Highlights
► We study PSAP protein interactions using PSAP-Bcl-XL chimeras. ► We demonstrate the role of the C-terminal TMD of Bcl-XL in oligomerization. ► We discuss the involvement of motif GXXXXG in this interaction.
Abbreviations: PSAP/Mtch1, presenilin 1-associated protein/mitochondrial carrier homolog 1; MOM, mitochondrial outer membrane; IMS, intermembrane space; MOMP, mitochondrial outer membrane permeabilization; TMD, transmembrane domain; mRFP, monomeric red fluorescent protein; VAMP2, vesicle-associated membrane protein 2
Keywords: Apoptosis, Mitochondria, Bcl-XL, Oligomerization, Transmembrane domain
Abstract
Bcl-XL is a pro-survival member of the Bcl-2 family that can be found in the outer mitochondrial membrane and in soluble cytosolic homodimers. Bcl-XL can bind pro-apoptotic members of this family preventing them from activating the execution phase of apoptosis. Bcl-XL has been shown to homodimerize in different ways, although most binding and structural assays have been carried out in the absence of its carboxyl terminal transmembrane domain. We show here that this domain can by itself direct protein oligomerization, which could be related to its previously reported role in mitochondrial morphology alterations and apoptosis inhibition.
Structured summary of protein interactions
Vamp2 physically interacts with Vamp2 by blue native page (View interaction)
Vamp2 physically interacts with Vamp2 by cross-linking study (View interaction)
Bcl-Xl physically interacts with Bcl-Xl by blue native page (View interaction)
Bcl-Xl physically interacts with Bcl-Xl by cross-linking study (View interaction)
1. Introduction
Apoptosis is a type of cell death characterized by the silent elimination of unnecessary or damaged cells, which is required for development and tissue homeostasis. Two major routes of apoptosis have been described: the extrinsic route is initiated at the cell surface and mediated by death receptors, like Fas-CD95 [1]; the intrinsic route is mediated by mitochondria [2], where many different signals are integrated and the final decision about the fate of the cell is made. Both routes are linked in some cells by Bid, a member of the Bcl-2 protein family [3]. Bcl-2 family members participate directly in the decision step [4]. Pro-apoptotic members of this family, Bax and Bak, have been shown to permeabilize the mitochondrial outer membrane (MOM), facilitating the release to the cytosol of intermembrane space (IMS) proteins like cytochrome c [5], AIF [6] or Smac/DIABLO [7], [8]. These proteins then activate a series of caspase-dependent or -independent mechanisms that dismantle the cell. A second mechanism for the release of apoptotic factors from the IMS involves permeability transition, which allows the entry of water in the matrix with subsequent burst of the MOM [9].
A complex interplay between pro-apoptotic and pro-survival Bcl-2 family members controls MOM permeabilization (MOMP), considered the point-of-no-return during apoptosis induction. Several models have been proposed to explain how Bcl-2 members regulate MOMP through interactions between pro-survival and pro-apoptotic proteins, both in solution and at the MOM [10]. Activation of these proteins involves conformational changes, specially relevant for Bax, which usually resides in the cytosol and requires a conformational change to expose hydrophobic domains required for membrane insertion, followed by further changes necessary for pore formation through oligomerization at the MOM [11].
Homodimerization of the pro-survival protein Bcl-XL in the cytosol involves a C-terminal membrane-targeting α helix from one monomer and a hydrophobic groove on the other monomer [12], although other dimerization modes have been described [13], [14], [15], [16]. Release of that helix from the hydrophobic groove allows its insertion into the MOM. Most interaction and structural studies carried out with this protein employed deletion mutants lacking the C-terminal helix. This mutant can also bind membranes through its N-terminal domain [17], and Bax, Bcl-2 and Bcl-XL have been reported to insert α-helices 5 and 6 into the MOM during apoptosis [18], [19], [20]. Many interaction and structural studies have been carried out in the absence of membranes, underestimating their role in protein interactions and conformational changes [17], [21].
Since the C-terminal helix (transmembrane domain or TMD) of Bcl-XL and flanking sequences contain the necessary information for MOM targeting and insertion in a C-in N-out orientation [22], we used this domain to target fragments of PSAP/Mtch1 to the outer mitochondrial membrane [23]. PSAP was first identified as a presenilin-1 associated protein with homology to inner membrane mitochondrial carriers, and therefore is also known as mitochondrial carrier homolog 1 (Mtch1) [24]. The importance of its closest homolog, Mtch2, in Bid-induced apoptosis has been reported recently [25]. We reported that PSAP is a MOM protein with two pro-apoptotic domains [23]. Since apoptosis induction by these domains could depend on interactions with other proteins, we sought to analyze PSAP interactions by crosslinking. These assays suggested that the TMD of Bcl-XL could be involved in oligomerization, which was confirmed analyzing fusions to monomeric red fluorescent protein (mRFP).
2. Materials and methods
2.1. General reagents
All reagents were of molecular biology grade. Restriction enzymes were from Roche, Stratagene, Invitrogen, Fermentas and New England Biolabs; Accuprime Pfx DNA polymerase, custom-made primers and T4 DNA Ligase were from Invitrogen; Pfu polymerase, from Stratagene.
2.2. Construction of expression vectors
Expression vectors containing PSAP or Mtch2 sequences have been previously described [23]. pJAC295, expressing myc-mRFP-TMD Bcl-XL (39.1 kDa), was constructed by amplifying the mRFP sequence from another vector by PCR with primers containing Eco RI (mRFPEcoF: 5′-CTAGGATCGAATTCGGATGGCCTCCTCCGAGGACGT-3′) and Hind III (mRFPHindR: 5′-CTAGGATCAAGCTTGGCGCCGGTGGAGTGGCGGC-3′) restriction sites. The digested PCR product was used to replace PSAP sequences in the vector expressing myc-PSAP65-112-Bcl (which contains the TMD of Bcl-XL) [23]. The resulting vector expresses mRFP preceded by a myc tag and followed by the sequence KLESRKGQERFNRWFLTGMTVAGVVLLGSLFSRK, where the first two amino acids (kl) correspond to the Hind III site and the remaining amino acids, to the TMD of Bcl-XL.
The same approach was used to construct vector pAOC2, expressing myc-mRFP-TMD VAMP2 (32.2 kDa), replacing PSAP sequences in a vector that expresses myc-PSAP39-168-Vamp (which contains the TMD of VAMP2) [23]. In this case, sequence KLLKRKYWWKNLKMMIILGVICAIILIIIIVYFSS followed mRFP. pAL2 (28.7 kDa), expressing myc-mRFP, was constructed by digesting pJAC295 with Hind III and Not I, to eliminate the TMD of Bcl-XL, blunting with Pfu polymerase, and re-ligating. Vectors were transformed into E. coli JM109 from Promega. Correct clones were confirmed by restriction digestion and sequencing. pAOC5, expressing myc-Bcl-XL (full length Bcl-XL preceded by a myc tag, 28.6 kDa), was constructed by carrying out a PCR on a vector containing the cDNA for Bcl-XL with primers BCLXLECOF (5′-CTAGGATCGAATTCGAATGTCTCAGAGCAACAACCGGGAGC-3′) and BLCLXLNOTR (5′-CTAGGATCGCGGCCGCTCATTTCCGACTGAAGAGTGAG-3′). The PCR product was digested with Eco RI and Not I and inserted into pCMVMyc. pAOC6, expressing myc-BclXL-ΔTMD (Bcl-XL without its carboxyl terminal transmembrane domain, preceded by a myc tag, 26.7 kDa), was constructed by PCR with primers BCLXLECOF and BCLXLWOTMXHOR (5′-CTAGGATCCTCGAGGGCTGCTGCATTGTTCCCATAG-3′). The product was digested with Eco RI and Xho I and cloned into pCMVMyc. pAOC7, expressing myc-BclXL-TMD2Pit2 (BclXL preceded by a myc tag and with its carboxyl terminal domain replaced by the second transmembrane domain of Pit 2, 27.3 kDa) was constructed by doing PCR on vector pVLG60 (pEGFpN1-PSAP-DTM1-Pit2TM2, see Lamarca et al. [26]) with primers Pit2TM2XhoF (5′-CTAGGATCCTCGAGAGGCAGGCATGCATTTTAGCTTC-3′) and Pit2TM2NotR (5′-CATGGTACGCGGCCGCTCATTTGGCGCCTAGTAACACGGAG), the product was digested with Xho I and Not I and used to replace the equivalent fragment in vector pAOC6. The sequence of the second transmembrane domain of Pit 2 (TMD2Pit2) used is RQACILASIFETTGSVLLGAK. pAOC8, which expresses myc-BclXL-TMD3Pit2 (BclXL preceded by a myc tag and with its carboxyl terminal domain replaced by the third transmembrane domain of Pit 2, 27.3 kDa), was generated by PCR with primers Pit2TM3XhoF (5′-CTAGGATCCTCGAGATGGCTGGGGAAGTTAGTGC-3′) and Pit2TM3NotR (5′-CTAGGATCGCGGCCGCTCACAGGAAGGAAGCAATCAGCT-3′) in the same way as done with pAOC7, but using vector pVLG61 (pEGFpN1-PSAP-DTMI-Pit2TM3, see Lamarca et al. [26]) as template for the PCR. The sequence of the third transmembrane domain of Pit2 (TMD3Pit2) used is MAGEVSAMVGSAVWQLIASFL.
2.3. Cell culture and transfection
HEK293 cells and HeLa cells were cultured as previously described [26] and transfected using GeneJuice Reagent (Novagen) at a confluence of 80% in 24-well plates.
2.4. Crosslinking and immunoblotting
12 h post-transfection, cells were washed twice with 500 μl phosphate-buffered saline (PBS) and resuspended in 180 μl crosslinking buffer (20 mM sodium phosphate, 150 mM NaCl, pH 7.5) containing 0.2 mM phenyl-methyl-sulfonyl-fluoride (PMSF). To detect de effect of expression of chimeras on Bax and Bak oligomerization, 12 h post-transfection cells were treated with 30 μM camptothecin for 24 h, and then processed as indicated. Samples were divided in two tubes, 10 μl of a 25 mM solution in DMSO of the amine-reactive membrane-permeant crosslinker BSOCOES (Bis[2-(succinimidooxycarbonyloxy)ethyl]sulfone) (Pierce) was added to one tube and 10 μl DMSO to the second tube, and incubated for 1 h or 20 min (for Bax and Bak) at room temperature with agitation. 5 μl quenching solution (1 M Tris Hcl, pH 7.5) was then added and samples agitated for 15 min at room temperature. Cells were recovered by centrifugation and analyzed using SDS–PAGE (Laemmli) followed by immunoblotting with anti-myc (Invitrogen), anti-PSAP MGAS [26], anti-Bax (Millipore) or anti-Bak (Millipore) antibodies after transfer onto polyvinylidene fluoride (PVDF) membranes (Invitrogen), as described [23]. Proteins were visualized by incubation with goat-anti mouse or goat anti-rabbit secondary antibodies conjugated to HRP and detected by ECL. Molecular mass markers were from Invitrogen (BenchMark Pre-Stained Protein Ladder) or Bio-Rad (Precision Plus Protein Standards).
2.5. Blue-native electrophoresis (BN-PAGE)
BN-PAGE was carried out with digitonin-treated cells using the NativePAGE™ Novex® Bis–Tris Gel System from Invitrogen, 4–16% gradient gels, following instructions supplied by the manufacturer. 10 μg protein was loaded per well. Native protein standards were from Invitrogen (NativeMark Unstained Protein Standard).
2.6. Cell viability and death assays
We used the Cell Proliferation Kit II (XTT) (Roche) as a quick method to measure cell death induced by camptothecin. HEK293 cells were cultured and transfected in 96-well plates, 12 h post-transfection cells were treated with 30 μM camptothecin or DMSO and 24 h later they were incubated with XTT. Two independent assays were carried out in triplicate.
For trypan blue exclusion assays, HEK293 cells were cultured in 24-well plates until they reached 80% confluency and then transfected and treated with camptothecin or DMSO as indicated above. 24 h post-transfection wells were washed with PBS, which was transferred to tubes to avoid loosing detached cells. Attached cells were trypsinized for 5 min and mixed with detached cells in PBS from the same well. Complete media, with serum, was added to inactivate trypsin. Cells were mixed with trypan blue (Roche) in a 1:1 ratio and counted using a hemacytometer. For each sample, four quadrants were scored twice, with cell numbers ranging between 18 and 35 per quadrant. For one of the experiments an automatic cell counter (Countess from Invitrogen) was used, obtaining similar results as with manual counts. Each experiment was carried out in triplicates three independent times.
2.7. Immunocytochemistry
For subcellular localization of Bcl-XL chimeras, cells grown on round coverslides in 24-well plates were first incubated with 25 nM Mitotracker Red CMX-Ros (Invitrogen) for 30 min at 37 °C, the culture media replaced with fresh media and incubated again for 30 min at 37 °C. Cells were then fixed with a solution containing 50% culture media and 50% of a 3.7% formaldehyde solution in PBS for 5 min, followed by a 15-min incubation with 3.7% formaldehyde solution in PBS, and then blocked and permeabilized with a solution containing 1% BSA and 0.1% TX-100 in PBS for 1 h. After three 10-min washes with PBS, cells were incubated with an anti-myc antibody from Invitrogen at a 1:700 dilution for 2 h, washed with PBS and incubated with an Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (Molecular Probes) for 1 h, washed again with PBS and mounted for microscopy using Fluoromount G (Sourthernbiotech).
Subcellular localization of mRFP chimeras was carried out as for Bcl-XL chimeras but using 100 nM Mitotracker Green.
2.8. Microscopy
We used a Leica DMI6000B inverted fluorescence microscope with structured illumination (Optigrid) and software Metamorph. For red fluorescent protein and Mitotracker red we used a BP560/40 excitation filter and a BP645/75 emission filter with a 595 nm dicroic, and for Mitotracker green and Alexa Fluor 488, we used a filter for GFP, excitation BP470/40, emission BP525/50 and 500 nm dicroic.
3. Results
In order to study PSAP function we generated several deletion mutants which were used to study mitochondrial import and induction of apoptosis by this protein [23]. We targeted PSAP fragments to the MOM by fusion to the TMD of Bcl-XL, demonstrating that MOM localization was required for apoptosis induction, since endoplasmic reticulum-targeting by fusion to the TMD of VAMP2 [27] did not induce apoptosis.
Since apoptosis induction by PSAP could be mediated by interactions with other proteins, we attempted to detect PSAP-interacting proteins by crosslinking, using some of the above-mentioned chimeras. When HEK293 cells expressing the first 168 residues of PSAP fused to the TMD of Bcl-XL (PSAP1–168TMD Bcl-XL, 21 kDa) were treated with BSOCOES, a band was observed by western blot suggesting dimerization of the protein (Fig. 1 , lane 2). Similar results were obtained with the protein containing the TMD of VAMP2 (PSAP1–168TMD VAMP2, 21.2 kDa). Since both proteins share the same PSAP sequences, we first assumed that PSAP was mediating dimerization. Nevertheless, a literature search indicated that the TMD of VAMP2 is responsible for its homo or heteromerization at the ER membrane [28], suggesting that the dimers observed in our experiments could depend on TMD instead of PSAP sequences. This interpretation could also explain a band migrating between those corresponding to the monomer (21.2 kDa) and dimer (42.4 kDa) in lanes 3 and 4 of Fig. 1, as a heterodimer between endogenous VAMP2 (12.6 kDa) and PSAP1–168TMD VAMP2 (expected molecular mass: 33.8 kDa, marked with an asterisk in Fig. 1). Note that some of the bands are also present in the absence of crosslinker, suggesting strong interactions.
Fig. 1.

Crosslinking of PSAP chimeras. Western blot of crosslinker-treated cells (+) or untreated (−) expressing PSAP1-168-TMD Bcl-XL or PSAP1-168-TMD VAMP2, separated on a 10% SDS gel and detected with anti-MGAS. Sizes of molecular mass markers are indicated on the left. Bands likely corresponding to heterodimers of PSAP1-168-TMD VAMP2 with endogenous VAMP2 are indicated with an asterisk on the right.
When similar constructs containing shorter PSAP fragments were used, oligomerization was observed. Fig. 2 shows the results obtained with a construct containing residues 65–112 preceded by a myc tag and followed by the TMD of Bcl-XL (11.2 kDa). Crosslinking of a protein chimera containing Mtch2 sequences (16 kDa) instead of PSAP sequences (lanes 3 and 4 in Fig. 2) also produced oligomers.
Fig. 2.
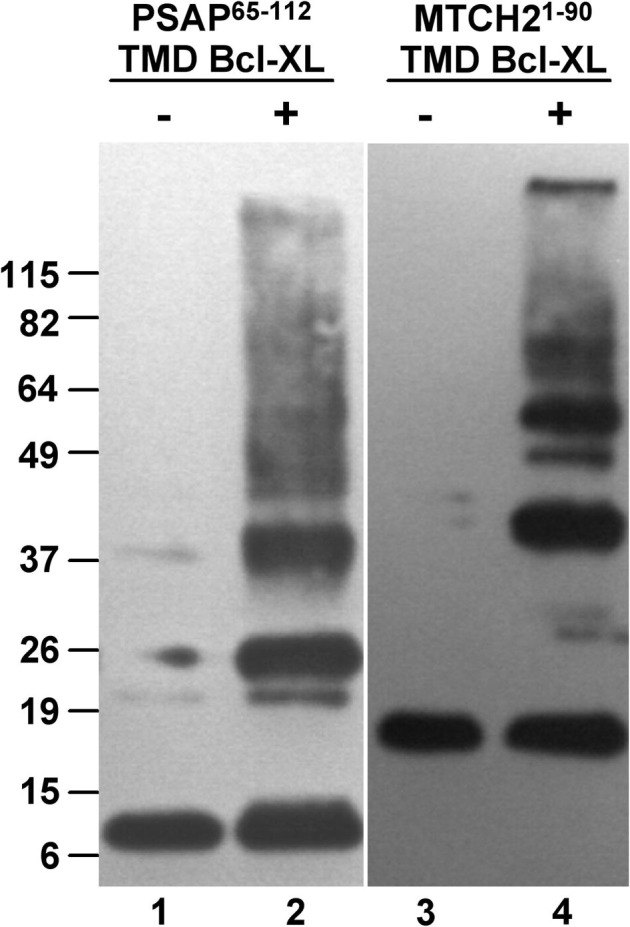
Crosslinking of PSAP and Mtch2 chimeras. Western blot of crosslinker-treated cells (+) or untreated (-) expressing myc-PSAP-65-112-TMD Bcl-XL or myc-Mtch2-1-90-TMD Bcl-XL, separated on a 10% SDS gel and detected with anti-myc. Sizes of molecular mass markers are indicated on the left.
Our constructs contained parts of PSAP or Mtch2 and the TMD of Bcl-XL, proteins that can all be inserted into the MOM, therefore we could not rule out a mixed effect due to different sequences from either protein. In fact, whether dimers or oligomers were observed depended on the sequences attached to the TMD (Fig. 1 and Fig. 2). In order to clarify this subject, we fused the TMD of Bcl-XL or that of VAMP2 to monomeric red fluorescent protein (mRFP) preceded by a myc tag for detection. We used myc-mRFP (28.7 kDa) as control. Crosslinking assays indicated that similar complexes could be detected when each TMD was fused to mRFP (Fig. 3 A), whereas myc-mRFP remained as a monomer, therefore indicating that the TMDs were responsible for the dimers observed. The contribution of PSAP sequences to dimers and oligomers shown in Fig. 1, Fig. 2 is under investigation and will be reported elsewhere.
Fig. 3.
Crosslinking and blue-native electrophoresis of mRFP chimeras. (A) Western blot of crosslinker-treated (+) or untreated (−) cells expressing myc-mRFP-TM Bcl-XL, myc-mRFP-TMD VAMP2 or myc-mRFP alone, separated on a 10% SDS gel and detected with anti-myc. Sizes of molecular mass markers are indicated on the left. Bands corresponding to dimers (63.7 and 64.4 kDa) are indicated with an asterisk on the right. (B) Blue-native electrophoresis analysis of mRFP chimeras. Digitonin-solubilized proteins from cells expressing myc-mRFP-TMD Bcl-XL, myc-mRFP-TMD VAMP2 or myc-mRFP alone were separated on native 4–16% gradient gels, blotted and detected with anti-myc. Sizes of native molecular mass markers are indicated on the left.
In order to analyze these interactions with a different technique, we used blue-native electrophoresis (Fig. 3B), confirming that fusion of mRFP to either the TMD of Bcl-XL or the TMD of VAMP2 induced oligomerization of the protein. The pattern obtained indicated the presence of oligomers up to pentamers, although less abundant (or less stable) than the dimers. These data clearly indicate the involvement of the TMD of Bcl-XL in self-association, an event that had not been reported previously. The fact that only dimers could be observed upon crosslinking could be due to the location of the reacting amino groups in the oligomers. Crosslinking covalently fixes those interactions where reacting groups (amino groups in this case) are localized in close proximity, and uncrosslinked molecules are separated later during denaturing electrophoresis. Blue native electrophoresis does not rely on specific reactive groups but on the overall stability of protein complexes where individual molecules are held together by non-covalent bonds. It could be that, in our complexes, reacting amino groups are only located closely between monomers of a single dimer, and dimers associate in oligomers in such a way that adjacent dimers cannot be crosslinked with each other. This could explain why dimers are mainly observed upon crosslinking whereas oligomers are observed after blue-native electrophoresis.
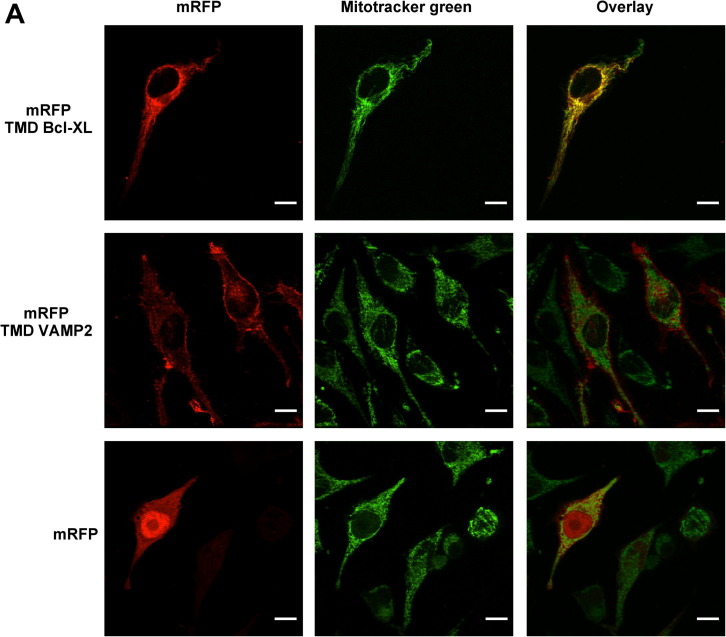
We analyzed the subcellular localization of our chimeras by fluorescence microscopy, using Mitotracker green to label mitochondria. As expected, mRFP-TMD Bcl-XL localized to mitochondria, mRFP-TMD VAMP2 localized to membranes and mRFP distributed evenly throughout the cell (Fig. 4 A).
Fig. 4.
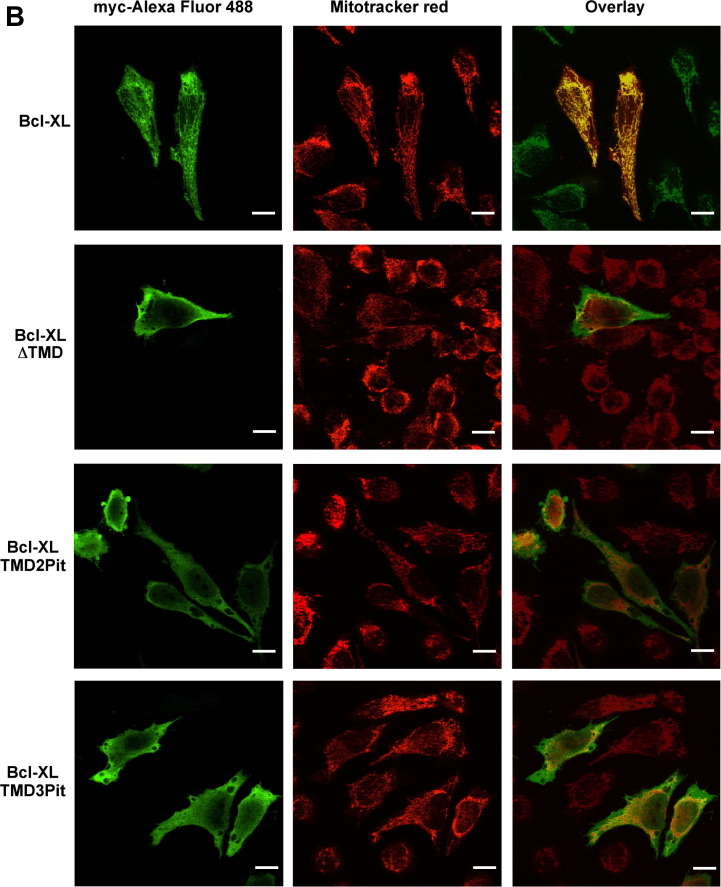
Subcellular localization of mRFP and Bcl-XL chimeras. (A) HeLa cells were grown on round coverslips, transfected with vectors encoding myc-mRFP-TMD Bcl-XL, myc-mRFP-TMD VAMP2 or myc-mRFP, incubated with Mitotracker green, fixed with formaldehyde and mounted for microscopy. (B) HeLa cells grown as in A and transfected with myc-BclXL, myc-BclXL-ΔTM, myc-BclXL-TMD2Pit or myc-BclXL-TMD3Pit, incubated with Mitotracker red CMX-Ros, with an anti-myc antibody, with goat anti-mouse Alexa fluor 488-conjugated antibody and mounted for microscopy. Images were acquired with a Leica DMI6000B inverted fluorescence microscope using structured illumination (Optigrid) with filters suitable for mRFP/Mitotracker Red and for Mitotracker green/Alexa fluor 488, and overlayed to show co-localization using software Metamorph. Bar is 10 μm.
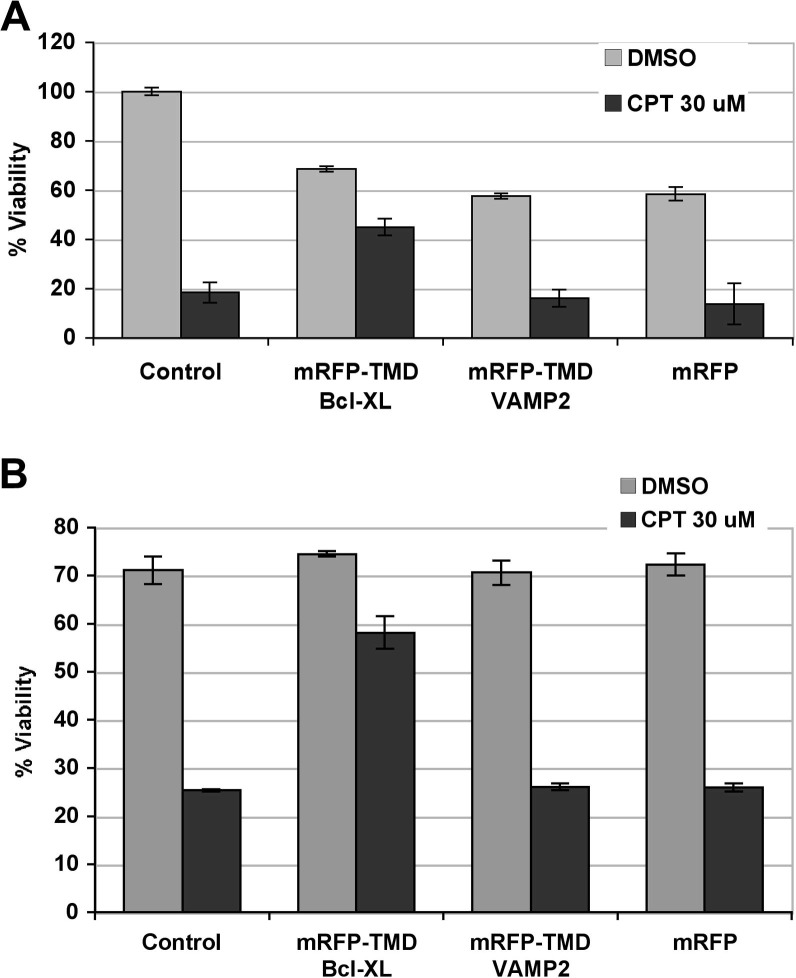
The TMD of Bcl-XL fused to yellow fluorescent protein has been reported to alter mitochondrial morphology and to moderately protect cells against staurosporine-induced apoptosis [29]. In order to find out if our mRFP-TMD Bcl-XL chimera also protected cells against camptothecin-induced cell death we carried out cell viability assays using XTT, which were later confirmed by trypan blue exclusion assays. The results, shown in Fig 5 , clearly indicated that the TMD of Bcl-XL fused to mRFP protected cells from death, whereas the TMD of VAMP2 fused to mRFP or mRFP alone did not.
Fig. 5.
Protection of mRFP-TMD Bcl-XL against camptothecin-induced cell death. HEK293 cells transfected with myc-mRFP-TMD Bcl-XL, myc-mRFP-TMD VAMP2 or myc-mRFP alone were treated with DMSO or with 30 μM camptothecin for 24 h starting 12 h post-transfection. (A) Cells were analyzed using the Cell Proliferation Kit II (XTT) (Roche). Results of two independent assays carried out in triplicates are shown. (B) Cells were analyzed by trypan blue exclusion. Results of three independent assays carried out in triplicates are shown. Error bars indicate standard deviation.
Since Bcl-XL can prevent Bax and Bak oligomerization, we analyzed the state of oligomerization of these two proapoptotic proteins upon transfection of cells with mRFP chimeras and induction of apoptosis with camptothecin. We did not see any effect on Bax and Bak oligomerization in cells transfected with either mRFP-TMD Bcl-XL, mRFP-TMD VAMP2 or mRFP alone (Fig 6 ).
Fig. 6.
Effect of overexpression of chimeras in Bax and Bak oligomerization. HEK293 cells were transfected with vectors encoding myc-mRFP-TMD Bcl-XL, myc-mRFP-TMD VAMP2 or myc-mRFP, treated with 30 μM camptothecin for 24 h and the effect on Bax and Bak oligomerization analyzed by crosslinking with BSOCOES. Fractions from each sample were analyzed by western blot using antibodies against Bax, Bak or myc (to detect overexpressed proteins). Sizes of molecular mass markers run alongside are indicated on the left in kDa. −, without BSOCOES; +, with BSOCOES.
Even though Bcl-XL is able to dimerize in absence of its TMD under some conditions, we analyzed how the absence of its TMD or the presence of a TMD from an unrelated protein affected its dimerization. For this, we constructed a new vector to express Bcl-XL without its TMD, as well as another vector encoding Bcl-XL with its TMD replaced by the second TMD of Pit 2, a plasma membrane sodium transporter [30]. We expressed these constructs in cells and carried out crosslinking assays using BSOCOES. Under these conditions, we could detect dimers of full-length Bcl-XL (Fig 7 , lane 2) and no dimers of Bcl-XL lacking its TMD (Fig 7, lane 4). Curiously, Bcl-XL with its TMD replaced by the second TMD of Pit2 was also able to dimerize (Fig 7, lane 6).
Fig. 7.
Effect of different TMDs in Bcl-XL dimerization. Full length Bcl-XL (Bcl-XL), Bcl-XL without its carboxyl terminal TMD (Bcl-XL ΔTMD), Bcl-XL with the second TMD of Pit 2 (Bcl-XL TMD2Pit) or Bcl-XL with the third TMD of Pit 2 (Bcl-XL TMD3Pit) all preceded by a myc tag, were overexpressed in HEK293 cells, treated with the crosslinker BSOCOES and analyzed by western blot with an anti-myc antibody. −, without BSOCOES; +, with BSOCOES.
Analysis of the sequences of the TMD of Bcl-XL and the second TMD of Pit 2 revealed the presence of the sequence motif GXXXXG (where G is glycine and X, any amino acid), in tandem in Bcl-XL (GMTVAGVVLLG) and alone in the TMD of Pit2 (GSVLLG). Note that five out of six amino acids are conserved between sequence GVVLLG in Bcl-Xl and sequence GSVLLG in the second TMD of Pit2. Since the sequence motif GXXXG had been reported to be involved in dimerization of the BH3-only Bcl-2 family member BNIP3 through its TMD, and also in other proteins [31], [32], [33], [34], [35], [36], [37], [38], which is very similar to the motifs found in the TMD of Bcl-XL and the second TMD of Pit2, we constructed a new vector where the TMD of Bcl-XL was replaced by the third TMD of Pit2, which lacks sequence GXXXXG. Crosslinking assays with this new chimera indicated that it was unable to dimerize (Fig 7, lane 8).
We analyzed the subcellular localization of these chimeras, with myc-BclXL localizing to mitochondria as expected and the remaining chimeras showing a diffuse localization in the cell (Fig 4B). This also indicates that sequences involved in dimerization and mitochondrial targeting within the TMD of Bcl-XL are different, since the second TMD of Pit2 is able to induce oligomerization but not mitochondrial localization.
4. Discussion
We have shown that the transmembrane C-terminal domain of Bcl-XL can directly participate in oligomerization. This is a mode of self-association unreported for Bcl-XL. Bcl-2 family members control their activities through a complex network of protein-protein interactions, posttranslational modifications, transcriptional control or protein degradation [11]. Conformational changes induced upon heteromeric or homomeric protein interactions are very important for their activities.
Bcl-XL has been shown to dimerize by different mechanisms under several conditions [12], [13], [14], [15], [16], [17]. Many of these studies, as well as structural studies, used Bcl-XL mutants lacking the C-terminal TMD under non-physiological assay conditions, although its presence is likely to affect the overall fold of the protein, conformational changes and protein interactions.
Bcl-XL has been reported to regulate apoptosis by heterodimerization-dependent and -independent mechanisms. The former implies binding to pro-apoptotic proteins; the latter, could depend, at least in part, on the formation of an ion channel that can counteract the effects of MOM permeabilization by proteins like Bax or Bak [39]. Interestingly, C-terminal cleavage products of Bcl-XL can form a pore large enough to allow cytochrome c release [40], changing a pro-survival protein into a pro-apoptotic one, what has been also described for Bcl-2 upon binding of a Nur77-derived peptide [41]. Zheng et al. [29] reported changes in mitochondrial morphology and protection against apoptosis mediated by a YFP-TM Bcl-XL fusion protein, suggesting that this TMD could be required for a bioenergetic function of Bcl-XL distinct from BH3 domain sequestration. Our results suggest that oligomerization mediated by the TMD of Bcl-XL could be involved in these activities.
The TM domains of other Bcl-2 family members have been also reported to be involved in protein dimerization, like the TMD of BH3-only protein BNIP3 [37] or the TMD of Bax [42]. In the case of BNIP3, the sequence GXXXG has been shown to directly participate in dimerization. Interestingly, a similar sequence is present in the TMD of Bcl-XL, but in tandem (GMTVAGVVLLG: GMTVAG and GVVLLG), with one more amino acid between the two glycines (GXXXXG). The presence of sequence GXXXG in tandem had been previously described in other proteins [35], [38]. Curiously, a very similar sequence containing that motif is also present in the second TMD of Pit2 (GSVLLG), which we used to replace the TMD of BclXL expecting to block dimerization (before we analyzed the TMD sequences), and Bcl-XL with that TMD also dimerizes. BclXL with its TMD replaced by the third TMD of Pit2, which does not contain that sequence motif, does not dimerize, strongly suggesting the importance of motif GXXXXG in dimerization.
Sequence GXXXG was first reported to be involved in TMD dimerization by Engelman and coworkers [34], [43]. Furthermore, within these motifs, residues V and L are also usually present close to G residues, as is the case for the TMD of Bcl-XL and the second TMD of Pit2. Sequence specificity in the dimerization of transmembrane alpha-helices was first reported by Lemmon et al. [31], reporting later that the pattern LIxxGVxxGVxxT is involved in dimerization [44]. Furthermore, the GXXXG motif has been involved in the formation of a membrane channel by the Helicobacter pylori vacuolating toxin [38], where this motif appears in three tandem repeats. This motif has also been found in several other proteins, like subunits e [45] and g [46] of yeast mitochondrial ATP synthase, APH-1 [47], ABCG2 [48], the SARS coronavirus spike protein (were it is involved in trimerization) [49], prion protein [50], amyloid precursor protein [51], the two-peptide bacteriocin lactococcin G [52], carnitine palmitoyltransferase 1A [53], ErbB [54], protein FliH and its Type III secretion homologue YscL [55], the Japanese encephalitis virus precursor membrane (prM) protein [56], the PufX polypeptide of Rhodobacter sphaeroides RC-LH1 photosynthetic complex [57], human organic anion transporter 1 (hOAT1) [58], VDAC1 [59] and the NS4B protein of hepatitis c virus [60]. A review about some of these proteins can be seen in Senes et al. [61].
Although in the TMD of Bcl-XL the two glycines are separated by four residues instead of 3 as in the GXXXG motif, our results strongly suggest that this motif is mediating interaction of Bcl-XL molecules through their TMDs, since the second TMD of Pit2, with 5 out of 6 conserved residues within that GXXXXG motif also allows dimerization of the protein, whereas the third TMD of Pit2, which lacks that motif, does not dimerize when fused to Bcl-XL.
In summary, we have shown that the TMD of Bcl-XL is involved in protein dimerization and that motif GXXXXG is very likely responsible for this interaction. This will most likely have important implications in Bcl-XL function.
Acknowledgements
We thank Dr. Ester Perales and Dr. Patricio Fernández-Silva for their help with blue-native electrophoresis. We thank Drs. Isabel Marzo, Javier Sancho and Ramón Hurtado for critical reading. We thank Rubén Calvo for his help with cell culture. This work was funded by grants BFU2006-07026 from the Spanish Ministry of Education, BFU2009-11800 from the Spanish Ministry of Science and Innovation and UZ2010 BIO-03 from the University of Zaragoza to J.A.C. A.O. is the recipient of a Ph.D. Fellowship from the Banco Santander/Universidad de Zaragoza. A.L.M. was a recipient of a short-term fellowship from the Universidad Nacional Autónoma de México-CONACYT.
References
- 1.Nagata S., Goldtein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G., Reed J.C. Mitochondrial control of cell death. Nat. Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 3.Li H., Zhu H., Xu C.J., Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 4.Cory S., Adams J.M. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 5.Liu X., Kim C.N., Yang J., Jemmerson R., Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 6.Susin S.A. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 7.Du C., Fang M., Li Y., Li L., Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Verhagen A.M. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 9.Crompton M., Virji S., Doyle V., Johnson N., Ward J.M. The mitochondrial permeability transition pore. Biochem. Soc. Symp. 1999;66:167–179. doi: 10.1042/bss0660167. [DOI] [PubMed] [Google Scholar]
- 10.Leber B., Lin J., Andrews D.W. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chipuk J.E., Moldoveanu T., Llambi F., Parsons M.J., Green D.R. The BCL-2 family reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong S.Y., Gaume B., Lee Y.J., Hsu Y.T., Ryu S.W., Yoon S.H., Youle R.J. Bcl-x(L) sequesters its C-terminal membrane anchor in soluble, cytosolic homodimers. Embo J. 2004;23:2146–2155. doi: 10.1038/sj.emboj.7600225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill J.W., Manion M.K., Maguire B., Hockenbery D.M. BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 2006;356:367–381. doi: 10.1016/j.jmb.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Denisov A.Y., Sprules T., Fraser J., Kozlov G., Gehring K. Heat-induced dimerization of BCL-xL through alpha-helix swapping. Biochemistry. 2007;46:734–740. doi: 10.1021/bi062080a. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y., Lin Z., Shen X., Chen K., Jiang H., Liu D. Bcl-xL forms two distinct homodimers at non-ionic detergents: implications in the dimerization of Bcl-2 family proteins. J. Biochem. 2008;143:243–252. doi: 10.1093/jb/mvm216. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Liu D., Shen X., Chen K., Jiang H. Structure assembly of Bcl-x(L) through alpha5-alpha5 and alpha6-alpha6 interhelix interactions in lipid membranes. Biochim. Biophys. Acta. 2009;1788:2389–2395. doi: 10.1016/j.bbamem.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Thuduppathy G.R., Terrones O., Craig J.W., Basanez G., Hill R.B. The N-terminal domain of Bcl-xL reversibly binds membranes in a pH-dependent manner. Biochemistry. 2006;45:14533–14542. doi: 10.1021/bi0616652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annis M.G., Soucie E.L., Dlugosz P.J., Cruz-Aguado J.A., Penn L.Z., Leber B., Andrews D.W. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. Embo J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billen L.P., Kokoski C.L., Lovell J.F., Leber B., Andrews D.W. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol. 2008;6:e147. doi: 10.1371/journal.pbio.0060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim P.K., Annis M.G., Dlugosz P.J., Leber B., Andrews D.W. During apoptosis bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol. Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Saez A.J., Ries J., Orzaez M., Perez-Paya E., Schwille P. Membrane promotes tBID interaction with BCL(XL) Nat. Struct. Mol. Biol. 2009;16:1178–1185. doi: 10.1038/nsmb.1671. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamarca V., Marzo I., Sanz-Clemente A., Carrodeguas J.A. Exposure of any of two proapoptotic domains of presenilin 1-associated protein/mitochondrial carrier homolog 1 on the surface of mitochondria is sufficient for induction of apoptosis in a Bax/Bak-independent manner. Eur. J. Cell Biol. 2008;87:325–334. doi: 10.1016/j.ejcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Xu X., Shi Y., Wu X., Gambetti P., Sui D., Cui M.Z. Identification of a novel PSD-95/Dlg/ZO-1 (PDZ)-like protein interacting with the C terminus of presenilin-1. J. Biol. Chem. 1999;274:32543–32546. doi: 10.1074/jbc.274.46.32543. [DOI] [PubMed] [Google Scholar]
- 25.Zaltsman Y. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat. Cell Biol. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamarca V., Sanz-Clemente A., Perez-Pe R., Martinez-Lorenzo M.J., Halaihel N., Muniesa P., Carrodeguas J.A. Two isoforms of PSAP/MTCH1 share two proapoptotic domains and multiple internal signals for import into the mitochondrial outer membrane. Am. J. Physiol. Cell Physiol. 2007;293:C1347–C1361. doi: 10.1152/ajpcell.00431.2006. [DOI] [PubMed] [Google Scholar]
- 27.Hanson P.I., Heuser J.E., Jahn R. Neurotransmitter release – four years of SNARE complexes. Curr. Opin. Neurobiol. 1997;7:310–315. doi: 10.1016/s0959-4388(97)80057-8. [DOI] [PubMed] [Google Scholar]
- 28.Laage R., Rohde J., Brosig B., Langosch D. A conserved membrane-spanning amino acid motif drives homomeric and supports heteromeric assembly of presynaptic SNARE proteins. J. Biol. Chem. 2000;275:17481–17487. doi: 10.1074/jbc.M910092199. [DOI] [PubMed] [Google Scholar]
- 29.Zheng J.Y., Tsai Y.C., Kadimcherla P., Zhang R., Shi J., Oyler G.A., Boustany N.N. The C-terminal transmembrane domain of Bcl-xL mediates changes in mitochondrial morphology. Biophys. J. 2008;94:286–297. doi: 10.1529/biophysj.107.104323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salaun C., Marechal V., Heard J.M. Transport-deficient Pit2 phosphate transporters still modify cell surface oligomers structure in response to inorganic phosphate. J. Mol. Biol. 2004;340:39–47. doi: 10.1016/j.jmb.2004.04.050. [DOI] [PubMed] [Google Scholar]
- 31.Lemmon M.A., Flanagan J.M., Treutlein H.R., Zhang J., Engelman D.M. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 32.MacKenzie K.R., Prestegard J.H., Engelman D.M. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 33.Brosig B., Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russ W.P., Engelman D.M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 35.McClain M.S., Cao P., Cover T.L. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect. Immun. 2001;69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendrola J.M., Berger M.B., King M.C., Lemmon M.A. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 37.Sulistijo E.S., Jaszewski T.M., MacKenzie K.R. Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J. Biol. Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 38.McClain M.S., Iwamoto H., Cao P., Vinion-Dubiel A.D., Li Y., Szabo G., Shao Z., Cover T.L. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J. Biol. Chem. 2003;278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 39.Minn A.J. Bcl-xL regulates apoptosis by heterodimerization-dependent and -independent mechanisms. Embo J. 1999;18:632–643. doi: 10.1093/emboj/18.3.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basanez G. Pro-apoptotic cleavage products of Bcl-xL form cytochrome c-conducting pores in pure lipid membranes. J. Biol. Chem. 2001;276:31083–31091. doi: 10.1074/jbc.M103879200. [DOI] [PubMed] [Google Scholar]
- 41.Kolluri S.K. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell. 2008;14:285–298. doi: 10.1016/j.ccr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Er E., Lalier L., Cartron P.F., Oliver L., Vallette F.M. Control of Bax homodimerization by its carboxyl terminus. J. Biol. Chem. 2007;282:24938–24947. doi: 10.1074/jbc.M703823200. [DOI] [PubMed] [Google Scholar]
- 43.Senes A., Gerstein M., Engelman D.M. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 44.Lemmon M.A., Treutlein H.R., Adams P.D., Brunger A.T., Engelman D.M. A dimerization motif for transmembrane alpha-helices. Nat. Struct. Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 45.Arselin G., Giraud M.F., Dautant A., Vaillier J., Brethes D., Coulary-Salin B., Schaeffer J., Velours J. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur. J. Biochem. 2003;270:1875–1884. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 46.Saddar S., Stuart R.A. The yeast F(1)F(0)-ATP synthase: analysis of the molecular organization of subunit g and the importance of a conserved GXXXG motif. J. Biol. Chem. 2005;280:24435–24442. doi: 10.1074/jbc.M502804200. [DOI] [PubMed] [Google Scholar]
- 47.Lee S.F. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the gamma-secretase complex. J. Biol. Chem. 2004;279:4144–4152. doi: 10.1074/jbc.M309745200. [DOI] [PubMed] [Google Scholar]
- 48.Polgar O. Mutational analysis of ABCG2: role of the GXXXG motif. Biochemistry. 2004;43:9448–9456. doi: 10.1021/bi0497953. [DOI] [PubMed] [Google Scholar]
- 49.Arbely E., Granot Z., Kass I., Orly J., Arkin I.T. A trimerizing GxxxG motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry. 2006;45:11349–11356. doi: 10.1021/bi060953v. [DOI] [PubMed] [Google Scholar]
- 50.Choi J.K. Generation of monoclonal antibody recognized by the GXXXG motif (glycine zipper) of prion protein. Hybridoma (Larchmt) 2006;25:271–277. doi: 10.1089/hyb.2006.25.271. [DOI] [PubMed] [Google Scholar]
- 51.Munter L.M. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. Embo J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oppegard C., Schmidt J., Kristiansen P.E., Nissen-Meyer J. Mutational analysis of putative helix-helix interacting GxxxG-motifs and tryptophan residues in the two-peptide bacteriocin lactococcin G. Biochemistry. 2008;47:5242–5249. doi: 10.1021/bi800289w. [DOI] [PubMed] [Google Scholar]
- 53.Jenei Z.A., Borthwick K., Zammit V.A., Dixon A.M. Self-association of transmembrane domain 2 (TM2), but not TM1, in carnitine palmitoyltransferase 1A: role of GXXXG(A) motifs. J. Biol. Chem. 2009;284:6988–6997. doi: 10.1074/jbc.M808487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escher C., Cymer F., Schneider D. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J. Mol. Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Trost B., Moore S.A. Statistical characterization of the GxxxG glycine repeats in the flagellar biosynthesis protein FliH and its Type III secretion homologue YscL. BMC Microbiol. 2009;9:72. doi: 10.1186/1471-2180-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y.J., Peng J.G., Wu S.C. Characterization of the GXXXG motif in the first transmembrane segment of Japanese encephalitis virus precursor membrane (prM) protein. J. Biomed. Sci. 2010;17:39. doi: 10.1186/1423-0127-17-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crouch L.I., Holden-Dye K., Jones M.R. Dimerisation of the Rhodobacter sphaeroides RC-LH1 photosynthetic complex is not facilitated by a GxxxG motif in the PufX polypeptide. Biochim. Biophys. Acta. 2010;1797:1812–1819. doi: 10.1016/j.bbabio.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Duan P., Wu J., You G. Mutational analysis of the role of GXXXG motif in the function of human organic anion transporter 1 (hOAT1) Int. J. Biochem. Mol. Biol. 2011;2:1–7. [PMC free article] [PubMed] [Google Scholar]
- 59.Thinnes F.P. Apoptogenic interactions of plasmalemmal type-1 VDAC and Abeta peptides via GxxxG motifs induce Alzheimer’s disease – a basic model of apoptosis? Wien Med. Wochenschr. 2011;161:274–276. doi: 10.1007/s10354-011-0887-5. [DOI] [PubMed] [Google Scholar]
- 60.Han Q. Conserved GXXXG- and S/T-Like Motifs in the Transmembrane Domains of NS4B Protein Are Required for Hepatitis C Virus Replication. J. Virol. 2011;85:6464–6479. doi: 10.1128/JVI.02298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senes A., Engel D.E., DeGrado W.F. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]