Abstract
Transmissible gastroenteritis virus (TGEV) is a swine enteropathogenic coronavirus that causes significant economic losses in swine industry. Current studies on TGEV internalization mainly focus on viral receptors, but the internalization mechanism is still unclear. In this study, we used single‐virus tracking to obtain the detailed insights into the dynamic events of the TGEV internalization and depict the whole sequential process. We observed that TGEVs could be internalized through clathrin‐ and caveolae‐mediated endocytosis, and the internalization of TGEVs was almost completed within ~2 minutes after TGEVs attached to the cell membrane. Furthermore, the interactions of TGEVs with actin and dynamin 2 in real time during the TGEV internalization were visualized. To our knowledge, this is the first report that single‐virus tracking technique is used to visualize the entire dynamic process of the TGEV internalization: before the TGEV internalization, with the assistance of actin, clathrin, and caveolin 1 would gather around the virus to form the vesicle containing the TGEV, and after ~60 seconds, dynamin 2 would be recruited to promote membrane fission. These results demonstrate that TGEVs enter ST cells via clathrin‐ and caveolae‐mediated endocytic, actin‐dependent, and dynamin 2‐dependent pathways.
Keywords: actin, caveolae, clathrin, dynamin 2, TGEV
Abbreviations
- a.u.
arbitrary units
- BSA
bovine serum albumin
- Cav1
caveolin 1
- CCPs
clathrin‐coated pits
- CCSs
clathrin‐coated structures
- CCVs
clathrin‐coated vesicles
- Clc
clathrin light chain B
- CPE
cytopathic effect
- CPZ
chlorpromazine
- CTB
cholera toxin beta subunit
- CytoD
cytochalasin D
- DIC
differential interference contrast
- DiD
1,1'‐dioctadecyl‐3,3,3',3'‐tetramethylindodicarbocyanine, 4‐chlorobenzenesulfonate salt
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- Dyn2
dynamin 2
- ECL
enhanced chemiluminescence
- EGFP
enhanced green fluorescent protein
- EMCCD
electron‐multiplying charge‐coupled device
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- HRP
horseradish peroxidase
- IFA
immunofluorescence assays
- IgG
immunoglobulin G
- K44A
dominant negative mutant of dynamin 2
- MOI
multiplicity of infection
- MSD
mean squared displacement
- MβCD
methyl‐β‐cyclodextrin
- NS
nonsignificant
- ORF
open reading frame
- pAPN
porcine aminopeptidase N
- PBS
phosphate‐buffered saline
- PFA
paraformaldehyde
- PMSF
phenylmethanesulfonyl fluoride
- P/S/C
penicillin‐streptomycin‐ciprofloxacin
- qPCR
quantitative real‐time PCR
- SDS‐PAGE
sodium dodecyl sulfate‐polyacrylamide gel electrophoresis
- ST cells
swine testis cells
- TCID50
tissue culture infective dose 50
- Tf
transferring
- TGE
transmissible gastroenteritis
- TGEV
transmissible gastroenteritis virus
- TGEV‐N
transmissible gastroenteritis nucleocapsid protein
- WT
wild‐type
1. INTRODUCTION
Transmissible gastroenteritis virus (TGEV) is an alphacoronavirus and the causative agent of transmissible gastroenteritis (TGE).1 TGEV is an enveloped virus with a nonsegmented positive‐stranded RNA genome that encodes four structural proteins: spike (S) glycoprotein, membrane (M) glycoprotein, envelope (E) protein, and nucleocapsid (N) protein.2 TGEVs cause severe vomiting, diarrhea, and rapid dehydration in sucking piglets, resulting in severe economic losses in the swine industry.1 In order to prevent and treat TGE, it is necessary to study the internalization mechanism of TGEVs. The swine testis (ST) cell line, which is highly susceptible to TGEV infection, provides a useful model to study the internalization mechanism of TGEVs.3
At present, it has been known that porcine aminopeptidase N (pAPN), also known as CD13, is a primary receptor of TGEV that can be used by virus for binding and internalizing into host cells,3 and the TGEV S glycoprotein plays a crucial role in the early steps of infection.4 Importantly, it has proved that TGEVs directly interact with pAPN on the cell membrane to form complex that promotes the TGEV internalization by receptor‐mediated endocytosis.5 It is reported that clathrin‐ and caveolae‐mediated endocytosis are both involved in the TGEV internalization.6 And studies have demonstrated that actin can gather around the cell membrane when TGEVs infect host cells and the disruption of actin can inhibit the TGEV internalization.7 In addition, depletion of cholesterol, which is present on viral envelope or cell membrane, can result in a reduction of the TGEV internalization.8 Furthermore, transferrin receptor 1 is a supplementary receptor that assists the TGEV infection and replication.9 Moreover, epidermal growth factor receptor plays a synergistic role with pAPN to promote the TGEV internalization.6
Before the broad application of single‐virus tracking technique, previous studies on virus internalization, which relied on fixed cell assays, could only provide entire information about the average state of the whole population of viruses and reflect discontinuous moments in the highly dynamic process of virus internalization.10 With the development of single‐virus tracking technique, it is feasible to reveal the transient but indispensable events during virus internalization and display a continuous and precise image of the interaction between virus and cellular components. In addition, the detailed kinetic information over the entire process could also be obtained from single‐virus tracking analysis.11, 12 This approach has already been used to reveal the internalization mechanism of many viruses such as influenza A virus, vesicular stomatitis virus, and human immunodeficiency virus.13, 14, 15 Therefore, single‐virus tracking provides previously unavailable information about the dynamics of endocytic processes and is particularly well suited to address how viruses are targeted to endocytic mechanism.
Although it is known that TGEVs enter host cells through endocytosis, but the specific pathway used by TGEVs is still unclear. Using single‐virus tracking technique combined with dual‐color fluorescence imaging, we dissected the dynamic process of the TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis in individual ST cells. During endocytosis, some essential molecules, including actin and dynamin, also play a vital role. Therefore, the dynamic interactions of TGEVs and actin or dynamin also were observed. These experiments demonstrate that TGEVs enter host cells via clathrin‐ and caveolae‐mediated endocytosis. In addition, the TGEV internalization is actin and dynamin 2 dependent.
2. MATERIALS AND METHODS
2.1. Cells, virus, antibodies, and reagents
Swine testis (ST) cells were maintained in our laboratory and cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS, Biological Industries, Kibbutz Beit Haemek, Israel) and 1% penicillin‐streptomycin‐ciprofloxacin (P/S/C, Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2. TGEVs (strain H16) adapted to ST cells were maintained in our laboratory. Rabbit anti‐dynamin 2 monoclonal antibody, rabbit anti‐β‐actin monoclonal antibody, fluorescein isothiocyanate (FITC)‐conjugated anti‐mouse immunoglobulin G (IgG), and horseradish peroxidase (HRP)‐conjugated anti‐rabbit IgG were purchased from HuaBio (Hangzhou, Zhejiang, China). Anti‐TGEV polyclonal antibody was obtained through immunization of mice with inactivated TGEVs. No phenol red DMEM (NPR‐DMEM) was purchased from Procell (Wuhan, Hubei, China). ProLong Live Antifade Reagent and 1,1'‐dioctadecyl‐3,3,3',3'‐tetramethylindodicarbocyanine, 4‐chlorobenzenesulfonate salt (DiD) were purchased from Invitrogen (Carlsbad, CA, USA). The endocytic pathway inhibitors as chlorpromazine (CPZ), nystatin, and cytochalasin D (CytoD) were purchased from Aladdin (Scottsdale, AZ, USA). Pitstop 2, methyl‐β‐cyclodextrin (MβCD), and dynasore were purchased from Sigma‐Aldrich (Saint Louis, MO, USA). CPZ, CytoD, pitstop 2 and dynasore were dissolved in dimethyl sulfoxide (DMSO), and nystatin and MβCD were dissolved in water. Alexa 568‐conjugated human transferrin (Tf‐568) was purchased from Invitrogen (Carlsbad, CA, USA). FITC‐conjugated cholera toxin beta subunit (CTB‐FITC) was purchased from Sigma‐Aldrich (Saint Louis, MO, USA).
2.2. Virus production, purification, and labeling
TGEVs were propagated by inoculating monolayers of ST cells in DMEM supplemented with 2% FBS at a multiplicity of infection (MOI) of 0.1. The cytopathic effect (CPE) of TGEVs was assessed at 2 days postinfection prior to the removal of cell debris by low‐speed centrifugation at 4°C. Two hundred milliliters supernatant were subsequently pelleted with ultracentrifuge (XPN‐100, Beckman, Brea, CA, US) at 30 000 rpm for 3 hours at 4°C. The TGEV pellet was resuspended overnight in HNE buffer (5 mM HEPES, 150 mM NaCl, 0.1 mM EDTA, pH = 7.4) and purified by ultracentrifugation on a sucrose density gradient (20%‐60% [w/v] in HNE buffer) at 30 000 rpm for 3 hours at 4°C.16 The 40%‐60% section containing TGEVs was harvested, aliquoted, and stored at −80°C. For labeling with the lipophilic fluorescent dye, the purified TGEVs were incubated with DiD for 3 hours at room temperature. Unbound dye was removed with 0.2‐μm pore size filters.17 DiD‐labeled TGEVs were stored at −80°C in the dark and avoided repeated freezing and thawing.
2.3. Virus titration
To investigate the influence of DiD on TGEV infectivity, the viral titers were determined by tissue culture infective dose 50 (TCID50) assay. First, 10‐fold serially diluted aliquots of DiD‐labeled and ‐unlabeled TGEVs were used to confluent monolayers of ST cells in 96‐well plates. After 1‐hour infection at 37°C, cells were washed with phosphate‐buffered saline (PBS) three times to remove the noninternalized TGEVs, and subsequently incubated in DMEM supplemented with 2% FBS at 37°C. The CPE was measured after 2 days. Each sample was titrated in triplicate. The TCID50 was calculated using Reed and Munch mathematical analysis.
2.4. Virus infection and drug administration
To test the effects of endocytic pathway inhibitors on TGEV infection, first, ST cells were seeded in 12‐well plates and cultured for 24 hours until they reached 80% confluence. Then, these ST cells were treated with the indicated concentrations of CPZ, nystatin, CytoD, or dynasore for 30 minutes at 37°C, respectively. Afterward, the ST cells were inoculated with TGEVs at a MOI of 2 and incubated for 2 hours in the presence of the corresponding inhibitors. Next, the ST cells were washed once with bicarbonate‐free DMEM buffer (pH = 4.7) and twice with PBS. Finally, the infected ST cells were collected to detect the levels of viral RNA by qPCR assays. All assays were performed in three replicates. The primer sequences used in qPCR assays were listed in Table 1.
Table 1.
Primer sequences for qPCR
| Name | Accession number | Sequence (5′‐3′) |
|---|---|---|
| TGEV‐N | FJ755618 | F: CATGGTGAAGGGCCAACGTA |
| R: GGCAACCCAGACAACTCCAT | ||
| GAPDH | NM_001206359 | F: TCGGAGTGAACGGATTTGGC |
| R: TGCCGTGGGTGGAATCATAC |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.5. Plasmid construction and transfection
For expression vectors, clathrin light chain B (Clc) and β‐actin sequences were cloned into pEGFP‐C3 (where EGFP is enhanced green fluorescent protein) vector (Addgene, Cambridge, MA, USA). Caveolin 1 (Cav1) and dynamin 2 (Dyn2) sequences were cloned into pEGFP‐N1 vector (Addgene, Cambridge, MA, USA) and Cav1 was cloned into pmKO2‐N1 vector (Addgene, Cambridge, MA, USA). The dominant negative mutant (K44A) of Dyn2 was constructed by point mutation. For RNA interference vector, Dyn2 shRNA target sequences were inserted into pLKO.1‐Puro (MiaoLingBio, Wuhan, Hubei, China). The primer sequences used for cloning and RNA interference target sequences are listed in Tables 2 and 3. All constructs were verified by DNA sequencing. ST cells grown to 80% confluence were transfected with 2 μg of plasmid DNA using LipoMax transfection reagent (Sudgen, Bellevue, WA, USA).
Table 2.
Primer sequences used to construct the plasmids
| Name | Accession number | Sequence (5′‐3′) |
|---|---|---|
| β‐Actin | NM_001101 | F: GGGTACCGATGGATGATGATATCGCCGC |
| R: GGGATCCCGCTAGAAGCATTTGCGGTGGAC | ||
| Cav1 | AB451284 | F: GCTCGAGATGTCTGGGGGCAAATACGTAG |
| R: GGAATTCGTATTTCTTTCTGCAAGTTGATGCG | ||
| Clc | NM_001834 | F: GGGTACCGATGGCTGATGACTTTGGCTTC |
| R: GGGATCCCGCTAGCGGGACAGTGGCGT | ||
| Dyn2 | BC054501 | F: GCTCGAGATGGGCAACCGCGGGATG |
| R: GGAATTCGGTCGAGCAGGGATGGCTCG | ||
| K44A | F: AGCGCCGGCGCGAGCTCGGTGCTGGAGAAC | |
| R: CACCGAGCTCGCGCCGGCGCTCTGGCCGCC |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 3.
RNA interference sequences
| Name | Accession number | Sequence (5′‐3′) |
|---|---|---|
| sh‐ctrl | KY765411 | TTCGGAAGCAAGAGGACCTCA |
| sh‐1 | GGATGTCCTGGAGAACAAGCT | |
| sh‐2 | GCTGGTGAAGATGGAGTTTGA | |
| sh‐3 | GGGAGATCAGCTATGCCATTA |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.6. Western blot
Cell samples were washed twice with PBS and lysed with NP‐40 lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitor phenylmethanesulfonyl fluoride (PMSF) (Beyotime, Shanghai, China). After centrifugation at 8000 rpm for 10 minutes at 4°C, the supernatants of the cell lysates were normalized for equal protein content using a BCA protein assay kit (GenStar, Beijing, China). Equal amounts of protein were subjected to sodium dodecyl sulfate‐polyacrylamide gel (10%) electrophoresis (SDS‐PAGE), followed by transferring onto 0.2‐μm nitrocellulose blotting membranes (GE Healthcare Life Sciences, Marlborough, MA, USA), and the membranes were blocked by 5% nonfat milk for 1 hour at room temperature. Afterward, the membranes were incubated overnight at 4°C with primary antibodies specific for each target protein. Then HRP‐conjugated secondary antibodies were used. Treated with enhanced chemiluminescence (ECL) buffer (Vazyme, Nanjing, Jiangsu, China), the bands were quantified with ImageJ software.
2.7. Immunofluorescence assays
First, ST cells were treated with the endocytic pathway inhibitors for 30 minutes, respectively. Subsequently, ST cells were inoculated with TGEVs and incubated at 4°C for 1 hours to bind the TGEVs to ST cells. Afterward, ST cells were incubated at 37°C for 5 minutes and 2 hours, respectively. Then, the ST cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes, permeabilized with 0.2% Triton X‐100 for 10 minutes, and blocked with 1% bovine serum albumin for 1 hour at room temperature. Next, the ST cells were incubated with the TGEV polyclonal antibody at 4°C overnight, and incubated with the FITC‐conjugated secondary antibody for 1 hour at 37°C. Finally, the ST cells and the infected TGEVs were observed by differential interference contrast and fluorescence microscope (Nikon A1, Tokyo, Japan), respectively.
2.8. Endocytic markers internalization assays
ST cells were initially pretreated with the endocytic pathway inhibitors as CPZ, pitstop 2, nystatin, and MβCD for 30 minutes, respectively. Then, ST cells were inoculated with the endocytic markers as 25 µg/mL Tf‐568 and 2 µg/mL CTB‐FITC at 4°C for 1 hour, respectively. Afterward, ST cells were incubated at 37°C for 15 minutes. Untreated ST cells inoculated with the same amount of the endocytic markers were used as the control. Finally, the ST cells were fixed with 4% PFA for 30 minutes for observation.
2.9. Fluorescence co‐localization assays
ST cells were transfected with Clc‐EGFP and Cav1‐EGFP, respectively. After 24 hours, ST cells were inoculated with DiD‐labeled TGEVs and incubated at 4°C for 1 hour to bind the TGEVs to ST cells. Then, ST cells were incubated at 37°C for 5 minutes. Finally, the cells were fixed for observation.
2.10. Live cell imaging and analysis
Live cell imaging was performed on a laser scanning confocal microscope (Nikon A1, Tokyo, Japan) at 37°C. The microscope was equipped with a heated environmental chamber, an electron‐multiplying charge‐coupled device (EMCCD) camera (Nikon, Tokyo, Japan) and a 60×/1.40 oil‐immersed micro‐objective (Nikon, Tokyo, Japan). To image TGEV internalization, first, the ST cells were cultured in glass bottom cell culture dishes (Nest, Wuxi, Jiangsu, China) and grown to 70% confluence prior to transfection. Then, after transfection of 24 hours, DiD‐labeled TGEVs were bound to cells on ice for 30 minutes and subsequently washed twice with prechilled PBS. Next, NPR‐DMEM containing 3 μg/mL trypsin and ProLong Live Antifade Reagent (1:100) was added to the cell culture dish. The microscope stage and the micro‐objective were maintained at 37°C within an environmental chamber, and the air above the cells was supplied with 5% CO2. Finally, dual‐color fluorescence images were recorded with 2‐3‐second intervals for 10 minutes.
Each captured frame was processed using the Gaussian spatial filter to remove the background and the noise. The fluorescence intensity was measured using the NIS‐Elements software. The trajectories were generated by pairing spots in each captured frame according to the similarity of viral fluorescence intensity. The instantaneous velocity was calculated based on the reconstructed trajectory using the ImageJ software. The mean squared displacement (MSD) was calculated via our built program using the Matlab software to distinguish their diffusion modes.18
3. RESULTS
3.1. Performances of DiD‐labeled TGEVs
In order to visualize individual TGEVs in live ST cells, TGEVs were labeled with DiD, a lipophilic fluorescent dye that spontaneously partitions into the TGEV envelope. To compare the infectivity of the DiD‐labeled and unlabeled TGEVs, virus titers were assessed by TCID50. The results in Figure 1A indicate that DiD labeling did not significantly reduce the TGEV infectivity. Moreover, in order to verify the DiD‐labeling performance, DiD‐labeled TGEVs were added to a glass bottom cell culture dish, and a fluorescence image of individual DiD‐labeled TGEVs was recorded as shown in Figure 1B. According to the statistical analysis on the fluorescence intensities of individual DiD‐labeled TGEVs in Figure 1C, it is found that DiD was evenly labeled on the TGEVs. Therefore, the DiD‐labeled TGEVs not only had undifferentiated infectivity compared to the unlabeled TGEVs, but also had evenly distributed fluorescence intensities, both supporting the single‐virus tracking.
Figure 1.
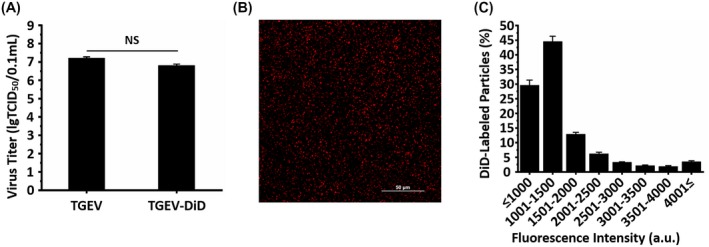
Performances of DiD‐labeled TGEVs. A, Virus titers of unlabeled and DiD‐labeled TGEVs. Data represent the results of three independent experiments implemented in triplicates. NS, nonsignificant. B, Representative image showing DiD‐labeled TGEVs visualized by fluorescence microscopy using a 647‐nm laser. Scale bar, 50 μm. C, Fluorescence intensity histogram of 198 062 individual DiD‐labeled TGEVs. The fluorescence intensity was analyzed using the NIS‐Elements software. a.u., arbitrary units. All graphs show mean ± SD
3.2. Live cell imaging of the TGEV internalization via clathrin‐mediated endocytosis
Clathrin‐mediated endocytosis is a key process in vesicular trafficking that transports a wide range of cargo molecules from the cell surface to the interior.19 Recent studies have shown that clathrin‐mediated endocytosis is associated with the TGEV internalization in the porcine intestinal columnar epithelial cells.6 To further confirm the function of clathrin‐mediated endocytosis on TGEV internalization into ST cells, which are highly permissive to TGEV infection,3 we investigated the effect of chlorpromazine (CPZ) as a cationic amphiphilic inhibitor on TGEV infectivity by Immunofluorescence assay (IFA).20 ST cells were treated with 10 µM CPZ for 30 minutes, and then inoculated with TGEVs for 2 hours at 37°C. Untreated ST cells infected with the same amount of TGEVs were used as the control. Finally, the cells were fixed and the infected TGEVs were stained with anti‐TGEV polyclonal antibody and FITC‐conjugated anti‐mouse IgG for observation. According to Figure 2A, less infected TGEVs were observed in the CPZ treated ST cells compared to the control ST cells, revealing that CPZ significantly but not completely inhibited the TGEV internalization into ST cells. In order to quantify the CPZ effect on TGEV infection, ST cells were treated with different CPZ concentrations of 1, 5, 10, 50 µM, and infected with the same amount of TGEVs. The relative TGEV levels were measured by qPCR in Figure 2B, indicating that higher CPZ concentration significantly reduced the TGEV infectivity. Moreover, pitstop 2, which is a selective inhibitor of clathrin‐mediated endocytosis that acts via blocking ligand access to the clathrin terminal domain,21 significantly inhibited the TGEV internalization (Figure S1A). Transferrin uptake assays indicated that CPZ and pitstop 2 successfully inhibited the clathrin‐mediated endocytosis (Figure S1B). In addition, the co‐localization experiments showed that the TGEV particles were markedly co‐localized with clathrin (Figure S1E,F), suggesting that clathrin‐mediated endocytosis is important for TGEV internalization. The above results further confirm that TGEVs can be internalized into the host cells via clathrin‐mediated endocytosis.
Figure 2.
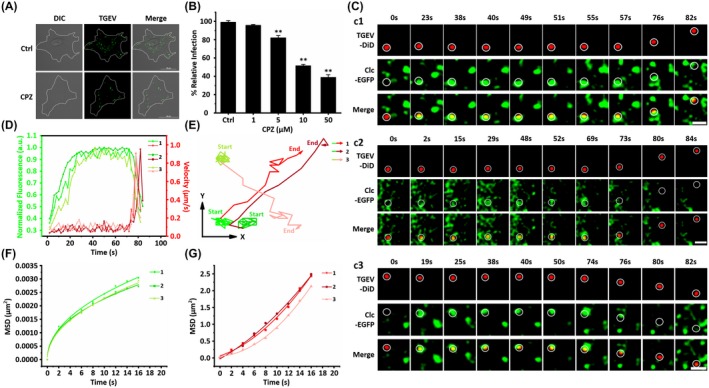
Live cell imaging of the TGEV internalization via clathrin‐mediated endocytosis. A, Representative immunofluorescence images of TGEVs in control and CPZ treated cells. The ST cells were treated or untreated with 10 μM CPZ for 30 min and subsequently inoculated with TGEVs for 2 h, and then the ST cells were fixed and the infected TGEVs were stained with the anti‐TGEV primary antibody and the fluorescent secondary antibody. Scale bar, 20 μm. B, Effects of CPZ with different concentrations on TGEV infection. The ST cells were treated or untreated with the indicated concentrations of CPZ for 30 min and inoculated with TGEVs for 2 h, subsequently, the relative TGEV levels were measured by qPCR. Data present as mean ± SD from three independent experiments. **P < .01 by t test. C, Time‐lapse images of the internalization of three TGEVs via clathrin‐mediated endocytosis shown in Videos S1‐S3. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. D, Fluorescence intensity curves of clathrin at the site of virus and velocity curves of viral diffusion corresponding to (C). E, Trajectories of viral diffusion corresponding to (C). F and G, MSD vs time plots of viral movement. The colors are in accordance with those in (E)
To further investigate the dynamic TGEV internalization via clathrin‐mediated endocytosis, single‐virus tracking was adopted using the fluorescence confocal microscope (Nikon A1, Tokyo, Japan). The TGEVs were labeled with DiD, and a fusion protein of clathrin light chain B and enhanced green fluorescent protein (Clc‐EGFP) was expressed in the ST cells to observe the clathrin‐coated structures (CCSs), which were generated by recruiting clathrin from cytoplasm on the cell membrane. Figure 2C depicts the typical dynamic motions of three individual TGEVs internalizing into the ST cells. All these time‐lapse images show that the TGEVs first moved slowly in local regions; afterward, they significantly accelerated and moved rapidly through large distances. Moreover, there was no clathrin around the TGEVs in the beginning, then the clathrin gradually appeared with the TGEVs, and finally the clathrin around the TGEVs disappeared. Furthermore, we define entry as the time point at which the particle velocity of TGEV begins to increase, that is, the virus leaves the cell membrane and enters the cell. Analyzed from 38 TGEVs entries via clathrin‐mediated endocytosis, the statistical results show that the time duration from the beginning of recruitment of Clc to TGEV entry is 86.42 ± 17.30 seconds. Meanwhile, according to Figures 2C and S2A, the TGEV internalization via clathrin‐mediated endocytosis could be completed within 2 minutes of warm‐up. In order to analyze these dynamic motions in details, both the TGEV velocities and the clathrin fluorescence intensities were extracted as shown in Figure 2D. The TGEVs moved slowly with the velocities of 0.076/0.079/0.108 μm/s, and their velocities rapidly increased to 0.907/0.952/0.912 μm/s. In addition, the clathrin fluorescence signals gradually increased and then nearly plateaued when the TGEV velocities were low, indicating the generation and the gradual maturation of the clathrin‐coated pits (CCPs). The subsequent drastic decline and the eventual disappearance of the clathrin fluorescence signals occurred with the TGEV acceleration, demonstrating that the TGEVs were successfully encapsulated into clathrin‐coated vesicles (CCVs) and internalized into the ST cells, followed by the rapid uncoating of the CCVs only within a few seconds. By analyzing the TGEV velocities and the clathrin fluorescence signals, the TGEV motions can be separated into two stages as shown in Figure 2E. Additionally, the TGEV motions were also studied using mean square displacement (MSD). It is found that the TGEVs first experienced anomalous diffusion during the assembly of CCPs on the cell membrane (Figure 2F); and then the rapid motions of TGEVs were in directed diffusion (Figure 2G), suggesting that the TGEVs were successfully internalized into the ST cells. Both the results in Figure 2E‐G illustrate that the TGEVs first recruited clathrin to form vesicles‐containing viruses and then entered into the ST cells.
These results prove that TGEVs could enter ST cells via clathrin‐mediated endocytosis. In addition, single‐virus tracking reveals that the TGEVs were first attached to the cell membrane and recruited clathrin to form the CCSs; after the CCSs containing TGEVs matured, the TGEVs were successfully internalized into the ST cells, and the CCSs finally disappeared. However, inhibiting clathrin‐mediated endocytosis did not completely block the TGEV internalization, indicating that there are still other infection pathways for TGEVs.
3.3. Live cell imaging of the TGEV internalization via caveolae‐mediated endocytosis
Caveolae‐mediated endocytosis is the most commonly reported clathrin‐independent pathway.22 And caveolae‐mediated endocytosis is also associated with the TGEV internalization in the porcine intestinal columnar epithelial cells.6 To further determine whether TGEVs can be internalized through caveolae‐mediated endocytosis, nystatin as a caveolae‐mediated endocytosis inhibitor was used in IFA.23 ST cells were treated with 10 µM nystatin for 30 minutes and inoculated with TGEVs for 2 hours at 37°C; and those untreated with nystatin were used as control. Afterward, the ST cells were fixed and the internalized TGEVs were stained with anti‐TGEV polyclonal antibody and FITC‐conjugated anti‐mouse IgG for observation as shown in Figure 3A. It is found that nystatin significantly inhibited the TGEV internalization compared to the control group. Furthermore, to quantify the nystatin effect on TGEV infection, ST cells were treated with different nystatin concentrations of 1, 5, 10, 50 µM, and infected with the same amount of TGEVs. The relative TGEV levels measured by qPCR in Figure 3B illustrate that the TGEV infection could be significantly reduced with higher nystatin concentrations. Moreover, methyl‐β‐cyclodextrin (MβCD), which can deplete cholesterol to inhibit caveolae‐mediated endocytosis,24 significantly inhibited the TGEV internalization (Figure S1C). Cholera toxin beta subunit (CTB) uptake assays indicated that nystatin and MβCD successfully inhibited the caveolae‐mediated endocytosis (Figure S1D). In addition, the co‐localization experiments showed that the TGEV particles were markedly co‐localized with Cav1 (Figure S1E,G), suggesting that caveolae‐mediated endocytosis is important for TGEV internalization. These results further prove that the TGEV internalization also depends on caveolae‐mediated endocytosis.
Figure 3.
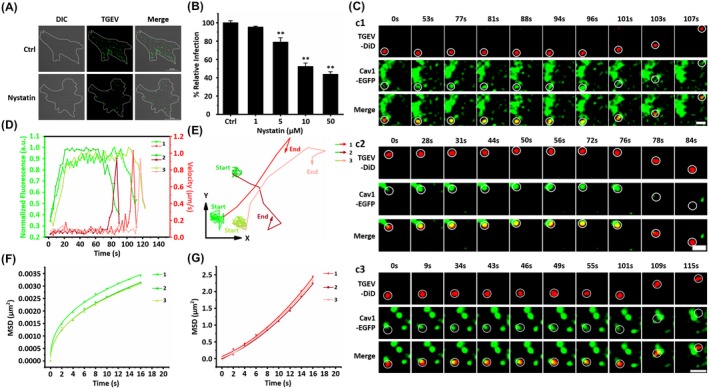
Live cell imaging of the TGEV internalization via caveolae‐mediated endocytosis. A, Representative immunofluorescence images of TGEVs in control and nystatin treated cells. The ST cells were treated or untreated with 10 μM nystatin for 30 min and subsequently inoculated with TGEVs for 2 h, and then the ST cells were fixed and the infected TGEVs were stained with the anti‐TGEV primary antibody and the fluorescent secondary antibody. Scale bar, 20 μm. B, Effects of nystatin with different concentrations on TGEV infection. The ST cells were treated or untreated with the indicated concentrations of nystatin for 30 min and inoculated with TGEVs for 2 h, subsequently, the relative TGEV levels were measured by qPCR. Data present as mean ± SD from three independent experiments. **P < .01 by t test. C, Time‐lapse images of the internalization of three TGEVs via caveolae‐mediated endocytosis shown in Video S4‐S6. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. D, Fluorescence intensity curves of Cav1 at the site of virus and velocity curves of viral diffusion corresponding to (C). E, Trajectories of viral diffusion corresponding to (C). F and G, MSD vs time plots of viral movement. The colors are in accordance with those in (E)
To reveal the dynamics of TGEV internalization via caveolae‐mediated endocytosis, we transfected ST cells with a vector that expressed Cav1 and EGFP fusion protein (Cav1‐EGFP) as well as labeled DiD on the TGEVs, and simultaneously tracked the Cav1‐EGFP and the DiD signals in the live ST cells. Figure 3C depicts the typical dynamics of three TGEVs internalizing into the ST cells. Furthermore, results of 17 TGEV entries via caveolae‐mediated endocytosis show that the average duration from the beginning of recruitment of Cav1 to TGEV entry is 88.56 ± 19.31 seconds, and according to Figures 3C and S2 B, the TGEV internalization via caveolae‐mediated endocytosis could be finished within 2 minutes of warm‐up. Figure 3D shows the TGEV velocities and the caveolae fluorescence signals extracted from Figure 3C. Similar to the case of TGEV internalization via clathrin‐mediated endocytosis, the TGEV velocities were rather low as 0.078/0.065/0.079 μm/s at first but significantly accelerated to 0.937/0.932/0.913 μm/s after 96/75/111 seconds. Moreover, according to the fluorescence signals, caveolae gradually increased and then maintained stable in the low TGEV velocity stage, but finally disappeared after the TGEVs reached fast velocities. Therefore, the TGEV internalization via caveolae‐mediated endocytosis can also be separated into two stages shown in Figure 3E as the trajectories and Figure 3F,G as the MSD. In the first stage, the TGEVs were attached to the cell membrane with rather low speed in anomalous motion mode accompanied with the generation and the gradual maturation of caveolae; while in the second stage, these TGEVs successfully entered into the ST cells with accelerated velocities in directed motion mode but with the disappearance of caveolae.
These results explicitly demonstrate that caveolae‐mediated endocytosis is involved in the TGEV internalization into the ST cells. In addition, single‐virus tracking reveals that the TGEVs gradually recruited Cav1 to form caveolae when TGEVs attached to the cell membrane, after the caveolae containing TGEVs matured, these TGEVs were internalized into the ST cells, followed by the rapid uncoating of caveolae. These results prove that TGEV internalization depends on both clathrin‐ and caveolae‐mediated endocytosis.
3.4. Proportion of TGEV internalized via two endocytic pathways
To determine the proportion of the TGEV internalization via clathrin‐ or caveolae‐mediated endocytosis, ST cells were transfected with two vectors that expressed Clc‐EGFP fusion protein and Cav1‐mKO2 fusion protein. The DiD‐labeled TGEVs were tracked in co‐transfected ST cells. Figure 4A depicts a typical dynamic motion of an internalized TGEV via clathrin‐mediated endocytosis in co‐transfected ST cells. The time‐lapse images show that the TGEV first moved slowly in local regions; afterward, the TGEV significantly accelerated and moved rapidly through large distances. Moreover, there was no clathrin around the TGEV in the beginning, then the clathrin gradually appeared with the TGEV, and finally the clathrin around the TGEVs disappeared, but there was no Cav1 around the TGEV all along. Similarly, Figure 4B depicts a typical dynamic motion of an internalized TGEV via caveolae‐mediated endocytosis in co‐transfected ST cells. The time‐lapse images show that the TGEV first moved slowly in local regions; afterward, the TGEV significantly accelerated and moved rapidly through large distances. Moreover, there was no Cav1 around the TGEV in the beginning, then the Cav1 gradually appeared with the TGEV, and finally the Cav1 around the TGEVs disappeared, but there was no clathrin around the TGEV all along. In addition, we observed 61 successfully internalized TGEVs in 20 co‐transfected ST cells, of which 37 were internalized via clathrin‐mediated endocytosis and 24 via caveolae‐mediated endocytosis. According to the pie chart, about 60.7% of TGEVs could be internalized via clathrin‐mediated endocytosis and 39.3% of TGEVs could be internalized via caveolae‐mediated endocytosis (Figure 4C).
Figure 4.
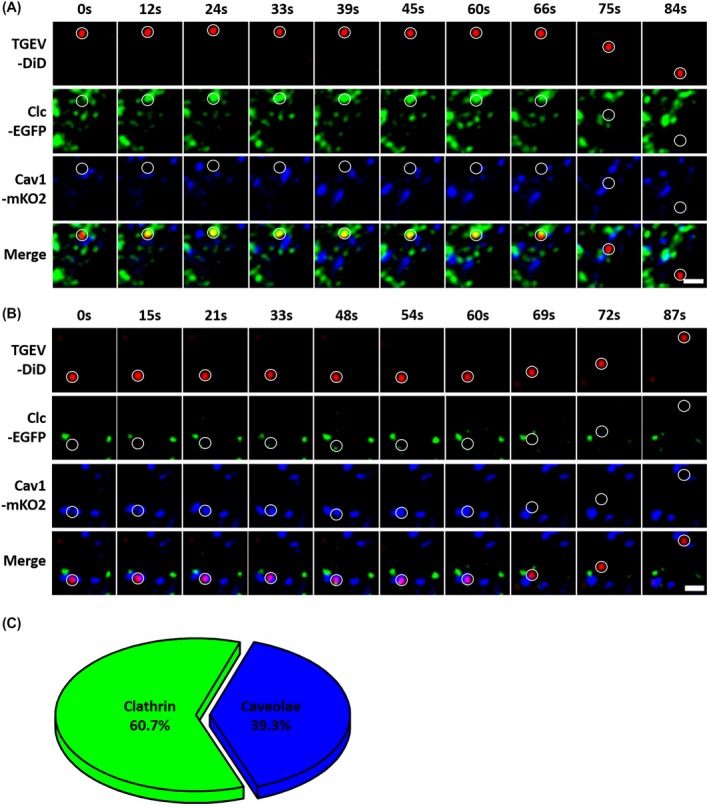
The proportion of the TGEV internalization via clathrin‐ or caveolae‐mediated endocytosis. A, Time‐lapse images of TGEV internalized via clathrin‐mediated endocytosis in co‐transfected ST cells. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. B, Time‐lapse images of TGEV internalized via caveolae‐mediated endocytosis in co‐transfected ST cells. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. C, The pie chart of the proportion of TGEV internalized via clathrin‐ or caveolae‐mediated endocytosis
3.5. Actin promotes TGEV internalization
Besides the TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis, the function of endocytosis‐related proteins on virus internalization was also studied. Actin was proved as an essential element of endocytosis.25 Moreover, it is reported that TGEV infection can stimulate actin gather around the cell membrane,7 but it is still unclear whether the accumulation of actin is directly involved in the TGEV internalization. To further confirm the actin function, CytoD as an actin monomer‐sequestering inhibitor was used in IFA.26 ST cells were treated with 1 μM CytoD and then infected with TGEVs. Untreated ST cells infected with the same amount of TGEVs were used as control. Finally, the ST cells were fixed and the infected TGEVs were stained with anti‐TGEV polyclonal antibody and FITC‐conjugated anti‐mouse IgG for observation. As shown in Figure 5A, the ST cells treated with CytoD exhibited significantly lower TGEV infection levels compared to the control group. Moreover, qPCR was also adopted to study the effects of different CytoD concentrations as 0.1, 0.5, 1, 5 μM on virus infection: with the increase of CytoD concentrations, the relative TGEV levels decreased as shown in Figure 5B. Both results in Figure 5A,B further prove that actin is involved in the TGEV internalization.
Figure 5.
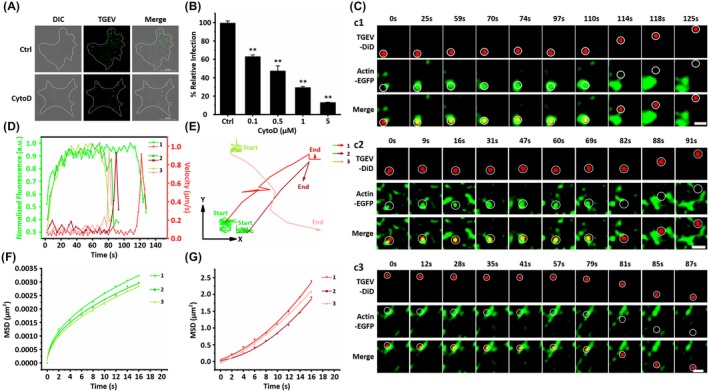
Actin promotes TGEV internalization. A, Representative immunofluorescence images of TGEVs in control and CytoD‐treated cells. The ST cells were treated or untreated with 1 μM CytoD for 30 min and subsequently inoculated with TGEVs for 2 h, and then the ST cells were fixed and the infected TGEVs were stained with the anti‐TGEV primary antibody and the fluorescent secondary antibody. Scale bar, 20 μm. B, Effects of CytoD with different concentrations on TGEV infection. The ST cells were treated or untreated with the indicated concentrations of CytoD for 30 min and inoculated with TGEVs for 2 h, subsequently, the relative TGEV levels were measured by qPCR. Data present as mean ± SD from three independent experiments. **P < .01 by t test. C, Time‐lapse images of the internalization of three TGEVs through actin‐dependent pathway shown in Videos S7‐S9. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. D, Fluorescence intensity curves of actin at the site of virus and velocity curves of viral diffusion corresponding to (C). E, Trajectories of viral diffusion corresponding to (C). F and G, MSD vs time plots of viral movement. The colors are in accordance with those in (E)
Single‐virus tracking was also implemented to investigate the dynamic TGEV‐actin interactions by tracking individual DiD‐labeled TGEVs internalizing into the ST cells expressing actin‐EGFP as shown in Figure 5C. Additionally, Figure 5D shows the TGEV velocities and the actin fluorescence signals extracted from Figure 5C. Actin around the TGEVs gradually increased and maintained with rather high levels when the TGEVs performed rather low velocities as 0.067/0.105/0.099 μm/s; after 114/81/63 seconds, actin around the TGEVs rapidly decreased as TGEVs accelerated to 0.913/0.931/0.956 μm/s, both synchronizing with the TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis. Therefore, it is speculated that actin could assist clathrin and caveolae endocytic pathways to promote the TGEV internalization. Similar to the conditions of TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis, the TGEV motions could also be divided into two stages shown in Figure 5E as the trajectories and Figure 5F and G as the MSD. The first stage describes the attached TGEVs recruited actin with low velocities in anomalous diffusion mode; and the second stage shows the TGEVs successfully entered into the ST cells with high velocities in directed diffusion mode.
In addition, the average duration from the beginning of recruitment of actin to TGEV entry is 87.11 ± 16.37 seconds according to 65 TGEV entries with actin, which is almost consistent with the above observed dynamics of clathrin‐ and caveolae‐mediated endocytosis. The results prove that actin can assist clathrin and caveolae endocytic pathways to promote the TGEV internalization from the beginning of the TGEV internalization.
3.6. Dynamin promotes TGEV internalization
Besides actin, dynamin also plays critical roles in several types of endocytosis.27 To study the dynamin function on TGEV internalization, dynasore as a cell permeable small molecule inhibitor of dynamin was used in IFA.28 ST cells were treated with 100 μM dynasore and infected with TGEVs. Untreated cells infected with the same amount of TGEVs were used as control. Afterward, the ST cells were fixed and the internalized TGEVs were stained with anti‐TGEV polyclonal antibody and FITC‐conjugated anti‐mouse IgG for observation as shown in Figure 6A. The ST cells treated with dynasore were resistant to TGEV internalization compared to those untreated cells. Moreover, the effects of different dynasore concentrations of 10, 50, 100, 500 μM on TGEV infection were also analyzed using qPCR as shown in Figure 6B, indicating that the relative TGEV levels decreased with higher dynasore concentrations. Both results prove that dynamin is involved in the TGEV internalization.
Figure 6.
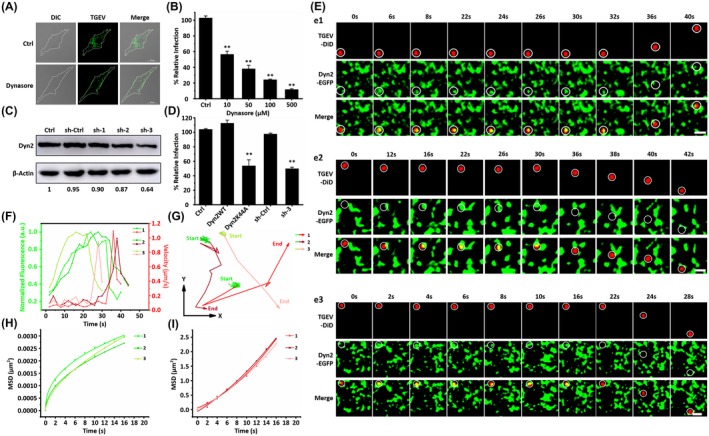
Dynamin 2 promotes TGEV internalization. A, Representative immunofluorescence images of TGEVs in control and dynasore‐treated cells. The ST cells were treated or untreated with 100 μM dynasore for 30 min and subsequently inoculated with TGEVs for 2 h, and then the ST cells were fixed and the infected TGEVs were stained with the anti‐TGEV primary antibody and the fluorescent secondary antibody. Scale bar, 20 μm. B, Effects of dynasore with different concentrations on TGEV infection. The ST cells were treated or untreated with the indicated concentrations of dynasore for 30 min and inoculated with TGEVs for 2 h, subsequently, the relative TGEV levels were measured by qPCR. Data present as mean ± SD from three independent experiments. **P < .01 by t test. C, Analysis on the expression of Dyn2 in the ST cells transfected with shRNAs. The ST cells were transfected with shRNAs for 24 h and subsequently lysed and collected to measure the expression level of Dyn2 by western blot. D, Effects of Dyn2WT, Dyn2K44A, sh‐ctrl, and sh‐3 on TGEV infection. The ST cells were transfected with corresponding plasmids and shRNAs for 24 h and inoculated with TGEVs for 2 h, subsequently, the relative TGEV levels were measured by qPCR. Data present as mean ± SD from three independent experiments. **P < .01 by t test. E, Time‐lapse images of the internalization of three TGEVs through Dyn2‐dependent pathway shown in Videos S10‐S12. Circles indicate the positions of the TGEVs in each panel. Scale bar, 2 μm. F, Fluorescence intensity curves of Dyn2 at the site of virus and velocity curves of viral diffusion corresponding to (E). G, Trajectories of viral diffusion corresponding to (E). H and I, MSD vs time plots of viral movement. The colors are in accordance with those in (G)
To further confirm that dynamin could contribute to the TGEV internalization, we knocked down intracellular dynamin 2 (Dyn2) expression using shRNAs. First, three shRNAs targeting the open reading frame (ORF) of Dyn2 were selected; then, shRNAs were transfected into the ST cells, and finally, the reduction of Dyn2 expression was quantified by immunoblotting with antibody directed against the Dyn2. As shown in Figure 6C, the Dyn2 band of the ST cells transfected with sh‐3 significantly reduced, whereas the actin band did not change; therefore, sh‐3 was used for gene silencing. Moreover, using a qPCR assay, the TGEV infectivity of the ST cells transfected with sh‐3 was reduced to about 50% as shown in Figure 6D. In addition, we also investigated the effect of the Dyn2 dominant negative mutant Dyn2K44A, which can decrease the GTPase activity thus resulting in reduced endocytosis.29 The TGEV infectivity was significantly reduced in the ST cells transfected with the Dyn2K44A vector compared to those ST cells transfected with wild‐type Dyn2 (Dyn2WT) in Figure 6D. All the results in Figure 6A‐D prove that Dyn2 is essential for the TGEV internalization.
In order to reveal the dynamic interactions between TGEVs and Dyn2, the DiD‐labeled TGEVs were tracked in live ST cells expressing Dyn2‐EGFP as shown in Figure 6E. Additionally, the corresponding TGEV velocities and the Dyn2 fluorescence signals were extracted in Figure 6F. At ~30 seconds before TGEV acceleration, the Dyn2 fluorescence signals just began to increase, and then rapidly reduced when the TGEV velocities rapidly increased. According to the trajectories in Figure 6G and the MSD in Figure 6H,I, Dyn2 increased around the TGEVs when they were in anomalous diffusion mode with low velocities, and decreased around the TGEVs when they were in directed diffusion mode with high velocities. Compared to the TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis as well as actin assistance, Dyn2 recruitment occurred in the late stage of the TGEV internalization.
Moreover, the statistical results showed that the average duration from the beginning of recruitment of Dyn2 to TGEV entry is 21.12 ± 4.41 seconds statistically obtained from 51 TGEV entries with Dyn2. Compared to the TGEV internalization via actin assistance, it is proved that Dyn2 can assist clathrin or caveolae endocytic pathways to promote the TGEV internalization in its late stage.
4. DISCUSSION
Although viruses are simple in structures and compositions, viral infections are complex processes. Viruses have evolved sophisticated mechanisms to hijack host cell machinery for initiating productive infections. Merely in internalization, viruses make use of a repertoire of cellular processes that involve hundreds of cellular proteins.30 In this work, we investigated the TGEV internalization in real time using single‐virus tracking, which could observe and analyze the mechanistic and kinetic events during TGEV internalization.
Based on electron microscope, previous study suggested that the TGEVs entered the host cells by endocytosis. Unfortunately, it is still unknown whether the TGEV endocytosis relies on clathrin dependent or independent pathway; however, the observed thickening of the membrane below some attached TGEVs in many cases indicated the involvement of the clathrin‐mediated endocytosis. Using single‐virus tracking, we observed that the TGEVs entered the ST cells via clathrin‐ and caveolae‐mediated endocytosis, and the entire TGEV internalization could be completed within ~2 minutes as shown in Figure 7B and S2. In addition, the endocytic pathway inhibitors CPZ, pitstop 2, nystatin, and MβCD significantly reduced TGEV internalization, further illustrating that the TGEVs could enter the ST cells via clathrin‐ and caveolae‐mediated endocytosis. According to the previous study, it was reported that the TGEV internalization reduced in the porcine intestinal columnar epithelial cells treated with shRNAs that targeting clathrin or caveolin; and the TGEVs could be clearly co‐localized with endocytic transferrin and cholera toxin beta subunit.6 Therefore, our conclusion obtained from single‐virus tracking that the TGEVs utilized clathrin‐ and caveolae‐mediated endocytosis for the internalization is in agreement with the previous studies.
Figure 7.
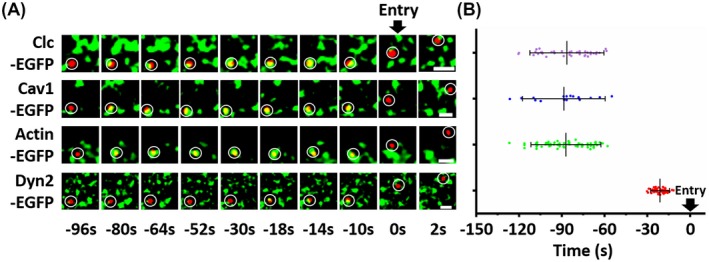
Average time duration from the beginning of recruitment of endocytic related proteins to TGEV entry. A, Four events of the TGEV internalization through clathrin‐ and caveolae‐mediated endocytosis, actin‐ and Dyn2‐dependent pathway, respectively. The time of each frame in the four events is unified. B, Statistical analysis on the average duration from the beginning of recruitment of endocytic related proteins to TGEV entry. Only the internalized TGEVs were used in the analysis. Each dot represents an individual TGEV. The numbers of virus particles are 38, 17, 65, and 51 corresponding to clathrin‐ and caveolae‐mediated endocytosis, actin‐ and Dyn2‐dependent pathways, respectively. Data present as mean ± SD
In the past decades, it has been proved that many viruses can be internalized via clathrin‐ and caveolae‐mediated endocytosis by different methods. By using specific inhibitors, RNA interference, and dominant negative mutants, it has been indicated that hepatitis C virus, African swine fever virus, hepatitis E virus, and porcine hemagglutinating encephalomyelitis virus enter their host cells via clathrin‐mediated endocytosis,31, 32, 33, 34 while grass carp reovirus and porcine sapelovirus enter their host cells via caveolae‐mediated endocytosis.35, 36 In addition to these conventional methods, single‐virus tracking provides many new insights into the study of the virus endocytic mechanism. Using this method, it has been successfully revealed the dynamic process of internalization of many viruses as influenza virus, dengue virus, vesicular stomatitis virus, and infectious hematopoietic necrosis virus.13, 14, 18, 37, 38 By comparing our data with those obtained on other viruses mentioned above, we found that the dynamic of TGEV internalization is consistent with these viruses. These viruses first move slowly in local regions; afterward, they significantly accelerate and move rapidly through large distances. In addition, the internalizations of these viruses all experience two diffusion modes, first anomalous diffusion mode and then directed diffusion mode.
Increasing evidences from different cell types reveal that actin plays an active and critical role during endocytosis. It was reported that actin was required for clathrin‐coated vesicle formation, and membrane tension determined the actin dependence of clathrin‐coated assembly.39, 40 Moreover, using single‐virus tracking for vesicular stomatitis virus, it was proved that clathrin‐coated vesicles depended upon the actin machinery during virus internalization.14 Furthermore, caveolae‐mediated endocytosis was proved to be dependent on the actin cytoskeleton, revealing that caveolae was dynamic structure that could be internalized into the cell, and the kinase activity regulated internalization relied on intact actin network.41 Additionally, using single‐virus tracking for simian virus 40, actin polymerization was required in virus internalization via caveolae‐mediated endocytosis.42 In this work, using single‐virus tracking, it is observed that the TGEVs recruited actin, and this process synchronized with the TGEV internalization via clathrin‐ and caveolae‐mediated endocytosis (Figure 7), suggesting that actin could assist endocytic pathways to promote the TGEV internalization from the beginning of the TGEV internalization. In addition, the actin monomer‐sequestering inhibitor CytoD significantly reduced TGEV internalization, further confirming that actin is important for the TGEV internalization.
The role of dynamin in endocytosis is considered as scission factor that pinches off membrane invaginations.27 Numerous studies have shown that most viruses utilize the dynamin‐dependent clathrin‐ and caveolae‐mediated endocytosis. Simian virus 40 entered CV‐1 cells via caveolae‐ and dynamin‐dependent endocytosis, and human immunodeficiency virus entered TZM‐bl cells via clathrin‐ and dynamin‐dependent endocytosis.42, 43 Using single‐virus tracking, we found that the TGEVs started to recruit Dyn2 often at 20 seconds before the TGEVs entered the ST cells as shown in Figure 7, suggesting that Dyn2 had the function of pinching off the formed vesicles containing TGEVs from the cell membrane. Furthermore, the inhibition, RNA interference, and dominant negative mutant assays further prove the Dyn2 function on the TGEV internalization in the late stage.
To our best knowledge, it is the first time that TGEV internalization was directly observed using single‐virus tracking. Moreover, the kinetics during TGEV internalization were analyzed in single‐virus level as shown in Figure 8. After the TGEV attaches to the cell membrane, the TGEV first recruits clathrin or Cav1 to form CCV or caveolae with the assistance of actin. When the vesicle containing the TGEV matures, Dyn2 is recruited in the late stage of TGEV internalization to induce membrane fission and pinch off the endocytic vesicle from the cell membrane. Finally, uncoating disassembles the endocytic proteins, and the nascent vesicle containing TGEV is released in the host cell. This direct visualization on TGEV internalization in ST cells provides insights into TGEV infection, and may help the development of inhibitors to block the TGEV internalization into ST cells for preventing and treating TGE.
Figure 8.
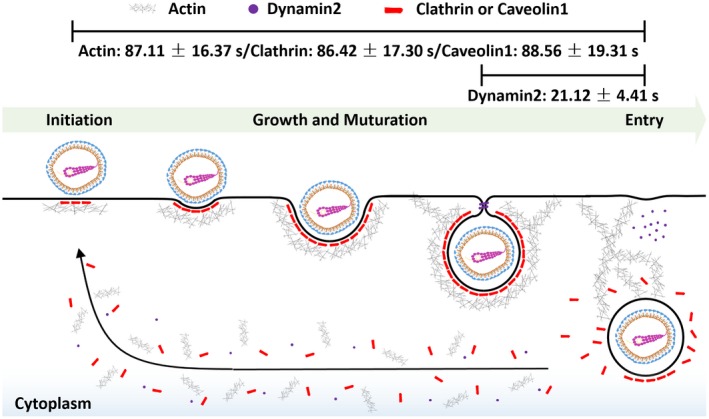
Scheme of TGEV internalization in the ST cell. TGEV enters ST cells via clathrin‐ and caveolae‐mediated endocytosis. The vesicle containing the TGEV is first initiated at the binding site of virus and grows with the assistance of actin. Dyn2 is subsequently recruited in the late stage of TGEV internalization to pinch off the vesicle from the cell membrane. Finally, virus is successfully internalized, and the endocytic proteins rapidly disassemble
CONFLICT OF INTEREST
The authors declared that they have no conflicts of interest to this work.
AUTHOR CONTRIBUTIONS
J. Wang, Y.‐Y. Li, and F. Liu conceived and designed the study; S.‐Y. Wang contributed analysis tools; J. Wang performed the experiments and analyzed the data; J. Wang and S.‐Y. Wang produced the Figures, compiled the tables; and J. Wang, S.‐Y. Wang, and F. Liu edited and revised the manuscript.
Supporting information
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program (2018YFD0500100); National Natural Science Foundation of China (31870154, 31522056, 61705092); Natural Science Foundation of Jiangsu Province of China (BK20170194); Jiangsu Key Research and Development Program (BE2018709); and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Wang J, Li Y, Wang S, Liu F. Dynamics of transmissible gastroenteritis virus internalization unraveled by single‐virus tracking in live cells. The FASEB Journal. 2020;34:4653–4669. 10.1096/fj.201902455R
REFERENCES
- 1. Laude H, Rasschaert D, Delmas B, Godet M, Gelfi J, Charley B. Molecular biology of transmissible gastroenteritis virus. Vet Microbiol. 1990;23:147‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salanueva IJ, Carrascosa JL, Risco C. Structural maturation of the transmissible gastroenteritis coronavirus. J Virol. 1999;73:7952‐7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delmas B, Gelfi J, L'Haridon R, et al. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwegmann‐Wessels C, Bauer S, Winter C, Enjuanes L, Laude H, Herrler G. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol J. 2011;8:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen GH, Delmas B, Besnardeau L, et al. The coronavirus transmissible gastroenteritis virus causes infection after receptor‐mediated endocytosis and acid‐dependent fusion with an intracellular compartment. J Virol. 1998;72:527‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu W, Zhang S, Shen Y, Yang Q. Epidermal growth factor receptor is a co‐factor for transmissible gastroenteritis virus entry. Virology. 2018;521:33‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu W, Zhu L, Yang X, Lin J, Yang Q. The epidermal growth factor receptor regulates cofilin activity and promotes transmissible gastroenteritis virus entry into intestinal epithelial cells. Oncotarget. 2016;7:12206‐12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ren X, Glende J, Yin J, Schwegmann‐Wessels C, Herrler G. Importance of cholesterol for infection of cells by transmissible gastroenteritis virus. Virus Res. 2008;137:220‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang S, Hu W, Yuan L, Yang Q. Transferrin receptor 1 is a supplementary receptor that assists transmissible gastroenteritis virus entry into porcine intestinal epithelium. Cell Commun Signal. 2018;16:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun E, He J, Zhuang X. Live cell imaging of viral entry. Curr Opin Virol. 2013;3:34‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brandenburg B, Zhuang X. Virus trafficking—learning from single‐virus tracking. Nat Rev Microbiol. 2007;5:197‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rust MJ, Lakadamyali M, Brandenburg B, Zhuang X. Single‐virus tracking in live cells. Cold Spring Harb Protoc. 2011;2011:p065623. [DOI] [PubMed] [Google Scholar]
- 13. Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Q, Li W, Yin W, et al. Single‐particle tracking of human immunodeficiency virus type 1 productive entry into human primary macrophages. ACS Nano. 2017;11:3890‐3903. [DOI] [PubMed] [Google Scholar]
- 16. Hoornweg TE, van Duijl‐Richter MKS, Ayala Nuñez NV, Albulescu IC, van Hemert MJ, Smit JM. Dynamics of chikungunya virus cell entry unraveled by single‐virus tracking in living cells. J Virol. 2016;90:4745‐4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A. 2003;100:9280‐9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun E, Liu A, Zhang Z, Liu S, Tian Z, Pang D. Real‐time dissection of distinct dynamin‐dependent endocytic routes of influenza A virus by quantum dot‐based single‐virus tracking. ACS Nano. 2017;11:4395‐4406. [DOI] [PubMed] [Google Scholar]
- 19. McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin‐mediated endocytosis. Nat Rev Mol Cell Bio. 2011;12:517‐533. [DOI] [PubMed] [Google Scholar]
- 20. Wang LH, Rothberg KG, Anderson RG. Mis‐assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dutta D, Williamson CD, Cole NB, Donaldson JG. Pitstop 2 is a potent inhibitor of clathrin‐independent endocytosis. PLoS One. 2012;7:e45799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lajoie P, Nabi IR. Lipid rafts, caveolae, and their endocytosis. Int Rev Cel Mol Bio. 2010;282:135‐163. [DOI] [PubMed] [Google Scholar]
- 23. Rothberg KG, Ying YS, Kamen BA, Anderson RG. Cholesterol controls the clustering of the glycophospholipid‐anchored membrane receptor for 5‐methyltetrahydrofolate. J Cell Biol. 1990;111:2931‐2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Danthi P, Chow M. Cholesterol removal by methyl‐beta‐cyclodextrin inhibits poliovirus entry. J Virol. 2004;78:33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661‐686. [DOI] [PubMed] [Google Scholar]
- 26. Sampath P, Pollard TD. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991;30:1973‐1980. [DOI] [PubMed] [Google Scholar]
- 27. Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell‐permeable inhibitor of dynamin. Dev Cell. 2006;10:839‐850. [DOI] [PubMed] [Google Scholar]
- 29. Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blanchard E, Belouzard S, Goueslain L, et al. Hepatitis C virus entry depends on clathrin‐mediated endocytosis. J Virol. 2006;80:6964‐6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hernaez B, Alonso C. Dynamin‐ and clathrin‐dependent endocytosis in African swine fever virus entry. J Virol. 2009;84:2100‐2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holla P, Ahmad I, Ahmed Z, Jameel S. Hepatitis E virus enters liver cells through a dynamin‐2, clathrin and membrane cholesterol‐dependent pathway. Traffic. 2015;16:398‐416. [DOI] [PubMed] [Google Scholar]
- 34. Li Z, Zhao K, Lan Y, et al. Porcine hemagglutinating encephalomyelitis virus enters Neuro‐2a cells via clathrin‐mediated endocytosis in a Rab5‐, cholesterol‐, and pH‐dependent manner. J Virol. 2017;91:e01083‐e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang F, Guo H, Zhang J, Chen Q, Fang Q. Identification of the caveolae/raft‐mediated endocytosis as the primary entry pathway for aquareovirus. Virology. 2018;513:195‐207. [DOI] [PubMed] [Google Scholar]
- 36. Zhao T, Cui L, Yu X, Zhang Z, Shen X, Hua X. Porcine sapelovirus enters PK‐15 cells via caveolae‐dependent endocytosis and requires Rab7 and Rab11. Virology. 2019;529:160‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Schaar HM, Rust MJ, Waarts BL, et al. Characterization of the early events in dengue virus cell entry by biochemical assays and single‐virus tracking. J Virol. 2007;81:12019‐12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu H, Liu Y, Liu S, Pang DW, Xiao G. Clathrin‐mediated endocytosis in living host cells visualized through quantum dot labeling of infectious hematopoietic necrosis virus. J Virol. 2011;85:6252‐6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yarar D, Waterman‐Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin‐mediated endocytosis. Mol Biol Cell. 2005;16:964‐975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boulant S, Kural C, Zeeh J, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin‐mediated endocytosis. Nat Cell Biol. 2011;13:1124‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parton RG, Joggerst B, Simons K. Regulated internalization of caveolae. J Cell Biol. 1994;127:1199‐1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pelkmans L, Püntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40‐induced internalization of caveolae. Science. 2002;296:535‐539. [DOI] [PubMed] [Google Scholar]
- 43. Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin‐dependent fusion with endosomes. Cell. 2009;137:433‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


