Abstract
A single membrane‐bound aminopeptidase N (APN) occurs in the pea aphid (Acyrthosiphon pisum Harris) midgut, with a pH optimum of 7.0, pI of 8.1 and molecular mass of 130 kDa. This enzyme accounts for more than 15.6% of the total gut proteins. After being solubilized in detergent, APN was purified to homogeneity. The enzyme is a glycoprotein rich in mannose residues, which binds the entomotoxic lectins of the concanavalin family. The internal sequence of APN is homologous with a conservative domain in APNs, and degenerated primers of highly conserved APN motifs were used to screen a gut cDNA library. The complete sequence of APN has standard residues involved in zinc co‐ordination and catalysis and a glycosyl‐phosphatidylinositol anchor, as in APNs from Lepidoptera. APN has a broad specificity towards N‐terminal amino acids, but does not hydrolyze acidic aminoacyl‐peptides, thus resembling the mammalian enzyme (EC 3.4.11.2). The k cat/K m ratios for different di‐, tri‐, tetra‐, and penta‐peptides suggest a preference for tripeptides, and that subsites S1, S2′ and S3′ are pockets able to bind bulky aminoacyl residues. Bestatin and amastatin bound APN in a rapidly reversible mode, with K i values of 1.8 µm and 0.6 µm, respectively. EDTA inactivates this APN (k obs 0.14 m −1·s−1, reaction order of 0.44) at a rate that is reduced by competitive inhibitors. In addition to oligopeptide digestion, APN is proposed to be associated with amino‐acid‐absorption processes which, in contrast with aminopeptidase activity, may be hampered on lectin binding.
Keywords: aminopeptidase N, Aphididae, glycosyl‐phosphatidylinositol (GPI) anchor, mannose lectin receptor, substrate specificity
Abbreviations
- APN
aminopeptidase N
- ConA
concanavalin A
- ConBr
concanavalin A ortholog from Canavalia brasiliensis
- EST
expressed sequence tag
- GPI
glycosyl‐phosphatidylinositol
- LeupNA
l‐leucine‐p‐nitroanilide
- WGA
wheat germ agglutinin
Aminopeptidase N (APN) is an exopeptidase that catalyzes the sequential release of N‐terminal amino acids of peptides (EC 3.4.11.2). It is found in the midgut of insect larvae either as soluble enzyme or associated with the microvillar membrane. Properties of APN preparations from midgut tissue have been described for at least six orders of insects [1, 2].
APNs are the major proteins in some insect midgut microvillar membranes. Probably linked to its abundance, APN is one of the targets of the insecticidal Bacillus thuringiensisδ‐endotoxins [3, 4, 5, 6]. These toxins, after binding to receptors such as APNs, form channels through which midgut cell contents leak, finally leading to insect death [7]. Also, in humans, APN is the binding site for coronavirus infection [8].
Such findings raised interest in this enzyme, leading to APN cloning from target insects in the Lepidoptera family [3, 6, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]. All these APNs are inserted into the midgut microvillar membrane by a C‐terminal glycosyl‐phosphatidylinositol (GPI) anchor. Sequence comparisons with vertebrate and fungal aminopeptidases showed that their most striking similarities were in the zinc‐binding motif, including residues His142, His146, and Glu166 (putative zinc ligands, numbering according to thermolysin), and Glu143 (catalytic active residue). This conserved motif classifies the enzymes as members of the M1 family of neutral zinc metallopeptidases [21]. In spite of these research efforts, there are few detailed studies on the substrate specificity of lepidopteran microvillar APNs [1, 2].
Kinetic data on a midgut APN from Coleoptera showed its similarity to mammalian APN, a family showing a broad specificity towards aminoacyl β‐naphthylamides. Chemical modification experiments revealed that a metal ion, a carboxylic group, and the lateral chains of His, Arg and Tyr are important for enzyme activity [22, 23].
APN sequences obtained so far are restricted to the Lepidoptera, although insect targets of B. thuringiensis toxins now include many Coleoptera (beetles) and Diptera (flies, mosquitoes). A homologue aminopeptidase has been found in the Drosophila genome [24], and several enzymes found in Rhynchosciara americana have been characterized [25, 26]. No membrane‐bound aminopeptidase from Hemiptera (bugs, aphids, whiteflies, scales) has been studied so far [2], nor has any truly hemipteran‐active B. thuringiensis toxin yet been identified. In fact, the Hemipteran Dysdercus peruvianus has a soluble aminopeptidase [27]. Although they are key components of trophic and toxic interactions involving insects, comparative structural and functional data on insect aminopeptidases are lacking.
In aphids, APN occurs in the apical network of lamellae, which in this insect replaces the usual regularly arranged microvilli [28]. Furthermore, Sauvion et al. [29] found strong interaction of the lectin concanavalin A (ConA) with putative glycosylated receptors at the cell surface. In this paper, we describe the purification to homogeneity of the midgut membrane‐bound APN from adult pea aphids Acyrthosiphon pisum (Hemiptera: Aphididae) and the cloning of its corresponding cDNA. The data show that this APN prefers tripeptides, has broad amino‐acid specificity, and is the most important mannose‐specific lectin‐binding site in midgut membranes.
Results
Solubilization of A. pisum membrane‐bound midgut APN
About 98% of APN midgut activity [l‐leucine‐p‐nitroanilide (LeupNA) as substrate] was found to be membrane‐bound. The soluble fraction was eluted as a single peak from a Mono Q column, with a retention time similar to that of the solubilized enzyme (data not shown). The soluble enzyme was disregarded in further studies.
Acyrthosiphon pisum membrane‐bound APN was well solubilized by all detergents tested (detergent concentration, % solubilization, % activity recovery): Chaps (32.7 mm, 90 ± 6%, 97 ± 8%), deoxycholate (7.3 mm, 91 ± 7%, 81 ± 9%), Triton X‐100 (9.7 mm, 96 ±5%, 116 ± 9%), Nonidet (9.7 mm, 91 ± 9%, 79 ±8%), Control (8 ± 1% solubilization, 100 ± 8% recovery). As the best yield (solubilization) and recovery of activity were found with Triton X‐100, this detergent was chosen for preparing the starting sample.
Purification of A. pisum midgut APN
The solubilized A. pisum APN was purified to homogeneity by one chromatographic step using a Mono Q column (Fig. 1A). From starting material consisting of 300 guts, with total activity 2.2 U and 343 µg protein, it was possible to recover 28 µg purified APN with specific activity 40.3 U·mg−1. The final yield was ≈ 50%, with a purification factor of 6.4. SDS/PAGE of purified APN resulted in a single 150‐kDa protein band (Fig. 1B). The enzyme was found in the midgut as a major protein band and was preferentially solubilized by Triton X‐100 (Fig. 1B, lane 2).
Figure 1.
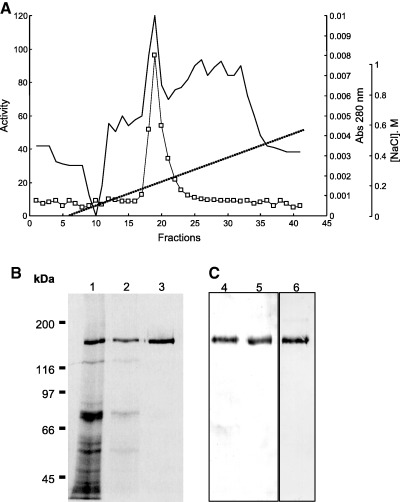
Chromatographic purification of midgut aminopeptidase from A. pisum. (A) Chromatography on Mono Q equilibrated with 20 mm Tris/HCl buffer (pH 7.0)/0.1% Triton X‐100. Elution was accomplished with a gradient of 0–600 mm NaCl gradient in the same Tris buffer (substrate used LeupNA). (B) SDS/PAGE of samples obtained after the steps from A. pisum APN purification (12% polyacrylamide slab gels, silver staining). Lane 1, midgut homogenate; lane 2, Triton X‐100‐released proteins from midgut cell membranes; lane 3, Mono Q eluate (purified aminopeptidase). (C) Glycoprotein detection (Dig Glycan detection kit), after western blots of proteins. Lane 4, midgut homogenate; lane 5, purified APN; lane 6, purified with the differentiation kit with the mannose‐specific lectin Galanthus nivalis agglutinin.
SDS/PAGE of proteins in fractions eluted from a gel‐filtration column showed a correspondence between eluted activity and band intensity in stained gels (not shown), indicating homogeneity of the purified enzyme. The molecular mass calculated from gel filtration was 200 ± 30 kDa, a little higher than that obtained from SDS/PAGE.
In addition, APN can be purified using a single chromatographic step in ConA–Br‐Sepharose (data not shown). The purified protein had the same mobility on SDS/PAGE and the same internal peptide sequence (see below) as APN purified on a Mono Q column.
Properties of the purified APN from A. pisum
Acyrthosiphon pisum APN is a glycoprotein (Fig. 1C) and seems to be the major and/or most glycosylated protein from aphid midgut extracts (Fig. 1C, lane 4). It binds specifically to the lectin (Galanthus nivalis agglutinin) that recognizes a mannose moiety (Fig. 1C, lane 6). This agrees with the APN pattern of elution from ConA–Br‐Sepharose columns (see above).
The APN purified from A. pisum had a pH optimum of 7.0 ± 0.5 (Fig. 2A) when assayed with LeupNA as substrate. Isoelectric focusing gave a single peak of pI 8.4 ± 0.2 (Fig. 2B), and density‐gradient ultracentrifugation produced a single peak of molecular mass 130 ± 20 kDa (Fig. 2C).
Figure 2.
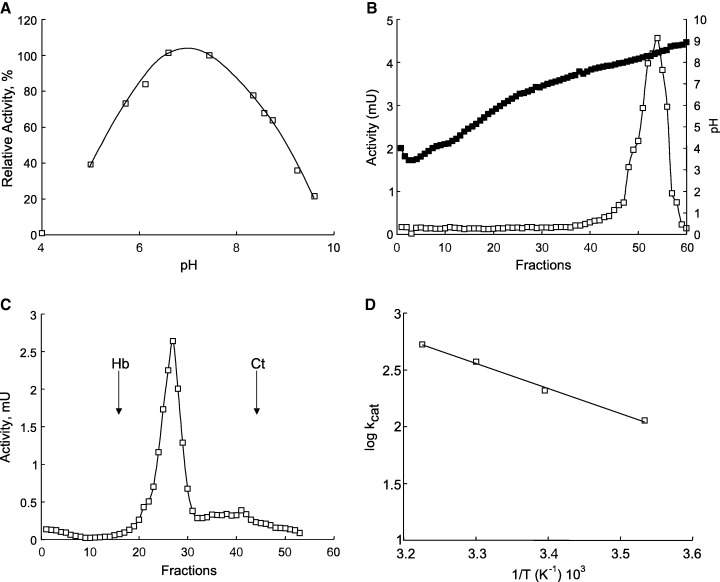
Properties of purified midgut APN from A. pisum. (A) Effect of pH on enzyme activity (optimal pH 7.0 ± 0.5). Buffers used: 100 mm sodium phosphate buffer (pH 5–7) and 100 mm Tris/HCl buffer (pH 7–9.5). (B) Isoelectric focusing (pI 8.4 ± 0.2). (C) Density‐gradient centrifugation. Hb, Haemoglobin; Ct, catalase. Molecular mass was calculated as 130 kDa. (D) Arrhenius plot. Activation energy was determined as E a = 42.2 kJ·mol−1.
The thermodynamic parameters of activation for A. pisum APN (Fig. 1D) were calculated by Arrhenius plot (plot of k cat against 1/T). From the slope of the line, the activation energy (E a) was determined to be 42.2 kJ·mol−1. Other thermodynamic parameters of activation were calculated using the relations of the transition state theory [23]. Thus, ΔS‡, ΔG‡ and ΔH‡at 25 °C were estimated to be −65.1 J·mol−1·K−1 (−15.5 cal·mol−1·K−1), 59.0 kJ·mol−1 (14 kcal·mol−1) and 39.7 kJ·mol−1 (9.5 kcal·mol−1), respectively.
Purified APN (Mono Q column) was submitted to MS sequencing. First, it was treated with trypsin. The digested protein was separated by Q‐ToF, and two of the resulting peptides were submitted to MS sequencing. The resulting sequences were (a) MDLLAIPDFR, (b) AGAMENWGMNTYK, and (c) NDSKITIYTYK. The same sequence as produced by peptide number 2 could be recovered from the protein purified by ConA–Br‐Sepharose, together with a series of more than 19 mass hits covering the entire sequence (25 p.p.m. precision cut‐off), including matching oxidized methionines. A Mowse score of 3.82E + 9 identified the purified and cloned sequences unambiguously (see below).
Kinetic parameters of A. pisum APN
The purified aphid APN showed a broad specificity towards N‐terminal aminoacyl residues, although it was unable to hydrolyze l‐aspartic acid α‐(β‐naphthylamide) (Table 1). The preferred substrates (higher k cat/K m values) were those bearing leucine or methionine at the N‐terminus, and the least preferable those presenting a proline at the N‐terminus (Table 1). There was a slight preference for tripeptides (Table 1), as judged by a comparison of k cat/K m values for peptides of the Leu‐(Gly)n series which differ only in the number of Gly residues (Table 1).
Table 1.
Kinectic parameters for purified APN from A. pisum. Relative values of k cat/K m were calculated using LeupNA as reference for synthetic substrate. For peptide subtrates, PheGlyGlyPhe was used as reference. The values were determined at least twice by 10 independent determinations with different substrate concentrations. SEM values were calculated by fitting data by a weighted linear regression using the software SigmaPlot®. AlaβNA, l‐alanine‐β‐naphthylamide; AlapNA, l‐alanine‐p‐nitroanilide; ArgpNA, l‐arginine‐p‐nitroanilide; MetβNA, l‐methionine‐β‐naphthylamide; MetpNA, l‐methionine‐p‐nitroanilide; ProβNA, l‐proline‐β‐naphthylamide.
| Substrate | K m (mm) | k cat (s−1) | k cat/K m (mm −1·s−1) | k cat/K m (relative) |
|---|---|---|---|---|
| LeupNA | 0.057 ± 0.007 | 119 ± 15 | 2090 ± 360 | 100 |
| MetpNA | 0.026 ± 0.008 | 45 ± 3 | 1720 ± 540 | 80.2 |
| ArgpNA | 0.19 ± 0.02 | 158 ± 31 | 832 ± 180 | 40.5 |
| AlapNA | 1.1 ± 0.04 | 222 ± 4 | 202 ± 9 | 9.9 |
| LeuβNA | 0.038 ± 0.003 | 105 ± 2 | 2760 ± 220 | 133 |
| ArgβNA | 0.058 ± 0.06 | 72 ± 2 | 1220 ± 130 | 59.0 |
| MetβNA | 0.043 ± 0.005 | 28 ± 1 | 642 ± 76 | 30.9 |
| AlaβNA | 0.28 ± 0.04 | 67 ± 2 | 240 ± 8 | 11.4 |
| ProβNA | 1.6 ± 0.2 | 6.8 ± 0.2 | 4.3 ± 0.5 | 0.2 |
| AspβNA | – | < 0,19 | ||
| LeuGly | 0.26 ± 0.08 | 5.6 ± 0.4 | 22 ± 7 | 11.1 |
| LeuGlyGly | 0.28 ± 0.06 | 26 ± 2 | 92 ± 20 | 48.9 |
| LeuGlyGlyGly | 0.9 ± 0.1 | 16 ± 1 | 19 ± 2 | 10 |
| LeuGlyGlyGlyGly | 1.1 ± 0.1 | 17 ± 1 | 15 ± 1 | 7.8 |
| LeuLeuLeu | 0.070 ± 0.02 | 6.4 ± 0.3 | 91 ± 23 | 47.8 |
| PheGly | 0.60 ± 0.08 | 13 ± 4 | 22 ± 8 | 11.1 |
| PheGlyGly | 1.1 ± 0.3 | 23 ± 2 | 22 ± 6 | 11.1 |
| PheGlyPheGly | 0.71 ± 0.07 | 92 ± 2 | 130 ± 13 | 68.8 |
| PheGlyGlyPhe | 0.27 ± 0.03 | 50 ± 2 | 188 ± 21 | 100 |
Leucine hydroxamate is a simple intersecting linear competitive inhibitor of APN (Fig. 3), with K i =5 ± 1 µm; the same is true for arginine hydroxamate (K i = 34 ± 7 µm). K i values for aminoacyl hydroxamates depend on the hydroxamate used, not on the substrate used (l‐leucine‐β‐naphthylamide or l‐arginine‐β‐naphthylamide) (not shown). This indicates that l‐leucine‐β‐naphthylamide and l‐arginine‐β‐naphthylamide are hydrolyzed at the same active site.
Figure 3.

Inhibition of purified A. pisum APN by leucine hydroxamate. Lineweaver–Burk plots of LeupNA‐hydrolyzing activity against different concentrations (mm) of leucine hydroxamate. Insert: replots of slopes calculated from Lineweaver–Burk plots against the concentration of leucine hydroxamate. K i = 5 ± 1 µm (n = 4).
Acyrthosiphon pisum APN inhibition by amastatin and bestatin are rapidly reversible by dilution (not shown), as observed with microsomal aminopeptidase [30]. Their pattern of inhibition is an intersecting, competitive, linear type, with K i = 1.8 µm for bestatin and K i = 0.6 µm for amastatin (not shown).
Inactivation of A. pisum APN by EDTA follows pseudo‐first‐order kinetics with k obs = 0.14 m −1·s−1, which is virtually completely suppressed by the competitive inhibitor arginine hydroxamate at a concentration corresponding to 25‐fold its K i value (Fig. 4). The reaction order with respect to EDTA was 0.44. As EDTA has two metal‐binding sites, the data support the conclusion that removal of only one metal ion is sufficient to inactivate the enzyme. In agreement with this, the partially EDTA‐inactivated enzyme has the same K m and pH optimum as native aminopeptidase (not shown).
Figure 4.
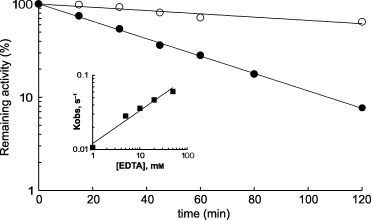
Inactivation of A. pisum APN by EDTA at 37 °C. Reaction mixtures contained different concentrations of EDTA in 100 mm Tris/HCl buffer, pH 7.0, containing 0.1% Triton X‐100. After different incubation times, the reaction was stopped by 100 times dilution. Inactivation by 50 mm EDTA in the absence (•) or presence (○) of 850 µm (25 × K i) arginine hydroxamate, which is a competitive inhibitor of aminopeptidase. Buffer used: 100 mm Tris/HCl, pH 7.0, containing 0.1% Triton X‐100. The insert shows a plot of the log of the observed first‐order rate kinetics of inactivation constant against log of EDTA concentration. n, the slope of the plot, was calculated as 0.44 and estimates the number of molecules of EDTA needed to inactivate each active site of the enzyme.
Carbohydrate and lectin interactions with A. pisum APN
The enzyme strongly interacts with lectins that bind mannose‐like Galanthus nivalis agglutinin and ConA, as observed in blotting assays (Fig. 1C) and in the purification steps (see above). The interaction with lectins was evaluated by density‐gradient ultracentrifugation (Fig. 5). After 30 min of preincubation of APN with the lectins, wheat germ agglutinin (WGA), which binds to sialic acid and N‐acetylglucosamine moieties, or ConA, which binds to glucose and mannose moieties, the samples were submitted to density‐gradient ultracentrifugation. APN with WGA results in a single peak (Fig. 5A), as observed in Fig. 2C, which corresponds to the APN without bound lectin (Fig. 5A). Mixing ConA with APN resulted in all the activity being at the bottom of the tube, meaning a molecular mass higher than 400 kDa (Fig. 5B), resulting from lectin binding and agglutination. When a competitive monosaccharide (α‐methyl mannoside) was added to the incubation mixture of ConA with APN at a concentration of 500 mm, a peak of intermediate molecular mass was observed (Fig. 5C), corresponding to the partially aggregated form of APN with ConA (the molecular mass of ConA is 73 kDa under the density‐gradient conditions).
Figure 5.
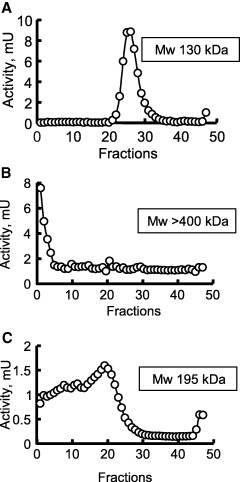
Density‐gradient ultracentrifugation of A. pisum APN in the presence of the glucose/mannose‐binding lectin ConA and WGA (sialic acid/N‐acetylglucosamine binding). (A) APN with WGA lectin; (B) APN with ConA; (C) APN with ConA and 500 mmα‐methyl mannoside, a competitive sugar. Note that the sedimentation of APN with WGA is closer to that in Fig. 2C.
The activity recovered from the density gradients was similar with and without lectins (data not shown). The kinetics parameters of APN (k cat and K m) associated with ConA were unaffected, indicating that the catalytic site and the mannosylated site(s) are quite far apart on the enzyme molecule.
Sequence coding the A. pisum APN
Full‐length cDNA was obtained for A. pisum APN with 3234 bp (GenBank accession number DQ440823). This sequence codes for a protein of 973 amino acids with residues 1–17 corresponding to the signal peptide predicted with signalp (http://www.cbs.dtu.dk/services/SignalP)[31]. The mature protein has a putative unglycosylated molecular mass of 109 011 Da and pI 5.30. The full‐length cDNA contains a short 5′‐UTR from 1 to 132 bp and 3′‐UTR from 3055 to 3234 bp.
The protein encoded by this cDNA contains the three peptide sequences obtained from the purified enzyme, showing identity between the purified enzyme and the cDNA sequence, as well as MS peaks with high Mowse score (see above) which unambiguously identified the cloned sequence.
The coding protein has high similarity to other aminopeptidases. It possesses the domain HEXXH + G, characteristic of gluzincins, and the domain GAMEN, found in many of these enzymes (Fig. 6). Putative N‐glucosylation sites (predicted at http://www.cbs.dtu.dk/services/NetNGlyc/) are assigned in Fig. 6, as well as the GPI anchor site in its C‐terminal domain, as predicted by the DGPI software [32]. The presence of the signal peptide and GPI anchor signal are consistent with the known characteristics of insect APNs. O‐glycosylation sites were predicted to be present in the region (close to the C‐terminus) of the APN using the NetOGlyc 3.1 server [33]. clustalw sequence alignment with other insect aminopeptidases (Fig. 7) showed that A. pisum APN has a weak similarity to class 2 aminopeptidases from Lepidoptera[20].
Figure 6.

cDNA coding sequence of A. pisum APN and its deduced sequence (GenBank accession number DQ440823). The predicted signal peptide is underlined, and the C‐terminal GPI cleavage signal sequence is dotted underlined. The characteristic zinc binding/gluzincin motif, HEXXH + E, and the gluzincin aminopeptidase motif, GAMEN, are highlighted in a bold/gray box. Peptides identified by MS analysis are double underlined. Boxed residues correspond to the MS sequenced peptides. Putative N‐glycosylated asparagine residues are dark‐shaded using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/), and putative O‐glycosylated threonines residues identified by the NetOGlyc 3.1 server (http://www.cbs.dtu.dk/services/NetOGlyc/) are also shaded.
Figure 7.
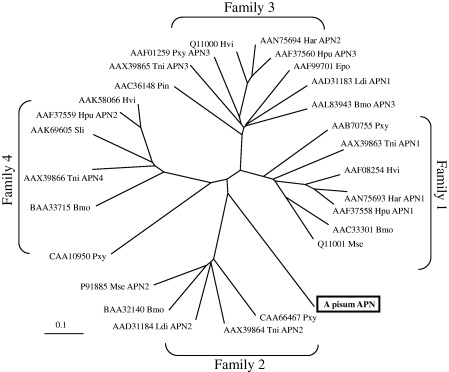
Sequence tree of Lepidoptera aminopeptidases and A. pisum APN. The tree was obtained using the clustalx alignment program. Families were numbered as described by Wang et al. [20]. Sequences used: Helicoverpa armigera, Har, APN1 (HaAPN1) (GenBank accession number AAN75693) and APN2 (HaAPN2) (GenBank accession number AAN75694) [19]; Helicoverpa puntigera, Hpu, APN1 (HpAPN1) (GenBank accession number AAF37558), APN2 (HpAPN2) (GenBank accession number AAF37559) and APN3 (H pAPN3) (GenBank accession number AAF37560) [15]; Heliothis virescens, Hvi, 110 kDa APN(HvAPN 110 kDa) (GenBank accession number AAK58066) [18], 120‐kDa APN(HvAPN 120 kDa) (GenBank accession number ACC46929) [9] and 170‐kDa APN(HvAPN 170 kDa) (GenBank accession number AAF08254) [11]; Plutella xylostella, Pxy, APNA (PxAPNA) (GenBank accession number AAB70755) [12], APN1 (PxAPN1) (GenBank accession number CAA66467) [5], APN3 (PxAPN3) (GenBank accession number AAF01259) [17] and APN4 (PxAPN4) (GenBank accession number CAA10950); Bombyx mori, Bmo, APN1 (BmAPN1) (GenBank accession number AAC33301) [6], APN2 (BmAPN2) (GenBank accession number BAA32140) [10], APN3 (BmAPN3) (GenBank accession number AAL83943) [17] and APN4 (BmAPN4) (GenBank accession number BAA33715); Epiphyas postvittana, Epo, APN(EpAPN) (GenBank accession number AAF99701) [13]; Lymantria dispar, Ldi, APN1 (LdAPN1) and APN2 (LdAPN2) (GenBank accession numbers AAD31183 and AAD31184) [64]; Plodia interpunctella, Pin, APN(PiAPN) (GenBank accession number AAC36148) [14]; Manduca sexta, Mse, APN1 (MsAPN1) (GenBank accession number CAA61452) [3] and APN2 (MsAPN2) (GenBank accession number CAA66466) [5]; Spodoptera litura, Sli, APN(SlAPN) (GenBank accession number AAK69605) [16].
Discussion
Occurrence, properties and sequence of A. pisum APN
Acyrthosiphon pisum has a membrane‐bound and a soluble aminopeptidase corresponding to 98% and 2% of the midgut aminopeptidase activity, respectively, when LeupNA is used as substrate. There is a single molecular species of A. pisum membrane‐bound aminopeptidase in this tissue, as judged by Mono Q chromatography after solubilization of almost 100% of its activity.
The membrane‐bound APN from A. pisum was purified to homogeneity, with a yield of 51.9% and specific activity of 40.3 U·mg−1. Taking into account that the specific activity of the homogenized midgut of this insect is 6.3 U·mg−1 per animal and that each midgut has 19 µg protein [27], it is possible to calculate that there are about 3 µg APN per midgut and that APN amounts to 15.6% of midgut protein. This is confirmed by SDS/PAGE of the midgut homogenate, where it is possible to recognize APN as a major protein in the preparation.
APN is a glycosylated protein of molecular mass 130 kDa (density‐gradient centrifugation) and pI 8.4. Molecular masses determined by SDS/PAGE (150 kDa) or gel filtration (200 kDa) are probably artifacts. The molecular mass of the unglycosylated protein is 109 kDa and pI 5.3 (predicted from the amino‐acid sequence). The data led us to conclude that ≈ 16% of the molecular mass of APN is carbohydrate.
Immunoblot for identification of glycosylated proteins recognized APN as the most abundant glycoprotein in the midgut and thus as an important target for lectins. Taking into account that more than one lectin can bind a single APN molecule (Fig. 5), and the abundance of APN in microvillar membrane, this enzyme is potentially the most important lectin‐binding site in aphid midgut. The calculated amount of aminopeptidase in microvillar membrane may explain the capacity of each aphid to feed on a diet containing lectins amounting to as much as 1 µg of ConA in 48 h [29]. Also immunohistochemical observations of lectin binding on the midgut demonstrated that the stomach (ventriculus 1) cell membranes are the primary target for ConA, followed by the intestine (remaining midgut chambers) cell membranes [29]. Activity measurements found APN along the midgut, and imunolocalization with APN antibodies showed that APN is associated with a specialized plasma membrane associated with the apical lamellae. These consist of a complex network of lamellae, linked one to another by trabecullae to resist the osmotic pressure caused by high‐sugar phloem‐sap ingestion [28]. The apical lamellae replace the regularly arranged microvilli observed in most midgut cells. APN localization data are in agreement with the lectin‐binding site found by Sauvion et al. [29]. However, as presented here, APN activity is not affected by glucose/mannose‐binding lectin.
Acyrthosiphon pisum APN has a broad specificity towards the N‐terminal amino‐acid residues of peptides, but it does not hydrolyze acidic aminoacyl‐peptides, and, although it is by no means proven, it appears to prefer peptides longer than dipeptides. Thus, A. pisum APN resembles the vertebrate enzyme (EC 3.4.11.2) [34] and the following insect midgut enzymes: microvillar membrane APN from Tenebrio molitor[22], soluble Tineola bisselliela (Lepidoptera) aminopeptidase [35, 36], soluble Attagenus megatoma (Coleoptera) aminopeptidase [37], soluble and microvillar R. americana (Diptera) aminopeptidases [25, 26], and membrane‐bound Spodoptera littoralis (Lepidoptera) aminopeptidase [38]. Another resemblance between A. pisum APN and the vertebrate enzyme are both inhibited by bestatin and amastatin, which in both cases is rapidly reversible [30]. It should be noted, however, that A. pisum APN has a K m value for peptides much smaller than those for T. molitor APN [22].
Substrates with different N‐terminal amino‐acid residues are hydrolyzed at the same site of A. pisum APN, as hydroxamate K i values do not depend on the aminoacyl β‐naphthylamide used as substrate.
Substrates with a bulky aminoacyl residue in position P1 (numbering of Schechter & Berger [39]) are better substrates for APN. The same is true for the P2′ position (compare Leu‐Gly‐Gly with Leu‐Leu‐Leu in Table 1), but not for the P1′ position (compare aminoacyl‐naphthylamide with aminoacyl‐p‐nitroanilide). This suggests that the subsites S1, S2′ and probably S3′ of the enzyme are pockets able to bind bulky aminoacyl residues, and this hypothesis agrees with the fact that amastatin is a better inhibitor of A. pisum APN than bestatin. Bestatin has bulky residues putatively able to interact with S1′ and S2′ of the enzyme (see above), and amastatin with S1, S1′ and S2′[40].
Acyrthosiphon pisum APN is the first insect digestive aminopeptidase that does not belong to the order Lepidoptera to be fully characterized and sequenced. A rapid survey of the A. pisum databank (http://urgi.infobiogen.fr/cgi-bin/annotation_form.pl?organism=apisum) allows the identification of more than 25 contigs with some relation to the word ‘aminopeptidase’, and similarly, a high number of ‘aminopeptidases’ are encoded in the Drosophila genome (http://flybase.org). A total of 29 A. pisum expressed sequence tags (ESTs) have full complementarities with the cloned APN in almost 60 000 ESTs. From these 29 ESTs, 22 belong to libraries from the digestive tract, and seven from libraries from whole insect (none from other tissue libraries), meaning that this aminopeptidase is potentially very specific to the aphid midgut.
The APN sequence has all identified residues essential for zinc binding and catalysis. In the sequence, it was easy to recognize the signal peptide, several potential glycosylation sites, as well as a GPI anchor at its C‐terminus. This anchor is possibly an adaptation to a phloem‐based diet, avoiding excretion of the enzyme into the honeydew, as the phylogenetically related Hemipteran Dysdercus peruvianus has a soluble aminopeptidase, in spite of the fact that, in this case, the enzyme is trapped between the microvillar and perimicrovillar membranes [27, 28].
Function of A. pisum APN and lectin toxicity
The role of the microvillar aminopeptidase is postulated to be hydrolysis of oligopeptides formed by the action of luminal proteinases [1, 2, 22]. In aphids, a cathepsin L was found to be partially associated with modified perimicrovillar membranes and is possibly involved in degradation of toxic proteins found in the phloem sap [28, 41]. APN is certainly responsible for the final digestion of peptides generated by cathepsin L. Another possibility is that APN is somehow associated with putative amino‐acid‐binding sites at the plasma membranes associated with the apical lamellae (modified perimicrovillar membranes). These are thought to increase the amino‐acid concentration (usually low in the aphid diet) [28], thus facilitating absorption. APN may also be directly linked to absorptive sites in apical lamellae, as has been suggested for Lepidoptera [42]. Finally, APN may serve as the primary digestive enzyme responsible for the assimilation of the phloem sap small peptide fraction, chemical components largely unexplored at the moment.
As B. thuringiensis is not effective in aphid control, lectins have been used as insecticidal agents against aphids [43]. The soluble protein, ferritin, is the snowdrop lectin‐binding protein in the planthopper Nilaparvata lugens[44]. The authors postulated that alteration of iron metabolism might be related to its lectin toxicity. Although ferritin was not the most abundant protein in midgut preparations, this protein was the most specifically recognized in N. lugens. However, none of the 2D PAGE protein spots observed in pea aphid homogenates as binding to ConBr was identified as corresponding to ferritin (F. A. Mendonca de Sousa & Y. Rahbé, unpublished), although the ferritin gene is largely transcribed in A. pisum midguts [45]. It is possible that the mechanism of toxicity found in planthopper is different from that found in aphids.
In A. pisum, the lectin, ConA, is a potent toxin affecting survival and growth, but WGA is relatively ineffective [46]. These data agree with the fact that aphids do not possess a peritrophic membrane [28]. Consequently, this toxicity must result from lectin binding to target proteins in the apical membranes from the midgut, although not related to the inhibition of APN activity. One explanation of this effect is a decrease in amino‐acid absorption caused by ConA binding to APN, with deleterious effects on the putative associated proteins thought to bind to amino acids (see above). Indeed, ConA‐intoxicated aphids have been shown to display altered hemolymph free amino‐acid profiles and modified excretion of asparagine in their honeydew [47]. It is still possible that a reduction in membrane protein lateral mobility or its resistance to phloem osmotic pressure is the major cause of lectin toxicity to aphids. These possibilities need to be evaluated.
Experimental procedures
Animals
Acyrthosiphon pisum Harris aphids (Hemiptera: Aphididae), clone Ap‐LL01, were maintained in the laboratory on broad bean seedlings (Vicia faba) in ventilated plexiglass cages (21 °C; 70% relative humidity; 16 h light/8 h darkness). For the experiments, a limited number of mass‐reared adults were allowed to lay eggs for 24 h on young Vicia plants, and the resulting apterous insects were used as 9‐day‐old adults.
Chemicals
Buffer salts, detergents, molecular‐mass markers, protein inhibitors, and most substrates were purchased from Sigma‐Aldrich (St Louis, MO, USA). Glycoprotein detection kits came from Boehringer‐Mannheim (Mannheim, Germany). The peptides Leu‐Gly‐Gly‐Gly and Leu‐Gly‐Gly‐Gly‐Gly were gifts from Dr L. Juliano (Unifesp, São Paulo, Brazil).
Preparation of samples
Adult apterous aphids were immobilized on a flat surface, using adhesive tape, and their guts were removed under a stereomicroscope in Yeager's physiological solution [48]. The midguts were separated and homogenized in double‐distilled water with the aid of a Potter‐Elvehjem homogenizer. The homogenates were labeled crude homogenate and stored. Crude homogenates were used to assay APN or were centrifuged at 100 000 g for 1 h at 4 °C, resulting in a supernatant (labeled midgut soluble fraction) and a pellet (midgut cell membranes). Washed midgut cell membranes were prepared by dispersing the midgut cell membranes in water, followed by three freezing and thawing cycles, and re‐centrifugation at 100 000 g for 1 h at 4 °C. All centrifugations were performed on a Hitachi Ultracentrifuge model Himac 70P‐72 with an RPS 40T rotor.
Protein determination and enzymatic assays
Protein was determined as described by Bradford [49] using ovalbumin as standard. When samples contained detergent, protein was determined by the method of Smith et al. [50], as modified by Morton & Evans [51], using BSA as standard.
Routine assays of APN were performed using 1 mm LeupNA as substrate (initially solubilized in dimethyl sulfoxide) in 100 mm Tris/HCl buffer, pH 7.0, at 30 °C. Unless otherwise specified, the same conditions were used for all other substrates. Naphthylamine liberated from aminoacyl‐β‐naphthylamides, nitroaniline from aminoacyl‐p‐nitroanilides, and phenylalanine and leucine from the different peptides were determined spectrophotometrically by the methods of Hopsu et al. [52], Erlanger et al. [53] and Nicholson & Kim [54], respectively. In each determination, incubations were continued for at least four different periods of time, and the initial rates were calculated. All assays were performed so that the measured activity was proportional to protein and incubation time. Controls without enzyme or without substrate were included. One enzyme unit (U) is defined as the amount that hydrolyzes 1 µmol substrate·min−1, at 30 °C.
Solubilization of APN by detergents
In order to evaluate the solubilizing efficiency of detergents, samples of 200 µL midgut homogenate at a concentration of 13 guts per mL (which contains ≈ 50 µg protein) of midgut cell membranes were suspended in 10 mm Hepes buffer, pH 7.4, in the presence and absence of several detergents. After 17 h at 4 °C with shaking, the suspensions were centrifuged at 100 000 g for 1 h at 4 °C, and supernatants were assayed for APN. APN activity was determined in the resulting supernatants and referred to the original preparation of cell membranes (as percentage solubilization). Recovery is the percentage of the sum of solubilized plus nonsolubilized activity referred to the original preparation of cell membranes. Data are mean ± SEM calculated from determinations carried out in three different preparations.
For routine solubilization of APN, midgut cell membranes were suspended in 10 mm Hepes buffer, pH 7.4, containing 10 mm Triton X‐100. After 1 h at 4 °C, the suspension was centrifuged at 25 000 g for 30 min at 4 °C, and the supernatant used as a source of enzyme.
Purification of detergent‐solubilized APN
Cell membranes corresponding to ≈ 300 A. pisum midguts (wet weight ≈ 6 mg) were solubilized with Triton X‐100 as described above, and applied to a Mono Q HR 5/5 column (0.5 cm internal diameter × 5 cm) equilibrated with 20 mm Tris/HCl buffer, pH 7.0, containing 0.1% Triton X‐100 in an FPLC system. Controls showed that protease inhibitors are not necessary. Elution was carried out with a gradient of 0–0.6 m NaCl in the same buffer. The flux was 1.0 mL·min−1, and fractions of 0.4 mL were collected. Fractions showing activity with LeupNA were pooled, and purification was checked by SDS/PAGE.
Alternatively, the APN was purified on a 3‐mL ConA–Br‐Sepharose column (6 × 30 mm). The column was washed with 15 mL 20 mm acetate buffer, pH 4.2, containing 0.5 m NaCl, then with 15 mL 100 mm citrate/phosphate buffer, pH 6.0, containing 2 mm CaCl2, before being equilibrated with 15 mL 20 mm Tris/HCl buffer, pH 7.0, with 0.1% Triton X‐100. The solubilized samples were applied to the column and eluted with 20 mm Tris/HCl buffer, pH 7.0, containing 0.1% Triton X‐100 and 0.5 mα‐methyl mannoside. Fractions of 1 mL were collected at a flow rate of 1 mL·min−1.
SDS/PAGE
Electrophoresis of A. pisum samples in denaturing conditions (SDS/PAGE) was carried out on 7.5% (w/v) polyacrylamide gels containing 0.1% (w/v) SDS, on a discontinuous pH system [55], using Mini Protean II cells (Bio‐Rad, Hercules, CA, USA). Samples were lyophilized and suspended in sample buffer containing 60 mm Tris/HCl buffer, pH 6.8, 2.0% (w/v) SDS, 5% (v/v) 2‐mercaptoethanol, 10% glycerol and 0.2% (w/v) bromophenol blue and heated for 3 min at 95 °C in a water bath before being loaded on to the gels. Electrophoresis was carried out at 200 V until the tracking dye reached the bottom of the gel. The gel was then silver‐stained [56] or stained using 0.1% (w/v) Coomassie Blue R in 10% acetic acid/40% methanol for 30 min. In the last case, destaining was achieved with several washes in a solution containing 40% methanol and 10% acetic acid.
Isoelectric focusing
Isoelectric focusing was performed as described by Terra et al. [57], in columns of 7.5% polyacrylamide gel containing 10% ampholytes pH 3–10 (Pharmalyte 3–10, Pharmacia, Uppsala, Sweden). Samples were applied after polymerization and prefocusing (30 min at 31 V·cm−1) on the top of the alkaline side of the gels. For samples in detergent, 0.1% Triton X‐100 was added to the gels and fractionation buffer. Recoveries of activities applied to the gels were 80–100%.
Density‐gradient centrifugation
Samples of purified APN (100 µL) containing ≈ 1 µg purified protein with or without 100 µg lectins were layered on the top of 10‐mL glycerol gradients (10–30%, w/v) made up in 100 mm Tris/HCl buffer, pH 7.0, in the presence or absence of 500 mmα‐methyl mannoside. Centrifugation and collection of fractions were performed as described previously [58]. The molecular mass of APN was calculated by the method of Martin & Ames [59], using the sedimentation rates of hemoglobin (64.5 kDa) and bovine liver catalase (232 kDa) as reference standards. Activity recovery was 80–100%.
Determination of molecular mass by gel filtration
Samples of 200 µL, containing 2–4 µg purified protein, were applied to a gel‐filtration column in an FPLC system (Pharmacia‐LKB Biotechnology, Uppsala, Sweden) by using a Superose HR 10/30 column (1.0 cm internal diameter × 30 cm) equilibrated and eluted in 100 mm Tris/HCl buffer, pH 7.0, containing 0.1% Triton X‐100. Fractions of 0.4 mL were collected at a flow rate of 0.5 mL·min−1. Molecular masses were calculated using the following proteins as standards: aprotinin (6.5 kDa), cytochrome c (12.4 kDa), ovalbumin (45 kDa), BSA (65 kDa), and β‐amylase (200 kDa). Recoveries were 80–100%.
Detection of carbohydrates in purified APN
Protein samples were blotted on to nitrocellulose sheets after SDS/PAGE [60]. Detection of carbohydrates was performed with the DIG Glycan Detection kit, and identification of carbohydrate moieties was accomplished with lectins by using the DIG Glycan Differentiation kit. The procedures followed the supplier's instructions (Boehringer Mannheim).
Kinetic studies
The effect of substrate concentration on the activity of purified APN was determined using at least 10 different substrate concentrations. K m and k cat values (mean ± SEM) were determined by a weighted linear regression using the software SigmaPlot® (Jandel Scientific, Systat Software Inc., Richmond, CA, USA). In the inhibition studies, purified APN was incubated with four different inhibitor concentrations in each of 10 different concentrations of substrate. K i values were calculated as described by Segel [61].
EDTA inactivation
Purified APN was incubated with EDTA (1–50 mm) for different times at 40 °C in 100 mm citrate/phosphate buffer pH 6. The EDTA‐inactivation reactions were stopped by 100‐fold dilution of reaction mixtures with 100 mm Tris/HCl buffer (pH 7.0)/0.1% Triton X‐100. The remaining activity was measured using LeupNA as substrate, under the conditions described above. Protection against inactivation by EDTA was investigated with aminoacyl hydroxamates, which are simple linear competitive inhibitors of A. pisum APN. The reaction order was determined by incubation of APN with different concentrations of EDTA.
Microsequencing of purified APN
Purified APN was electroblotted on to poly(vinylidene difluoride) membranes after SDS/PAGE [62]. The membranes were stained for protein using 0.1% Coomassie Blue R‐250 in a 50% (v/v) methanol, and were destained with 50% methanol. Dried poly(vinylidene difluoride) membranes were submitted to tryptic digestion, and the resulting peptides (two peptides) were submitted to MS sequencing at the sequencing facility of the Pasteur Institute (Paris).
Alternatively, in‐gel digestion was performed for protein identification: spots were excised from preparative gels using pipette tips. The spots were washed with 100 µL 25 mm NH4HCO3 for 30 min, twice destained for 30 min with 100 µL 25 mm NH4HCO3/acetonitrile (v/v), and dehydrated in acetonitrile. Gel spots were completely dried using a vacuum centrifuge before trypsin digestion. The dried gel volume was evaluated, and 3 vol. trypsin (V5111; Promega, Madison, WI, USA; 10 ng·µL−1 in 25 mm NH4HCO3) was added. Digestion was performed at 37 °C over 5 h. The gel pieces were centrifuged, and 8–12 µL acetonitrile (depending on gel volume) was added to extracted peptides. The mixture was sonicated for 5 min and centrifuged. For MALDI‐TOF MS analysis, 1 µL supernatant was loaded directly on to the MALDI target. The matrix solution (5 mg·mL−1α‐cyano‐4‐hydroxycinnamic acid in 50% acetonitrile containing 0.1% trifluoroacetic acid) was added immediately and allowed to dry at room temperature. A Voyager DE‐Pro model MALDI‐TOF mass spectrometer (Perseptive BioSystems, Farmingham, MA, USA) was used in positive‐ion reflector mode for peptide mass fingerprinting. External calibration was performed with a standard peptide solution (Proteomix; LaserBio Laboratories, Sophia‐Antipolis, France). Internal calibration was performed using peptides resulting from autodigestion of porcine trypsin. Monoisotopic peptide masses were assigned and used from NCBI database searches (plus A. pisum APN sequence) with the ‘ms‐fit’ software.
Cloning of APN from A. pisum
Total RNA was extracted from midgut epithelium of A. pisum with Trizol following the instructions of the manufacturer, Invitrogen, which are based on those of Chomczynski & Sacchi [63]. mRNA was purified with Qiagen mRNA purification kit, and a cDNA library was constructed with the kit Smart (Clontech, Mountain View, CA, USA), following the instructions of the manufacturer. A partial sequence of a cDNA coding for APN was amplified using degenerated primers for the APN consensus sequence that contains the peptide AGAMENWGM identified in MS analysis (primers APN‐U538: 5′‐TTYCCITGYTIYGAYGARCC‐3′, based on peptide ‘TGLYRSS’; APN‐L1051: 5′‐RTTICCRAACCACWKRTG‐5′, based on peptide ‘THQWFGN’). PCR was performed using Taq DNA polymerase (Invitrogen) using the standard method. The PCR product was cloned in pGEM‐T Easy Vector (Promega), sequenced, and the identified fragment sequence had high identity with that of APN.
This sequenced cDNA fragment contains the two identified peptides sequenced from purified protein, and was blasted (blastn) against the A. pisum ESTs deposited at NCBI [45]. The recovered fragments were clustered and blasted again against the A. pisum ESTs until the N‐terminus was completed with signal peptide and 5′‐UTR sequence. Putative 3′ reads were recovered from A. pisum ESTs using blast (blastn) with the best full‐length blast hit as driver (Apis mellifera, access number XP 366261). Also, ESTs corresponding to A. pisum APN can be recovered along the sequence using MS peaks with Mascot engine (http://www.matrixscience.com/). Reads were clustered into two contigs covering the 3′ region and the 5′ region. The final gap between these contigs was recovered using PCR with forward primers 5′‐GGCATGGTGAGGACTAGTTGGCCG‐3′ combined with reverse primer 5′‐GCCATGCCGCCGTCTCGTTGATGG‐3′, and the complete sequence was obtained and deposited at GenBank with the accession number DQ440823.
Acknowledgements
This work was supported by the Brazilian research agencies FAPESP, CAPES/COFECUB and CNPq (PRONEX program). We are indebted to Dr L. Juliano (Medical School, UNIFESP) for the synthesis of several peptides used as substrates, to Dr C. Ferreira for helpful discussion, and to Mrs L.Y. Nakabayashi for technical assistance. We thank Mrs L. Duportest and C. Deraison for their help in obtaining the A. pisum cDNA library, and G. Duport for her invaluable skills in aphid dissection. FAMS, YR and PTC were given grants for exchanges between USP, UFC and INRA‐INSA through a French–Brazilian contract from CAPES/COFECUB (contract 261/98, co‐ordinated by Drs S. Grenier and J. R. Parra). WRT is a research fellow of CNPq. Many thanks go to Dr Christophe Chambon (INRA Clermont‐Ferrand Theix Proteomic Facility) for his help with MALDI‐TOF analysis of the purified enzyme.
Database The sequence described here has been deposited in the GenBank database with the accession number DQ440823
References
- 1. Terra WR & Ferreira C (1994) Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol 109B, 1–62. [Google Scholar]
- 2. Terra WR & Ferreira C (2005) Biochemistry of digestion In Comprehensive Molecular Insect Science (Gilbert LI, Iatrou K & Gill SS, eds), Vol. 4, pp. 171–224. Elsevier, Oxford. [Google Scholar]
- 3. Knight PJ, Knowles BH & Ellar DJ (1995) Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA (c) toxin. J Biol Chem 270, 17765–17770. [DOI] [PubMed] [Google Scholar]
- 4. Valaitis AP, Lee MK, Rajamohan F & Dean DH (1995) Brush border membrane aminopeptidase‐N in the midgut of the gypsy moth serves as the receptor for the CryIA (c) delta‐endotoxin of Bacillus thuringiensis . Insect Biochem Mol Biol 25, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 5. Denolf P, Hendrickx K, Vandamme J, Jansens S, Peferoen M, Degheele D & VanRie J (1997) Cloning and characterization of Manduca sexta and Plutella xylostella midgut aminopeptidase N enzymes related to Bacillus thuringiensis toxin‐binding proteins. Eur J Biochem 248, 748–761. [DOI] [PubMed] [Google Scholar]
- 6. Yaoi K, Nakanishi K, Kadotani T, Imamura M, Koizumi N, Iwahana H & Sato R (1999) cDNA cloning and expression of Bacillus thuringiensis Cry1Aa toxin binding 120 kDa aminopeptidase N from Bombyx mori . Biochim Biophys Acta 1444, 131–137. [DOI] [PubMed] [Google Scholar]
- 7. Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR & Dean DH (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62, 775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT & Holmes KV (1992) Human aminopeptidase N is a receptor for human Coronavirus 229E. Nature 357 (6377), 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gill SS, Cowles EA & Francis V (1995) Identification, isolation and cloning of a Bacillus thuringiensis Cry1Ac toxin‐binding protein from the midgut of the lepidopteran insect Heliothis virescens . J Biol Chem 270, 27277–27282. [DOI] [PubMed] [Google Scholar]
- 10. Hua G, Tsukamoto K & Ikezawa H (1998) Cloning and sequence analysis of the aminopeptidase N isozyme (APN2) from Bombyx mori midgut. Comp Biochem Physiol 121B, 213–222. [DOI] [PubMed] [Google Scholar]
- 11. Oltean DI, Pullikuth AK, Lee HK & Gill SS (1999) Partial purification and characterization of Bacillus thuringiensis Cry1A toxin receptor A from Heliothis virescens and cloning of the corresponding cDNA. Appl Environ Microbiol 65, 4760–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang WX, Gahan LJ, Tabashnik BE & Heckel DG (1999) A new aminopeptidase from diamondback moth provides evidence for a gene duplication event in Lepidoptera. Insect Mol Biol 8, 171–177. [DOI] [PubMed] [Google Scholar]
- 13. Simpson RM & Newcomb RD (2000) Binding of Bacillus thuringiensis delta‐endotoxins Cry1Ac and Cry1Ba to a 120‐kDa aminopeptidase‐N of Epiphyas postvittana purified from both brush border membrane vesicles and baculovirus‐infected Sf9 cells. Insect Biochem Mol Biol 30, 1069–1078. [DOI] [PubMed] [Google Scholar]
- 14. Zhu YC, Kramer KJ, Oppert B & Dowdy AK (2000) cDNAs of aminopeptidase‐like protein genes from Plodia interpunctella strains with different susceptibilities to Bacillus thuringiensis toxins. Insect Biochem Mol Biol 30, 215–224. [DOI] [PubMed] [Google Scholar]
- 15. Emmerling M, Chandler D & Sandeman M (2001) Molecular cloning of three cDNAs encoding aminopeptidases from the midgut of Helicoverpa punctigera, the Australian native budworm. Insect Biochem Mol Biol 31, 899–907. [DOI] [PubMed] [Google Scholar]
- 16. Agrawal N, Malhotra P & Bhatnagar RK (2002) Interaction of gene‐cloned and insect cell‐expressed aminopeptidase N of Spodoptera litura with insecticidal crystal protein Cry1C. Appl Environ Microbiol 68, 4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakanishi K, Yaoi K, Nagino Y, Hara H, Kitami M, Atsumi S, Muira N & Sato R (2002) Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella: their classification and the factors that determine their binding specificity of Bacillus thuringiensis Cry1Ac toxin. FEBS Lett 519, 215–220. [DOI] [PubMed] [Google Scholar]
- 18. Banks DJ, Hua G & Adang MJ (2003) Cloning of a Heliothis virescens 110 kDa aminopeptidase N and expression in Drosophila S2 cells. Insect Biochem Mol Biol 33, 499–508. [DOI] [PubMed] [Google Scholar]
- 19. Rajagopal R, Agrawal N, Selvapandiyan A, Sivakumar S, Ahmad S & Bhatnagar RK (2003) Recombinantly expressed isoenzymic aminopeptidases from Helicoverpa armigera (American cotton bollworm) midgut display differential interaction with closely related Bacillus thuringiensis insecticidal proteins. Biochem J 370, 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P, Zhang X & Zhang J (2005) Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni . Insect Biochem Mol Biol 35, 611–620. [DOI] [PubMed] [Google Scholar]
- 21. Rawlings ND & Barrett AJ (1995) Evolutionary families of metallopeptidases. Methods Enzymol 248, 183–228. [DOI] [PubMed] [Google Scholar]
- 22. Cristofoletti PT & Terra WR (1999) Specificity, anchoring, and subsites in the active center of a microvillar aminopeptidase purified from Tenebrio molitor (Coleoptera) midgut cells. Insect Biochem Mol Biol 29, 807–819. [Google Scholar]
- 23. Cristofoletti PT & Terra WR (2000) The role of amino acid residues in the active site of a midgut microvillar aminopeptidase from the beetle Tenebrio molitor . Biochim Biophys Acta 1479, 185–195. [DOI] [PubMed] [Google Scholar]
- 24. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA & Galle RF (2000) The genome sequence of Drosophila melanogaster . Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 25. Ferreira C & Terra WR (1984) Soluble aminopeptidases from cytosol and luminal contents of Rhynchosciara americana midgut caeca. Properties and phenanthroline inhibition. Insect Biochem 14, 145–150. [Google Scholar]
- 26. Ferreira C & Terra WR (1986) Substrate specificity and binding loci for inhibitors in an aminopeptidase purified from the plasma membrane of midgut cells of an insect (Rhynchosciara americana). Arch Biochem Biophys 244, 478–485. [DOI] [PubMed] [Google Scholar]
- 27. Silva CP, Ribeiro AF & Terra WR (1996) Enzyme markers and isolation of the microvillar and perimicrovillar membranes of Dysdercus peruvianus (Hemiptera: Pyrrhocoridae) midgut cells. Insect Biochem Mol Biol 26, 1011–1018. [Google Scholar]
- 28. Cristofoletti PT, Ribeiro AF, Deraison C, Rahbé Y & Terra WR (2003) Midgut adaptation and digestive enzyme distribution in a phloem feeding insect, the pea aphid Acyrthosiphon pisum . J Insect Physiol 49, 11–24. [DOI] [PubMed] [Google Scholar]
- 29. Sauvion N, Nardon C, Febvay G, Gatehouse AM & Rahbé Y (2004) Binding of the insecticidal lectin Concanavalin A in pea aphid, Acyrthosiphon pisum (Harris) and induced effects on the structure of midgut epithelial cells. J Insect Physiol 50, 1137–1150. [DOI] [PubMed] [Google Scholar]
- 30. Wilkes SH & Prescott J (1985) The slow, tight binding of bestatin and amastatin to aminopeptidases. J Biol Chem 25, 13154–13162. [PubMed] [Google Scholar]
- 31. Bendtsen JD, Nielsen H, von Heijne G & Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795. [DOI] [PubMed] [Google Scholar]
- 32. Kronegg J & Buloz D (1999) Detection/prediction of GPI cleavage site (GPI‐anchor) in a protein (DGPI). http://129.194.185.165/dgpi/ .
- 33. Julenius K, Molgaard A, Gupta R & Brunak S (2005) Prediction, conservation analysis and structural characterization of mammalian mucin‐type O‐glycosylation sites. Glycobiology 15, 153–164. [DOI] [PubMed] [Google Scholar]
- 34. Norén O, Sjostrom H, Danielsen EM, Cowell GM & Skovbjerg H (1986) The enzymes of the enterocyte plasma membrane In Molecular and Cellular Basis of Digestion (Desnuelle P, Sjostrom H. & Norén O, eds). Elsevier, Amsterdam. [Google Scholar]
- 35. Ward CW (1975a) Aminopeptidases in webbing clothes moth larvae. Properties and specificities of enzymes of highest electrophoretic mobility. Aust J Biol Sci 28, 447–455. [DOI] [PubMed] [Google Scholar]
- 36. Ward CW (1975b) Aminopeptidases in webbing clothes moth larvae. Properties and specificities of the enzymes of intermediate electrophoretic mobility. Biochim Biophys Acta 410, 361–369. [DOI] [PubMed] [Google Scholar]
- 37. Baker JE & Woo SM (1981) Properties and specificities of a digestive aminopeptidase from larvae of Attagenus megatoma (Coleoptera: Dermestidae). Comp Biochem Physiol 69B, 189–193. [Google Scholar]
- 38. Lee MJ & Anstee JH (1995) Characterization of midgut exopeptidase activity from larval Spodoptera littoralis . Insect Biochem Mol Biol 25, 63–71. [Google Scholar]
- 39. Schechter I & Berger A (1967) On the size of the active site in proteases. Biochem Biophys Res Commun 27, 157–162. [DOI] [PubMed] [Google Scholar]
- 40. Rich DH, Moon BJ & Harbeson S (1984) Inhibition of aminopeptidases by amastatin and bestatin derivatives. Effect of inhibitor structure on slow‐binding processes. J Med Chem 27, 417–422. [DOI] [PubMed] [Google Scholar]
- 41. Deraison C, Darboux I, Duportets L, Gorojankina T, Rahbé Y & Jouanin L (2004) Cloning and characterization of a gut‐specific cathepsin L from the aphid Aphis gossypii . Insect Mol Biol 13, 165–177. [DOI] [PubMed] [Google Scholar]
- 42. Parenti P, Morandi P, Mcgivan JD, Consonnic P, Leonardi G & Giordana B (1997) Properties of the aminopeptidase N from the silkworm midgut (Bombyx mori). Insect Biochem Mol Biol 27, 397–403. [Google Scholar]
- 43. Gatehouse AMR & Gatehouse JA (1996) Effects of lectins on insects In Effects of Antinutrients on the Nutritional Value of Legume Diets:9th World Conference of Food Science and Technology, COST 98 Action, Budapest (Bardocz S, Gelencser E. & Pusztai A, eds), pp. 14–21. COST publications, Brussels. [Google Scholar]
- 44. Du JP, Foissac X, Carss A, Gatehouse AMR & Gatehouse JA (2000) Ferritin acts as the most abundant binding protein for snowdrop lectin in the midgut of rice brown planthoppers (Nilaparvata lugens). Insect Biochem Mol Biol 30, 297–305. [DOI] [PubMed] [Google Scholar]
- 45. Sabater‐Munoz B, Legeai F, Rispe C, Bonhomme J, Dearden P, Dossat C, Duclert A, Gauthier JP, Ducray DG, Hunter W, et al. (2006) Large‐scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera). Genome Biol 7, R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rahbé Y & Febvay G (1993) Protein toxicity to aphids: an in vitro test on Acyrthosiphon pisum . Entomologia Expis Applicata 67, 149–160. [Google Scholar]
- 47. Sauvion N (1995) Effects and mechanisms of toxicity of two lectins of the glucose‐mannose group towards the pea aphid, Acyrthosiphon pisum (Harris). Potential use of plant lectins for creating transgenic plant resistant to aphids. PhD Thesis, INSA‐Lyon: http://tel.ccsd.cnrs.fr/documents/archives0/00/00/70/06/tel-00007006-00/tel-00007006.pdf .
- 48. Dreux P (1963) Modification de la solution de Robert Levy et Yeager permettant le fonctionnement du vaisseau dorsal de la chenille de Galleria mellonella en millieu artificiel. C R Seances Soc Biol Fil 157, 1000–1005. [Google Scholar]
- 49. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 50. Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ & Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150, 76–85. [DOI] [PubMed] [Google Scholar]
- 51. Morton RE & Evans TA (1992) Modification of the bicinchoninic acid protein assay to eliminate lipid interference in determining lipoprotein protein content. Anal Biochem 204, 332–334. [DOI] [PubMed] [Google Scholar]
- 52. Hopsu UK, Mäkinen KK & Glenner GG (1966) Purification of a mammalian peptidase selective for N‐terminal arginine and lysine residues: aminopeptidase B. Arch Biochem Biophysics 114, 557–566. [DOI] [PubMed] [Google Scholar]
- 53. Erlanger BF, Kokowsky N & Cohen W (1961) The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophysics 95, 271–278. [DOI] [PubMed] [Google Scholar]
- 54. Nicholson JA & Kim YS (1975) An one‐step 1‐amino acid oxidase assay for intestinal peptide hydrolase activity. Anal Biochem 63, 110–117. [Google Scholar]
- 55. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 56. Blum H, Beier H & Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide. Electrophoresis 8, 93–99. [Google Scholar]
- 57. Terra WR, Ferreira C & De Bianchi AG (1978) Physical properties and Tris inhibition of an insect trehalase and a thermodynamic approach to the nature of its active site. Biochim Biophys Acta 524, 131–141. [DOI] [PubMed] [Google Scholar]
- 58. Terra WR & Ferreira C (1983) Further evidence that enzymes involved in the final stages of digestion by Rhynchosciara americana do not enter the endoperitrophic space. Insect Biochem 13, 143–150. [Google Scholar]
- 59. Martin RG & Ames BN (1961) A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem 236, 1372–1379. [PubMed] [Google Scholar]
- 60. Towbin H, Staehelin T & Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Segel IH (1975) Enzyme Kinetics. Behavior and Analysis of Rapid Equilibrium and Steady‐State Enzyme Systems. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 62. Matsudaira P (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 262, 10035–10038. [PubMed] [Google Scholar]
- 63. Chomczynski P & Sacchi N (1987) Single‐step method of RNA isolation by acid guanidinium triocyanate–phenol–chloroform extraction. Anal Biochem 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 64. Garner KJ, Hiremath S, Lehtoma K & Valaitis AP (1999) Cloning and complete sequence characterization of two gypsy moth aminopeptidase‐N cDNAs, including the receptor for Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol 29, 527–535. [DOI] [PubMed] [Google Scholar]


