related lectins not only to liberated agalactosylated glycans but also to agalactosylated glycoproteins, we performed glycoconjugate microarray analyses [34]. Cell‐culture supernatants containing DC‐SIGN–, DC‐SIGNR– or LSECtin–Fc were pre‐incubated with Cy3‐conjugated anti‐human IgG, and the resulting complexes were applied to the glycoconjugate array, as previously described [34]. Culture supernatants derived from parental Chinese hamster ovary (CHO) cells were used as controls. Binding signals were detected using an evanescent‐field fluorescence‐assisted scanner (relevant data only are shown in Fig. 4A and full data are shown in Fig. S3). DC‐SIGN–Fc exhibited substantial binding to agalactosylated α1‐acid glycoprotein (αAGP) and transferrin (TF). The binding of DC‐SIGN–Fc to agalactosylated αAGP and TF is not a result of its specificity to Lewis‐related glycans, because it showed no detectable affinity for their intact (sialylated) and galactosylated forms. These data support the above results, obtained by FAC, that DC‐SIGN–Fc shows specificity to agalactosylated N‐glycans. The binding of DC‐SIGN–Fc was abolished in the presence of 10 mm EDTA, indicating that the binding occurs via the C‐type CRD. Weak signals on intact, galactosylated and agalactosylated TF are caused by the nonspecific reactivity of anti‐human IgG used as a secondary antibody (Fig. S3).
Figure 4.
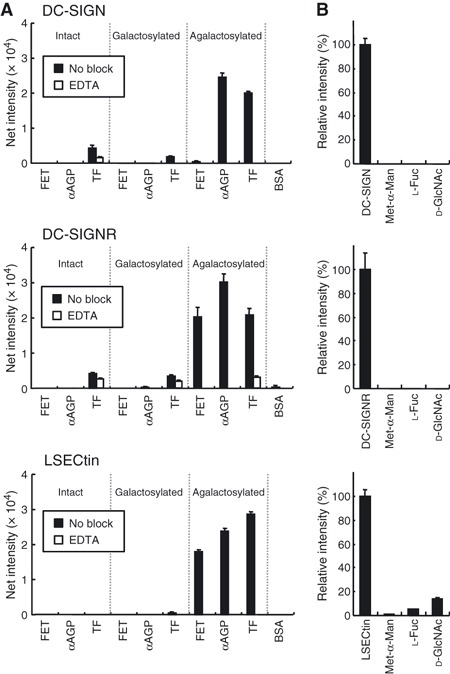
Binding of DC‐SIGN‐related lectin–Fc chimeras to agalactosylated glycoproteins. (A) Culture supernatants derived from CHO cells transfected with vectors expressing DC‐SIGN–Fc, DC‐SIGNR–Fc and LSECtin–Fc were precomplexed with Cy3‐conjugated anti‐human IgG and then applied to each well of slide glasses in the presence or absence of 10 mm EDTA. Fluorescently labeled proteins were detected using an evanescent‐field fluorescence‐assisted scanner. (B) Carbohydrate‐inhibition assay. Media were pre‐incubated with 50 mm monosaccharides (Met‐α‐Man, l‐Fuc and d‐GlcNAc) before assay.
DC‐SIGNR–Fc showed substantial affinity to a series of agalactosylated glycoproteins, including fetuin, but not to their sialylated (intact) and galactosylated forms. In all cases, the binding of DC‐SIGNR–Fc to these agalactosylated glycoproteins was completely abolished in the presence of 10 mm EDTA. LSECtin–Fc bound exclusively to a panel of agalactosylated glycoproteins (fetuin, αAGP and TF). The binding was also abrogated in the presence of 10 mm EDTA. Although these DC‐SIGN‐related lectin–Fc chimeras showed substantial binding to agalactosylated glycoproteins, they showed no detectable affinity to GlcNAc‐containing O‐glycans, such as core 2, 3, 4 and 6, and chitobiose (GlcNAcβ1‐4GlcNAc) (Fig. S3), suggesting that their primary targets are N‐glycans.
To examine whether the binding is carbohydrate‐dependent, we performed inhibition assays using three monosaccharide competitors: Met‐α‐Man, L‐Fuc and D‐GlcNAc (Fig. 4B). Data were expressed as the ratio of fluorescence intensity relative to that obtained for agalactosylated αAGP in the absence of competitors. In the presence of either of these monosaccharide inhibitors, binding of DC‐SIGN‐related lectin–Fc chimeras to agalactosylated αAGP was inhibited. These results indicate that the DC‐SIGN‐related lectin–Fc chimeras bind to glycoproteins containing agalactosylated complex‐type N‐glycans in a C‐type CRD‐dependent manner.
DC‐SIGN‐related lectins bind to agalactosylated glycoproteins expressed on cell surfaces
To verify binding of the DC‐SIGN‐related lectins to the agalactosylated N‐glycans of glycoproteins expressed on cell surfaces, we next examined their binding to CHO cells and their glycosylation‐deficient mutants, Lec1 and Lec8 cells, by flow cytometry. CHO cells are known to express complex‐, hybrid‐ and high‐mannose‐type N‐glycans, and O‐glycans, such as core 1 [35], whereas Lec1, a GnT‐I‐deficient mutant cell line, lacks both complex‐ and hybrid‐type N‐glycans and thus is dominated by high‐mannose‐type N‐glycans [36]. Lec8 cells have a deletion mutation in the Golgi uridine diphosphate‐Gal transporter, and thus express much reduced levels of galactosylated glycoconjugates [37]. Fc‐fusion protein chimeras were purified, precomplexed with Cy3‐labeled anti‐human IgG, and incubated with the Lec1, Lec8 and CHO cell lines (Fig. 5A). DC‐SIGN–Fc bound strongly to Lec8 cells as well as to Lec1 cells (Fig. S4), but did not bind to parental CHO cells. Similarly, DC‐SIGNR–Fc bound strongly to Lec8 and Lec1 cells, but not to CHO cells. By contrast, LSECtin–Fc bound only to Lec8 cells (and not to Lec1 or CHO cells). In the presence of 20 mm EDTA, the binding of Fc‐fusion proteins to Lec8 cells was abolished.
Figure 5.

Binding of DC‐SIGN‐related lectin–Fc chimeras to Lec8 cells. (A) DC‐SIGN–, DC‐SIGNR– and LSECtin–Fc precomplexed with Cy3‐conjugated anti‐human IgG (20 μg·mL−1) were incubated with Lec8 cells (filled histogram). Negative controls represent staining obtained using Cy3‐conjugated anti‐human IgG (dotted line). For the chelating assay, Lec8 cells were incubated with the Fc–fusion protein chimeras in the presence of 20 mm EDTA (thin line). Parental CHO cells were used as controls. After incubation on ice for 1 h, cells were analyzed by flow cytometry. (B) For the inhibition assay, Lec8 cells (2 × 105) were pre‐incubated with 1 mg·mL−1 of PVL (GlcNAc‐binding lectin) on ice for 1 h. MFI, mean fluorescence intensity.
We then performed inhibition tests using a GlcNAc‐binding lectin from Psathyrella velutina (PVL). When PVL (1 mg·mL−1) was pre‐incubated with Lec8 cells, binding of DC‐SIGN–, DC‐SIGNR– and LSECtin–Fc was reduced to 30–40% (Fig. 5B). These results, together with FAC and glycoconjugate microarray analysis, indicate that DC‐SIGN–, DC‐SIGNR– and LSECtin–Fc bind to agalactosylated N‐glycans of glycoproteins displayed on cell surfaces in a Ca2+‐dependent manner.
Adhesion of CHO cells, expressing DC‐SIGN‐, DC‐SIGNR‐ and LSECtin, to Lec8 cells
It is known that the DC‐SIGN‐related lectins have the functional ability to mediate cellular adhesion in a carbohydrate‐binding manner. To confirm the cellular interaction of the DC‐SIGN‐related lectins with agalactosylated cells, we performed cell‐adhesion assays using Lec8 cells and lectin‐transfected CHO cells. CHO cell lines stably expressing DC‐SIGN, DC‐SIGNR or LSECtin were generated, and their levels of expression were analyzed with the aid of specific antibodies. Flow cytometric analysis indicated that DC‐SIGN and DC‐SIGNR were apparently overexpressed on the surface of CHO cells, whereas LSECtin was expressed less strongly (Fig. 6A). By contrast, no reactivity was observed for untransfected CHO cells (data not shown).
Figure 6.

Adhesion of CHO cells expressing DC‐SIGN, DC‐SIGNR and LSECtin to Lec8 cells. (A) CHO cells stably expressing DC‐SIGN, DC‐SIGNR and LSECtin were prepared as described in the Materials and methods. Surface expression of the DC‐SIGN‐related lectins was detected by flow cytometry using monoclonal anti‐DC‐SIGN, monoclonal anti‐DC‐SIGNR and polyclonal anti‐LSECtin, followed by PE‐conjugated anti‐mouse and FITC‐conjugated anti‐goat IgGs, respectively (filled histogram). Isotype‐control antibodies were used as negative controls (dotted histogram). (B) CMRA‐labeled Lec8 cells (5 × 104 cells) were incubated with CHO cells expressing DC‐SIGN, DC‐SIGNR and LSECtin, at 4 °C for the indicated time. (C) CMRA‐labeled Lec8 cells were incubated with parental Flp‐In‐CHO cells and with Flp‐In‐CHO cells expressing DC‐SIGN, DC‐SIGNR and LSECtin in the presence or absence of 2 mm CaCl2 for 30 min at 4 °C. After gentle washing, cell–cell adhesion was determined using a microplate reader.
These transfected cells were incubated in each well of 96‐well plates for 2 days. After washing, the cells were co‐cultured on ice with CMRA‐labeled Lec8 cells (5 × 104). After removal of unbound Lec8 cells by gentle washing, adherent cells were detected directly using a microplate reader. As shown in Fig. 6B, all three transfectants showed increased adhesion to Lec8 cells in a time‐dependent manner. In the absence of 2 mm CaCl2, adhesion of these transfected CHO cells to Lec8 cells was reduced to the level of control CHO cells (Fig. 6C). These results, together with the results of the glycoconjugate microarray, indicate that DC‐SIGN, DC‐SIGNR and LSECtin mediate intercellular interaction with agalactosylated cells via C‐type CRDs in a Ca2+‐dependent manner.
DC‐SIGN‐related lectins internalize agalactosylated αAGP into cells
Previous studies have shown that DC‐SIGN, DC‐SIGNR and LSECtin could internalize exogenous ligands, such as bacterial and viral glycoproteins/glycolipids, into cells. We examined whether agalactosylated glycoproteins are internalized into cells expressing these C‐type lectin receptors. As a model ligand, we chose agalactosylated αAGP, which was recognized by DC‐SIGN‐related lectin–Fc chimeras on a glycoconjugate microarray. αAGP was pretreated with both sialidase and β‐galactosidase, and the resulting agalactosylated αAGP was biotinylated. DC‐SIGN‐, DC‐SIGNR‐ and LSECtin‐expressing CHO cells were then incubated on ice for 1 h with the biotin‐labeled agalactosylated αAGP precomplexed with phycoerythrin (PE)‐conjugated streptavidin (10 μg·mL−1), and the temperature was raised to 37 °C to trigger internalization. The internalized fluorescence was detected by flow cytometry. As shown in Fig. 7A, agalactosylated αAGP was found to be internalized into all of the DC‐SIGN‐, DC‐SIGNR‐ and LSECtin‐expressing CHO cells, whereas the internalization was not observed for its intact (extensively sialylated) form. In the absence of CaCl2, no internalization was observed. Neither intact nor agalactosylated αAGP were internalized by parental CHO cells. When the transfected cell lines were incubated at 37 °C for prolonged periods of time (up to 120 min), the internalization levels of agalactosylated αAGP were found to increase over the incubation period (Fig. 7B). These results clearly demonstrate that DC‐SIGN‐, DC‐SIGNR‐ and LSECtin‐expressing cells internalize agalactosylated, but not intact, αAGP in a Ca2+‐dependent manner.
Figure 7.

Uptake of agalactosylated αAGP by CHO cells stably expressing DC‐SIGN, DC‐SIGNR and LSECtin. (A) CHO cells stably expressing DC‐SIGN, DC‐SIGNR and LSECtin were incubated with 10 μg·mL−1 of biotin‐labeled agalactosylated αAGP (blue line) and its intact form (green line) precomplexed with PE‐conjugated streptavidin on ice for 30 min, and allowed to internalize at 37 °C for 1 h in the presence or absence (orange line) of 2 mm CaCl2. Negative controls represent staining obtained using PE‐conjugated streptavidin (red line). Cells were analyzed by flow cytometry. Parental untransfected CHO cells were used as mock cells. (B) CHO cells expressing DC‐SIGN, DC‐SIGNR and LSECtin cells were internalized at 37 °C for the times shown with 10 μg·mL−1 of biotin‐labeled agalactosylated αAGP precomplexed with PE‐conjugated streptavidin.
Adhesion and uptake by cells expressing endogenous DC‐SIGN and LSECtin
We examined the endocytic and adhesive functions of the DC‐SIGN‐related lectins using cell lines endogenously expressing the receptors: differentiated THP‐1 cells (dTHP‐1 cells), treated with 4β‐phorbol 12‐myristate 13‐acetate expressing DC‐SIGN (Fig. 8A); and HL‐60 cells expressing LSECtin (Fig. 8B).
Figure 8.

Cell adhesion and uptake by cells expressing endogenous DC‐SIGN and LSECtin. Flow cytometry histograms obtained after immunofluorescence staining of dTHP‐1 (A) and HL‐60 cells (B) with anti‐DC‐SIGN and anti‐LSECtin mAbs followed by labeling with FITC‐ and PE‐conjugated anti‐mouse IgG (black line), respectively. Negative controls represent staining obtained using isotype‐control antibody (dotted). Cells were analyzed by flow cytometry. (C) dTHP‐1 cells were incubated with CMRA‐labeled Lec8 cells (2 × 104 cells). (D) CMRA‐labeled HL‐60 cells (1 × 105 cells) were incubated with Lec8 cells. After incubation at 4 °C for 30 min followed by gentle washing, bound cells were determined by analysis on a microplate reader. dTHP‐1 (E) and HL‐60 cells (F) were incubated with 10 μg·mL−1 of FITC‐conjugated agalactosylated αAGP on ice for 30 min, and were allowed to internalize at 37 °C for 2 h. Cells were analyzed by flow cytometry. For analysis in the inhibition assay, these cells were pre‐incubated, at 37 °C for 30 min, with mAbs specific for either DC‐SIGN or LSECtin.
Lec8 cells were incubated with the above dTHP‐1 and HL‐60 cells expressing endogenous DC‐SIGN and LSECtin, respectively, at 4 °C for 30 min. As shown in Figs. 8C and D, dTHP‐1 and HL‐60 cells adhered to Lec8 cells. Cell adhesion was specifically blocked by pretreatment with mAbs specific for DC‐SIGN and LSECtin (by approximately 30% for dTHP‐1 cells and by approximately 70% for HL‐60 cells, respectively).
We next investigated the endocytic activity of DC‐SIGN and LSECtin. Cells were first incubated with fluorescein isothiocyanate (FITC)‐conjugated, agalactosylated αAGP on ice for 30 min, and then warmed to 37 °C for 120 min to trigger internalization. As shown in Figs 8E and F, FITC‐conjugated agalactosylated αAGP was internalized into the dTHP‐1 and HL‐60 cells expressing endogenous DC‐SIGN and LSECtin, respectively. The internalization was inhibited by pretreatment with the blocking mAbs (by approximately 30% for dTHP‐1 cells and by approximately 65% for HL‐60 cells, respectively). These results indicate that endogenous DC‐SIGN and LSECtin expressed on immune‐related cells can mediate both intercellular interaction with agalactosylated cells and internalization of an agalactosylated glycoprotein.
Identification of agalactosylated glycoprotein ligands for DC‐SIGN in human serum
In order to identify agalactosylated glycoprotein ligands for DC‐SIGN, DC‐SIGN–Fc protein‐immobilized gel was incubated with serum and bound proteins were eluted with EDTA. The eluate was resolved by SDS/PAGE and blotted with biotin‐labeled PVL, which is specific for GlcNAc‐containing glycans. As shown in Fig. 9A, three major bands at approximately 160, 75 and 55 kDa were detected, indicating that agalactosylated glycoproteins recognized by DC‐SIGN are indeed present in human serum. No band was detected in the absence of DC‐SIGN–Fc. As shown in Fig. 9B, the three major bands (i.e. 1, of 160 kDa; 2, of 75 kDa; and 4, of 55 kDa) were present on a silver‐stained gel, as well as an extra band (band 3, of 65 kDa), which corresponded to serum albumin, probably as a contaminant. Protein identification by MS revealed that bands 1, 2 and 4 corresponded to α2‐macroglobulin, serotransferrin and IgG heavy chain, respectively (Table 2). These results suggest that α2‐macroglobulin, serotransferrin and IgG heavy chain might have agalactosylated bi‐antennary N‐glycans, and thus are candidate molecules for DC‐SIGN ligands in human serum.
Figure 9.

Identification of agalactosylated ligands for DC‐SIGN in human serum. The DC‐SIGN‐immobilized gel was incubated with human serum at 4 °C overnight. After washing, bound glycoproteins were eluted with EDTA. The eluate was resolved by SDS/PAGE, and was detected by PVL‐blotting (A) and silver‐staining (B).
Table 2.
MS identification of serum agalactosylated glycoproteins bound by DC‐SIGN.
Discussion
Through the detailed analysis of DC‐SIGN‐related lectins by quantitative FAC, we identified a common recognition unit for agalactosylated N‐glycans, namely the terminal GlcNAc residue at the GnT‐I position on the trimannosyl core structure. However, each lectin showed distinct affinities to a variety of agalactosylated N‐glycans. We used C‐type CRDs fused with the Fc region of IgG, because the standard procedure, in which lectins are covalently immobilized on N‐hydroxysuccinimide‐activated Sepharose through primary amine groups [31], gave insufficient interaction for the DC‐SIGN‐related lectins tested. By contrast, the present procedure, using Fc–chimera conjugates, yielded good results with sufficient availabilities of the immobilized ligands (i.e. 10–60%). However, there is still room for discussion about K d/K a values determined in this work, which were basically defined for the disulfide‐linked homodimer of each C‐type CRD fused with an IgG Fc portion. The DC‐SIGN‐related lectins are thought to form relatively large multimers at the cell surface [6, 30]. In addition, the K d values are highly dependent on the experimental conditions, including temperature (in this work, 25 °C), flow rate (0.125 mL·min−1), lectin density (various) and lectin‐immobilization method (Fc chimera). In this study, the lectin densities were considerably different from one another. Hence, the affinity was expressed as ‘apparent K d/K a’ throughout this manuscript. Nevertheless, the major conclusion reached in this work was that DC‐SIGN, DC‐SIGNR and LSECtin exhibited common binding affinity to mono‐ and/or bi‐antennary agalactosylated N‐glycans. Although affinity enhancement by multivalent lectin–carbohydrate interaction is well documented, it is assumed that a simple 1 : 1 interaction, rather than multiple interactions, occurs under the FAC conditions used, because the oligosaccharide concentrations used for the analysis were low (2.5–5 nm). Moreover, accumulated evidence from crystallography suggests that multiple lectin–oligosaccharide interactions, leading to affinity enhancement, does not seem to occur in the cases of DC‐SIGN and DC‐SIGNR and their counterpart glycan ligands (see below).
In the crystallographic analysis of DC‐SIGN with GlcNAc2Man3 (PDB code, 1k9i) [38], the trimannosyl core structure was found to form hydrogen bonds via Ca2+‐coordination with Glu347 and Asn349 in an EPN motif, as well as via van der Waals contact with Phe313. The terminal β1‐2GlcNAc residue on the α1‐3Man branch also forms both a hydrogen bond with Asn349 and van der Waals contact with Val351. In the structure of DC‐SIGNR (1k9j) [38], Asn361 in the EPN motif and Ser363 (Val351 in DC‐SIGN) makes hydrogen bonds with the terminal β1‐2GlcNAc residue. These observations support the present result that β1‐2GlcNAc at the GnT‐I position is critical for recognition by DC‐SIGN and DC‐SIGNR (Fig. S5). Crystallographic studies also revealed that the terminal β1‐2GlcNAc residue of the α1‐6Man branch forms hydrogen bonds and/or van der Waals contacts (Asn311 and Phe313 of DC‐SIGN; Asn323 and Phe325 of DC‐SIGNR). In the case of LSECtin, for which neither crystallographic analyses nor NMR studies have been reported, we used a modeling structure available from MODBASE published by Pieper et al. [39] (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi). Comparison between LSECtin and DC‐SIGN CRDs (1k9i) indicated that the terminal β1‐2GlcNAc residue on the α1‐3Man branch would interact with Trp259, in addition to Ca2+‐coordinated hydrogen bonds to the α1‐3Man residue (Fig. S5). Trp259 of LSECtin corresponds to Val351 in DC‐SIGN and to Ser363 in DC‐SIGNR, indicating that Trp259 would be involved in the binding to the terminal β1‐2GlcNAc residue. Phe313 in DC‐SIGN and Phe325 in DC‐SIGNR also correspond to Arg219 in LSECtin, which probably interacts with the β1‐2GlcNAc residue on the α1‐6Man branch. In the present FAC analysis, highly branched agalactosylated N‐glycans were not recognized by the DC‐SIGN‐related lectins, presumably for reasons of steric hindrance. Indeed, there is no structural space to accommodate the terminal β1‐4GlcNAc residue on the α1‐3Man branch according to their reported crystal and modeling structures. The DC‐SIGN‐related lectins are unlikely to accommodate O‐glycans because of steric hindrance caused by the presence of an attached polypeptide.
In addition to agalactosylated N‐glycans, mannosylated glycans are also ligands for DC‐SIGN and DC‐SIGNR (Fig. 2, 004‐015, 913‐915). The characteristic features are (a) DC‐SIGN apparently shows higher affinity to high‐mannose‐type N‐glycans than to DC‐SIGNR, (b) the binding affinities of both DC‐SIGN and DC‐SIGNR are enhanced when the number of αMan structures increases, consistent with previous results [4] and (c) DC‐SIGNR shows higher affinity to mannosylated glycans (913‐915) than to high‐mannose‐type N‐glycans (005‐014), whereas DC‐SIGN shows the opposite. A significant difference was also observed in the binding affinities of the ligands for Fuc‐containing glycans. An earlier report by Appelmelk et al. [40] indicated that DC‐SIGN showed strong recognition of a series of Lewis antigen‐immobilized polyacrylamides (i.e. Lewisa/b/x/y) when analyzed using an ELISA. Subsequently, Guo et al. [4] demonstrated different aspects of specificities between DC‐SIGN and DC‐SIGNR CRDs using glycan microarray analyses, and further discussed their binding mechanisms based on crystallographic analyses with lacto‐N‐fucopentaose III. They found that the Lewis antigens were high‐affinity ligands for DC‐SIGN, but not for DC‐SIGNR. In fact, DC‐SIGN Val351 is involved in tight van der Waals contact with 2‐OH of Fuc on Lewisx antigen in addition to recognition of 3‐ and 4‐OH in a Ca2+‐dependent binding manner (PDB code, 1sl5), whereas DC‐SIGNR Ser363 excludes the contact with 2‐OH of Fuc (1sl6) [4]. van Liempt et al. [23] also supported different binding modes between DC‐SIGN and DC‐SIGNR, regarding Lewisa/x tri‐saccharides, using modeling and docking simulation combined with a mutagenesis study.
Glycan‐binding receptors expressed on animal cells play key roles in endogenous cellular‐adhesion events [23]. One the well‐characterized example of this is leukocyte–endothelial cell adhesion mediated by selectins. Like selectins, DC‐SIGN and DC‐SIGNR may also function as adhesion receptors for endogenous cells, such as T cells [2], endothelial cells [42] and neutrophils [8]. Herein, we provided evidence that DC‐SIGN, DC‐SIGNR and LSECtin serve as cellular adhesion receptors for mammalian agalactosylated CHO cells (Lec8 cells). This finding suggests that these DC‐SIGN‐related lectins can mediate cellular adhesion events through recognition of agalactosylated N‐glycoproteins expressed on endogenous cells. Considering the fact that agalactosylated glycoproteins are abundant in mouse brain [43], and vessel‐associated DC‐SIGN+ cells are present in human brain tissue [44], interaction of DC‐SIGN with agalactosylated cells might be involved in cell–cell adhesion events in the brain.
Ligand clearance by specific C‐type lectin receptors has been shown experimentally. Grewal et al. [45] provided sophisticated evidence that Ashwell receptor (asialoglycoprotein receptors 1 and 2) mediates clearance for asialo‐types of endogenous von Willebrand factor and platelets, and thus modulates homeostasis in blood coagulation. In this article, we demonstrated that agalactosylated αAGP, an acute‐phase serum glycoprotein produced in liver, was internalized in cells expressing DC‐SIGN, DC‐SIGNR and LSECtin. We also identified candidate agalactosylated glycoproteins (α‐2‐macroglobulin, serotransferrin and IgG heavy chain) for DC‐SIGN in human serum. As all of the DC‐SIGN‐related lectins have been found to be expressed in LSECs, they are probably involved in the clearance of agalactosylated glycoproteins in serum.
The cell wall of Gram‐negative bacteria contains lipopolysaccharide (LPS) consisting of three domains: lipid A, core saccharide and O‐antigen. Recently, Steeghs et al. [18] reported that DC‐SIGN expressed on DCs mediates cell adhesion and internalization of Neisseria meningitidis through the recognition of GlcNAc, a repeating unit in the core saccharide region of LPS. Similarly, Zhang et al. [46] showed that DC‐SIGN on HeLa cells binds to several Gram‐negative bacteria (Escherichia coli, Salmonella typhimurium, Neisseria gonorrhoeae and Haemophilus ducreyi) through GlcNAc in the core region of LPS. By contrast, LSECtin has been reported to bind to Ebola virus glycoprotein and SARS virus spike protein in a Ca2+‐dependent manner [28, 29]. Subsequently, Powlesland et al. [30] suggested the presence of bi‐antennary agalactosylated N‐glycans, as well as high‐mannose‐type N‐glycans, on Ebola virus glycoprotein by MS analysis. Similar, but detailed, data have also been obtained for SARS virus spike protein [47]. These findings support the fact that LSECtin binds to these viral glycoproteins in a β1‐2GlcNAc‐binding manner. Consistently, both DC‐SIGN and DC‐SIGNR would act as pathogen‐recognition receptors for Man/GlcNAc‐containing glycans presented in Ebola and SARS viruses [10, 48, 49], illustrating that the three lectins are also involved in interacting with such viruses. In this regard, an EPN motif may function as a bridging player towards nonself (pathogen) and undesired self. Further investigations of natural agalactosylated glycan ligands for these DC‐SIGN‐related lectins remain to be made for understanding their physiological functions.
Materials and methods
Materials
Mouse monoclonal anti‐DC‐SIGN (clone 120507), mouse monoclonal anti‐DC‐SIGNR (120604), polyclonal goat anti‐LSECtin, and mouse and goat isotype IgGs were purchased from R&D Systems. FITC‐conjugated rabbit anti‐goat IgG, PE‐conjugated goat anti‐mouse IgG, Cy3‐conjugated goat anti‐human IgG and PE‐conjugated streptavidin were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Mouse monoclonal anti‐LSECtin IgG (clone SOTO‐1) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). PVL was purchased from Wako (Osaka, Japan). Human αAGP was purchased from Sigma (Tokyo, Japan). Agalactosylated αAGP was prepared by treatment with Arthrobacter ureafaciens sialidase (Roche, Tokyo, Japan) and Streptococcus 6646K β‐galactosidase (Seikagaku, Tokyo, Japan). Degalactosylation of αAGP was analyzed using a lectin microarray (Fig. S6) [50].
Plasmids
The coding sequences of DC‐SIGN CRD (251‐404 amino acids), DC‐SIGNR CRD (240‐376) and LSECtin CRD (160‐293) were amplified by PCR using specific primer sets (forward and reverse, respectively: 5′‐CGCCTGTGCCACCCCTGTCCCTGGGAATG‐3′ and 5′‐CGCAGGAGGGGGGTTTGGGGTGGCAGGG‐3′ for DC‐SIGN CRD; 5′‐CGCCTGTGCCGCCACTGTCCCAAGGACTG‐3′ and 5′‐TTCGTCTCTGAAGCAGGCTGCGGGCTTTTT‐3′ for DC‐SIGNR CRD; and 5′‐AACTCCTGCGAGCCTTGCCCCACGTC‐3′ and 5′‐GCAGTTGTGCCTTTTCTCACAGATC‐3′ for LSECtin CRD). An amplified fragment was purified and ligated into a pSecTag/FRT/V5‐His vector (Invitrogen, Tokyo, Japan). A gene encoding the Fc region of human IgG was inserted into the vector via AgeI and PmeI sites. Full‐length (FL) cDNAs encoding DC‐SIGN, DC‐SIGNR and LSECtin were amplified by PCR using specific primer sets (forward and reverse, respectively; 5′‐ACCATGAGTGACTCCAAGGAACCAAGACT‐3′ and 5′‐CGCAGGAGGGGGGTTTGGGGTGGCAGGG‐3′ for DC‐SIGN FL; 5′‐ACCATGAGTGACTCCAAGGAACCAAGG‐3′ and 5′‐TTCGTCTCTGAAGCAGGCTGCGGGCTTTTT‐3′ for DC‐SIGNR FL; and 5′‐ATAATGGACACCACAAGGTACAGCAAGTA‐3′ and 5′‐GCAGTTGTGCCTTTTCTCACAGATC‐3′ for LSECtin FL). The derived fragment was ligated into a pcDNA5/FRT/V5‐His vector (Invitrogen).
Cell culture
CHO cells, their glycosylation‐deficient mutants (Lec1 and Lec8 cells) and HL‐60 cells were cultured, at 37 °C and 5% CO2, in RPMI 1640 supplemented with 5% fetal bovine serum (Invitrogen), 100 U·mL−1 of penicillin and 100 μg·mL−1 of streptomycin. For maintenance of Lec8 cells, proline (20 mg·L−1) was added to the complete medium. THP‐1 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U·mL−1 penicillin and 100 μg·mL−1 streptomycin. To induce differentiation, THP‐1 cells (1 × 106 cells) were treated with 50 ng·mL−1 of 4β‐phorbol 12‐myristate 13‐acetate (Sigma). After incubation for 96 h, cells were used for uptake and adhesion experiments. To examine the surface expression of DC‐SIGN on dTHP‐1 cells and of LSECtin on HL‐60 cells, the cells suspended in NaCl/Pi (2.7 mm KCl, 1.5 mm KH2PO4, 137 mm NaCl and 8.1 mm Na2HPO4) containing 1% (w/v) BSA (NaCl/Pi/BSA) were incubated with 10 μg·mL−1 of primary antibodies (monoclonal anti‐DC‐SIGN, monoclonal anti‐LSECtin and isotype IgGs) for 30 min on ice. After washing with NaCl/Pi/BSA, the cells were incubated with 10 μg·mL−1 of a secondary antibody (PE‐conjugated anti‐mouse IgG) for 30 min on ice. After washing with NaCl/Pi/BSA the cells were analyzed using flow cytometry (FACSCantoII; BD Biosciences, San Jose, CA, USA). The data obtained were analyzed using flowjo software (FlowJo, Ashland, OR, USA).
Construction of stably expressing CHO cell lines
Plasmids were transfected into Flp‐In‐CHO cells (Invitrogen) by Lipofectamine LTX (Invitrogen) in accordance with the manufacturer’s procedure. Stably expressing CHO cell clones were selected in Ham’s F12 Medium supplemented with 5% fetal bovine serum or 5% low IgG‐fetal bovine serum (Invitrogen), 100 U·mL−1 of penicillin, 100 μg·mL−1 of streptomycin and 0.5 mg·mL−1 of hygromycin B (Invitrogen). To examine the surface expression of DC‐SIGN‐related lectins on CHO cells, 1 × 106 cells in NaCl/Pi/BSA were incubated with 10 μg·mL−1 of primary antibodies (monoclonal anti‐DC‐SIGN, monoclonal anti‐DC‐SIGNR, monoclonal anti‐LSECtin mAb and isotype IgGs) for 30 min on ice. After washing with NaCl/Pi/BSA, the cells were incubated, for 30 min on ice, with 10 μg·mL−1 of appropriate secondary antibodies (FITC‐conjugated anti‐goat or PE‐conjugated anti‐mouse IgGs). After washing with NaCl/Pi/BSA, the cells were analyzed by flow cytometry.
Preparation of DC‐SIGN‐related lectin‐Fc chimeras
CHO cells stably expressing DC‐SIGN‐related lectin–Fc chimeras were cultured for 15 days with medium exchanges every 3 days. Collected culture supernatants containing DC‐SIGN‐related lectin–Fc chimeras were purified by affinity chromatography on Protein A–Sepharose (Amersham, Tokyo, Japan), as described previously [51].
FAC
FAC was performed essentially as described previously [31, 32]. Briefly, DC‐SIGN‐related lectin–Fc chimeras were coupled to Protein A–Sepharose at a concentration of 12–15 mg·mL−1. Flow rate and column temperature were kept at 0.125 mL·min−1 and 25 °C, respectively, and an excess volume of each glycan in NaCl/Tris [10 mm Tris/HCl, pH 7.4, containing 0.8% (w/v) NaCl] containing 1 mm CaCl2, was successively injected into the columns. Elution of PA‐labeled glycans (2.5–5 nm) was monitored using a fluorescence detector (excitation/emission wavelengths: 310/380 nm; see Fig. S1). The elution front relative to that of an appropriate standard oligosaccharide (i.e. V−V0) was determined. The app K d value was obtained from V−V0 and Bt (effective ligand content of the column), according to the basic equation of FAC, app K d = Bt/(V−V0) − [A]0. For concentration‐dependence analysis, varying concentrations ([A]0) of glycan derivatives (Man9GlcNAc2‐methotrexate (MTX), lactose‐β‐p‐nitrophenyl, Manα1‐3Man‐PA, lactose‐PA and NGA2‐Fmoc, see Fig. S2) were successively injected into the columns, and the elution was monitored by fluorescence (excitation/emission wavelengths: 270/380 nm for PA) and UV detectors (304 nm for methotrexate, 280 nm for p‐nitrophenyl and 274 nm for Fmoc), respectively. Woolf–Hofstee‐type plots, (V−V0) versus (V−V0)[A]0, were constructed to determine Bt and app K d values from the intercept of the axis and the slope of the fitted curves, respectively.
Glycoconjugate microarray
The glycoconjugate microarray system was generated as described previously [34]. The glycan structures of modified glycoproteins were analyzed using a lectin microarray [50]. Binding of DC‐SIGN‐related lectin–Fc chimeras to glycoproteins was analyzed using the glycoconjugate microarray system. Briefly, culture supernatants containing DC‐SIGN‐related lectin–Fc chimeras were pre‐incubated with Cy3‐conjugated anti‐human IgG at room temperature for 30 min. The resulting culture supernatants were applied to each well of slide glasses, and incubated at 20 °C overnight. The fluorescence images of the slides were detected using an evanescent‐field fluorescence‐assisted GlycoStation ReaderTM (GP Biosciences, Yokohama, Japan). The data obtained were analyzed using the arraypro AnalyzerTM version 4.5 (Media Cybernetics, Bethesda, MD, USA). For the inhibition assay, 10 mm EDTA or 50 mm monosaccharides [methyl‐α‐mannoside (Met‐α‐Man), L‐Fuc and D‐GlcNAc] were pre‐incubated in the precomplex solutions at room temperature for 30 min.
Binding of DC‐SIGN‐related lectin–Fc chimeras to cells
Cells (1 × 106) in TSA buffer [NaCl/Tris containing 2 mm CaCl2, 2 mm MgCl2 and 0.5% (w/v) BSA] were incubated with 20 μg·mL−1 of DC‐SIGN‐related lectin–Fc chimeras precomplexed with Cy3‐conjugated anti‐human IgG, in the presence or absence of 20 mm EDTA, for 1 h on ice. After washing with TSA buffer, cells were analyzed by flow cytometry. For use in the inhibition assay, 2 × 105 Lec8 cells were pre‐incubated with 1 mg·mL−1 of PVL on ice for 1 h.
Cell adhesion assay
Cells (5 × 103) were cultured in 96‐well plates for 2 days at 37 °C. For labeling, cells were incubated for 30 min at 37 °C in RPMI 1640 containing 10 μm Cell‐Tracker CMRA (Invitrogen), in accordance with the manufacturer’s procedure [52]. After washing with NaCl/Tris containing 2 mm MgCl2, with or without 2 mm CaCl2, the CMRA‐labeled cells were added to each well (2–10 × 104 cells per 100 μL), and incubated at 4 °C for the indicated period of time. After removal of nonadherent cells by gravity washing in cold NaCl/Tris containing 2 mm MgCl2, either with or without 2 mm CaCl2, adherent cells were detected using a microplate reader with excitation/emission wavelengths of 548/576 nm (SpectraMax M5; Molecular Devices, Sunnyvale, CA, USA). For blocking studies, dTHP‐1 and HL‐60 cells were pre‐incubated for 30 min at 37 °C with 10 μg·mL−1 of mAbs specific for either DC‐SIGN or LSECtin.
Internalization assay
Biotin‐conjugated glycoproteins were prepared using N‐hydroxysuccinimide‐PEG4‐biotin (Pierce, Rockford, IL, USA), according to the manufacturer’s instructions. Parental Flp‐In‐CHO cells, or Flp‐In‐CHO cells expressing DC‐SIGN, DC‐SIGNR or LSECtin (2 × 105 cells), were cultured in six‐well plates for 2 days at 37 °C. After washing with TSA buffer, either with or without 2 mm CaCl2, the cells were incubated for 30 min on ice with 10 μg·mL−1 of biotin‐labeled glycoproteins precomplexed with PE‐conjugated streptavidin followed by incubation at 37 °C for various time‐points up to 120 min. After tryptic digestion, the cells were analyzed by flow cytometry. For assay of internalization with dTHP‐1 and HL‐60 cells, agalactosylated αAGPs were conjugated with FITC (Sigma) according to the manufacturer’s instructions. dTHP‐1 and HL‐60 cells were treated for 30 min on ice with 10 μg·mL−1 of FITC‐conjugated agalactosylated αAGP in TSA buffer. The cells were then incubated at 37 °C for 2 h. For blocking studies, dTHP‐1 and HL‐60 cells were pre‐incubated for 30 min at 37 °C with 10 μg·mL−1 of mAbs specific for either DC‐SIGN or LSECtin.
Lectin blotting
Human serum was precleared by incubation with Protein A–Sepharose at 4 °C for 3 h. The serum was then incubated with DC‐SIGN–Fc‐immobilized gel at 4 °C overnight. After washing with NaCl/Tris containing 2 mm CaCl2 and 2 mm MgCl2, bound proteins were eluted with NaCl/Tris containing 5 mm EDTA. The eluate was run on SDS/PAGE and electroblotted onto a polyvinylidene difluoride (PVDF) membrane. After blocking with NaCl/Tris containing Tween 20 (NaCl/Tris‐T), the membrane was incubated with 1 μg·mL−1 of biotinylated PVL for 1 h at room temperature. After washing with NaCl/Tris‐T, the membrane was incubated with streptavidin‐conjugated alkaline phosphatase (Promega, Tokyo, Japan) for 30 min at room temperature. After washing with NaCl/Tris‐T, the membrane was visualized with Western Blue‐stabilized substrate for alkaline phosphatase (Promega).
In‐gel digestion with trypsin and MS
SDS/PAGE gels were silver‐stained, as described previously [53]. In‐gel digestion with trypsin (Promega) was performed as described previously [54]. The peptide mass was analyzed using an Ultraflex MALDI‐TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The spectra obtained were analyzed using the flex Analysis software, version 2.0. Protein identification was performed using the ms‐fit software, version 5.0.0 (http://jpsl.ludwig.edu.au/).
Supporting information
Table S1. appK d values (μM) of the DC‐SIGN‐related lectins for PA‐glycans
Fig. S1. Schematic representation of 157 oligosaccharide structures used for FAC analysis.
Fig. S2. Structural formulae of Man9GlcNAc2‐MTX, Manα1‐3Man‐PA and NGA2‐Fmoc.
Fig. S3. Glycoconjugate microarray analysis of DC‐SIGN‐related lectin‐Fc chimeras.
Fig. S4. Binding of DC‐SIGN‐related lectin‐Fc chimeras to Lec1 cells.
Fig. S5. Structural analysis of binding sites of DC‐SIGN‐related lectin CRDs.
Fig. S6. Generation of agalactosylated αAGP.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer‐reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting info item
Acknowledgements
We thank N. Uchiyama, Y. Kubo, J. Murakami and M. Fukumura for technical assistance, Dr H. Matsuzaki for technical support for mass spectrometric analysis and Dr T. Angata for providing the pcDNA/EK‐Fc plasmid. This work was supported, in part, by a grant from the New Energy and Industrial Technology Development Organization (NEDO) and Grants‐in‐Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan (to H.T.).
References
- 1. Soilleux EJ, Morris LS, Leslie G, Chehimi J, Luo Q, Levroney E, Trowsdale J, Montaner LJ, Doms RW, Weissman D et al. (2002) Constitutive and induced expression of DC‐SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J Leukoc Biol 71, 445–457. [PubMed] [Google Scholar]
- 2. Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y & Figdor CG (2000) Identification of DC‐SIGN, a novel dendritic cell‐specific ICAM‐3 receptor that supports primary immune responses. Cell 100, 575–585. [DOI] [PubMed] [Google Scholar]
- 3. Curtis BM, Scharnowske S & Watson AJ (1992) Sequence and expression of a membrane‐associated C‐type lectin that exhibits CD4‐independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc Natl Acad Sci USA 89, 8356–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI & Drickamer K (2004) Structural basis for distinct ligand‐binding and targeting properties of the receptors DC‐SIGN and DC‐SIGNR. Nat Struct Mol Biol 11, 591–598. [DOI] [PubMed] [Google Scholar]
- 5. van Liempt E, Bank CM, Mehta P, Garcia‐Vallejo JJ, Kawar ZS, Geyer R, Alvarez RA, Cummings RD, Kooyk Y & van Die I (2006) Specificity of DC‐SIGN for mannose‐ and fucose‐containing glycans. FEBS Lett 580, 6123–6131. [DOI] [PubMed] [Google Scholar]
- 6. Mitchell DA, Fadden AJ & Drickamer K (2001) A novel mechanism of carbohydrate recognition by the C‐type lectins DC‐SIGN and DC‐SIGNR. Subunit organization and binding to multivalent ligands. J Biol Chem 276, 28939–28945. [DOI] [PubMed] [Google Scholar]
- 7. Bogoevska V, Nollau P, Lucka L, Grunow D, Klampe B, Uotila LM, Samsen A, Gahmberg CG & Wagener C (2007) DC‐SIGN binds ICAM‐3 isolated from peripheral human leukocytes through Lewis x residues. Glycobiology 17, 324–333. [DOI] [PubMed] [Google Scholar]
- 8. van Gisbergen KP, Sanchez‐Hernandez M, Geijtenbeek TB & van Kooyk Y (2005) Neutrophils mediate immune modulation of dendritic cells through glycosylation‐dependent interactions between Mac‐1 and DC‐SIGN. J Exp Med 201, 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR et al. (2000) DC‐SIGN, a dendritic cell‐specific HIV‐1‐binding protein that enhances trans‐infection of T cells. Cell 100, 587–597. [DOI] [PubMed] [Google Scholar]
- 10. Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL & Delgado R (2002) C‐type lectins DC‐SIGN and L‐SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 76, 6841–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tassaneetrithep B, Burgess TH, Granelli‐Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL et al. (2003) DC‐SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med 197, 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez‐Hernandez M, Vandenbroucke‐Grauls CM, Appelmelk B & Van Kooyk Y (2003) Mycobacteria target DC‐SIGN to suppress dendritic cell function. J Exp Med 197, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tailleux L, Schwartz O, Herrmann JL, Pivert E, Jackson M, Amara A, Legres L, Dreher D, Nicod LP, Gluckman JC et al. (2003) DC‐SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med 197, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cambi A, Gijzen K, de Vries JM, Torensma R, Joosten B, Adema GJ, Netea MG, Kullberg BJ, Romani L & Figdor CG (2003) The C‐type lectin DC‐SIGN (CD209) is an antigen‐uptake receptor for Candida albicans on dendritic cells. Eur J Immunol 33, 532–538. [DOI] [PubMed] [Google Scholar]
- 15. Serrano‐Gomez D, Dominguez‐Soto A, Ancochea J, Jimenez‐Heffernan JA, Leal JA & Corbi AL (2004) Dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin mediates binding and internalization of Aspergillus fumigatus conidia by dendritic cells and macrophages. J Immunol 173, 5635–5643. [DOI] [PubMed] [Google Scholar]
- 16. Colmenares M, Puig‐Kroger A, Pello OM, Corbi AL & Rivas L (2002) Dendritic cell (DC)‐specific intercellular adhesion molecule 3 (ICAM‐3)‐grabbing nonintegrin (DC‐SIGN, CD209), a C‐type surface lectin in human DCs, is a receptor for Leishmania amastigotes. J Biol Chem 277, 36766–36769. [DOI] [PubMed] [Google Scholar]
- 17. van Die I, van Vliet SJ, Nyame AK, Cummings RD, Bank CM, Appelmelk B, Geijtenbeek TB & van Kooyk Y (2003) The dendritic cell‐specific C‐type lectin DC‐SIGN is a receptor for Schistosoma mansoni egg antigens and recognizes the glycan antigen Lewis x. Glycobiology 13, 471–478. [DOI] [PubMed] [Google Scholar]
- 18. Steeghs L, van Vliet SJ, Uronen‐Hansson H, van Mourik A, Engering A, Sanchez‐Hernandez M, Klein N, Callard R, van Putten JP, van der Ley P et al. (2006) Neisseria meningitidis expressing lgtB lipopolysaccharide targets DC‐SIGN and modulates dendritic cell function. Cell Microbiol 8, 316–325. [DOI] [PubMed] [Google Scholar]
- 19. van Gisbergen KP, Aarnoudse CA, Meijer GA, Geijtenbeek TB & van Kooyk Y (2005) Dendritic cells recognize tumor‐specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing nonintegrin. Cancer Res 65, 5935–5944. [DOI] [PubMed] [Google Scholar]
- 20. Nonaka M, Ma BY, Murai R, Nakamura N, Baba M, Kawasaki N, Hodohara K, Asano S & Kawasaki T (2008) Glycosylation‐dependent interactions of C‐type lectin DC‐SIGN with colorectal tumor‐associated lewis glycans impair the function and differentiation of monocyte‐derived dendritic cells. J Immunol 180, 3347–3356. [DOI] [PubMed] [Google Scholar]
- 21. Soilleux EJ, Barten R & Trowsdale J (2000) DC‐SIGN; a related gene, DC‐SIGNR; and CD23 form a cluster on 19p13. J Immunol 165, 2937–2942. [DOI] [PubMed] [Google Scholar]
- 22. Bashirova AA, Geijtenbeek TB, van Duijnhoven GC, van Vliet SJ, Eilering JB, Martin MP, Wu L, Martin TD, Viebig N, Knolle PA et al. (2001) A dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin (DC‐SIGN)‐related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV‐1 infection. J Exp Med 193, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Liempt E, Imberty A, Bank CM, Van Vliet SJ, Van Kooyk Y, Geijtenbeek TB & Van Die I (2004) Molecular basis of the differences in binding properties of the highly related C‐type lectins DC‐SIGN and L‐SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J Biol Chem 279, 33161–33167. [DOI] [PubMed] [Google Scholar]
- 24. Snyder GA, Ford J, Torabi‐Parizi P, Arthos JA, Schuck P, Colonna M & Sun PD (2005) Characterization of DC‐SIGN/R interaction with human immunodeficiency virus type 1 gp120 and ICAM molecules favors the receptor’s role as an antigen‐capturing rather than an adhesion receptor. J Virol 79, 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu W, Tang L, Zhang G, Wei H, Cui Y, Guo L, Gou Z, Chen X, Jiang D, Zhu Y et al. (2004) Characterization of a novel C‐type lectin‐like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem 279, 18748–18758. [DOI] [PubMed] [Google Scholar]
- 26. Dominguez‐Soto A, Aragoneses‐Fenoll L, Martin‐Gayo E, Martinez‐Prats L, Colmenares M, Naranjo‐Gomez M, Borras FE, Munoz P, Zubiaur M, Toribio ML et al. (2007) The DC‐SIGN‐related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood 109, 5337–5345. [DOI] [PubMed] [Google Scholar]
- 27. Dominguez‐Soto A, Aragoneses‐Fenoll L, Gomez‐Aguado F, Corcuera MT, Claria J, Garcia‐Monzon C, Bustos M & Corbi AL (2009) The pathogen receptor liver and lymph node sinusoidal endotelial cell C‐type lectin is expressed in human Kupffer cells and regulated by PU.1. Hepatology 49, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gramberg T, Hofmann H, Moller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH et al. (2005) LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 340, 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gramberg T, Soilleux E, Fisch T, Lalor PF, Hofmann H, Wheeldon S, Cotterill A, Wegele A, Winkler T, Adams DH et al. (2008) Interactions of LSECtin and DC‐SIGN/DC‐SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology 373, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Powlesland AS, Fisch T, Taylor ME, Smith DF, Tissot B, Dell A, Pohlmann S & Drickamer K (2008) A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem 283, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tateno H, Nakamura‐Tsuruta S & Hirabayashi J (2007) Frontal affinity chromatography: sugar‐protein interactions. Nat Protoc 2, 2529–2537. [DOI] [PubMed] [Google Scholar]
- 32. Nakamura S, Yagi F, Totani K, Ito Y & Hirabayashi J (2005) Comparative analysis of carbohydrate‐binding properties of two tandem repeat‐type Jacalin‐related lectins, Castanea crenata agglutinin and Cycas revoluta leaf lectin. FEBS J 272, 2784–2799. [DOI] [PubMed] [Google Scholar]
- 33. Nakamura‐Tsuruta S, Kominami J, Kamei M, Koyama Y, Suzuki T, Isemura M & Hirabayashi J (2006) Comparative analysis by frontal affinity chromatography of oligosaccharide specificity of GlcNAc‐binding lectins, Griffonia simplicifolia lectin‐II (GSL‐II) and Boletopsis leucomelas lectin (BLL). J Biochem 140, 285–291. [DOI] [PubMed] [Google Scholar]
- 34. Tateno H, Mori A, Uchiyama N, Yabe R, Iwaki J, Shikanai T, Angata T, Narimatsu H & Hirabayashi J (2008) Glycoconjugate microarray based on an evanescent‐field fluorescence‐assisted detection principle for investigation of glycan‐binding proteins. Glycobiology 18, 789–798. [DOI] [PubMed] [Google Scholar]
- 35. Patnaik SK, Potvin B, Carlsson S, Sturm D, Leffler H & Stanley P (2006) Complex N‐glycans are the major ligands for galectin‐1, ‐3, and ‐8 on Chinese hamster ovary cells. Glycobiology 16, 305–317. [DOI] [PubMed] [Google Scholar]
- 36. Chaney W & Stanley P (1986) Lec1A Chinese hamster ovary cell mutants appear to arise from a structural alteration in N‐acetylglucosaminyltransferase I. J Biol Chem 261, 10551–10557. [PubMed] [Google Scholar]
- 37. Oelmann S, Stanley P & Gerardy‐Schahn R (2001) Point mutations identified in Lec8 Chinese hamster ovary glycosylation mutants that inactivate both the UDP‐galactose and CMP‐sialic acid transporters. J Biol Chem 276, 26291–26300. [DOI] [PubMed] [Google Scholar]
- 38. Feinberg H, Mitchell DA, Drickamer K & Weis WI (2001) Structural basis for selective recognition of oligosaccharides by DC‐SIGN and DC‐SIGNR. Science 294, 2163–2166. [DOI] [PubMed] [Google Scholar]
- 39. Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti‐Renom MA et al. (2009) MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 37, D347–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Appelmelk BJ, van Die I, van Vliet SJ, Vandenbroucke‐Grauls CM, Geijtenbeek TB & van Kooyk Y (2003) Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell‐specific ICAM‐3‐grabbing nonintegrin on dendritic cells. J Immunol 170, 1635–1639. [DOI] [PubMed] [Google Scholar]
- 41. Taylor ME & Drickamer K (2007) Paradigms for glycan‐binding receptors in cell adhesion. Curr Opin Cell Biol 19, 572–577. [DOI] [PubMed] [Google Scholar]
- 42. Geijtenbeek TB, Krooshoop DJ, Bleijs DA, van Vliet SJ, van Duijnhoven GC, Grabovsky V, Alon R, Figdor CG & van Kooyk Y (2000) DC‐SIGN‐ICAM‐2 interaction mediates dendritic cell trafficking. Nat Immunol 1, 353–357. [DOI] [PubMed] [Google Scholar]
- 43. Shimizu H, Ochiai K, Ikenaka K, Mikoshiba K & Hase S (1993) Structures of N‐linked sugar chains expressed mainly in mouse brain. J Biochem 114, 334–338. [DOI] [PubMed] [Google Scholar]
- 44. Greter M, Heppner FL, Lemos MP, Odermatt BM, Goebels N, Laufer T, Noelle RJ & Becher B (2005) Dendritic cells permit immune invasion of the CNS in an animal model of multiple sclerosis. Nat Med 11, 328–334. [DOI] [PubMed] [Google Scholar]
- 45. Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V & Marth JD (2008) The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med 14, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang P, Snyder S, Feng P, Azadi P, Zhang S, Bulgheresi S, Sanderson KE, He J, Klena J & Chen T (2006) Role of N‐acetylglucosamine within core lipopolysaccharide of several species of gram‐negative bacteria in targeting the DC‐SIGN (CD209). J Immunol 177, 4002–4011. [DOI] [PubMed] [Google Scholar]
- 47. Krokhin O, Li Y, Andonov A, Feldmann H, Flick R, Jones S, Stroeher U, Bastien N, Dasuri KV, Cheng K et al. (2003) Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol Cell Proteomics 2, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K & Nabel GJ (2004) pH‐dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC‐SIGN. J Virol 78, 5642–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marzi A, Gramberg T, Simmons G, Moller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B et al. (2004) DC‐SIGN and DC‐SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol 78, 12090–12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuno A, Uchiyama N, Koseki‐Kuno S, Ebe Y, Takashima S, Yamada M & Hirabayashi J (2005) Evanescent‐field fluorescence‐assisted lectin microarray: a new strategy for glycan profiling. Nat Methods 2, 851–856. [DOI] [PubMed] [Google Scholar]
- 51. Angata T, Hingorani R, Varki NM & Varki A (2001) Cloning and characterization of a novel mouse Siglec, mSiglec‐F: differential evolution of the mouse and human (CD33) Siglec‐3‐related gene clusters. J Biol Chem 276, 45128–45136. [DOI] [PubMed] [Google Scholar]
- 52. Tateno H, Uchiyama N, Kuno A, Togayachi A, Sato T, Narimatsu H & Hirabayashi J (2007) A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology 17, 1138–1146. [DOI] [PubMed] [Google Scholar]
- 53. Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS & Mische SM (1999) Mass spectrometric identification of proteins from silver‐stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- 54. Shevchenko A, Wilm M, Vorm O & Mann M (1996) Mass spectrometric sequencing of proteins silver‐stained polyacrylamide gels. Anal Chem 68, 850–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. appK d values (μM) of the DC‐SIGN‐related lectins for PA‐glycans
Fig. S1. Schematic representation of 157 oligosaccharide structures used for FAC analysis.
Fig. S2. Structural formulae of Man9GlcNAc2‐MTX, Manα1‐3Man‐PA and NGA2‐Fmoc.
Fig. S3. Glycoconjugate microarray analysis of DC‐SIGN‐related lectin‐Fc chimeras.
Fig. S4. Binding of DC‐SIGN‐related lectin‐Fc chimeras to Lec1 cells.
Fig. S5. Structural analysis of binding sites of DC‐SIGN‐related lectin CRDs.
Fig. S6. Generation of agalactosylated αAGP.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer‐reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting info item


