Abstract
Endoplasmic reticulum (ER) stress triggers the integrated ER‐stress response (IERSR) that ensures cellular survival of ER stress and represents a primordial form of innate immunity. We investigated the role of IERSR during Leishmania amazonensis infection. Treatment of RAW 264.7 infected macrophages with the ER stress‐inducing agent thapsigargin (TG; 1 μM) increased L. amazonensis infectivity in an IFN1‐α receptor (IFNAR)‐dependent manner. In Western blot assays, we showed that L. amazonensis activates the inositol‐requiring enzyme (IRE1)/ X‐box binding protein (XBP)‐1‐splicing arms of the IERSR in host cells. In chromatin immunoprecipitation (ChIP) assays, we showed an increased occupancy of enhancer and promoter sequences for the Ifnb gene by XBP1 in infected RAW 264.7 cells. Knocking down XBP1 expression by transducing RAW 264.7 cells with the short hairpin XBP1 lentiviral vector significantly reduced the parasite proliferation associated with impaired translocation of phosphorylated IFN regulatory transcription factor (IRF)‐3 to the nucleus and a decrease in IFN1‐β expression. Knocking down XBP1 expression also increased NO concentration, as determined by Griess reaction and reduced the expression of antioxidant genes, such as heme oxygenase (HO)‐1, that protect parasites from oxidative stress. We conclude that L. amazonensis activation of XBP1 plays a critical role in infection by protecting the parasites from oxidative stress and increasing IFN1‐β expression.—Dias‐Teixeira, K. L., Calegari‐Silva, T. C., Dos Santos, G. R. R. M., Vitorino dos Santos, J., Lima, C., Medina, J. M., Aktas, B. H., Lopes, U. G. The integrated endoplasmic reticulum stress response in Leishmania amazonensis macrophage infection: the role of X‐box binding protein 1 transcription factor. FASEB J. 30, 1557–1565 (2016). http://www.fasebj.org
Keywords: ER stress, XBP1, thapsigargin IRE1, IFN1‐β, oxidative stress
Abbreviations
- ChIP
chromatin immunoprecipitation
- ERAD
endoplasmic reticulum‐associated degradation
- FBS
fetal bovine serum
- HEK
human embryonic kidney
- HO
heme oxygenase
- IERSR
integrated endoplasmic reticulum stress response
- IFNAR
IFN1‐α receptor
- IRE
inositol‐requiring enzyme
- IRF
IFN regulatory transcription factor
- KO
knockout
- NAC
N‐acetyl‐L‐cysteine
- PKR
RNA‐dependent protein kinase
- qPCR
quantitative PCR
- ROS
reactive oxygen species
- shRNA
short hairpin RNA
- TG
thapsigargin
- SOD
superoxide dismutase
- sXBP
spliced X‐box binding protein
- uXBP
unspliced XBP
- WT
wild‐type
Leishmaniasis is a family of diseases caused by the protozoan parasite Leishmania. The infection is endemic in >98 countries and territories. Approximately 200,000–400,000 new visceral leishmaniasis cases and 700,000–1,200,000 new cutaneous leishmaniasis cases occur each year worldwide (1). Leishmania amazonensis belongs to the Leishmania mexicana complex and causes localized cutaneous and diffuse cutaneous leishmaniasis (2). These parasites often hijack the host's innate immune response to establish a successful infection, particularly in macrophages. We have shown that L. amazonensis parasites induce IFN1‐β expression via RNA‐dependent protein kinase (PKR) in a TLR2‐dependent manner (3) and that IFN1‐β favors parasite growth. In addition, IFN1‐α receptor (IFNAR) signaling favors L. amazonensis infection in mice (4). Another component of host immune defense subverted by L. amazonensis is the reactive oxygen species (ROS) defense mechanism. We have shown that L. amazonensis suppresses NO production by repressing the iNOS expression via the NF‐kB transcription factor, which is essential for progression of infection (5). Finally, we have demonstrated that L. amazonensis infection induces superoxide dismutase 1 (SOD‐1), which favors parasite growth (3).
The integrated endoplasmic reticulum (ER) stress response (IERSR) is an evolutionarily conserved mechanism that restores cellular homeostasis and ensures cell survival during ER stress. In mammalian cells, IERSR consists of 3 signaling pathways: activation of transcription factor (ATF)‐6, inositol‐requiring enzyme (IRE)‐1, and PKR‐like endoplasmic reticulum kinase (PERK) (6). Activated IRE1, through the endonuclease domain in the cytosol, removes a 26‐nucleotide intron from cytoplasmic unspliced X‐box binding protein (uXBP)‐1 mRNA. The splicing results in a frame shift at the splice junction that also extends the open reading frame and gives rise to an activated form of XBP1 called spliced (s)XBP1. sXBP1 is an active transcription factor that induces transcription of a cluster of genes to expand the folding capacity of ER, promote ER‐associated retrograde transport and degradation (ERAD) of unfolded proteins, and phospholipid synthesis, thereby promoting cell survival (7–9).
XBP1 was characterized in 1990 as a basic leucine zipper domain (bZIP) transcription factor in B cells (10). Activation of XBP1 in response to ER stress is well established. An unexpected finding was that XBP1 was also activated by TLR4 and ‐2 in macrophages in absence of ER stress. Toll‐like receptor—mediated XBP1 activation leads to an increase in IL‐6, IL‐1β, and IFN‐1β production (11). In addition, XBP1 increases IFN‐1β expression in cells under ER stress in a TLR4‐ and ‐3‐dependent manner (12). Activated sXBP1 recruits the IFN regulatory transcription factor (IRF)‐3, and the histone acetylase complex cAMP‐responsive element‐binding (CREB) binding protein (CBP/p300) to the Ifnb enhancer promoter during ER stress, which is further enhanced by cotreatment with LPS (13). Another important role for XBP1 is the prevention of oxidative stress. sXBP1 induces expression of an tioxidant molecules such as SOD‐1 and catalase; consistently XBP1‐deficient cells produce more ROS (14).
It is well known that viral and bacterial infection can activate the IRE1/XBP1‐splicing arms of the IERSR. For example, Chikungunya virus promotes partial splicing of XBP1, although this effect does not appear to result in nuclear translocation (15). Coronavirus similarly favors expression of sXBP1 to protect infected cells from apoptosis (16). Finally, XBP1 can be activated in some bacterial infections. For example, Listeria monocytogenes induces XBP1 splicing, which reduces the number of intracellular bacterial (17). Pseudomonas aeruginosa likewise triggers the IERSR and induces sXBP1 expression, to facilitate survival of the infected host cell (18).
Despite the well‐known role of XBP1 in cellular homeostasis and its documented role in response to viral and bacterial infection, little is known about the role of XBP1 in the pathogenesis of the protozoan parasite. In this study, we found that ER stress renders macrophages more susceptible to L. amazonensis infection in an IFNAR‐dependent manner. L. amazonensis infection induces XBP1 splicing, its nuclear translocation, and its occupancy of the Ifnb enhancer and proximal promoter in macrophages. Knocking down XBP1 expression reduces L. amazonensis infection through reduction of IFN1‐β expression associated with impaired phospho‐IRF3 nuclear translocation. XBP1 knockdown also reduces heme oxygenase (HO)‐1 expression, and increases intracellular NO concentration. Our data demonstrate that L. amazonensis induces the IRE1a/XBP1 branch of the IERSR to escape cellular defense, particularly oxidative stress, and increases expression of IFN1‐β, an important cytokine that favors L. amazonensis infection.
MATERIAL AND METHODS
Cell culture and treatment
The mouse macrophage cell line RAW 264.7 (ATCC: TIB‐71; American Type Culture Collection, Manassas, VA, USA) and human embryonic kidney cell line HEK‐293FT (Thermo Scientific—Life Technologies, Waltham, MA, USA) were cultured in DMEM (Thermo Scientific‐Gibco, Grand Island, NY, USA) supplemented with 10% heat‐inactivated fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in 5% CO2. The HEK‐293FT cells were maintained in the same medium supplemented with 500 μg/ml geneticin. Murine primary macrophages derived from wild‐type (WT) or IFNAR‐knockout (KO) were elicited with thioglycollate, removed from C57BL/6 mice by peritoneal washing, and placed into 24‐well plates (2 × 105 per well; 1 h at 37°C) to allow the cells to adhere. Nonadherent cells were removed by washing with DMEM without serum. The adherent cells were incubated for 24 h in DMEM with 10% FBS for subsequent L. amazonensis infection assays. To determine the infection index in an ER stress condition, we treated the cells with vehicle or 1 μM of TG (Sigma‐Aldrich, St. Louis, MO, USA). To determine the role of NO in the L. amazonensis infection, we treated the cells with 20 mM N‐acetyl‐l‐cysteine (NAC) (Sigma‐Aldrich) for 30 min, infected them with L. amazonensis for 24 h, and then treated them again with 20 mM NAC for an additional 48 h. The cells were treated with 1 μg/ml LPS (Sigma‐Aldrich) as a positive control for NO measure.
Parasites, culture conditions, and infection
L. amazonensis (WHOM/R/75/Josefa) was cultivated in vitro in Schneider Insect Medium (Sigma‐Aldrich) supplemented with 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin. Stationary promastigote forms were transferred to fresh medium at a density of 10 parasites/ml, at 26°C. Macrophages were infected with stationary promastigote forms 5 d after inoculation of the culture at a parasite‐to‐cell ratio of 10:1. The infection index was calculated by multiplying the percentage of infected macrophages by the average number of parasites per macrophage on Giemsa‐stained slides.
Western blot analysis
RAW 264.7 cells were plated in 6‐well polystyrene plates (106) 1 d before infection. Total and nuclear protein extracts were prepared from infected cells 4 and 8 h after infection. For preparation of total cellular extracts, macrophages were washed 3 times with PBS and then lysed in 100 μl lyses buffer [50 mM Tris‐HCl (pH 7.5), 5 mM EDTA, 10 mM EGTA, 50 mM NaF, 20 mM β‐glycerophosphate, 250 mM NaCl, 0.1% Triton X‐100, and 1 μg/ml bovine serum albumin) supplemented with 1:100 dilution of protease inhibitor cocktail (Sigma‐Aldrich). Nuclear extracts were obtained as has been described (5). Total protein (50 μg) and nuclear protein (20 μg) were separated by SDS‐PAGE. The proteins were transferred to PVDF membranes (Bio‐Rad, Hercules, CA, USA) and blotted with antibodies against XBP1 (Santa Cruz Biotechnology, Dallas, TX, USA), lamin A/C (Santa Cruz Biotechnology), α‐tubulin (Santa Cruz Biotechnology), β‐actin (Sigma‐Aldrich), HSP70 (Santa Cruz Biotechnology), or HO‐1 (HO‐1 Assay Designs; Stressgen Biotechnologies, Victoria, BC, Canada) overnight. Membranes were incubated with horseradish peroxidase–conjugated IgG (1:4000) for 1 h in room temperature and washed extensively with Tris‐buffered saline‐Tween (TBST). Antibody antigen complexes were detected by the ECL detection system (GE Health Care, Pittsburgh, PA, USA).
Quantitative real‐time PCR
Total RNA was extracted from RAW 264.7 cells (1 × 106) with RNeasy Plus (Qiagen, Hilden, Germany), and 1 μg total RNA was reverse transcribed into the first‐strand cDNA with ImProm (Promega, Madison WI, USA) and oligo (dT)12–18 primer, according to the manufacturer's instructions. Forward 5′‐CCGCAGCACTCAGACTATG‐3′ and reverse 5′‐GGGTCCAACTTGTCCAGAAT‐3′ pairs were used to amplify uXBP1 mRNA; forward 5′‐CTGAGTCCGCAGCAGGT‐3'and reverse 5′‐AAACATGACAGGGTCCAACTT‐3′ primers were used to amplify sXBP1 mRNA; forward: 5′‐TCCAAGAAAGGACGAACATTC‐3′ and reverse 5′‐TGAGGACATCTCCCACGTCAA‐3′ primers were used to amplify IFN1‐β; and forward 5′‐GTGTCCGTCGTGGATCTG‐3′ and reverse 5′‐TGCTTCACCACCTTCTTGA‐3′ primers were used to amplify GAPDH. GAPDH was used for normalizing all other transcripts. The amplicon specificity was verified by the presence of a single melting temperature peak in dissociation curves run after realtime PCR. Real‐time quantitative (q) PCR was performed with the ABI 7500 detection system (Thermo Scientific‐Applied Biosystems, Foster City, CA, USA) using Go Taq qPCR Master Mix (Promega). All qPCR experiments were performed at least 3 times. All expression ratios were computed via the ΔΔ Ct method.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) analysis was performed according to the Simple ChIP Enzymatic Chromatin IP Kit protocol (Cell Signaling Technology, Danvers, MA, USA). After infection, RAW 264.7 cells (4 × 107) were subjected to the ChIP assay (3). The chromatin was immunoprecipitated with anti‐XBP1 (Santa Cruz Biotechnology) and anti‐[3H]histone Lys‐Ac (Santa Cruz Biotechnology) antibodies at 4° C on a rotator for 16 h. The DNA isolated from immunoprecipitated material was amplified by real‐time PCR with Go Taq qPCR Master Mix (Promega) for the Ifnb proximal promoter: forward 5′‐AACTGAAAGGGAGAACTGAAAG ‐3′ and reverse: 5′‐GCAAGATGAGGCAAAGGC‐3′. The primers used for the +6 kb site were forward 5′‐CGAAGGGAAAGAGAAATGTG‐3′ andreverse‐5′‐CTGGAGGTAACTGGTTGC‐3′.Asacontrol,1/50th of digested input chromatin was similarly processed and analyzed without immunoprecipitation.
Lentiviral transduction
XBP1 expression was knocked down in RAW 264.7 cells by transducing the cells with lentiviral XBP1 shRNA expression vector (19). The mouse XBP1 shRNA plasmids were obtained from the Dana Farber Cancer Institute/Broad Institute shRNA Consortium (Boston, MA, USA). The pMD2.G envelope plasmid, PSPAX2 packaging plasmid, and pLKO plasmid were obtained from Addgene (Cambridge, MA, USA). Three days after transduction, the RAW 264.7 cells were selected with 4 μg/ml puromycin for 7 d.
Nitrite concentration
The Griess reaction was used to analyze the nitrite (NO2 –) content as an indicator of NO production in the supernatant of RAW 264.7 (2 × 105) cultures. The absorbance at 540 nm was measured after incubation of 50 μl of supernatants with 50 μl of the solution containing N‐[naphthyl]ethylenediamine dihydrochloride (NEED; 1 mg/ml), sulfanilamide (10 mg/ml), and 5% phosphoric acid.
Statistical analyses
Data were analyzed by Student's t test for independent samples or 1‐way ANOVA using Prism 5 software (GraphPad, San Diego, CA, USA). Data are expressed as the average of 3 independent experimental, and significant differences are indicated for P < 0.05.
RESULTS
IERSR enhances L. amazonensis infection in a type 1 IFN–dependent manner
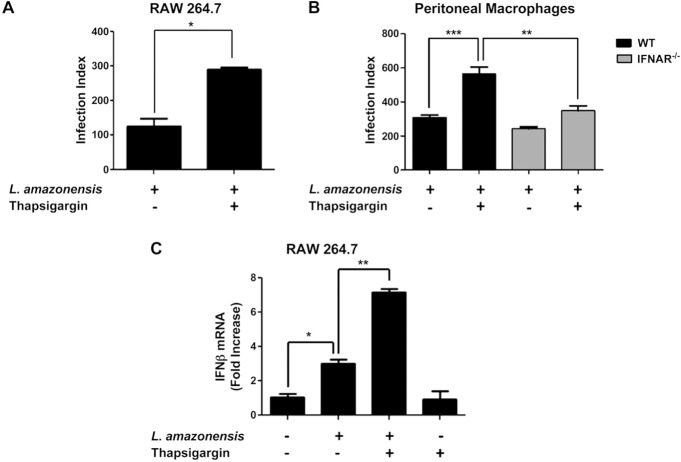
To test the hypothesis that activation of IERSR enhances L. amazonensis infection, we treated RAW 264.7 macrophages for 1 h with thapsigargin (TG), a well‐known inhibitor of sarcoendoreticulum calcium ATPase (SERCA) channels that causes ER stress by depleting ER‐Ca2+ stores, followed by infection with L. amazonensis for 48 h. The infection index showed that activation of ER stress increased the parasite burden compared with that in non treated cells (Fig. 1 A). This finding suggests that 1 or more components of the IERSR enhance parasite infection. To elucidate the molecular effectors of IERSR that enhance L. amazonensis infection, we first investigated the possible involvement of IFN1, because our earlier work had demonstrated that L. amazonensis induces IFN1‐β expression and that this cytokine plays an important role in L. amazonensis growth (3) and because of the known role of XBP1 in IFN‐1b expression (12, 13). Our results indicated that enhanced parasite growth in TG‐treated macrophages was dependent on type I IFNAR (Fig. 1 B). Primary macrophages with IFNAR deletion showed reduced L. amazonensis infection compared with that in WT counterparts after treatment with TG. We also observed a cooperative effect of TG treatment and L. amazonensis infection on IFN1‐β expression in a qPCR assay (Fig. 1 C). Finally, TG treatment did not enhance the infection in TLR2‐KO macrophages (Supplemental Fig. 1), suggesting that this agent enhances parasite infectivity in a TLR2‐dependent manner.
Figure 1. The induction of ER stress by TG favors L. amazonensis infection via IFN1 expression. A) RAW264.7 cells were treated with TG followed 1 h later by infection with L. amazonensis (10:1). After 48 h of infection, the cells were fixed with methanol and stained with Giemsa, and the infection index was calculated (percent L. amazonensis infected cells 3 number of amastigotes per cell). B) WT or IFNAR‐KO mouse—derived macrophages were infected with L. amazonensis as in (A) and the infection index was calculated. C) RAW264.7 cells were treated with TG or vehicle, and 1 h later, half the cells were infected with L. amazonensis (10:1) for 4 h. IFN‐1β transcript levels were measured with qPCR and normalized to GAPDH transcripts. Results are representative of 3 experiments. ∗P < 0.05; ∗∗P < 0.005; ∗∗∗P = 0.0001.
L. amazonensis infection induces XBP1 activation
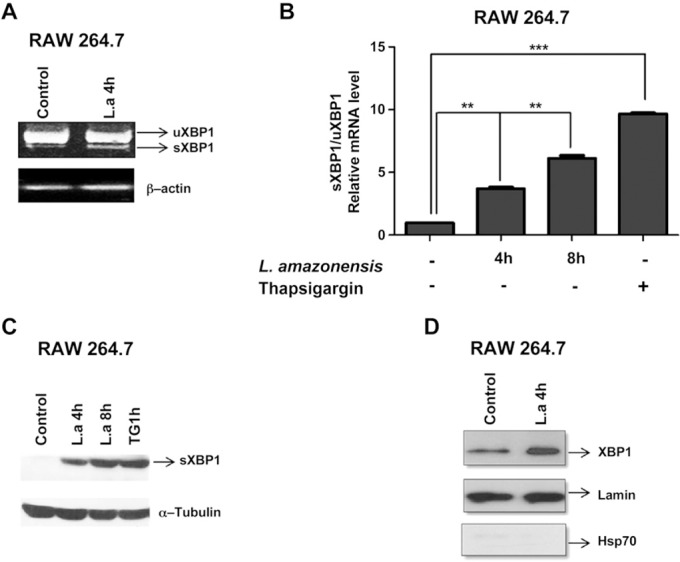
The IREIα/XBP1 arm of the IERSR plays an important role in cell survival during ER stress. We therefore sought to determine whether L. amazonensis activates this pathway to escape cellular defenses. Our semiquantitative and quantitative real‐time PCR assays indicated that L. amazonensis infection induced XBP1 splicing (Fig. 2 A, B). Our data demonstrated further that infection with L. amazonensis induced expression of sXBP‐1 protein (Fig. 2 C) and the nuclear translocation of XBP1 (Fig. 2 D), which is essential for the activity of this transcription factor. These data suggest that the L. amazonensis infection activates XBP1 and that this transcription factor play a role in the parasite infection.
Figure 2. L. amazonensis induces XBP1 splicing and nuclear translocation. A) RAW264.7 cells were infected with L. amazonensis (10:1) for 4 h, and sXBP1 transcript levels were measured with semi‐quantitative PCR and normalized to β‐actin. B) RAW264.7 cells were infected with L. amazonensis (10:1) for 4 or 8 h, and sXBP1 transcript levels were measured with qPCR normalized to uXBP1 transcript levels. RAW 264.7 cells treated with TG for 1 h were used as the control. ∗∗P = 0.0017; ∗∗∗P < 0.0001. C) Total protein extract of RAW 264.7 cells infected with L. amazonensis (10:1) for 4 or 8 h were analyzed by Western blot with anti‐XBP1 or α‐tubulin antibodies. RAW 264.7 cells treated with TG for 1 h were the positive control. D) Nuclear extract of RAW 264.7 cells infected with L. amazonensis (10:1) for 4 h were analyzed by Western blot with antibodies specific for XBP1 or lamin A/C. Purity of nuclear extracts was verified by Western blot analysis with an anti‐HSP70 antibody. Results are representative of 3 experiments.
L. amazonensis infection increases XBP1 occupancy of the Ifnb promoter and +6 kb enhancer site
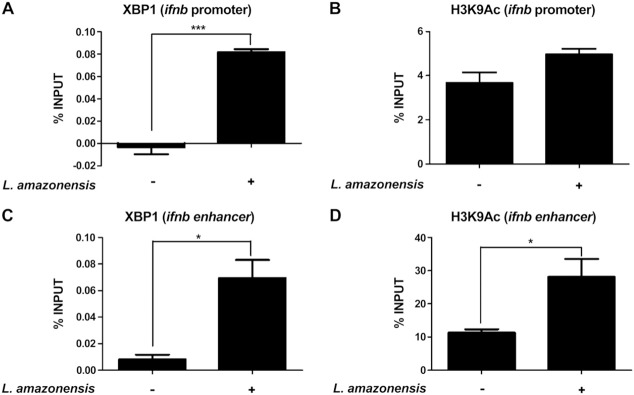
To determine whether and how XBP1 plays a role in the pathogenesis of L. amazonensis we first sought to determine whether L. amazonensis infection induces XBP1 binding to the Ifnb promoter. Our data showed that infection of L. amazonensis increased the occupancy by XBP1 of Ifnb promoter in RAW 264.7 cells compared with that in non‐infected cells (Fig. 3 A) without any effect on the epigenetic modification of [3H]histone (acetylation on Lys residue) associated to the Ifnb proximal promoter (Fig. 3 B). L. amazonensis infection also induced occupancy by XBP1 of the +6 kb enhancer site for Ifnb gene expression (Fig. 3 C), and this effect was associated with an increase in modification of [3H]histone (acetylation on Lys9 residue) (Fig. 3 D). These data suggest that XBP1 plays a critical role in IFN‐1β expression during L. amazonensis infection.
Figure 3. L. amazonensis leads to XBP1 occupancy of the Ifnb promoter and +6 kb enhancer sequences. A, B) RAW 264.7 cells were infected with L. amazonensis (10:1) for 4 h. Relative occupancy of XBP1 in the Ifnb promoter (A) and acetylation of histone 3 at Lys (B) were determined by ChIP followed by qPCR compared with that in the input sample. Occupancy of control IgG ChIP was combined for all experiments. C, D) RAW 264.7 cells were infected with L. amazonensis (10:1) for 4 h. Relative occupancy of XBP1 in the +6 kb enhancer site (C) and acetylation of histone 3 at Lys (D) were determined by ChIP and qPCR by normalizing to that in the input sample. Control IgG ChIP was combined from all experiments and served as the baseline. Results are representative of 3 experiments. ∗P < 0.05; ∗∗∗P= 0.0003.
XBP1 is necessary for L. amazonensis growth in infected macrophages
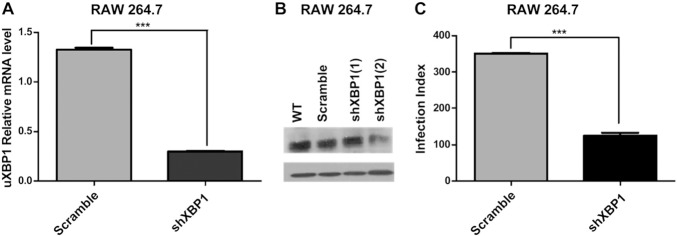
To evaluate the cause—effect relationship between XBP1 activation and the pathogenesis of L. amazonensis, we knocked down expression of XBP1 by transducing mouse macrophages with 3 independent lentiviral short hairpin (sh)RNA expression vectors targeting XBP1 and generated stably transduced cells. We evaluated these transduced cells for XBP1 silencing with a qPCR assay (Fig. 4 A). We further confirmed qPCR results by Western blot analysis of transduced cell extracts (Fig. 4 B). Based on these results, we selected cells transduced with an shXBP1 lentivirus that yielded the highest level of XBP1silencing (~90%) for evaluating the role of XBP1 in L. amazonensis infection. Our infection index assays demonstrated that shXBP1 knockdown cells significantly reduced the L. amazonensis burden when compared with their scrambled shRNA–transduced counterparts similarly infected with this parasite (Fig. 4 C). These data demonstrate clearly that XBP1 plays a critical role in the infection of macrophages by L. amazonensis.
Figure 4. XBP1 knockdown decreases L. amazonensis infection. A) RAW 264.7 macrophages transduced with scrambled (pLKO) or XBP1 (shXBP1) lentiviral shRNA expression vectors were treated with TG for 1 h. XBP1 mRNA levels were determined by qPCR and normalized to uXBP1 transcript levels. ∗∗∗P= 0.0004. B) RAW 264.7 macrophages transduced with lentiviral shRNA expression vectors as described in (A) were treated with TG for 1 h. XBP‐1 or β‐actin expression was detected by Western blot. C) Scrambled shRNA and XBP1 knockdown cells were infected with L. amazonensis (10:1) for 48 h. fixed with methanol, and stained with Giemsa, and the infection index was calculated as in ∗∗∗P < 0.0001. Results are representative of 3 independent experiments.
Promotion of L. amazonensis infection by XBP1 is dependent on type 1 IFN
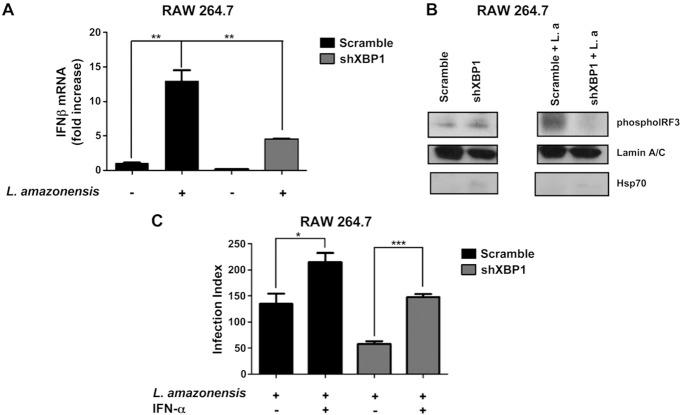
To test the prediction that XBP1 plays a critical role in L. amazonensis‐induced IFN‐1β expression, we determined IFN‐1β expression by qPCR in parasite‐infected, shXBP1‐silenced macrophages (Fig. 5 A). Induction of IFN‐1β mRNA by L. amazonensis infection was impaired in XBP1 knockdown cells. This effect was accompanied by reduced IRF3 nuclear translocation (Fig. 5 B). To provide further support for our contention that activation of XBP1 leads to IFN1 expression, thereby favoring the infection, we added recombinant IFN‐α to parasite‐infected, shXBP‐transduced macrophages and measured the parasite growth. As expected, addition of recombinant IFN‐1α significantly increased the levels of the parasites in XBP1‐silenced macrophages (Fig. 5 C). These data demonstrate conclusively that type 1 IFN signaling is a critical effector of XBP1 in promoting L. amazonensis infection.
Figure 5. XBP1 knockdown decreases expression of IFN‐1β in infected macrophages. A) Scrambled shRNA and XBP1 knockdown cells were infected with L. amazonensis (10:1) for 4 h, and relative IFN‐1β mRNA expression was determined by qPCR. ∗∗P = 0.0011. B) Nuclear extracts of RAW 264.7 cells infected with L. amazonensis (10:1) for 4 h analyzed by Western blot with antibodies specific for phosphorylated IRF3 or lamin A/C. The purity of nuclear extracts was verified by Western blot analysis with an anti‐HSP70 antibody. C) RAW264.7 cells were infected with L. amazonensis (10:1) for 24 h and then treated with recombinant IFN‐α for 48 h. The cells were fixed with methanol and stained with Giemsa, and the infection index was calculated as in ∗P < 0.05; ∗∗∗P = 0.0003. Results are representative of 3 independent experiments.
XBP1 protects macrophages infected with L. amazonensis against oxidative stress
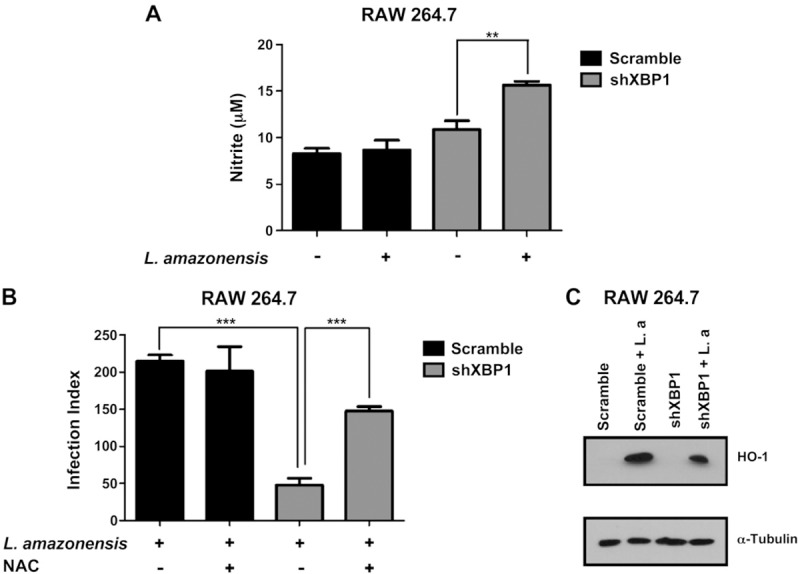
Another well‐established role of XBP1 is the protection of cells against oxidative stress by regulating the expression of antioxidant genes such SOD‐1 and catalase (14). The phagocytosis of parasites during infection creates an oxidative burst and induces the formation of ROS that act as a cellular defense against the parasite (20). However, L. amazonensis parasites subvert these defense mechanisms to survive inside macrophages. To determine whether XBP‐1 plays a role in subversion of host antioxidant defenses, we first determined the NO concentration in shXBP1‐ or scrambled shRNA–transduced cells. In these studies we observed that knocking down XBP1 expression increased nitrite concentration during L. amazonensis infection (Fig. 6 A). To further support the emerging hypothesis that sXBP1 protects cells from oxidative stress during L. amazonensis infection, we added NAC, an antioxidant agent, to shXBP1 cells infected with L. amazonensis and measured parasite growth. As expected, the addition of the antioxidant NAC significantly increased the parasite burden in shXBP1 macrophages (Fig. 6 B). Consistently, the shXBP1 transduced cells also showed a decrease in HO‐1 protein levels compared with scrambled shRNA–transduced cells during L. amazonensis infection. These data further support our hypothesis that XBP1 plays an important role in regulating HO‐1 expression and ROS levels during parasite infection (Fig. 6 C). Taken together, our data demonstrate conclusively that XBP1 plays a critical role in controlling the oxidative stress in L. amazonensis–infected cells, thereby promoting parasite survival and ensuring progression of infection.
Figure 6. XBP1 knockdown increases NO concentration and decreases HO‐1 expression in infected macrophages. A) Scrambled shRNA and XBP1 knockdown cells were infected with L. amazonensis (10:1) for 48 h, and nitrite concentrations were measured in the supernatant by the Griess reaction. ∗∗P = 0.0094. B) RAW264.7 cells were pretreated with NAC followed 30 min later by infection with L. amazonensis (10:1) for 24 h. Cells were treated with NAC for additional 48 h, fixed with methanol, and stained with Giemsa, and the infection index was calculated as in ∗∗∗P < 0.001: left, P= 0.0002; right, P= 0.0007. C) Total protein extract of scrambled shRNA and XBP1 knockdown cells infected with L. amazonensis (10:1) for 8 h were analyzed by Western blot with specific antibodies for HO‐1 or α‐tubulin. Results are representative of 3 independent experiments.
DISCUSSION
Higher eukaryotes have developed an array of immune defense mechanisms for protection against pathogenic organisms. In response, the pathogens have developed sophisticated approaches for evading hosts' defenses. In this never‐ending arms race between the host and the pathogen, pathogens very often hijack host defenses for survival. Leishmania is a pathogenic protozoan that successfully infects ~1 million people a year in Central and South America, Africa, and the Indian subcontinent. Several studies have shown that this parasite may subvert elements of innate immunity for survival and proliferation in mammalian hosts (21, 22). For example L. amazonensis induces IFN‐1β expression in a TLR2‐dependent manner (3), which appears to favor parasite growth. Similarly, IFNAR signaling favors L. amazonensis (4). L. amazonensis also subverts the ROS defense mechanism. Our group also showed that L. amazonensis suppresses NO production through iNOS repression in an NF‐kB transcription‐factor—dependent manner (5). Moreover, L. amazonensis infection induces SOD‐1, thus favoring parasite growth by reducing ROS accumulation (3). How L. amazonensis induces such responses to evade host defenses in a coordinated manner remains poorly understood.
Recent studies suggest that the transcription factor XBP1 is capable of activating many pathways that are obstructed by L. amazonensis. XBP1 is a critical component of the IERSR, which induces the expression of ER chaperones and components of ERAD. Furthermore, sXBP1 is involved in processes not directly related to the IERSR response, such as gluconeogenesis, glycolysis, and lipid metabolism (8). Finally, XBP1 is activated byTLR4 and ‐2 in macrophages, with an increase in IL‐6, IL‐1β, and IFN‐1β production in the absence of overt ER stress (11). XBP1 increases IFN‐1β expression after sequential treatment with TG followed by LPS, in a TLR4‐ and ‐3‐dependent manner (12). Induction of IFN‐1β by sXBP1 and the well‐established role of IFN signaling in L. amazonensis pathogenesis suggest that sXBP1 plays a role in this parasite's evasion of host defenses. Recent demonstration that Plasmodium infection of hepatocytes is associated with ER stress (23) and that XBP1 is critically important for parasite development in these cells (24) supports this hypothesis.
In our experiments, L. amazonensis infection was associated with induction of IERSR, including splicing of XBP1 and the nuclear translocation of this transcription factor (Fig. 2). We demonstrated further that chemical induction of ER stress with TG enhances parasite growth in a type I IFN‐dependent manner (Fig. 1). In further studies, we demonstrated that L. amazonensis infection induces XBP1 binding to the Ifnb promoter with no epigenetic modifications of [3H]histone (acetylation). Furthermore, we observed the occupancy of the Ifnb enhancer site by XBP1, which is associated with Lys9 acetylation of [3H]histone on this site, suggesting that the enhancer site plays a critical role in Ifnb expression during parasite infection (Fig. 3). These studies confirm and expand findings in previous reports that treatment of RAW 264.7 macrophages with TG and LPS induce the occupancy of a newly described enhancer site, +6 kb, for Ifnb gene expression; however, those studies did not report Ifnb promoter occupancy by XBP1 (13). Although suggestive, these results do not prove a cause–effect relationship between activation of XBP1 and parasite infection.
To determine a cause–effect relationship between L. amazonensis infection and XBP1 activation, we undertook functional genomic studies by knocking down XBP‐1 expression by transducing cells with an shXBP1 lentiviral expression vector. Knocking down XBP1 expression in macrophages significantly reduced L. amazonensis growth, demonstrating unequivocally that activation of XBP1 plays a critical role in L. amazonensis infection (Fig. 4). The XBP1 knockdown cells expressed significantly less IFN‐1β, most likely because of impaired translocation of the IRF3 transcription factor to the nucleus. Addition of recombinant type 1 IFNα to XBP1 knockdown cells restored L. amazonensis growth, demonstrating conclusively that XBP1 activation plays a critical role in L. amazonensis infection in a type 1 IFN‐dependent manner (Fig. 5).
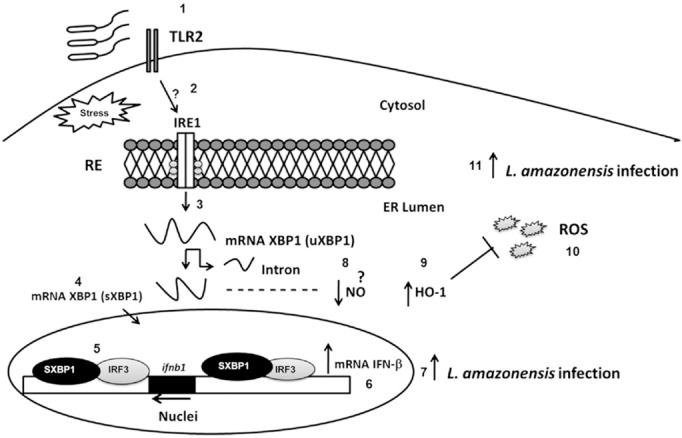
Knocking down XBP1 also restored the oxidative stress response suppressed by L. amazonensis. Specifically, shXBP1 cells infected with L. amazonensis showed high NO concentrations compared to scrambled shRNA‐transduced control cells, demonstrating that host antioxidant defenses are restored in XBP1 knockdown cells. Addition of the antioxidant agent NAC to XBP1 knockdown cells restored L. amazonensis growth, demonstrating conclusively that XBP1 activation plays a critical role in the L. amazonensis infection by preventing host cells from mounting an effective oxidative stress response (Fig. 6). L. amazonensis–infected XBP1 knockdown cells also showed lower HO‐1 expression, suggesting a role for XBP1 in HO‐1 expression (Fig. 6). These data demonstrate that XBP1 plays an important role in regulating oxidative stress during L. amazonensis infection. Down‐regulation of antioxidant defenses by sXBP1 promotes parasite growth and the progression of infection. These data are consistent with previous reports that sXBP1 plays important roles in regulating the oxidative stress response, most likely because of its effects on expression of SOD‐1 and catalase, 2 critical antioxidant genes (14). Figure. 7 explains the role of XBP1 during L. amazonensis infection. The coordinated induction of type IIFN and suppression of oxidative stress response by XBP1 cooperatively facilitate parasite proliferation and play a critical role in L. amazonensis pathogenicity. We showed that TLR2 is necessary for a TG‐induced parasite burden. The data suggest a scenario in which TLR2 and perhaps other TLRs are involved in XBP1 's effect on the infection.
Figure 7. Model depicting the role of XBP1 in L. amazonensis infection. L. amazonensis activates the IRE1α/XBP1 pathway (2) in a TLR2‐dependent manner (1). After XBP1 splicing, which leads to sXBP1 expression (3), this transcription factor translocates to the nucleus (4) and binds to the Ifnb gene promoter and enhancer site (5). XBP1 bound to the enhancer site recruits the transcription factor IRF3 and other chromatin modifiers, leading to an increase in IFN‐1β mRNA expression (6), thereby favoring L. amazonensis infection (7). In addition, through a poorly understood mechanism, XBP1 reduces the NO concentration (8), most likely by suppressing HO‐1 expression (9), leading to an inhibition of ROS production (10) that controls the oxidative stress and favors L. amazonensis infection (11).
Molecular mechanisms by which L. amazonensis activates XBP1 are poorly understood. It is conceivable that the formation of the large parasitophorous vacuole, originated by the fusion of membranes from cell compartments including ER, may activate the IERSR in infected cells.
In summary, our data demonstrate conclusively that XBP1 has a critical role in L. amazonensis infection, via a type I IFN–dependent mechanism. The results highlight the importance of XBP1 in L. amazonensis infection and suggest that IERSR inducers, such as viral coinfections may potentiate the parasite infection. This hypothesis is currently under investigation by our group. The molecular mechanism by which L. amazonensis activates XBP1 and sXBP1 in turn suppresses oxidative stress in parasite‐infected cells should be investigated further.
The authors declare no conflicts of interest.
Supporting information
Supplementary Material
Dias‐Teixeira, K. L. , Calegari‐Silva, T. C. , Dos Santos, G. R. R. M. , Vitorino dos Santos, J. , Lima, C. , Medina, J. M. , Aktas, B. H. , Lopes, U. G. The integrated endoplasmic reticulum stress response in Leishmania amazonensis macrophage infection: the role of X‐box binding protein 1 transcription factor. FASEB J. 30, 1557–1565 (2016). http://www.fasebj.org
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- World Health Organization/Tropical Disease Researchers (2015) Leishmaniasis: situation and trends. In Global Health Observatory (GHO) Data. Accessed September 15, 2015, at http://www.who.int/gho/neglected_diseases/leishmaniasis/en/
- Silveira, F. T. , Lainson, R. , and Corbett, C. E. (2004) Clinical and immunopathological spectrum of American cutaneous leishmaniasis with special reference to the disease in Amazonian Brazil: a review. Mem. Inst. Oswaldo Cruz 99, 239–251 [DOI] [PubMed] [Google Scholar]
- Vivarini, A. C. , Pereira, R. M. , Teixeira, K. L. D. , Calegari‐Silva, T. C. , Bellio, M. , Laurenti, M. D. , Corbett, C. E. P. , Gomes, C. M. C. , Soares, R. P. , Silva, A. M. , Silveira, F. T. , and Lopes, U. G. (2011) Human cutaneous leishmaniasis: interferon‐dependent expression of double‐stranded RNA‐dependent protein kinase (PKR) via TLR2. FASEB J. 25, 4162–4173 [DOI] [PubMed] [Google Scholar]
- Xin, L. , Vargas‐Inchaustegui, D. A. , Raimer, S. S. , Kelly, B. C. , Hu, J. , Zhu, L. , Sun, J. , and Soong, L. (2010) Type I IFN receptor regulates neutrophil functions and innate immunity to Leishmania parasites. J. Immunol. 184, 7047–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calegari‐Silva, T. C. , Pereira, R. M. , De‐Melo, L. D. , Saraiva, E. M. , Soares, D. C. , Bellio, M. , and Lopes, U. G. (2009) NF‐kappaB‐mediated repression of iNOS expression in Leishmania amazonensis macrophage infection. Immunol. Lett. 127, 19–26 [DOI] [PubMed] [Google Scholar]
- Walter, P. , and Ron, D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- Lee, A. H. , Iwakoshi, N. N. , and Glimcher, L. H. (2003) XBP‐1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta‐Alvear, D. , Zhou, Y. , Blais, A. , Tsikitis, M. , Lents, N. H. , Arias, C. , Lennon, C. J. , Kluger, Y. , and Dynlacht, B. D. (2007) XBP1 controls diverse cell type‐ and condition‐specific transcriptional regulatory networks. Mol. Cell 27, 53–66 [DOI] [PubMed] [Google Scholar]
- Hetz, C. , Martinon, F. , Rodriguez, D. , and Glimcher, L. H. (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1a. Physiol. Rev. 91, 1219–1243 [DOI] [PubMed] [Google Scholar]
- Liou, H. C. , Boothby, M. R. , Finn, P. W. , Davidon, R. , Nabavi, N. , Zeleznik‐Le, N. J. , Ting, J. P. , and Glimcher, L. H. (1990) A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247, 1581–1584 [DOI] [PubMed] [Google Scholar]
- Martinon, F. , Chen, X. , Lee, A. H. , and Glimcher, L. H. (2010) TLR activation of the transcription factorXBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. A. , Turner, M. J. , De Lay, M. L. , Klenk, E. I. , Sowders, D. P. , and Colbert, R. A. (2008) Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN‐beta induction via X‐box binding protein 1. Eur.J. Immunol. 38, 1194–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Liu, Y. P. , Sha, H. , Chen, H. , Qi, L. , and Smith, J. A. (2010) XBP‐1 couples endoplasmic reticulum stress to augmented IFN‐beta induction via a cis‐acting enhancer in macrophages. J. Immunol. 185, 2324–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Adachi, M. , Zhao, S. , Hareyama, M. , Koong, A. C. , Luo, D. , Rando, T. A. , Imai, K. , and Shinomura, Y. (2009) Preventing oxidative stress: a new role for XBP1. Cell Death Differ. 16, 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fros, J.J. , Major, L. D. , Scholte, F. E. , Gardner, J. , van Hemert, M.J. , Suhrbier, A. , and Pijlman, G. P. (2015) Chikungunya virus non‐structural protein 2‐mediated host shut‐off disables the unfolded protein response. J. Gen. Virol. 96, 580–589 [DOI] [PubMed] [Google Scholar]
- Fung, T.S. , Liao, Y. , and Liu, D.X. (2014) The endoplasmic reticulum stress sensor IRE1a protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. J. Virol. 88, 12752–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillich, H. , Loose, M. , Zimmer, K. P. , and Chakraborty, T. (2012) Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 14, 949–964 [DOI] [PubMed] [Google Scholar]
- Van 't Wout, E. F. , van Schadewijk, A. , van Boxtel, R. , Dalton, L. E. , Clarke, H. J. , Tommassen, J. , Marciniak, S. J. , and Hiemstra, P. S. (2015) Virulence factors of Pseudomonas aeruginosa induce both the unfolded protein and integrated stress responses in airway epithelial cells. PLOS Pathog. 11, e1004946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Ozel, D. , Qiao, Y. , Harbinski, F. , Chen, L. , Denoyelle, S. , He, X. , Zvereva, N. , Supko, J. G. , Chorev, M. , Halperin, J. A. , and Aktas, B.H. (2011) Chemical genetics identify eIF2α kinase heme‐regulated inhibitor as an anticancer target. Nat. Chem. Biol. 7, 610–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford, J. L. , Neumann, N. F. , and Belosevic, M. (2002) Macrophage‐mediated innate host defense against protozoan parasites. Crit. Rev. Microbiol. 28, 187–248 [DOI] [PubMed] [Google Scholar]
- Gregory, D.J. , and Olivier, M. (2005) Subversion of host cell signalling by the protozoan parasite Leishmania . Parasitology 130(Suppl), S27–S35 [DOI] [PubMed] [Google Scholar]
- Olivier, M. , Gregory, D.J. , and Forget, G. (2005) Subversion mechanisms by which Leishmania parasites can escape the host immune response: a signaling point of view. Clin. Microbiol. Rev. 18, 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio, P. , Zuzarte‐Luís, V. , Ruivo, M. T. , Falkard, B. , Nagaraj, N. , Rooijers, K. , Mann, M. , Mair, G. , Fidock, D. A. , and Mota, M. M. (2015) Parasite‐induced ER stress response in hepatocytes facilitates Plasmodium liver stage infection. EMBO Rep. 16, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky, A. , and Kappe, S. H. (2015) Host ER stress during malaria parasite infection. EMBO Rep. 16, 883–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material