Abstract
The intestinal microbiota is essential for nutrient acquisition, immune development, and exclusion of invading pathogens. The upper respiratory tract (URT) microbiota is less well studied and does not appear to abide by many of the paradigms of the gastrointestinal tract. Decades of carriage studies in children have demonstrated that microbe–microbe competition and collusion occurs in the URT. Whether colonization with common pathogens (e.g., Staphylococcus aureus and Streptococcus pneumoniae) alters immune development or susceptibility to respiratory conditions is just beginning to be understood. Herein, we discuss the biogeography of the URT microbiota, the succession and evolution of the microbiota through the life course, and discuss the evidence for microbe–microbe interactions in colonization and infection.
Keywords: Bacteria–bacteria interactions, microbiota, upper respiratory tract
Abbreviations
COPD, chronic obstructive pulmonary disease
Esp, extracellular serine protease
MRSA, methicillin‐resistant Staphylococcus aureus
MSSA, methicillin‐sensitive Staphylococcus aureus
OTUs, operational taxonomic units
TNF, tumor necrosis factor
URT, upper respiratory tract
The study of microbiota began with the discoveries by Antoine van Leeuwenhoek centuries ago with the identification of different bacteria from dental plaque. Early observations indicated that microbial communities influenced the development of the intestinal tract, including the stimulation of antibody production by potential beneficial bacteria, or probiotics 1. More recent studies have explored the complex, dynamic interactions and influences by external factors such as diet and environment on the gastrointestinal microbiota and its implications on health 2, 3. Some of the earliest studies also indicated that there were general patterns of succession which influenced how communities form and are altered via invasion of new members or destabilization (e.g., by antibiotics) 4, 5. We now know that the microbiota contributes to nutrient production and acquisition, as well as resistance to colonization by pathogens. Interestingly studies in germ‐free mice have demonstrated that the microbiota is required for normal physiological development of the organs such as the gut, lung, and brain 6. The gut microbiota drives the development of the mucosal immune system, including the initiation of secretion of immunoglobulins 7, the development of T‐cell populations 8, 9, and stimulating the production of antimicrobial peptides 10. The respiratory tract microbiota is less well studied, but it is believed that it also contributes to local immune education and the development of respiratory diseases, including asthma and allergy.
The communities of the upper respiratory tract (URT) microbiota (i.e., sinuses, nares, oropharyngeal) are distinct from one another and have unique patterns of colonization and succession. From early infancy, the URT is colonized by several pathogens, including Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae. These and other bacteria can also be referred to as ‘pathobionts’, which generally colonize the host asymptomatically, but have the potential to cause disease if they spread from the site of colonization 11. In fact, some pathobionts such as the Streptococcus anginosus group are believed to be the major cause of complicated lung infections such as pleural empyema 12, 13. The composition of the URT microbiota, including the presence of pathogens, changes rapidly during infancy, the process of which is called ecological succession. Ecological succession describes changes within the microbial community over time, which is influenced by both changing habitats (e.g., the developing human tissue) and exposure to new community members (i.e., colonizing bacteria). The progression of the URT microbiota has been shown to impact airway disease later in childhood 14. Understanding how microbial succession and colonization increases or reduces susceptibility to respiratory diseases such as asthma and allergy and how it promotes or prevents invasion by pathogens may inform novel interventions. This review describes the current understanding of how the URT microbiota influences health, infection, and inflammation.
Biogeography of the URT microbiota
The major functions of the URT are to heat, humidify, and filter air through several compartments (i.e., nares/vestibules, nasal cavity/meatus, nasopharynx, oropharynx, and pharynx) before the air reaches the lungs 15. These different compartments allow for colonization of pathogens, as differences in temperature, mucus secretion, and relative oxygen concentration throughout the URT may regulate colonization, as it does in the gastrointestinal tract 16, 17. The vestibules contain stratified squamous epithelial cells that transition into pseudostratified epithelial cells, as well as hairs that are important for trapping large particulate matter 15. Inspired air is heated and humidified within the turbinates of the nasal cavity, which also dehumidifies expired air. The nasal cavity has three turbinates that increase the surface area of the nose, and heat airflow entering the nasopharynx 18. Glands and goblet cells are important for secreting the hydrated mucus layer, which contributes to humidifying incoming air and traps microparticles and microbes entering the URT from the environment. Finally, the humidified air enters the nasopharynx, which moves into the lungs for respiration within the alveolar space. Ciliated epithelial cells direct mucus flow toward the esophagus to remove trapped particles, which is known as mucociliary clearance 19. Thus, the URT protects the lower airways from temperature or particle‐induced cellular damage. Air inspired into the URT impacts the inner ear since the Eustachian tubes contain air that is required for equalization of air pressure near the eardrum. Infections of the Eustachian tubes, known as acute or chronic otitis media, occur when pathogens evade physiological and immunological barriers of the URT 20, 21. Characterizing the microbiota of the deeper regions of the nasal cavity, including the sinuses, is challenging in routine clinical diagnostics; however, the more accessible regions of the nasal cavity have distinct microbiota profiles within an individual 22.
While the URT microbiota has long been recognized as a reservoir of pathogens, the lower respiratory tract has been thought to be sterile in healthy individuals. Studies from the 1960's failed to recover viable bacteria in the lower airways of healthy individuals in bronchial swabs 23. When bacteria or bacterial DNA were recovered from the airways it was believed to be due to sampling contamination by the upper airways 24. Consequently, the lungs were one of the few body sites which were not sampled as part of the Human Microbiome Project 25. Culture‐independent approaches identified the presence of microbial DNA that originated primarily from the oral cavity 26, 27. The question of whether there was a unique microbial community in healthy adults was thoroughly investigated in studies that used extremely careful sampling techniques 27, 28, 29, 30. Neutral community modeling, which hypothesizes that the bacteria found in the lungs should match those found in the URT if it was contaminated, suggested that the microbial community of the lungs was distinct from that of the URT 30. In chronic lower airway diseases such as cystic fibrosis, the lower airways are colonized by microbial communities clearly distinct from the URT 31.
An elegant study compared the regional colonization of bacteria in the oral cavity and established that there were three major niches in the mouth: the gingiva, buccal mucosa, and hard palate; saliva, tongue, tonsils, and throat; and the sub‐ and supragingival plaques 32. The different locations had similar bacterial members but at different ratios: Streptococcus species were at the highest abundance in the gingiva, buccal mucosa, and hard palate, whereas the tonsils and throat contained the highest abundance of Prevotella. Dental plaques have low turnover rates compared to tissue surfaces with sloughing epithelial cells, promoting the growth of biofilms containing anaerobic bacteria including Corynebacterium species 32.
Bacterial spread from or within individuals is facilitated through air flow, nasal secretions, or saliva. Infections of the lower respiratory tract often originate from bacteria in the oral cavity, in a process called ‘microaspiration’ 33, 34. Salivary excretions, which can be up to a liter a day, move bacteria from the tongue and throat/oropharynx toward the lower respiratory tract. Microaspiration is a frequent event, although contaminating commensal bacteria are cleared in healthy adults, and likely only cause disease in those with impaired lung or immune function 35.
The URT microbiota is also an important reservoir of potential lung pathogens, and determining if biogeographical niches exist for these pathogens may assist with disease prevention. Investigative studies that assess bacterial interactions between potential pathogens and other naturally colonizing members of the microbial community may lead to nonantibiotic‐based methods of clearing pathogens. A recent study by Yan et al. elegantly assessed different regions of the nasal cavity that would be exposed to the external environment (anterior nares), as well as drainage from the sinuses (middle meatus and sphenoethmoidal recess) in six St. aureus carriers and six noncarriers 22. Comparing different regions along the nasal cavity, all sites contained similar relative abundance of Firmicutes, Bacteroidetes, and Actinobacteria, but the relative abundance of Proteobacteria varied throughout the URT. The anterior nares had the lower microbial diversity compared to the middle meatus and sphenoethmoidal recess but the three sites selected had several unique operational taxonomic units (OTUs). When the St. aureus OTU was removed from the microbiota sequences, carriers could not be distinguished from noncarriers based on overall diversity. However, St. aureus carriers had high rates of Corynebacterium accolens, whereas noncarriers had high rates of Corynebacterium pseudodiphtheriticum, suggesting that some levels of niche competition could be important for colonization by St. aureus 22. This was corroborated with in vitro data showing that C. accolens and St. aureus supported each other's growth, whereas C. pseudodiphtheriticum inhibited the growth of St. aureus.
Culture‐based monitoring of URT microbiota
Nasopharyngeal carriage is a major contributor to infection, so the relationship between carriage rates, disease incidence, or antibiotic resistance is monitored by swabbing and culturing using conditions that enrich for specific pathogens. For example, St. aureus is often detected in greater than 30% of individuals, but rates of carriage of methicillin‐resistant St. aureus (MRSA) vary widely from 3 to 30% 36, 37, 38, 39. The distinction between MRSA and methicillin‐sensitive St. aureus (MSSA) is important, as those colonized by MRSA are at an increased risk for invasive disease. A study by Davis et al. demonstrated that upon hospital admission, 21% of patients were MSSA carriers, and 3.4% of patients carried MRSA 40; however, a much higher percentage of the MRSA carriers (19%) developed invasive staphylococcal disease than MSSA carriers (1.5%). Monitoring colonization may assist in the prevention of infection 40.
Nasal swabs have also been used to measure the efficacy of vaccination and to understand the interplay between age and environmental risk factors in carriage and disease risk 41, 42. For example, as a result of vaccination against S. pneumoniae, previously common serotypes which are included in the vaccine are being replaced with nonvaccine serotypes, and overall S. pneumoniae colonization has gone unchanged 43. Surveillance studies have found that vaccination against S. pneumoniae also alters carriage of St. aureus and H. influenzae 43.
The limitation of these carriage studies is that they provide detailed data about specific pathogens but are less helpful in understanding the microbial community as a whole. Recently, a more in‐depth culture‐based study of the nasal microbiota assessed four locations along the length of the nose: the anterior and posterior vestibules, and the middle and inferior meatus 44. In this study, 141 taxa, dominated by Staphylococcus, Corynebacterium, and Propionibacterium species were found in four different sites in 34 surgical patients. The majority of species were found in all regions of the nose, except Corynebacterium simulans, which was not present in the posterior vestibule, and Acinetobacter lwoffii, which was not present in the anterior vestibule and inferior meatus. There were distinct pairs of species that were generally cultured together. For example, Dolosigranulum pigrum was cultured in samples that had Corynebacterium propinquum and Staphylococcus epidermidis was cultured in samples that had Propionibacterium acnes. This suggests that these pairs have a mutualistic relationship that may promote colonization or facilitate survival within the URT. Although this study provided the most extensive culture‐based dataset on the composition of the URT microbiome, next‐generation sequencing on a subset of samples, identified 113 bacterial phylotypes that were not recovered by culture. This highlighted that the extensive culturing methods used did not identify all the bacteria as they were able to detect more bacterial groups via next‐generation sequencing 44. However, culture‐independent methods do not distinguish viable from non‐viable bacteria, and some of the ‘uncultured’ organisms may not be viable in the samples. Nonetheless, sequencing of the microbiota has become an important methodology for advanced bacterial composition assessment.
Culture‐independent study of URT microbiota
The most common method for assessing the composition of the bacterial community is by sequencing the16S rRNA gene. This gene has nine variable regions (V1–V9), differences in which allow for taxonomic identification of bacteria, which are classified as operational taxonomic units, or OTUs 45. In general, targeting one or more of these variable regions is sufficient to identify members of the microbiota at the genus level, however, the specific 16S variable regions used may vary due to tissue type or particular bacterial families present 46. While OTUs help quantitate the overall diversity of the microbiota, identification beyond the genera level can pose a significant challenge for determining exact community composition of the URT. As an example, the URT microbiota contains many members of the genus Streptococcus 47. The high degree of 16S sequence similarity between this genus means that it is not possible to resolve the relative abundances of members of these genera 48, 49. Nonetheless, culture‐independent techniques have been extremely useful in characterizing the composition and diversity of the URT microbiota. Sequencing of swabs of the middle turbinate, nasopharynx or anterior nares for nasal microbiota, and tongue or buccal mucosa for oral cavity sampling have identified that Corynebacteriaceae, Staphylococcaceae, and Propionibacteriaceae dominate in nasal cavity of healthy adults, although there is considerable compositional variability between the individuals 50. The oral cavity in these healthy adults was dominated by Streptococcaceae and Veillonellaceae. The microbial communities from the three sampling sites were distinct from each other, signifying important local niches for colonization. Interestingly, a separate study determined that swabs are representative of invasive (surgical) sampling, suggesting that less invasive measures are adequate for nasal microbial study 51. A summary of the most commonly found bacteria in healthy individuals are presented in Table 1.
Table 1.
Composition of the bacterial microbiota at different locations in the URT
| Population | Study | Sample site | Actinobacteria | Bacteroidetes | Firmicutes | Proteobacteria |
|---|---|---|---|---|---|---|
| Adult | 22 | Anterior nares | Corynebacterium, Propionibacterium | Prevotella | Dolosigranulum, Staphylococcus, Streptococcus | Moraxella, Escherichia–Shigella |
| Middle Meatus | Corynebacterium, Propionibacterium | Prevotella | Dolosigranulum, Staphylococcus, Streptococcus | Moraxella, Escherichia–Shigella | ||
| Adult | 101 | Sinus | Propionibacterium, Corynebacterium | Prevotella | Staphylococcus, Anaerococcus, Peptoniphilus | Ralstonia |
| Children | 49 | Nasopharynx | Corynebacterium, Propionibacterium, Bifidobacterium | Bacteroides | Staphylococcus, Faecalibacterium, Streptococcus | Moraxella |
| Oropharynx | Rothia, Corynebacterium | Prevotella, Porphyromonas | Streptococcus, Veillonella | Haemophilus, Moraxella | ||
| Adult | Nasopharynx | Corynebacterium, Propionibacterium, Bifidobacterium | Prevotella, Sphingobacterium | Staphylococcus, Faecalibacterium, Streptococcus | Pseudomonas, Haemophilus | |
| Oropharynx | Rothia, Corynebacterium | Prevotella, Porphyromonas | Streptococcus, Veillonella | Haemophilus, Moraxella | ||
| Elderly | 67 | Anterior nares | Propionibacterium, Corynebacterium, Bifidobacterium | Prevotella, Bacteroides | Streptococcus, Staphylococcus, Veillonella | Moraxella, Pseudomonas |
| Oropharynx | Propionibacterium, Corynebacterium, Bifidobacterium | Prevotella | Streptococcus, Staphylococcus, Veillonella | Moraxella, Pseudomonas | ||
| Adult | 32 | Gingiva | Actinomyces, Rothia | Porphyromonas, Prevotella | Streptococcus, Veillonella, Gemella | Haemophilus, Neisseria, Actinobacillus |
| Tongue | Actinomyces, Rothia | Prevotella, Porphyromonas | Streptococcus, Veillonella, Granulicatella | Neisseria, Haemophilus | ||
| Supragingival plaque | Actinomyces, Corynebacterium, Rothia | Porphyromonas, Prevotella, Capnocytophaga | Streptococcus, Gemella, Veillonella | Haemophilus, Neisseria, Actinobacillus |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Bacterial colonization, succession, and evolution in youth
Colonization of the URT begins at birth and, importantly, early colonization events impact respiratory health throughout life. Correlations between the method of delivery (i.e., vaginal or cesarean) and breastfeeding with susceptibility to respiratory infections, asthma and allergy have been observed for decades 52, 53, 54; it is now believed that this is due in part to the establishment of the URT microbiome. Recently, a study by Bosch et al. demonstrated that infants born via cesarean section were more likely to have reduced colonization levels of protective bacteria, including Corynebacterium and Dolosigranulum species 55. Breastfeeding improves infant health in part because it facilitates the transfer of maternal antibodies which are then found in the infant's nasal secretions 56, 57. A recent study established the role of exclusive breastfeeding versus exclusive formula feeding from 6 weeks of age up to an age of 6 months of life on the URT microbiota 58. Breastfed children had increased levels of Corynebacterium and Dolosigranulum species that may play an important role in protection against upper respiratory infections 14. Furthermore, it was noted at 6 weeks, but not 6 months of age, that there was a negative correlation between wheezing and the relative abundance of Dolosigranulum. Infants colonized by H. influenzae, Moraxella catarrhalis, and S. pneumoniae within the first month of life were more likely to demonstrate wheezing, compared to those that were not colonized, but colonization status of these pathogens at 1 year of life did not correlate with wheezing 59. A prospective cohort study involving 234 children demonstrated that nasopharyngeal colonization by Streptococcus at ~ 2 months of age was a strong predictor for asthma later in life 60. This study also demonstrated that antibiotic use within 4 weeks of sampling increased the likelihood of colonization by Streptococcus, Haemophilus, and Moraxella 60. The importance of the composition of colonizing bacteria in the first few months of life has been confirmed in mouse models of allergic asthma 61. Mice treated with antibiotics prior to weaning had more severe allergic asthma, but antibiotic treatment after weaning had no effect 61.
Studies of microbial succession have been performed in order to understand how the development of the URT microbiota influences susceptibility to allergic airway disease. A recent landmark study by Biesbroek et al. investigated the bacterial succession patterns in infants 14. The group followed a cohort of 60 infants over the first 2 years of life, sampling from 1.5 months of age to 24 months, and validated the findings with a cross‐sectional study of 140 children per age group. The relative bacterial load and α‐diversity did not change over time, however, the composition of the microbiota did change over time 14. At 1.5 months of age, five clusters were observed that were dominated by either Streptococcus, Moraxella, Staphylococcus, Corynebacterium, or Corynebacterium/Dolosigranulum. In the Streptococcus and Staphylococcus clusters, Corynebacterium and Dolosigranulum were found at very low abundance. As the infants progressed from 1.5 months to 24 months of age, those whose microbiota were dominated by Moraxella were likely to remain dominated by Moraxella, and infants who were colonized with Corynebacterium/Dolosigranulum were likely to become dominated by Moraxella. In contrast, microbial communities dominated by Streptococcus and Staphylococcus were not stable and would often have a completely different profile in subsequent samples, with no defined pattern 14. Interestingly, the Corynebacterium/Dolosigranulum cluster was highly correlated with breastfeeding. Furthermore, the children with a Corynebacterium‐ and/or Dolosigranulum‐dominated URT microbiota were less likely to have a URT infection compared with all other microbial profiles. This study demonstrated a repeatable pattern of ecological succession, and potentially identifies groups of infants that may be at risk for respiratory infections.
Another cross‐sectional study assessed composition of the nasopharyngeal and oral microbiota in 51 young children and 19 parents by both culture and 16S rRNA sequencing 49. Stearns et al. demonstrated that the oropharyngeal swabs had similar microbial communities regardless of age, and were dominated by Streptococcus, Prevotella and Veillonella. The adult nasopharyngeal samples were dominated by Firmicutes (including Lachnospiraceae, Staphylococcus, and Streptococcus species), Bacteroidetes (Sphingobacterium and Prevotella), and Actinobacteria (Corynebacterium, Bifidobacterium, Rothia, and Propionibacterium). In contrast, children were dominated by Moraxella, Haemophilus, Enterobacteriaceae, and Enterococcus. Targeted cultivation of these bacteria allowed for species identification, including Staphylococcus (St. epidermidis), Haemophilus (H. influenzae and Haemophilus parainfluenzae), D. pigrum, and Corynebacterium (C. durum and C. mucifaciens) 49. Furthermore, bacterial culture identified an increased bacterial density in the nasopharynx of children compared to healthy adults. This study demonstrated there is a dramatic difference between the infant and adult URT microbiota, suggesting progression to a more diverse, yet less dense, community.
Although the role of the URT microbiota in the immune system development is not as well described as the gut, there is some mechanistic evidence indicating that the microbiota in early life contributes to susceptibility to colonization by pathogens. A recent study demonstrated that macrophage recruitment to the URT during pneumococcal colonization was decreased in infant mice due to the inability to create a chemokine gradient within the nasal cavity 62. In this model, the neonatal microbiota contributed to the high baseline chemokine expression, as antibiotic treatment decreased baseline chemokine expression, which allowed for a chemokine gradient to form upon pneumococcal exposure. Understanding the mechanisms by which the URT microbiota drives early innate immune development and susceptibility to colonization by pathogens will be an exciting avenue for future research.
Bacterial colonization and evolution in age
There are not many studies describing how the URT microbiota changes in older adults, but there is some evidence that age‐related changes may contribute to the increased susceptibility to respiratory infections 63. As an example, in children, colonization by S. pneumoniae occurs frequently and is generally asymptomatic; however, when colonization is not appropriately controlled, dissemination from the nasopharynx may result in pneumonia, meningitis, or septicemia 64. In young adults, colonization is less frequent and of shorter duration due to adequate immune control, and consequently, disease is rare unless there are complicating comorbidities or influenza infection 65. The dynamics of carriage in the elderly are not as well studied; however, as in adults, carriage rates are low 66, 67. The combination of low colonization rates and high incidence of pneumonia implies that colonization is brief and proceeds swiftly to infection 64. In support of this, peaks of invasive pneumococcal disease in the elderly occur during Christmas holidays when contact with grandchildren, the major reservoir for S. pneumoniae, is presumed to occur 68. Furthermore, mouse models indicate during S. pneumoniae colonization the URT microbiota is profoundly distinct in aged mice, failing to return to a microbiota composition similar to prepneumococcal exposure, which younger mice were able to do 69, 70. Whether age‐related changes in the microbiota contribute to permissiveness to infection is not clear, although preliminary studies support this hypothesis.
The nasopharyngeal microbiota of older adults appears to undergo profound changes. The anterior nares of adults (18–40 years old) is distinct from that of the oropharynx, but this distinction is lost in the elderly (> 65 years old) who become dominated by Streptococcus, Prevotella, and Veillonella 67. This suggests the structure of the microbial community may degrade during aging, which facilitates an expansion of streptococcal species. As an example, Shannon diversity increases in elderly patients suffering from pneumonia compared to healthy elderly people 63. The Shannon diversity Index is a metric which accounts for the abundance of different bacteria, with a low Shannon diversity suggesting the microbiota is dominated by a few bacterial species, whereas a high Shannon diversity suggests many different taxa of even relative abundance 71. Pneumonia patients had a distinct decrease in anaerobic bacteria, including Prevotella, and decreased lactic acid bacteria Leptotrichia, and an increased viral load. A human experimental pneumococcal carriage model has identified that increased Shannon diversity in the URT microbiota correlates with a colonization permissive phenotype 72. Mouse models of pneumonia have also implicated a role for dysbiosis, however, distinct microbial interactions that could be occurring have yet to be fully elucidated 69, 70. Whether specific bacterial species or overall microbial community dynamics are important for susceptibility to disease in the elderly is still unknown (Fig. 1).
Figure 1.
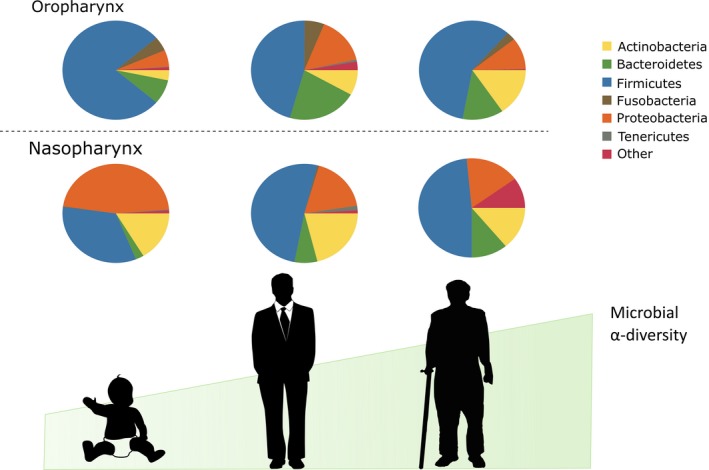
The evolution of the upper respiratory tract microbiota during aging. The oral and nasal microbiome at birth and infancy is influenced by environmental exposures, including breastfeeding. The nasal and oral tissue sites provide unique niches for bacteria to grow and evolve, but by late age the nasal and oral microbiome become quite similar, revealing potential breakdown of important host mechanisms in microbiota community composition. This is highlighted by increased α‐diversity (or microbial diversity within a host) during aging, indicating lack of community regulation.
Immunosenenescence, which is defined as age‐related changes in immune function, is a major contributing factor to the increased incidence of respiratory infections in people over 65 years old 73. It is unclear how immunosenescence affects the composition and maintenance of the URT microbiota, although there are associations with the immune status and dysbiosis of the intestinal microbiota 74, 75. Impaired innate immunity in the URT has been described in elderly mouse models 76, 77. For example, TLR‐1 expression, murine cathelicidin, and a decreased recruitment of macrophages in response to pneumococcal colonization contribute to decreased clearance of the bacteria in elderly mice 76. More recently, tumor necrosis factor (TNF)‐α was shown to be elevated in elderly mice, which promoted premature monocyte egress from the bone marrow and impaired bacterial clearance upon reaching the nasopharynx 77. These data correlate well with human data, demonstrating that elevated levels of circulating TNF‐α and IL‐6 are associated with increased risk for community‐acquired pneumonia 78.
Factors affecting URT microbiota
Seasons
Seasonal patterns of respiratory infections are well documented. Community‐acquired pneumonia is more prevalent during the winter and early spring months of the northern hemisphere 79. Infants have the highest prevalence of S. pneumoniae nasopharyngeal colonization during cooler and drier months worldwide 80. A number of factors have been proposed which may contribute to increased infection risk in the winter, including increased crowding or decreased immunity when exposed to colder air 81, 82. A study assessing children < 7 years old found that seasonality affected both pneumococcal pneumonia and invasive pneumococcal disease 83. Invasive pneumococcal disease increased in autumn, correlating with going back to school, whereas pneumonia cases increased in the winter 83. Furthermore, these children carried a higher density of S. pneumoniae during the winter months. Health care professionals have asymptomatic carriage of various pathogens which differed between winter and summer, with high M. catarrhalis and Coronavirus in the winter and high Klebsiella pneumoniae in the summer 84.
Whether seasonal changes in the URT microbiota also contribute to infection risk is unclear. Bogaert et al. found an increased relative abundance of Proteobacteria during winter months and increased carriers of Fusobacteria and Cyanobacteria, although it is unknown whether this affected respiratory infections 85. Infants with respiratory infections had increased carriage rates of Haemophilus in the spring–summer period and decreased Moraxella carriage during the autumn–winter period 60. Monitoring the changing dynamics within the URT microbiota may elucidate important interactions that occur in different humidity and temperature conditions.
Cigarette smoke
Exposure to cigarette smoke, whether by active or passive exposure, contributes to both chronic respiratory disease [e.g., asthma, chronic obstructive pulmonary disease (COPD)], and acute respiratory infections 86, 87, 88. Cigarette smoke increases infiltration of inflammatory immune cells, and decreases mucociliary clearance in the upper and lower respiratory tracts, which ultimately leads to bacterial colonization and infection 86. Smokers have increased microbial diversity compared to nonsmokers 89. Smokers have an increased likelihood of carrying pathogens, including S. pneumoniae, Streptococcus pyogenes, H. influenzae, and M. catarrhalis 90, 91. Consistent with this, smokers had less carriage of nonpathogenic Streptococcus, Prevotella, and Peptostreptococcus species, which have been shown to be inversely correlated with the presence of pathogens 90. Smokers had a large increase in carriage of several gram‐positive bacteria, including those associated with endocarditis and URT infections in the nasopharynx, which suggests that their increased risk of invasive disease is due to increased carriage of pathogenic bacteria 89. Additionally, mice exposed to cigarette smoke concurrently with S. pneumoniae nasopharyngeal colonization had heightened invasive pneumococcal disease compared to room air‐exposed mice, which was correlated with impaired innate immune responses 92. This supports that the impairment of innate immunity and colonization resistance likely contribute to exacerbations of COPD. The oral microbiota has also been shown to be affected by cigarette smoke 29. Smoking increases the relative abundance of Megasphaera species, associated with periodontitis, and decreases Peptostreptococcus, which has been shown to inhibit the growth of various URT pathogens in vitro 89, 91. Cigarette smoke has been demonstrated to affect bacterial adherence to oral tissue 93.
Cigarette smoke may also affect carriage and infection by altering bacteria directly. For example, biofilm formation and immune evasion increased in cigarette‐exposed St. aureus 94, 95. Cationic antimicrobial peptides, including cathelicidin, rely on membrane charge to target and form pores through bacterial membranes 96. Cigarette smoke extracts modulate the surface charge of St. aureus, thus reducing cathelicidin‐induced lysis 95. This effect is not limited to traditional cigarette smoke, as electronic cigarette smoke vapor increased virulence and biofilm formation, decreased bacterial killing, and impaired epithelial and alveolar macrophage‐induced killing of St. aureus 97. Collectively, smoking has been demonstrated to affect multiple facets of the URT microbiota, including host defense, bacterial adherence and colonization, as well as pathogen virulence.
Chronic URT disease
Chronic rhinosinusitis is an inflammatory disorder of the sinuses which lasts for greater than 12 weeks. Microbial dysbiosis, defined as alterations of the microbial community believed to be associated with disease susceptibility or pathology, is a feature of chronic rhinosinusitis 98. In contrast to other conditions (e.g., smoking, aging) in which there is an increase in microbial diversity, decreased microbial diversity has been reported in chronic rhinosinusitis patients 99, 100, 101. This decreased diversity may be due to increased prevalence of anaerobic bacteria that are believed to thrive as a result of increased prevalence of anaerobic pockets which occur during biofilm formation 102, 103, 104. The presence of Corynebacterium species correlates with optimal surgical outcomes, providing further evidence that Corynebacterium are beneficial members of the URT microbiota 101.
Genetic diseases affecting mucociliary clearance, such as cystic fibrosis and primary ciliary dyskinesia, affect the URT microbiota. Two recent studies have identified that infants with cystic fibrosis, which results in a thickened mucus layer, have an altered URT microbiota development compared to healthy infants including increased relative abundance of Staphylococcus species and decreased abundance of potential beneficial bacteria 105, 106. Many people with primary ciliary dyskinesia, a combination of diseases resulting in decreased ciliary action to clear mucus, also suffer from chronic rhinosinusitis, and there is a high reported rate of URT manifestations of disease, contributing to morbidity 107. While no studies have been completed to assess the URT microbiota of these patients, the increased number of infections and antibiotic exposure are likely to alter the composition. Monitoring the URT microbiota of patients with impaired mucociliary clearance may result in decreased or prevention of lung infections in this susceptible population.
Intramicrobiota interactions
In addition to environmental factors and the immune status of the host, intramicrobiota interactions also influence the composition of the microbial community. The majority of our understanding of competition comes from studies of pathogens, St. aureus and S. pneumoniae, which have a rich bactericidal arsenal which allows them to eliminate competitors in order to occupy specific niches. Although we have less information about how nonpathogenic members of the microbiota influence carriage, it is believed that they influence succession patterns, diversity, and infection risk 14. Below we summarize some of the best characterized bacterial interactions which alter carriage dynamics in the URT, although we recognize that viruses and fungal species could also be playing a role in the URT microbiome.
Staphylococcus aureus and Streptococcus pneumoniae
The strongest evidence for interspecies competition between St. aureus and S. pneumoniae in vivo comes from studies of URT microbiota changes after pneumococcal vaccination. These studies demonstrate that as S. pneumoniae carriage in the nasopharynx decreases, carriage rates of St. aureus increase 43. This is due in part to the bactericidal components produced by S. pneumoniae, such as hydrogen peroxide 65. Hydrogen peroxide production induces DNA damage in St. aureus which triggers the release of resident prophages, resulting in bacteriophage‐driven death of St. aureus 108. Some strains of St. aureus produce catalase or carotenoids, such as staphyloxanthin, which reduce the damaging effects of peroxides and neutrophil killing 65, 109. These compensatory defense mechanisms and the fact that not all strains of St. aureus carry lysogenic phage, may explain why not all studies find an inverse relationship between St. aureus and S. pneumoniae. The immune status of the host also contributes to the apparent inverse relationship in carriage. Mouse models have shown that antibodies against S. pneumoniae dehydrogenase have cross‐specificity for St. aureus which blocks subsequent St. aureus colonization 110. Similarly, the reciprocal relationship between S. pneumoniae and St. aureus do not occur in HIV‐positive children, which carry equal levels of both pathogens simultaneously, which implies that a fully competent immune system influences carriage 111.
Staphylococcus aureus and URT bacterial microbiota interactions
Staphylococcus aureus and H. influenzae have a positive correlation with each other. Margolis et al. demonstrated that levels of H. influenzae were higher when St. aureus was the initial colonizer of rat nasopharynx, possibly due to the liberation of nutrients by toxins released by St. aureus disrupting erythrocytes, including NAD and hemin 112, 113, 114. However, St. aureus has an inverse relationship with several microbial community members 115. After describing an inverse correlation between Corynebacterium species and St. aureus, Uehara et al. applied Corynebacterium to the nares of 17 St. aureus carriers. This resulted in the eradication of St. aureus in 71% of carriers, significantly more than those that received St. epidermidis or salt solutions, in a bacteriocin‐independent mechanism 115.
Staphylococcus epidermidis, Corynebacterium species, and St. aureus occupy similar niches in the URT. Niche competition is controlled by the production of peptides, including bacteriocins, by different species which either impact viability or biofilm formation. Bacteriocins are normally pore‐forming peptides produced by bacteria that target other, usually similar, bacteria. Some St. epidermidis strains produce an extracellular serine protease (Esp) that inhibits the biofilm formation and nasal colonization of St. aureus 116. St. epidermidis Esp disrupts biofilm formation of several different strains of St. aureus, including those in coculture for over 1 year, suggesting that resistance does not develop. Esp‐positive and ‐negative strains were placed in nasal cavities of St. aureus carriers, and only those colonized with Esp‐positive strains saw reduction or eradication of St. aureus. Furthermore, Esp‐induced disruption of St. aureus biofilms facilitated killing by human beta‐defensin 2 in previously resistant strains 116. The competition between St. epidermidis and St. aureus clearly involves both bacterial and host interactions.
Streptococcus pneumoniae and URT bacterial microbiota interactions
Intense intraspecies competition is a feature of the Streptococcus genera, and is mediated in large part by bacteriocins. These peptides target similar bacteria, but usually are coexpressed with an immunity peptide to protect the producing bacteria 117. A recent study assessed over 4000 S. pneumoniae genomes and found that there were over 250 unique combinations of bacteriocins across different strains, suggesting a very dynamic, complex interaction between strains 118. Production of hydrogen peroxide by S. pneumoniae allows antagonism of many other bacteria, however, bacterial members of the URT possess other pathways to compete with S. pneumoniae. H. influenzae and S. pneumoniae cocolonize the nasopharynx in healthy children, but competition is rampant between the two species. H. influenzae caused a rapid decrease in S. pneumoniae carriage within a day of cocolonization in immunocompromised mice 119. Interestingly, recruited neutrophils were activated by peptidoglycan from H. influenzae to enhance killing of S. pneumoniae 119. S. pneumoniae can impair fitness of H. influenzae and Neisseria meningitidis by producing neuraminidases that cleave sialic acids which these bacteria use to evade detection by the immune system 120.
In vitro studies have also suggested different bacteria can support or hinder S. pneumoniae growth. S. pneumoniae has the ability to respond to peptides from nasopharyngeal bacteria, including Prevotella species, which activate transcription of important colonization factors 121. In contrast, Corynebacterium species seem to impair the persistence of S. pneumoniae 122. Recently, it has been demonstrated that C. accolens have the ability to promote the killing of S. pneumoniae by metabolizing triacylglycerols, which are present on URT epithelium, to produce oleic and linoleic acids that are toxic to S. pneumoniae 123.
A human S. pneumoniae carriage model has been developed to assess relevant factors involved in colonization. Approximately 50% of those intranasally exposed to S. pneumoniae have successful colonization 124. Establishment of pneumococcal carriage was not correlated with carriage of St. aureus, M. catarrhalis, or H. influenzae, although there were subtle decreases in St. aureus carriage over time 125. Increased Shannon diversity increased likelihood of colonization, while St. aureus‐dominated nasopharyngeal samples were less likely to be colonized 125. Furthermore, successful colonization has been linked to increased viral carriage 126.
Conclusion
The URT is colonized by many different potential pathogens, and is constantly exposed to environmental bacteria, which compete for a niche for survival. Pathogens can survive and thrive in both the infant and elderly nasal and oral cavities, likely due to synergism between loss of colonization resistance and altered innate and adaptive immunity. Early colonization events, including birth route, breastfeeding, and exposure to other children influence the acquisition and succession of the URT microbiota, with Corynebacterium and Dolosigranulum species repeatedly demonstrated as protective profiles against disease 14. Future work establishing early colonization events through culture‐dependent and ‐independent methods, will help in the prevention of acute and chronic airway diseases throughout the lifespan. Limited mechanistic studies have been completed to describe beneficial microbial colonization and interaction with the immune system in the URT. Recent work has identified that specific bacterial groups in the nasal cavity are associated with efficacy of vaccination against influenza, suggesting that nasal immunity may be regulated by the nasal microbiota 127. Exploring host–bacterial interactions that have been uncovered in the gut, including adaptive immune development and pathogen clearance, could greatly decrease respiratory disease morbidity. Fecal transplants, which have been used to combat recurrent Clostridium difficile infections and diarrhea, have been identified to boost colonization resistance against the pathogen by altering the microbial community 128. Colonization resistance can be caused by limiting nutrients essential for pathogen growth, disrupting the niche required by the pathogen, or enhancing the immune response by the body. Taking advantage of colonization resistance is only beginning to be explored in the URT. Bacterial and viral pathogens of the URT require unique niches, thus various microbial communities may be required to resist different pathogens. Development of human and animal models that assess the intramicrobiota competition and host–microbiota interactions are an exciting avenue in the development of targeted, URT‐specific probiotics. The future of combating respiratory pathogens will require a thorough understanding of the dynamics of the URT microbiota and its’ interaction with the host.
Author contributions
LPS, MGS, and DMEB all contributed to the writing and critical review of this manuscript.
Acknowledgements
LPS is supported by a Canada Graduate Scholarship from the Canadian Institutes of Health Research (CIHR). MGS and DMEB are supported by the CIHR and hold Canada Research Chairs. Work in the Bowdish laboratory is supported by the McMaster Immunology Research Centre (MIRC) and the M.G. DeGroote Institute for Infectious Disease Research (IIDR).
Edited by Wilhelm Just
References
- 1. Yasui H, Nagaoka N, Mike A, Hayakawa K and Ohwaki M (1992) Detection of Bifidobacterium strains that induce large quantities of IgA. Microb Ecol Health Dis 5, 155–162. [Google Scholar]
- 2. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R et al (2010) Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA 107, 7503–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT and Ley RE (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108 (Suppl 1), 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ and Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yun Y, Srinivas G, Kuenzel S, Linnenbrink M, Alnahas S, Bruce KD, Steinhoff U, Baines JF and Schaible UE (2014) Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS One 9, e113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB and Gordon JI (2016) Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K et al (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. [DOI] [PubMed] [Google Scholar]
- 9. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV et al (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE and Backhed F (2012) Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Round JL and Mazmanian SK (2009) The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shinzato T and Saito A (1995) The Streptococcus milleri group as a cause of pulmonary infections. Clin Infect Dis 21 (Suppl 3), S238–S243. [DOI] [PubMed] [Google Scholar]
- 13. Ripley RT, Cothren CC, Moore EE, Long J, Johnson JL and Haenel JB (2006) Streptococcus milleri infections of the pleural space: operative management predominates. Am J Surg 192, 817–821. [DOI] [PubMed] [Google Scholar]
- 14. Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ and Bogaert D (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190, 1283–1292. [DOI] [PubMed] [Google Scholar]
- 15. Sahin‐Yilmaz A and Naclerio RM (2011) Anatomy and physiology of the upper airway. Proc Am Thorac Soc 8, 31–39. [DOI] [PubMed] [Google Scholar]
- 16. Siegel SJ and Weiser JN (2015) Mechanisms of bacterial colonization of the respiratory tract. Annu Rev Microbiol 69, 425–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rigottier‐Gois L (2013) Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu S, Liu Y, Sun X and Li S (2008) Influence of nasal structure on the distribution of airflow in nasal cavity. Rhinology 46, 137–143. [PubMed] [Google Scholar]
- 19. Wanner A, Salathe M and O'Riordan TG (1996) Mucociliary clearance in the airways. Am J Respir Crit Care Med 154, 1868–1902. [DOI] [PubMed] [Google Scholar]
- 20. Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D and Tung Y (1997) Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics. J Infect Dis 175, 1440–1445. [DOI] [PubMed] [Google Scholar]
- 21. Verhoeven D, Nesselbush M and Pichichero ME (2013) Lower nasopharyngeal epithelial cell repair and diminished innate inflammation responses contribute to the onset of acute otitis media in otitis‐prone children. Med Microbiol Immunol 202, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S and Relman DA (2013) Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laurenzi GA, Potter RT and Kass EH (1961) Bacteriologic flora of the lower respiratory tract. N Engl J Med 265, 1273–1278. [DOI] [PubMed] [Google Scholar]
- 24. Dickson RP, Erb‐Downward JR, Martinez FJ and Huffnagle GB (2016) The microbiome and the respiratory tract. Annu Rev Physiol 78, 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassis CM, Erb‐Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL and Huffnagle GB (2015) Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, Chen H, Berger KI, Goldring RM, Rom WN et al (2013) Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L et al (2010) Disordered microbial communities in asthmatic airways. PLoS One 5, e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L et al (2013) Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 187, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB and Schmidt TM (2015) Application of a neutral community model to assess structuring of the human lung microbiome. MBio 6, e02284–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Kehagia V, Connett GJ and Bruce KD (2006) Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J Clin Microbiol 44, 2601–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C and Izard J (2012) Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 13, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dickson RP, Erb‐Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB and Curtis JL (2015) Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc 12, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huxley EJ, Viroslav J, Gray WR and Pierce AK (1978) Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med 64, 564–568. [DOI] [PubMed] [Google Scholar]
- 35. Marik PE and Kaplan D (2003) Aspiration pneumonia and dysphagia in the elderly. Chest 124, 328–336. [DOI] [PubMed] [Google Scholar]
- 36. Jimenez‐Truque N, Saye EJ, Soper N, Saville BR, Thomsen I, Edwards KM and Creech CB (2016) Longitudinal assessment of colonization with Staphylococcus aureus in healthy collegiate athletes. J Pediatric Infect Dis Soc 5, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaw AG, Vento TJ, Mende K, Kreft RE, Ehrlich GD, Wenke JC, Spirk T, Landrum ML, Zera W, Cheatle KA et al (2013) Detection of methicillin‐resistant and methicillin‐susceptible Staphylococcus aureus colonization of healthy military personnel by traditional culture, PCR, and mass spectrometry. Scand J Infect Dis 45, 752–759. [DOI] [PubMed] [Google Scholar]
- 38. Kitti T, Boonyonying K and Sitthisak S (2011) Prevalence of methicillin‐resistant Staphylococcus aureus among university students in Thailand. Southeast Asian J Trop Med Public Health 42, 1498–1504. [PubMed] [Google Scholar]
- 39. Du J, Chen C, Ding B, Tu J, Qin Z, Parsons C, Salgado C, Cai Q, Song Y, Bao Q et al (2011) Molecular characterization and antimicrobial susceptibility of nasal Staphylococcus aureus isolates from a Chinese Medical College campus. PLoS One 6, e27328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis KA, Stewart JJ, Crouch HK, Florez CE and Hospenthal DR (2004) Methicillin‐resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clini Infect Dis 39, 776–782. [DOI] [PubMed] [Google Scholar]
- 41. Demczuk WH, Martin I, Griffith A, Lefebvre B, McGeer A, Shane A, Zhanel GG, Tyrrell GJ, Gilmour MW, Toronto Invasive Bacterial Diseases Network et al (2012) Serotype distribution of invasive Streptococcus pneumoniae in Canada during the introduction of the 13‐valent pneumococcal conjugate vaccine, 2010. Can J Microbiol 58, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 42. Waight PA, Andrews NJ, Ladhani SN, Sheppard CL, Slack MP and Miller E (2015) Effect of the 13‐valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet Infect Dis 15, 535–543. [DOI] [PubMed] [Google Scholar]
- 43. Bosch AA, van Houten MA, Bruin JP, Wijmenga‐Monsuur AJ, Trzcinski K, Bogaert D, Rots NY and Sanders EA (2016) Nasopharyngeal carriage of Streptococcus pneumoniae and other bacteria in the 7th year after implementation of the pneumococcal conjugate vaccine in the Netherlands. Vaccine 34, 531–539. [DOI] [PubMed] [Google Scholar]
- 44. Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, Wos‐Oxley M and Becker K (2016) The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol 18, 2130–2142. [DOI] [PubMed] [Google Scholar]
- 45. Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R and Rossello‐Mora R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12, 635–645. [DOI] [PubMed] [Google Scholar]
- 46. Claesson MJ, Wang Q, O'Sullivan O, Greene‐Diniz R, Cole JR, Ross RP and O'Toole PW (2010) Comparison of two next‐generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frandsen EV, Pedrazzoli V and Kilian M (1991) Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol 6, 129–133. [DOI] [PubMed] [Google Scholar]
- 48. Doern CD and Burnham CA (2010) It's not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J Clin Microbiol 48, 3829–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, Bowdish DM, Kellner JD and Surette MG (2015) Culture and molecular‐based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J 9, 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bassis CM, Tang AL, Young VB and Pynnonen MA (2014) The nasal cavity microbiota of healthy adults. Microbiome 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bassiouni A, Cleland EJ, Psaltis AJ, Vreugde S and Wormald PJ (2015) Sinonasal microbiome sampling: a comparison of techniques. PLoS One 10, e0123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I et al (2011) Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 128, 948–955 e1–3. [DOI] [PubMed] [Google Scholar]
- 53. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist‐Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL et al (2015) Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7, 307ra152. [DOI] [PubMed] [Google Scholar]
- 54. Dogaru CM, Nyffenegger D, Pescatore AM, Spycher BD and Kuehni CE (2014) Breastfeeding and childhood asthma: systematic review and meta‐analysis. Am J Epidemiol 179, 1153–1167. [DOI] [PubMed] [Google Scholar]
- 55. Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, de Steenhuijsen Piters WA, de Groot PK, Pernet P, Keijser BJ et al (2016) Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9, 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roberts SA and Freed DL (1977) Neonatal IgA secretion enhanced by breast feeding. Lancet 2, 1131. [DOI] [PubMed] [Google Scholar]
- 57. Taylor CE and Toms GL (1984) Immunoglobulin concentrations in nasopharyngeal secretions. Arch Dis Child 59, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA and Bogaert D (2014) The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 190, 298–308. [DOI] [PubMed] [Google Scholar]
- 59. von Linstow ML, Schonning K, Hoegh AM, Sevelsted A, Vissing NH and Bisgaard H (2013) Neonatal airway colonization is associated with troublesome lung symptoms in infants. Am J Respir Crit Care Med 188, 1041–1042. [DOI] [PubMed] [Google Scholar]
- 60. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E et al (2015) The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM et al (2012) Early life antibiotic‐driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 13, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siegel SJ, Tamashiro E and Weiser JN (2015) Clearance of pneumococcal colonization in infants is delayed through altered macrophage trafficking. PLoS Pathog 11, e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, Wang X, Trzcinski K, Bonten MJ, Rossen JW et al (2016) Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J 10, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kellner JD, Vanderkooi OG, MacDonald J, Church DL, Tyrrell GJ and Scheifele DW (2009) Changing epidemiology of invasive pneumococcal disease in Canada, 1998–2007: update from the Calgary‐area Streptococcus pneumoniae research (CASPER) study. Clin Infect Dis 49, 205–212. [DOI] [PubMed] [Google Scholar]
- 65. Regev‐Yochay G, Trzcinski K, Thompson CM, Malley R and Lipsitch M (2006) Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide‐mediated killing by Streptococcus pneumoniae . J Bacteriol 188, 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ridda I, Macintyre CR, Lindley R, McIntyre PB, Brown M, Oftadeh S, Sullivan J and Gilbert GL (2010) Lack of pneumococcal carriage in the hospitalised elderly. Vaccine 28, 3902–3904. [DOI] [PubMed] [Google Scholar]
- 67. Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG and Bowdish DM (2014) The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc 11, 513–521. [DOI] [PubMed] [Google Scholar]
- 68. Walter ND, Taylor TH Jr, Dowell SF, Mathis S, Moore MR and Active Bacterial Core Surveillance System Team (2009) Holiday spikes in pneumococcal disease among older adults. N Engl J Med 361, 2584–2585. [DOI] [PubMed] [Google Scholar]
- 69. Krone CL, Biesbroek G, Trzcinski K, Sanders EA and Bogaert D (2014) Respiratory microbiota dynamics following Streptococcus pneumoniae acquisition in young and elderly mice. Infect Immun 82, 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thevaranjan N, Whelan FJ, Puchta A, Ashu E, Rossi L, Surette MG and Bowdish DM (2016) Streptococcus pneumoniae colonization disrupts the microbial community within the upper respiratory tract of aging mice. Infect Immun 84, 906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jost L (2007) Partitioning diversity into independent alpha and beta components. Ecology 88, 2427–2439. [DOI] [PubMed] [Google Scholar]
- 72. Cremers AJ, Zomer AL, Gritzfeld JF, Ferwerda G, van Hijum SA, Ferreira DM, Shak JR, Klugman KP, Boekhorst J, Timmerman HM et al (2014) The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome 2, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chalmers JD, Campling J, Dicker A, Woodhead M and Madhava H (2016) A systematic review of the burden of vaccine preventable pneumococcal disease in UK adults. BMC Pulm Med 16, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O et al (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184. [DOI] [PubMed] [Google Scholar]
- 75. Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, O'Toole PW, Spector TD and Steves CJ (2016) Signatures of early frailty in the gut microbiota. Genome Med 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Krone CL, Trzcinski K, Zborowski T, Sanders EA and Bogaert D (2013) Impaired innate mucosal immunity in aged mice permits prolonged Streptococcus pneumoniae colonization. Infect Immun 81, 4615–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Puchta A, Naidoo A, Verschoor CP, Loukov D, Thevaranjan N, Mandur TS, Nguyen PS, Jordana M, Loeb M, Xing Z et al (2016) TNF drives monocyte dysfunction with age and results in impaired anti‐pneumococcal immunity. PLoS Pathog 12, e1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yende S, Tuomanen EI, Wunderink R, Kanaya A, Newman AB, Harris T, de Rekeneire N and Kritchevsky SB (2005) Preinfection systemic inflammatory markers and risk of hospitalization due to pneumonia. Am J Respir Crit Care Med 172, 1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murdoch KM, Mitra B, Lambert S and Erbas B (2014) What is the seasonal distribution of community acquired pneumonia over time? A systematic review Australas Emerg Nurs J 17, 30–42. [DOI] [PubMed] [Google Scholar]
- 80. Numminen E, Chewapreecha C, Turner C, Goldblatt D, Nosten F, Bentley SD, Turner P and Corander J (2015) Climate induces seasonality in pneumococcal transmission. Sci Rep 5, 11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Eccles R (2002) An explanation for the seasonality of acute upper respiratory tract viral infections. Acta Otolaryngol 122, 183–191. [DOI] [PubMed] [Google Scholar]
- 82. Makinen TM, Juvonen R, Jokelainen J, Harju TH, Peitso A, Bloigu A, Silvennoinen‐Kassinen S, Leinonen M and Hassi J (2009) Cold temperature and low humidity are associated with increased occurrence of respiratory tract infections. Respir Med 103, 456–462. [DOI] [PubMed] [Google Scholar]
- 83. Weinberger DM, Grant LR, Steiner CA, Weatherholtz R, Santosham M, Viboud C and O'Brien KL (2014) Seasonal drivers of pneumococcal disease incidence: impact of bacterial carriage and viral activity. Clin Infect Dis 58, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hassoun A, Huff MD, Weisman D, Chahal K, Asis E, Stalons D, Grigorenko E, Green J, Malone LL, Clemmons S et al (2015) Seasonal variation of respiratory pathogen colonization in asymptomatic health care professionals: a single‐center, cross‐sectional, 2‐season observational study. Am J Infect Control 43, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M and Sanders E (2011) Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6, e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taylor JD (2010) COPD and the response of the lung to tobacco smoke exposure. Pulm Pharmacol Ther 23, 376–383. [DOI] [PubMed] [Google Scholar]
- 87. Arcavi L and Benowitz NL (2004) Cigarette smoking and infection. Arch Intern Med 164, 2206–2216. [DOI] [PubMed] [Google Scholar]
- 88. Jones LL, Hashim A, McKeever T, Cook DG, Britton J and Leonardi‐Bee J (2011) Parental and household smoking and the increased risk of bronchitis, bronchiolitis and other lower respiratory infections in infancy: systematic review and meta‐analysis. Respir Res 12, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Charlson ES, Chen J, Custers‐Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD and Collman RG (2010) Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5, e15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Brook I and Gober AE (2005) Recovery of potential pathogens and interfering bacteria in the nasopharynx of otitis media‐prone children and their smoking and nonsmoking parents. Arch Otolaryngol Head Neck Surg 131, 509–512. [DOI] [PubMed] [Google Scholar]
- 91. Brook I and Gober AE (2005) Recovery of potential pathogens and interfering bacteria in the nasopharynx of smokers and nonsmokers. Chest 127, 2072–2075. [DOI] [PubMed] [Google Scholar]
- 92. Shen P, Morissette MC, Vanderstocken G, Gao Y, Hassan M, Roos A, Thayaparan D, Merlano M, Dorrington MG, Nikota JK et al (2016) Cigarette smoke attenuates the nasal host response to Streptococcus pneumoniae and predisposes to invasive pneumococcal disease in mice. Infect Immun 84, 1536–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. El Ahmer OR, Essery SD, Saadi AT, Raza MW, Ogilvie MM, Weir DM and Blackwell CC (1999) The effect of cigarette smoke on adherence of respiratory pathogens to buccal epithelial cells. FEMS Immunol Med Microbiol 23, 27–36. [DOI] [PubMed] [Google Scholar]
- 94. Kulkarni R, Antala S, Wang A, Amaral FE, Rampersaud R, Larussa SJ, Planet PJ and Ratner AJ (2012) Cigarette smoke increases Staphylococcus aureus biofilm formation via oxidative stress. Infect Immun 80, 3804–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McEachern EK, Hwang JH, Sladewski KM, Nicatia S, Dewitz C, Mathew DP, Nizet V and Crotty Alexander LE (2015) Analysis of the effects of cigarette smoke on staphylococcal virulence phenotypes. Infect Immun 83, 2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hancock RE and Diamond G (2000) The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol 8, 402–410. [DOI] [PubMed] [Google Scholar]
- 97. Hwang JH, Lyes M, Sladewski K, Enany S, McEachern E, Mathew DP, Das S, Moshensky A, Bapat S, Pride DT et al (2016) Electronic cigarette inhalation alters innate immunity and airway cytokines while increasing the virulence of colonizing bacteria. J Mol Med (Berl) 94, 667–679. [DOI] [PubMed] [Google Scholar]
- 98. Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN and Lynch SV (2012) Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 4, 151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R and Sanford T (2013) Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg 139, 1328–1338. [DOI] [PubMed] [Google Scholar]
- 100. Choi EB, Hong SW, Kim DK, Jeon SG, Kim KR, Cho SH, Gho YS, Jee YK and Kim YK (2014) Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy 69, 517–526. [DOI] [PubMed] [Google Scholar]
- 101. Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE and Frank DN (2015) Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol 136, 334–342 e1. [DOI] [PubMed] [Google Scholar]
- 102. Dlugaszewska J, Leszczynska M, Lenkowski M, Tatarska A, Pastusiak T and Szyfter W (2016) The pathophysiological role of bacterial biofilms in chronic sinusitis. Eur Arch Otorhinolaryngol 273, 1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Finegold SM, Flynn MJ, Rose FV, Jousimies‐Somer H, Jakielaszek C, McTeague M, Wexler HM, Berkowitz E and Wynne B (2002) Bacteriologic findings associated with chronic bacterial maxillary sinusitis in adults. Clin Infect Dis 35, 428–433. [DOI] [PubMed] [Google Scholar]
- 104. Stephenson MF, Mfuna L, Dowd SE, Wolcott RD, Barbeau J, Poisson M, James G and Desrosiers M (2010) Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J Otolaryngol Head Neck Surg 39, 182–187. [PubMed] [Google Scholar]
- 105. Mika M, Korten I, Qi W, Regamey N, Frey U, Casaulta C, Latzin P, Hilty M and SCILD Study Group (2016) The nasal microbiota in infants with cystic fibrosis in the first year of life: a prospective cohort study. Lancet Respir Med 4, 627–635. [DOI] [PubMed] [Google Scholar]
- 106. Prevaes SM, de Winter‐de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper‐Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK et al (2016) Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med 193, 504–515. [DOI] [PubMed] [Google Scholar]
- 107. Sommer JU, Schafer K, Omran H, Olbrich H, Wallmeier J, Blum A, Hormann K and Stuck BA (2011) ENT manifestations in patients with primary ciliary dyskinesia: prevalence and significance of otorhinolaryngologic co‐morbidities. Eur Arch Otorhinolaryngol 268, 383–388. [DOI] [PubMed] [Google Scholar]
- 108. Selva L, Viana D, Regev‐Yochay G, Trzcinski K, Corpa JM, Lasa I, Novick RP and Penades JR (2009) Killing niche competitors by remote‐control bacteriophage induction. Proc Natl Acad Sci USA 106, 1234–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Clauditz A, Resch A, Wieland KP, Peschel A and Gotz F (2006) Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74, 4950–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lijek RS, Luque SL, Liu Q, Parker D, Bae T and Weiser JN (2012) Protection from the acquisition of Staphylococcus aureus nasal carriage by cross‐reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci USA 109, 13823–13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. McNally LM, Jeena PM, Gajee K, Sturm AW, Tomkins AM, Coovadia HM and Goldblatt D (2006) Lack of association between the nasopharyngeal carriage of Streptococcus pneumoniae and Staphylococcus aureus in HIV‐1‐infected South African children. J Infect Dis 194, 385–390. [DOI] [PubMed] [Google Scholar]
- 112. Margolis E, Yates A and Levin BR (2010) The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host's immune response. BMC Microbiol 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Artman M, Domenech E and Weiner M (1983) Growth of Haemophilus influenzae in simulated blood cultures supplemented with hemin and NAD. J Clin Microbiol 18, 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nilsson IM, Hartford O, Foster T and Tarkowski A (1999) Alpha‐toxin and gamma‐toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect Immun 67, 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS and Maruchi N (2000) Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect 44, 127–133. [DOI] [PubMed] [Google Scholar]
- 116. Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T and Mizunoe Y (2010) Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. [DOI] [PubMed] [Google Scholar]
- 117. Dawid S, Roche AM and Weiser JN (2007) The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 75, 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Miller EL, Abrudan MI, Roberts IS and Rozen DE (2016) Diverse ecological strategies are encoded by Streptococcus pneumoniae bacteriocin‐like peptides. Genome Biol Evol 8, 1072–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lysenko ES, Ratner AJ, Nelson AL and Weiser JN (2005) The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 1, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shakhnovich EA, King SJ and Weiser JN (2002) Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect Immun 70, 7161–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hathaway LJ, Battig P, Reber S, Rotzetter JU, Aebi S, Hauser C, Heller M, Kadioglu A and Muhlemann K (2014) Streptococcus pneumoniae detects and responds to foreign bacterial peptide fragments in its environment. Open Biol 4, 130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP and Metlay JP (2012) Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78, 6262–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bomar L, Brugger SD, Yost BH, Davies SS and Lemon KP (2016) Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio 7, e01725‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gritzfeld JF, Wright AD, Collins AM, Pennington SH, Wright AK, Kadioglu A, Ferreira DM and Gordon SB (2013) Experimental human pneumococcal carriage. J Vis Exp 72, e50115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shak JR, Cremers AJ, Gritzfeld JF, de Jonge MI, Hermans PW, Vidal JE, Klugman KP and Gordon SB (2014) Impact of experimental human pneumococcal carriage on nasopharyngeal bacterial densities in healthy adults. PLoS One 9, e98829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Glennie S, Gritzfeld JF, Pennington SH, Garner‐Jones M, Coombes N, Hopkins MJ, Vadesilho CF, Miyaji EN, Wang D, Wright AD et al (2016) Modulation of nasopharyngeal innate defenses by viral coinfection predisposes individuals to experimental pneumococcal carriage. Mucosal Immunol 9, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Salk HM, Simon WL, Lambert ND, Kennedy RB, Grill DE, Kabat BF and Poland GA (2016) Taxa of the nasal microbiome are associated with influenza‐specific IgA response to live attenuated influenza vaccine. PLoS One 11, e0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Schenck LP, Beck PL and MacDonald JA (2015) Gastrointestinal dysbiosis and the use of fecal microbial transplantation in Clostridium difficile infection. World J Gastrointest Pathophysiol 6, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]


