Abstract
It has emerged that DDX3 plays a role in antiviral innate immunity. However, the exact mechanism by which DDX3 functions in antiviral innate immunity remains to be determined. We found that the expression of the protein activator of the interferon‐induced protein kinase (PACT) was regulated by DDX3 in human cells. PACT acts as a cellular activator of retinoic acid‐inducible gene‐I‐like receptors in the sensing of viral RNAs. DDX3 facilitated the translation of PACT mRNA that may contain a structured 5′ UTR. Knockdown of DDX3 decreased the viral RNA detection sensitivity of the cells. PACT partially rescued defects of interferon‐β1 and chemokine (C‐C motif) ligand 5/RANTES (regulated on activation normal T cell expressed and secreted) induction in DDX3‐knockdown HEK293 cells. Therefore, DDX3 may participate in antiviral innate immunity, at least in part, by translational control of PACT. Moreover, we show that overexpression of the hepatitis C virus (HCV) core protein inhibited the translation of a reporter mRNA harboring the PACT 5′ UTR. The HCV core protein was associated and colocalized with DDX3 in cytoplasmic stress granules, suggesting that the HCV core may abrogate the function of DDX3 by sequestering DDX3 in stress granules. The perturbation of DDX3 by viral proteins delineates a critical role for DDX3 in antiviral host defense.
Keywords: antiviral innate immunity, DDX3, hepatitis C virus, PACT, translational control
We here propose a model of DDX3‐mediated translational control during HCV infection. HCV genomic RNA (ssRNA) stimulates RIG‐I‐mediated antiviral innate immune response in the presence of PACT during viral replication. Interestingly, the HCV core protein abrogates the RIG‐I‐mediated antiviral innate immune response by direct binding of DDX3. Inactivation of DDX3 down‐regulates the expression of PACT and thus attenuates the viral RNA detection sensitivity of the cells.

Abbreviations
- CCL5
chemokine (C‐C motif) ligand 5
- dsRNA
double‐stranded RNA
- HCV
hepatitis C virus
- IFN
interferon
- IKKε
IκB kinase ε
- LPS
lipopolysaccharide
- MAVS
mitochondrial antiviral signaling protein
- NCL
nucleolin
- NF‐κB
nuclear factor kappa B
- PACT
protein activator of the interferon‐induced protein kinase
- PAMP
pathogen‐associated molecular pattern
- PKR
protein kinase R
- PRR
pattern recognition receptor
- RANTES
regulated on activation normal T cell expressed and secreted
- RIG‐I
retinoic acid‐inducible gene‐I
- RLR
RIG‐I‐like receptor
- RNAi
RNA interference
- SG
stress granule
- TBK1
TANK‐binding kinase 1
Introduction
RNA helicases of the DEAD‐box protein family play multiple roles in almost all aspects of RNA metabolism 1, 2. They use the energy released by ATP hydrolysis to unwind RNA‐duplex structures or remodel RNA–protein complexes 1, 2. Human DDX3 is a member of the DEAD‐box protein family. DDX3 and its yeast homolog Ded1 have been implicated in various RNA metabolism, including transcription 3, 4, pre‐mRNA splicing 5, 6, 7, mRNA transport 8, 9 and translation initiation 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20. The role of DDX3 in translation initiation appears to be evolutionarily conserved from yeast to humans 21. In the fission yeast Schizosaccharomyces pombe, Ded1 is involved in the translational control of two B‐type cyclins: Cig2 and Cdc13 22. Notably, Cig2 mRNA has an unusually long 5′ UTR, whereas the 5′ UTR of Cdc13 mRNA is predicted to contain a complex RNA secondary structure. Consistent with this notion, we found that human DDX3 is required for efficient translation of mRNAs that contain a long or structured 5′ UTR in mammalian cells 13, 14, 15. Given that the RNA helicase activity of DDX3 is required for its function in translational control 13, 14, 15, we propose that DDX3 may facilitate ribosome scanning by resolving secondary structures in the 5′ UTR of selected mRNAs during translation initiation.
It has been reported that DDX3 participates in a wide variety of cellular functions, such as cell cycle progression 15, 20, 23, 24, germ line development 25, viral replication 8, 13, 26, 27, cancer progression 3, 4, 28 and antiviral innate immunity 29, 30, 31, 32, 33. Recently, it has emerged that DDX3 plays a mediator role in antiviral innate immune signaling pathways 33. Viral infections are mainly detected by the host's pattern recognition receptors (PRRs), including the Toll‐like receptors and the retinoic acid‐inducible gene‐I (RIG‐I)‐like receptors (RLRs), which recognize different classes of viral nucleic acids and metabolites at the surface of cells, within endosomes or in the cytoplasm 34. The sensing of viral infections through PRRs leads to the production of inflammatory cytokines, chemokines and type I interferons (IFN‐α/β), which are required for the elimination of viruses. The activation of PRRs triggers intracellular signaling cascades that in turn up‐regulate transcription factors nuclear factor kappa B (NF‐κB) and interferon regulatory factors 3 and 7 (IRF3/7) 35. NF‐κB controls many genes of inflammatory cytokines, whereas IRF3/7 are mainly responsible for the expression of type I IFNs.
Many viruses have evolved a variety of strategies to evade antiviral innate immunity, either by avoiding PRR recognition or by interfering with IFN production. For example, hepatitis C virus (HCV) infection is sensed by cytoplasmic RLRs that recognize pathogen‐associated molecular patterns (PAMPs) in the viral RNA genome 36, 37. However, most people infected with HCV eventually develop a persistent or chronic infection. HCV can circumvent the host's antiviral innate immunity via the actions of the viral NS3/4A protease 38 and the HCV core protein 30. The HCV NS3/4A protease cleaves the mitochondrial antiviral signaling protein (MAVS) to prevent signal transduction of the RIG‐I signaling pathway 38. The HCV core protein suppresses type I IFN induction via its interactions with STAT1 39 and DDX3 30. Similarly, vaccinia virus protein K7 and hepatitis B virus polymerase also inhibit IFN‐β induction by targeting host DDX3 31, 40. The finding of DDX3 as a target of viral proteins for immune evasion highlights the importance of DDX3 in antiviral innate immunity.
DDX3 was recently reported to be a transcriptional regulator of the IFN‐β promoter 32, a signal transducer downstream of TANK‐binding kinase 1 (TBK1), as well as IκB kinase ε (IKKε) 29, 31, and a viral RNA sensor through interactions with RIG‐I and MAVS 41. However, the cause of such multiple acts of DDX3 remains ambiguous. DDX3 has been identified as a phosphorylation target of TBK1 32. Phosphorylated DDX3 can be recruited to the IFN‐β promoter, suggesting that DDX3 acts as a transcriptional regulator 32. Concurrently, DDX3 was shown to interact with IKKε as a scaffold protein, which facilitates the phosphorylation of IRF3/7 by IKKε and leads to IFN‐β induction 29, 31. Furthermore, DDX3 interacts with MAVS 41, an adaptor triggering RIG‐I‐mediated type I IFN induction, suggesting that DDX3 acts as a sensor of viral RNAs and placed DDX3 upstream of TBK1/IKKε. However, the exact mechanism by which DDX3 participates in antiviral innate immunity is still not fully understood.
PACT is a double‐stranded RNA (dsRNA)‐binding protein, which functions as a cellular activator of RIG‐I to facilitate the sensing of viral RNAs 42. Some viruses avoid immune detection by direct targeting of PACT 43, 44, 45, 46, 47. The perturbation of PACT‐induced activation of RIG‐I by a variety of viral proteins suggests a critical role for PACT in antiviral host defense. In the present study, we show that DDX3 was required for efficient translation of PACT mRNA in human cells. Knockdown of DDX3 resulted in impaired antiviral innate immune responses. PACT partially rescued defects of IFN‐β and chemokine (C‐C motif) ligand 5 (CCL5)/RANTES (regulated on activation normal T cell expressed and secreted) induction in DDX3‐knockdown cells. Taken together, it is shown that DDX3 may participate in antiviral innate immunity, at least in part, by translational control of PACT.
Results
DDX3 regulates the expression of PACT
We previously performed polysome fractionation of DDX3‐depleted HeLa cell extracts followed by cDNA microarray analysis of cellular mRNAs whose translation was impeded 15. Among these mRNA targets of DDX3, we found that several candidates are involved in antiviral innate immunity, including PACT. To confirm whether PACT is regulated by DDX3, we examined the expression of PACT in DDX3‐knockdown cells and mock‐treated cells. Immunoblotting analysis showed that the protein level of DDX3 was significantly reduced in HeLa and HEK293 cells transduced with either of DDX3‐targeting shRNAs (Fig. 1A,B, lanes 2–3) of which shDDX3‐2 was more effective than shDDX3‐1 in inhibiting DDX3 expression. Cyclin E1 has been identified as a translational target of DDX3 and thus serves as a positive control 15. As observed with cyclin E1, the level of PACT protein considerably declined in DDX3‐knockdown cells (Fig. 1A,B), suggesting that DDX3 may control the expression of PACT protein. We also evaluated PACT and cyclin E1 protein levels in DDX3‐overexpressing HeLa cells. Both PACT and cyclin E1 protein levels were up‐regulated by DDX3 in a dose‐dependent manner (Fig. 1C). PACT and cyclin E1 protein levels were increased by up to four‐ and five‐fold in accordance with the levels of DDX3 overexpression in HeLa cells using α‐tubulin as a normalization factor (Fig. 1C). The results confirmed our notion that PACT is regulated by DDX3.
Figure 1.
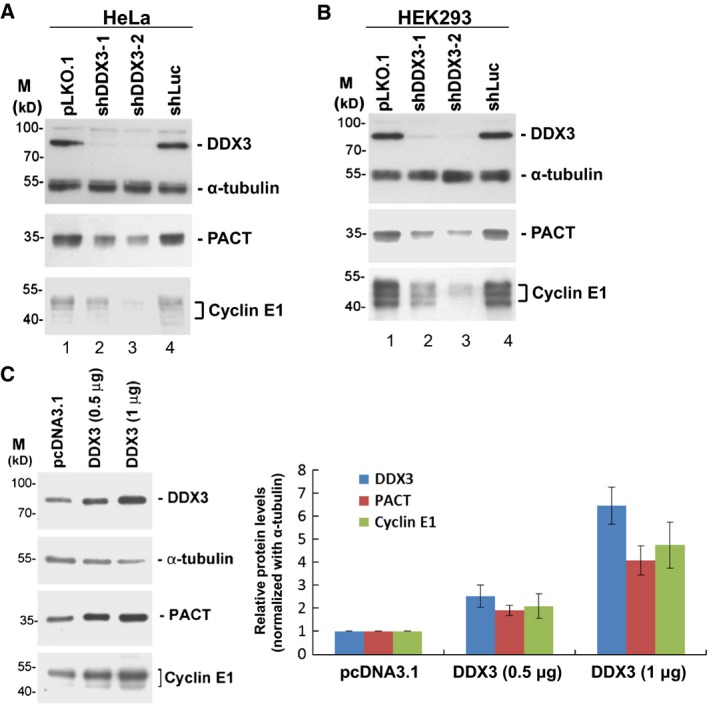
DDX3 regulates the expression of PACT. (A) HeLa cells were transduced with the empty lentiviral vector (lane 1, pLKO.1) or the pLKO.1 vector expressing the indicated shRNAs (lanes 2–4). After 24 h, puromycin was added to the culture medium for selection. Cells were harvested for analysis at 3 days post‐transduction. Immunoblotting was performed using antibodies against DDX3, α‐tubulin, PACT and cyclin E1. Detection of α‐tubulin was used as a loading control. (B) HEK293 cells were transduced with the lentiviral vectors as described in (A), except that puromycin was omitted. (C) HeLa cells were transfected with the pcDNA3.1 vector or the DDX3‐expressing pcDNA3.1 vector (0.5 and 1 μg). After 48 h, cells were harvested for immunoblotting analysis using antibodies against DDX3, α‐tubulin, PACT and cyclin E1. The bar graph shows the relative expression levels of DDX3, PACT and cyclin E1 normalized to α‐tubulin as the mean ± SEM of at least three independent experiments.
DDX3 regulates PACT expression at the protein level
To examine whether DDX3 affects the expression level of PACT mRNA, we detected both protein and mRNA expression levels in DDX3‐knockdown cells and mock‐treated cells. Immunoblotting analysis showed that knockdown of DDX3 in HeLa cells by either DDX3‐targeting shRNA caused an apparent reduction in the level of PACT protein (Fig. 2A), although the level of PACT mRNA was not affected by DDX3 knockdown (Fig. 2B). Quantitative immunoblotting analysis showed that the PACT protein levels were significantly reduced by up to 47% and 72% by shDDX3‐1 and shDDX3‐2, respectively (Fig. 2C). PACT mRNA levels were further measured by quantitative real‐time RT‐PCR and normalized to the levels of β‐actin mRNA. However, there was no significant change in the PACT mRNA level in DDX3‐knockdown cells compared to mock‐treated cells (Fig. 2C). Because DDX3 only affects PACT expression at the protein level, it is possible that DDX3 regulates the translation of PACT mRNA.
Figure 2.
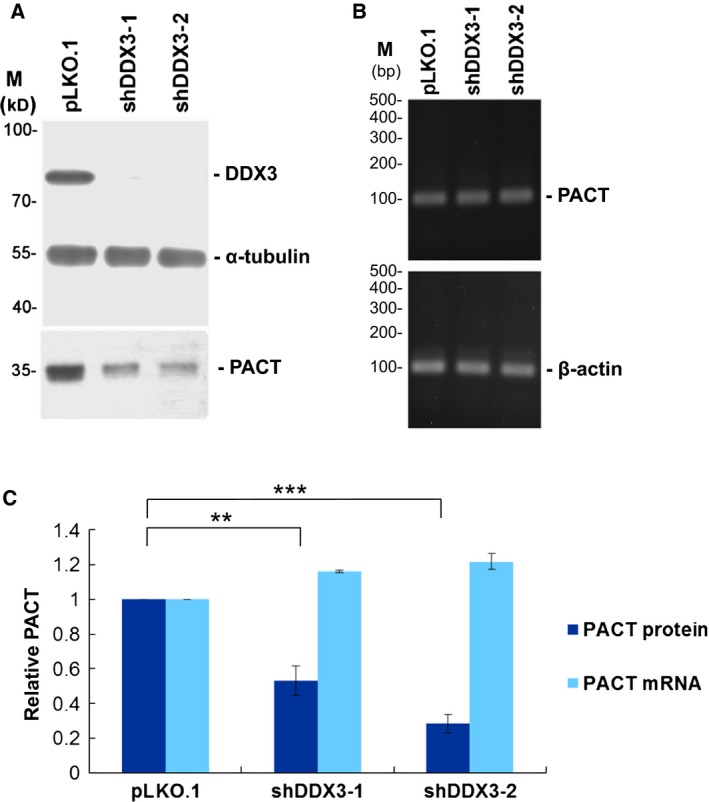
Knockdown of DDX3 inhibits PACT expression at the protein level. (A) HeLa cells were transduced with the empty lentiviral vector (pLKO.1) or the pLKO.1 vector expressing DDX3 shRNAs (shDDX3‐1 and shDDX3‐2). After 24 h, puromycin was added to the culture for selection. Cells were harvested for analysis at 3 days post‐transduction. Immunoblotting was performed using antibodies against DDX3, α‐tubulin and PACT. Detection of α‐tubulin was used as a loading control. (B) RNA extracted from mock‐treated (pLKO.1) or DDX3‐knockdown (shDDX3‐1 and shDDX3‐2) HeLa cells was analyzed by conventional RT‐PCR using specific primers for PACT and β‐actin mRNAs. PCR products were resolved by agarose gel electrophoresis. (C) The protein level of PACT relative to α‐tubulin in mock‐treated (pLKO.1) and DDX3‐knockdown (shDDX3‐1 and shDDX3‐2) HeLa cells were quantitatively analyzed. The levels of PACT and β‐actin mRNAs were detected by quantitative real‐time RT‐PCR. The bar graph shows the relative expression levels of PACT protein and mRNA as the mean ± SEM of at least three independent experiments (**P < 0.01, ***P < 0.001).
DDX3 is required for efficient translation of PACT mRNA
To confirm whether DDX3 affects the translation of PACT mRNA, polysome distribution of PACT mRNA was evaluated using a sucrose gradient sedimentation and the mRNA level was analyzed by quantitative real‐time RT‐PCR in DDX3‐knockdown and mock‐treated HeLa cells. Knockdown of DDX3 by shDDX3‐1 and shDDX3‐2 did not cause a detectable change in the polysome profile (Fig. 3A, top), which is consistent with the view that DDX3 is dispensable for general mRNA translation 14, 15. Accordingly, no visible change was observed in different density gradient fractions of 18S and 28S ribosomal RNAs (Fig. 3A, bottom). However, the distribution of PACT mRNA in polysome fractions was evidently changed in DDX3‐knockdown HeLa cells compared to mock‐treated cells (Fig. 3A, middle). Knockdown of DDX3 by shDDX3‐1 and shDDX3‐2 resulted in an accumulation of PACT and cyclin E1 mRNAs in the 40S ribosomal fraction (Fig. 3A, fraction 3), indicating a stalled translation initiation complex. By contrast, polysome distribution of β‐actin mRNA was not significantly affected by DDX3 knockdown.
Figure 3.
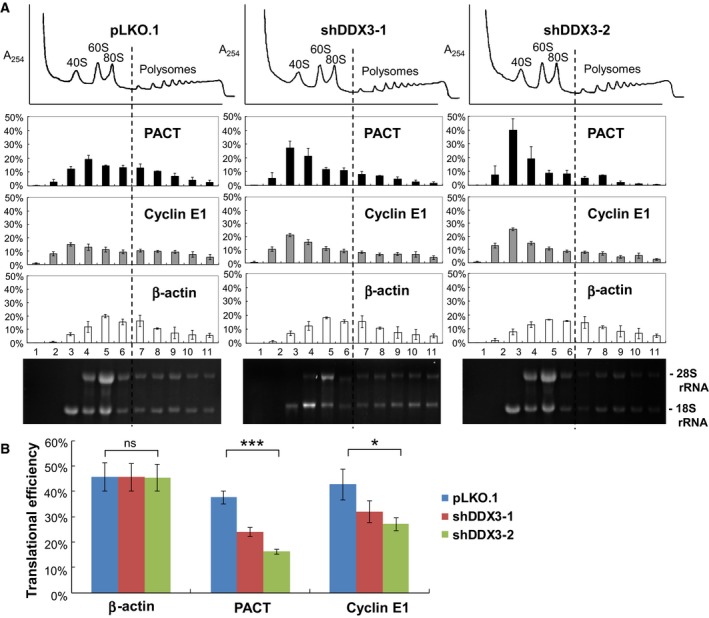
Knockdown of DDX3 inhibits translation initiation of PACT mRNA. (A) HeLa cells were transduced with the empty lentiviral vector (pLKO.1) or the pLKO.1 vector expressing DDX3 shRNAs (shDDX3‐1 and shDDX3‐2). Cells were harvested for analysis at 3 days post‐transduction. Cytoplasmic extracts were loaded on a linear 15–40% sucrose gradient ultracentrifugation. After centrifugation, the polysome profile was plotted by A 254 values (top). Total RNA was extracted from each fraction for analysis. The purified RNA was resolved on a 1% formaldehyde/agarose gel, and rRNAs were visualized by ethidium bromide staining (bottom). The levels of mRNA were analyzed by quantitative real‐time RT‐PCR using specific primers for PACT, cyclin E1 and β‐actin mRNAs (middle). (B) Translational efficiency of β‐actin, PACT and cyclin E1 mRNAs in mock‐treated (pLKO.1) and DDX3‐knockdown (shDDX3‐1 and shDDX3‐2) HeLa cells was calculated and shown as a percentage. The bar graph shows the changes of translational efficiency as the mean ± SEM of at least three independent experiments (ns, not significant; ***P < 0.001, *P < 0.05).
The distribution of an mRNA within the polysome fractions is reflective of its translational efficiency. Quantitative analysis of mRNAs by real‐time RT‐PCR showed that approximately 40% of PACT mRNA was associated with polysomes in mock‐treated cells, whereas only 24% (shDDX3‐1) and 16% (shDDX3‐2) of PACT mRNA remained associated with polysomes in DDX3‐knockdown cells (Fig. 3B), suggesting that the translational efficiency of PACT but not β‐actin mRNA was decreased by DDX3 knockdown (Fig. 3B). The translational efficiency of cyclin E1 mRNA was also down‐regulated in DDX3‐knockdown cells (Fig. 3B). Therefore, we conclude that DDX3 is required for efficient translation of PACT and cyclin E1 mRNAs in HeLa cells.
DDX3 facilitates the translation of PACT mRNA via its 5′ UTR
We have previously proposed that DDX3 is required for the translation of mRNAs that contain a long or structured 5′ UTR 13, 14, 15. The 201 nucleotides of 5′ UTR of PACT mRNA contain a GC‐rich (74.6% GC) sequence and are predicted to form a stable secondary structure with an estimated free energy of –109.50 kcal·mol−1 (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (Fig. 4A). To investigate the molecular mechanism by which DDX3 functions in the translation of PACT mRNA, we established a firefly luciferase (Fluc) reporter containing the 5′ UTR of PACT mRNA and performed a dual‐luciferase reporter assay. A Renilla luciferase (Rluc) reporter with an unstructured 5′ UTR was co‐transfected with the Fluc reporter as an internal control. Changes in the translation of reporter mRNAs were assessed by the relative Fluc/Rluc activity in DDX3‐knockdown cells compared to mock‐treated cells. A significant reduction (23.3% and 41.8% by shDDX3‐1 and shDDX3‐2, respectively) of the relative Fluc/Rluc activity was observed in DDX3‐knockdown cells without changing the level of the reporter mRNAs (Fig. 4B). The above data suggest that DDX3 participates in translational control of PACT mRNA, at least in part, via the structured PACT 5′ UTR.
Figure 4.
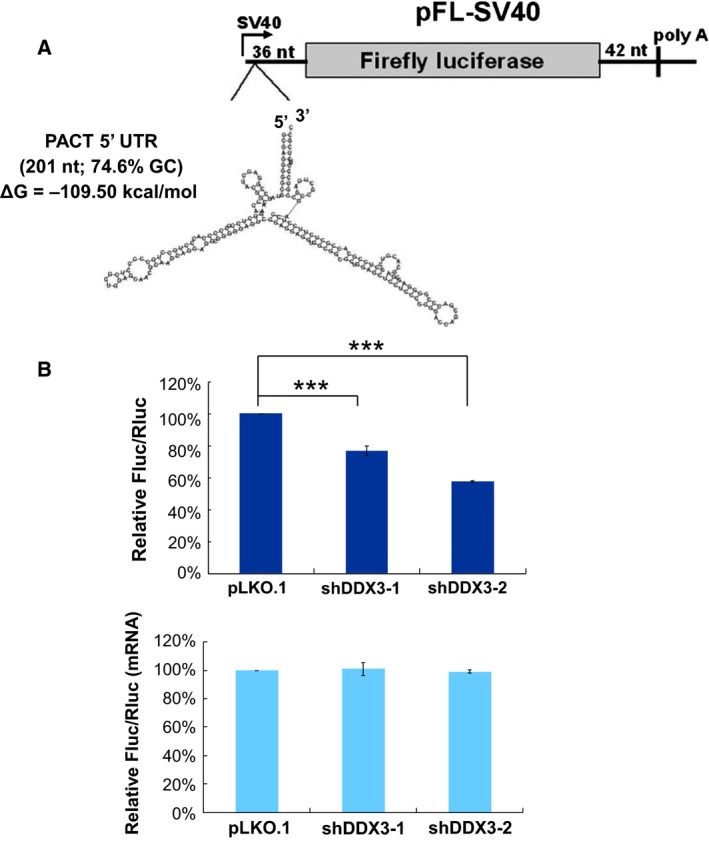
DDX3 facilitates the translation of reporter mRNAs containing the PACT 5′ UTR. (A) Schematic representation of the firefly luciferase (Fluc) reporter (pFL‐SV40) with the human PACT 5′ UTR (201 nucleotides), for which the predicted secondary structure is shown. (B) HeLa cells were transduced with the empty lentiviral vector (pLKO.1) or the pLKO.1 vector expressing DDX3 shRNAs (shDDX3‐1 and shDDX3‐2). After 48 h, HeLa cells were co‐transfected with the pFL‐SV40 reporter containing the PACT 5′ UTR and the control pRL‐SV40 vector encoding the Renilla luciferase (Rluc). Cells were lysed for analysis at 24 h post‐transfection. For each transfectant, the Fluc activity was normalized to that of the Rluc control. The bar graph shows the relative Fluc/Rluc activities in DDX3‐knockdown cells compared to mock‐treated cells (top). Fluc and Rluc mRNAs were analyzed by quantitative real‐time RT‐PCR (bottom). Data are shown as the mean ± SEM from at least three independent experiments (***P < 0.001).
PACT can partially rescue the antiviral innate immune defects caused by DDX3 knockdown
Because PACT functions in antiviral innate immunity, DDX3‐depleted cells may have compromised antiviral capacity. We therefore addressed the effects of DDX3 knockdown on the type I IFN and inflammatory chemokine induction. To mimic viral infection, we first stimulated HEK293 cells with the synthetic analog of dsRNA poly(I:C), as well as the HCV PAMP RNA, a polyU/UC sequence in the 3′ region of the HCV genome. Transfected poly(I:C) and the HCV PAMP RNA function as ligands for cytoplasmic RLRs to induce antiviral innate immune response 36. As expected, poly(I:C) induced IFN‐β1 and CCL5/RANTES expression in a dose‐dependent manner, and this was impaired in DDX3‐knockdown HEK293 cells (Fig. 5A, lanes 1–6). Similar results were obtained by transfection with the HCV PAMP RNA into HEK293 cells to induce IFN‐β1 and CCL5/RANTES expression (Fig. 5A, lanes 7–8). To mimic bacterial infection, we also stimulated HEK293 cells with lipopolysaccharide (LPS), a major constituent of the cell wall of most Gram‐negative bacteria. LPS‐induced IFN‐β1 and CCL5/RANTES expression was also impaired in DDX3‐knockdown HEK293 cells (Fig. 5A, lanes 9–10). By contrast, expression of the cytokine TNF‐α was not significantly affected by DDX3 knockdown in HEK293 cells (Fig. 5A). This result supported a role of DDX3 in innate immunity.
Figure 5.
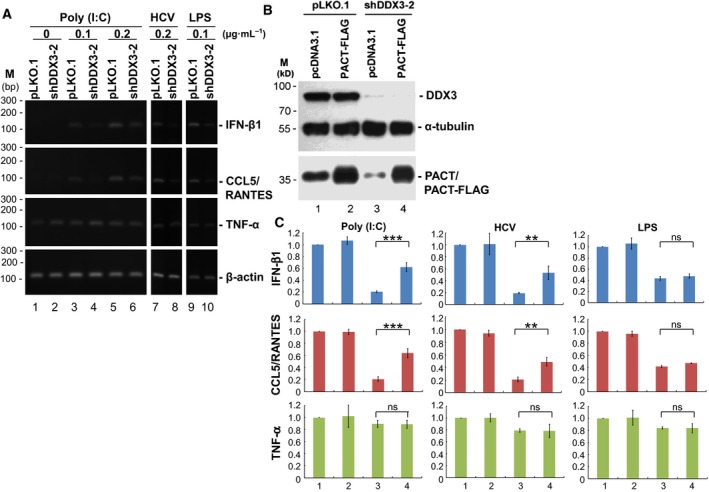
PACT partially rescued the antiviral innate immune defects caused by DDX3 knockdown. HEK293 cells were transduced with the empty lentiviral vector (pLKO.1) or the pLKO.1 vector expressing DDX3 shRNA (shDDX3‐2) for subsequent functional assessment. (A) At 3 days post‐transduction, HEK293 cells were transfected with poly(I:C) or the HCV PAMP RNA or treated with bacterial LPS for 4 h at different concentrations as indicated (μg·mL−1). Total RNA extracted from HEK293 cells was analyzed by conventional RT‐PCR using specific primers for IFN‐β1, CCL5/RANTES, TNF‐α, and β‐actin mRNAs. PCR products were resolved by 1.5% agarose gel electrophoresis. (B) At 48 h post‐transduction, cells were further transfected with the pcDNA3.1 vector or the FLAG‐tagged PACT‐expressing pcDNA3.1 vector (PACT‐FLAG) to assess whether PACT can be rescued in DDX3‐knockdown cells. Cell lysates were subjected to immunoblotting analysis using antibodies against DDX3, α‐tubulin and PACT. Detection of α‐tubulin was used as a loading control. (C) Under a similar experimental setting to that described in (B), HEK293 transfectants at 3 days post‐transduction were further transfected with 0.1 μg·mL−1 of poly(I:C) or 0.2 μg·mL−1 of the HCV PAMP RNA, or treated with 0.1 μg·mL−1 of bacterial LPS for 4 h to induce antiviral innate immune response. Total RNA extracted from HEK293 cells was analyzed by quantitative real‐time RT‐PCR using specific primers for IFN‐β1, CCL5/RANTES, TNF‐α and β‐actin mRNAs. The bar graph shows the relative mRNA levels of IFN‐β1, CCL5/RANTES and TNF‐α normalized to β‐actin as the mean ± SEM from at least three independent experiments (***P < 0.001, **P < 0.01; ns, not significant).
Next, we evaluated whether PACT can rescue the defect of impaired IFN‐β1 and CCL5/RANTES induction caused by DDX3 knockdown. Immunoblotting analysis showed that knockdown of DDX3 by shDDX3‐2 down‐regulated PACT expression in HEK293 cells (Fig. 5B, lane 3). Transient expression of FLAG‐tagged PACT restored PACT level in DDX3‐knockdown HEK293 cells (Fig. 5B, lane 4). To test whether PACT can rescue the defect of IFN‐β1 and CCL5/RANTES induction in DDX3‐knockdown cells, we transfected poly(I:C) or the HCV PAMP RNA into HEK293 cells to induce antiviral innate immunity. The production of IFN‐β1, CCL5/RANTES and TNF‐α mRNAs was measured by quantitative real‐time RT‐PCR using β‐actin for normalization (Fig. 5C). Knockdown of DDX3 showed an approximate 80% reduction in the IFN‐β1 and CCL5/RANTES induction in HEK293 cells transfected with poly(I:C) or the HCV PAMP RNA, whereas PACT partially rescued their expression (Fig. 5C, lanes 3–4). By contrast, transient expression of FLAG‐tagged PACT did not rescue impaired IFN‐β1 and CCL5/RANTES induction in DDX3‐knockdown HEK293 cells treated with bacterial LPS (Fig. 5C, lanes 3–4). These results indicated that DDX3 participates in antiviral immune response, at least partly, through translational control of PACT protein expression.
HCV core protein binds to DDX3 and abrogates its function in translational control
The HCV core protein interacts with DDX3 48, 49, 50 and interferes with DDX3‐mediated IFN‐β induction 30. However, the molecular mechanism underlying the impaired IFN‐β induction by the HCV core protein has not yet been fully defined. We assumed that the HCV core protein may bind to DDX3 and abrogate its function in antiviral innate immunity through translational control of PACT. To address this possibility, we examined the effects of HCV core on the translation of a reporter mRNA containing the PACT 5′ UTR by a dual‐luciferase reporter assay. Overexpression of GFP‐tagged HCV core, a matured form of HCV core from the HCV‐1b genotype 51, reduced the relative Fluc/Rluc activity without changing the level of the reporter mRNAs (Fig. 6A). We also demonstrated that GFP‐tagged HCV core associates with endogenous DDX3 in HEK293 cells by immunoprecipitation (Fig. 6B). This suggests that the HCV core protein may inhibit the translation of PACT mRNA by binding to DDX3. According to our previous study 14, DDX3 is predominantly localized in the cytoplasm and recruited to cytoplasmic stress granules (SGs) under stress conditions. We also observed that GFP‐tagged HCV core was mostly colocalized with DDX3 in cytoplasmic SGs (Fig. 6C), which accumulate stalled translation initiation complexes. Consistent with this notion, colocalization of GFP‐tagged HCV core with SG marker proteins eIF4A and HuR was observed in cytoplasmic SGs (Fig. 6C), suggesting that the HCV core protein may inhibit the function of DDX3 in antiviral innate immunity through a translational mechanism. In addition to cytoplasmic SGs, colocalization of GFP‐tagged HCV core with nucleolin (NCL) in the nucleolus was also observed (Fig. 6C).
Figure 6.
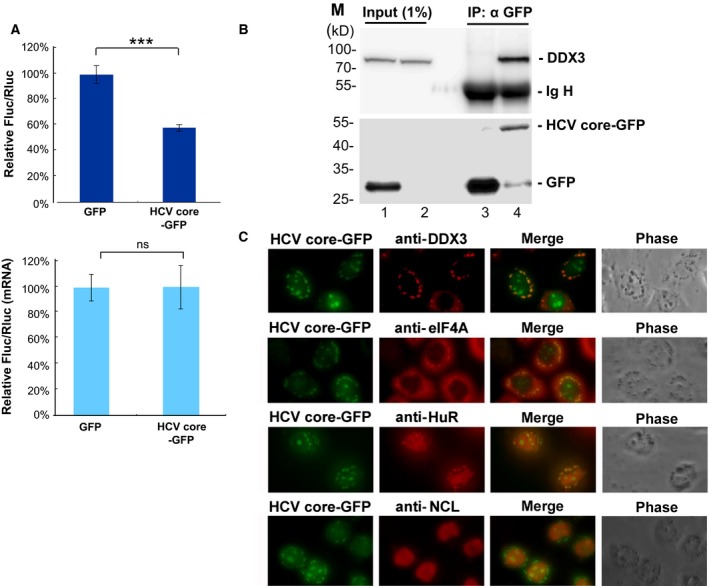
HCV core protein inhibits PACT translation by direct binding and redistribution of DDX3 into cytoplasmic SGs. (A) HeLa cells were co‐transfected with the pFL‐SV40 reporter containing the PACT 5′ UTR and the control pRL‐SV40 vector in combination with the pEGFP‐N1 vector encoding GFP or HCV core‐GFP. Cells were lysed for analysis at 48 h post‐transfection. For each transfectant, the Fluc activity was normalized to that of the Rluc control. The bar graph shows the relative Fluc/Rluc activities in HCV core‐overexpressing (HCV core‐GFP) cells compared to control (GFP) cells (top). Fluc and Rluc mRNAs were analyzed by quantitative real‐time RT‐PCR (bottom). Data are shown as the mean ± SEM from at least three independent experiments (***P < 0.001; ns, not significant). (B) HEK293 cells were transfected with the pEGFP‐N1 vector encoding GFP or HCV core‐GFP. Cells were lysed for analysis at 48 h post‐transfection. Immunoprecipitation was performed using anti‐GFP antibody coupled to protein A sepharose beads. Immunoprecipitates were treated with 1 mg·mL−1 RNase A at 37 °C for 30 min. Bound proteins were eluted and subjected to immunoblotting with anti‐DDX3 antibody (top) or anti‐GFP antibody (bottom). Ig H represents the immunoglobulin heavy chain. (C) HeLa cells were transfected with the pEGFP‐N1 vector encoding HCV core‐GFP for 24 h. Immunofluorescent staining of HeLa cells was carried out using antibodies against DDX3, eIF4A, HuR and nucleolin (NCL). Co‐localization of HCV core‐GFP (green) and DDX3, eIF4A or HuR (red) in cytoplasmic SGs was observed under a fluorescence microscope.
In the present study, we propose a model of DDX3‐mediated translational control during HCV infection (Fig. 7). HCV genomic RNAs can stimulate a RIG‐I‐mediated antiviral innate immune response in the presence of PACT during viral replication 36. However, the HCV core protein interferes with the antiviral innate immune response by abrogating DDX3‐mediated translational activation of PACT. Loss of the PACT protein attenuates the detection sensitivity of RIG‐I to the HCV genomic RNAs and, in turn, affects IFN‐β induction in HCV‐infected cells (Fig. 7). Taken together, HCV can evade antiviral innate immunity through the DDX3/PACT axis to establish persistent or chronic infection.
Figure 7.
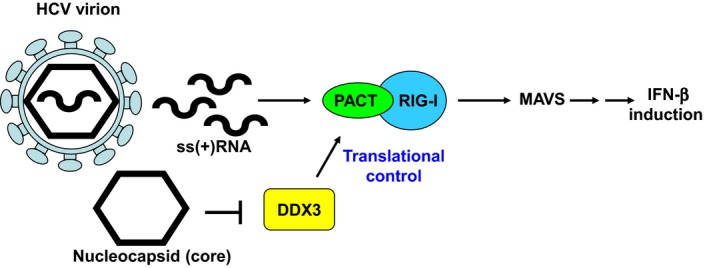
A model of DDX3‐mediated translational control of PACT in antiviral innate immune response to HCV infection. The HCV genomic RNA (ssRNA) contains specific PAMPs, which can be detected by RIG‐I to trigger antiviral innate immunity (IFN‐β induction). The dsRNA binding protein PACT is a binding partner of RIG‐I and functions as an activator of RIG‐I in the sensing of HCV RNAs. Interestingly, the HCV core protein abrogates the RIG‐I‐mediated antiviral innate immune response by direct binding of DDX3. Inactivation of DDX3 down‐regulates the expression of PACT and thus attenuates the viral RNA detection sensitivity of the cells.
Discussion
Previous studies have indicated that DDX3 is involved in antiviral innate immunity 31, 32, although the underlying molecular mechanisms are still not fully understood. In the present study, we show, for the first time, that DDX3 regulates PACT expression by promoting the translation of PACT mRNA, which likely harbors a structured 5′ UTR (Figs 1, 2, 3, 4). Our results thus provide another layer of evidence indictaing that DDX3 participates in antiviral innate immunity through regulation of PACT protein level. The accumulation of PACT mRNA in the fractions containing 40S ribosomal subunits in DDX3‐knockdown cells (Fig. 3) supports the view that DDX3 participates in ribosome scanning of PACT mRNA during translation initiation.
We also showed that DDX3 is required for IFN‐β1 and CCL5/RANTES induction when cells were transfected with viral RNAs (Fig. 5), supporting its role in antiviral innate immunity. Our results indicate that the impairment of innate immunity observed in DDX3‐knockdown cells was likely a result of the down‐regulation of PACT. PACT can partially rescue the impaired innate immunity caused by DDX3 depletion (Fig. 5). In agreement with a previous study 41, we suggest that DDX3 functions in the sensing of viral infection. Many viruses have evolved various strategies for evading detection by both innate and adaptive immune responses. HCV can utilize the core protein to counteract the antiviral effects of DDX3. We provide evidence that HCV core results in a decrease in the translation of reporter mRNA containing the PACT 5′ UTR (Fig. 6). The interaction between HCV core and DDX3 appears to play an important role in attenuating antiviral immune responses. The redistribution of DDX3 into cytoplasmic SGs was frequently observed in HCV core‐overexpressing cells (Fig. 6). Thus, the HCV core protein may inhibit DDX3 function by direct binding and/or sequestration into SGs to interfere with the expression of PACT, which is required for activation of RIG‐I‐mediated virus detection (Fig. 7). In addition to HCV core, vaccinia virus protein K7 31 and hepatitis B virus polymerase 40 proteins also abrogate IFN induction by targeting DDX3. It is likely that these viruses have developed a common strategy for evading or suppressing the host's antiviral innate immunity. Notably, PACT also serves as a target of a variety of viruses. It has been reported that viral proteins, including the influenza A virus NS1 protein 44, the Ebola virus VP35 protein 43, 46, the herpes simplex virus US11 protein 45 and the middle east respiratory syndrome coronavirus 4a protein 47, can abrogate RIG‐I‐mediated IFN induction by direct interaction with the PACT. By contrast, DDX3‐mediated translational control of PACT expression is a novel strategy for counteracting antiviral innate immunity.
Notably, PACT also serves as an activator of the protein kinase R (PKR) 52, 53, which is activated by viral dsRNAs to inhibit translation in infected cells. Activated PKR is able to phosphorylate the eukaryotic translation initiation factor eIF2α, thereby inhibiting translation initiation of cellular and viral mRNAs. PACT‐mediated activation of PKR plays a key role in host defense against viral infections. It is obvious that DDX3 can modulate PKR activity through translational control of PACT. Moreover, PACT also functions as a binding partner of Dicer in RNA interference (RNAi) 54, which is a gene silencing process triggered by dsRNAs 55, 56. RNAi also plays an antiviral role in infected plants 57, invertebrates 58 and mammals 59. Dicer requires PACT to facilitate substrate specificity during small interfering RNA and microRNA biogenesis. The loss of PACT strongly reduces the accumulation of mature microRNA 60. Therefore, it would be interesting to study the importance of DDX3‐mediated translational control of PACT in the RNAi pathway.
Experimental procedures
Cell culture and transfection
HeLa and HEK293 cells were grown at 37 °C in DMEM supplemented with 10% FBS, 100 U·mL−1 penicillin, and 100 μg·mL−1 streptomycin. Cell transfection was performed using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA), essentially in accordance with the manufacturer's instructions.
Plasmid constructs
The plasmid expressing FLAG‐tagged DDX3 has been described previously 4. For the in vivo translation assay, the PACT 5′ UTR fragment obtained by RT‐PCR using total RNA from HEK293 cells as the template was inserted into an unique HindIII site upstream of the firefly luciferase coding region in the pFL‐SV40 vector 14. The control reporter pRL‐SV40 (Promega, Madison, WI, USA) has been described previously 14. To generate a construct encoding the HCV PAMP RNA, the HCV PAMP fragment (223 nucleotides) obtained by PCR using HCV subgenomic replicon HCV‐EV71I‐Luc 61 as the template was inserted into yT&A cloning vector (Yeastern Biotech, Taipei, Taiwan). To generate a construct expressing FLAG‐tagged PACT protein, the ORF of PACT obtained by RT‐PCR using total RNA from HEK293 cells as the template was inserted into pcDNA3.1 (Life Technologies). To generate a construct expressing HCV core‐GFP fusion protein, the HCV core DNA fragment was subcloned from pFS‐HCVc174 51 into NheI and XhoI sites of pEGFP‐N1 (Clontech, Palo Alto, CA, USA). All constructs were confirmed by direct sequencing.
Lentivirus‐mediated RNAi knockdown
All of the plasmids required for lentivirus production were provided by the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The two pLKO.1‐shRNA vectors used to knockdown DDX3 were: TRCN0000000002 (shDDX3‐1) and TRCN0000000004 (shDDX3‐2). The control plasmid was TRCN0000231693 (shLuc). The transfection reagent Lipofectamine® 2000 (Life Technologies) was used for lentiviral production in 293T cells with a packaging construct (pCMV ΔR8.91), an envelope construct (pMD.G) and pLKO.1‐shRNA. To knockdown endogenous DDX3, HeLa cells were transduced with shRNA‐expressing lentivirus at a multiplicity of infection of 10 virus particles·cell−1 in growth medium containing 8 μg·mL−1 polybrene at 37 °C, 5% CO2. After 24 h, puromycin (2 μg·mL−1) was added to the medium for selecting infected cells. Cells were harvested 3 days post‐transduction for analysis.
Immunoblotting and immunoprecipitation
For immunoblot analysis, proteins were transferred onto a PVDF Transfer Membrane (Perkin Elmer, Waltham, MA, USA). Protein blots were blocked with 3% skim milk in TBST buffer (100 mm Tris‐HCl, pH 7.6, 150 mm NaCl and 0.1% Tween 20) at room temperature for 1 h. The primary antibodies used included affinity‐purified rabbit anti‐DDX3 (0.1 μg·mL−1) 14, rabbit anti‐α‐tubulin (dilution 1 : 2000; Cell Signaling, Beverly, MA, USA), rabbit anti‐PACT (dilution 1 : 1000; Cell Signaling), mouse anti‐cyclin E (0.4 μg·mL−1; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti‐GFP (0.2 μg·mL−1; Abcam, Cambridge, MA, USA). Blots were incubated with primary antibodies in blocking buffer at room temperature for 2 h, followed by incubation with horseradish peroxidase‐conjugated secondary antibodies at room temperature for 2 h. Detection was performed using Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA) for X‐ray film exposure.
To immunoprecipitate HCV core‐GFP, 2 μL of anti‐GFP antibody (Abcam) were coupled to protein A‐sepharose beads in NET‐2 buffer (50 mm Tris‐HCl, pH 7.4, 150 mm NaCl and 0.05% NP‐40). Transfected HEK293 cells (107 cells) were lysed in NET‐2 buffer containing 1% NP‐40 and 1× protease inhibitor cocktail (Thermo Scientific, Waltham, MA, USA). Cell lysates were incubated with the antibody‐beads conjugate at 4 °C with gentle agitation for 4 h. The beads were washed four times with 1 mL of cold NET‐2 buffer to remove unbound proteins. Immunoprecipitates were treated with 1 mg·mL−1 RNase A at 37 °C for 30 min. Bound proteins were eluted with 1× SDS sample buffer and resolved by 10% SDS/PAGE.
RT‐PCR and quantitative real‐time PCR
RT‐PCR was used to detect the mRNA expression level. Extracted total RNA was reverse‐transcribed into cDNA using the High‐Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer's instructions. The resulting cDNA was subjected to conventional PCR or quantitative real‐time PCR analysis. Conventional PCR was performed using GoTaq DNA polymerase (Promega) and the forward and reverse primers: PACT [forward primer (FP): 5′‐CCAGGGAAAACACC GATTCA‐3′ and reverse primer (RP): 5′‐GCACGTGTATTTGCACATCAGA‐3′], β‐actin (FP: 5′‐GCCCTGAGGCACTCTTCCA‐3′ and RP: 5′‐CGGATGTCCACGTCACACTT‐3′), cyclin E1 (FP: 5′‐TGCTTCGGCCTTGTATCATTT‐3′ and RP: 5′‐TGGAACCATCCACTTGACACA‐3′), IFN‐β1 (FP: 5′‐CAGAAGCTCCTGTGGCAATTG‐3′ and RP: 5′‐GGAACTGCTGCAGCTGCTTA‐3′), CCL5/RANTES (FP: 5′‐CAGCCCTCGCTGTCATCCT‐3′ and RP: 5′‐GGGCAATGTAGGCAAAGCA‐3′) and TNF‐α (FP: 5′‐TGAGGCCAAGCCCTGGTAT‐3′ and RP: 5′‐GAGATAGTCGGGCCGATTGA‐3′).
Quantitative real‐time PCR was performed using StepOnePlus™ Real‐Time PCR Systems (Life Technologies) in accordance with the manufacturer's instructions. The levels of mRNAs were detected with Fast SYBR® Green Master Mix (Applied Biosystems). In addition, quantitative real‐time PCR of Fluc and Rluc was performed using the Custom TaqMan Assay Design Tool (Applied Biosystems). Quantitative analysis was performed by the measurement of C t values during the exponential phase of amplification. Relative quantitation values were calculated using the 2−ΔΔCt method.
Sucrose gradient sedimentation and polysome profiling
Cells were collected in cold PBS containing 100 μg·mL−1 cycloheximide. All subsequent steps were performed at 4 °C. Cell pellets were resuspended in RSB‐150 (10 mm Tris‐HCl, pH 7.4, 3 mm MgCl2 and 150 mm NaCl) containing 100 μg·mL−1 cycloheximide, 40 μg·mL−1 digitonin (Calbiochem, San Diego, CA, USA), 20 U·mL−1 RNasin (Promega) and 1× protease inhibitor cocktail (Thermo Scientific). After incubation on ice for 5 min, cells were disrupted by passage through a 26‐gauge needle five times. Cytoplasmic extracts were collected by centrifugation at 3000 g for 2 min, and clarified by further centrifugation at 11 000 g for 15 min. The samples were loaded on a linear 15–40% sucrose gradient and centrifuged at 178 000 g for 3 h. After centrifugation, total RNA was extracted from each fraction using phenol/chloroform extraction in the presence of 1% SDS and 0.25 m NaCl, followed by ethanol precipitation. For polysome profile analysis, the gradients were monitored at 254 nm using an ISCO fractionation system (Teledyne ISCO Inc., Lincoln, NE, USA).
In vivo translation assay
DDX3‐knockdown and mock‐treated HeLa cells were transfected with a pFL‐SV40 derived reporter (0.2 μg) and the control pRL‐SV40 vector (0.2 μg). At 24 h post‐transfection, cells were lysed in 1× Passive Lysis Buffer (Promega). For the HCV core overexpression assay, 1 μg of plasmids encoding GFP or HCV core‐GFP were co‐transfected with a pFL‐SV40 derived reporter (0.2 μg) and the control pRL‐SV40 vector (0.2 μg) into HeLa cells (2 × 105 cells well−1). At 48 h post‐transfection, cells were lysed in 1× Passive Lysis Buffer (Promega). The activities of firefly luciferase and Renilla luciferase were measured using the Dual‐Luciferase Reporter Assay System (Promega).
Immunofluorescent staining
HCV core‐GFP was transiently expressed in HeLa cells for 24 h. Immunofluorescent staining of HeLa cells was performed as described previously 14. Briefly, cells grown on cover slips were rinsed with cold PBS and fixed with 3% formaldehyde in PBS for 30 min, followed by permeabilization with 0.5% Triton X‐100 in PBS for 10 min. After washing with PBS, cells were blocked with 1% BSA in PBS for 30 min and then incubated with the relevant primary antibodies in PBS at RT for 1 h. The primary antibodies used included mouse anti‐DDX3 (0.4 μg·mL−1; Santa Cruz Biotechnology), rabbit anti‐eIF4A1 (1 μg·mL−1; Abcam), mouse anti‐HuR (1 μg·mL−1; Santa Cruz Biotechnology) and mouse anti‐NCL (0.4 μg·mL−1; Santa Cruz Biotechnology). After washing with PBS twice, cells were incubated with appropriate secondary antibodies, including Alexa Fluor® 555 goat anti‐mouse IgG (2 μg·mL−1; Invitrogen) and Alexa Fluor® 555 goat anti‐rabbit IgG (2 μg·mL−1; Invitrogen), in PBS at RT for 1 h. After extensive washing with PBS, the specimens were mounted immediately and observed using an inverted fluorescence microscope (TE2000‐U; Nikon Eclipse, Tokyo, Japan) equipped with a charge‐coupled device camera.
Statistical analysis
All data are reported as the mean ± SE and were analyzed using prism, version 4.0 (GraphPad Software Inc., La Jolla, CA, USA). Statistical analysis was performed using an unpaired t‐test. P < 0.05 was considered statistically significant.
Author contributions
MCL performed most of the experiments. MCL and WYT planned the experiments. MCL analyzed the data. MCL, HSS, SWW and WYT wrote the paper.
Acknowledgements
We would like to thank Drs Yan‐Hwa Wu Lee and King‐Song Jeng for the plasmids. This work was supported by grants from Ministry of Science and Technology, Taiwan (NSC100‐2320‐B‐006‐021‐MY3 and 104‐2320‐B‐182‐037) and Chang Gung Memorial Hospital, Taiwan (CMRPD3E0011).
References
- 1. Cordin O, Banroques J, Tanner NK & Linder P (2006) The DEAD‐box protein family of RNA helicases. Gene 367, 17–37. [DOI] [PubMed] [Google Scholar]
- 2. Rocak S & Linder P (2004) DEAD‐box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5, 232–241. [DOI] [PubMed] [Google Scholar]
- 3. Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P et al (2008) Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27, 3912–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP & Lee YH (2006) DDX3, a DEAD box RNA helicase with tumor growth‐suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 66, 6579–6588. [DOI] [PubMed] [Google Scholar]
- 5. Jamieson DJ, Rahe B, Pringle J & Beggs JD (1991) A suppressor of a yeast splicing mutation (prp8‐1) encodes a putative ATP‐dependent RNA helicase. Nature 349, 715–717. [DOI] [PubMed] [Google Scholar]
- 6. Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD & Abelson J (2002) Composition and functional characterization of the yeast spliceosomal penta‐snRNP. Mol Cell 9, 31–44. [DOI] [PubMed] [Google Scholar]
- 7. Merz C, Urlaub H, Will CL & Luhrmann R (2007) Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13, 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yedavalli VS, Neuveut C, Chi YH, Kleiman L & Jeang KT (2004) Requirement of DDX3 DEAD box RNA helicase for HIV‐1 Rev‐RRE export function. Cell 119, 381–392. [DOI] [PubMed] [Google Scholar]
- 9. Kanai Y, Dohmae N & Hirokawa N (2004) Kinesin transports RNA: isolation and characterization of an RNA‐transporting granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- 10. Chuang RY, Weaver PL, Liu Z & Chang TH (1997) Requirement of the DEAD‐Box protein ded1p for messenger RNA translation. Science 275, 1468–1471. [DOI] [PubMed] [Google Scholar]
- 11. Soto‐Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D & Ohlmann T (2012) DEAD‐box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hilliker A, Gao Z, Jankowsky E & Parker R (2011) The DEAD‐box protein Ded1 modulates translation by the formation and resolution of an eIF4F‐mRNA complex. Mol Cell 43, 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lai MC, Wang SW, Cheng L, Tarn WY, Tsai SJ & Sun HS (2013) Human DDX3 interacts with the HIV‐1 Tat protein to facilitate viral mRNA translation. PLoS One 8, e68665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lai MC, Lee YH & Tarn WY (2008) The DEAD‐box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip‐associated protein and participates in translational control. Mol Biol Cell 19, 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai MC, Chang WC, Shieh SY & Tarn WY (2010) DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol 30, 5444–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL & Reed R (2008) Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36, 4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK & Wu Lee YH (2012) Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441, 119–129. [DOI] [PubMed] [Google Scholar]
- 18. Soto‐Rifo R, Rubilar PS & Ohlmann T (2013) The DEAD‐box helicase DDX3 substitutes for the cap‐binding protein eIF4E to promote compartmentalized translation initiation of the HIV‐1 genomic RNA. Nucleic Acids Res 41, 6286–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Senissar M, Saux AL, Belgareh‐Touze N, Adam C, Banroques J & Tanner NK (2014) The DEAD‐box helicase Ded1 from yeast is an mRNP cap‐associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res 42, 10005–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen HH, Yu HI, Cho WC & Tarn WY (2015) DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1‐mediated signaling pathway. Oncogene 34, 2790–2800. [DOI] [PubMed] [Google Scholar]
- 21. Tarn WY & Chang TH (2009) The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol 6, 17–20. [DOI] [PubMed] [Google Scholar]
- 22. Grallert B, Kearsey SE, Lenhard M, Carlson CR, Nurse P, Boye E & Labib K (2000) A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J Cell Sci 113 (Pt 8), 1447–1458. [DOI] [PubMed] [Google Scholar]
- 23. Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC & Wu Lee YH (2006) DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus‐associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 25, 1991–2003. [DOI] [PubMed] [Google Scholar]
- 24. Li Q, Zhang P, Zhang C, Wang Y, Wan R, Yang Y, Guo X, Huo R, Lin M, Zhou Z et al (2014) DDX3X regulates cell survival and cell cycle during mouse early embryonic development. J Biomed Res 28, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnstone O, Deuring R, Bock R, Linder P, Fuller MT & Lasko P (2005) Belle is a Drosophila DEAD‐box protein required for viability and in the germ line. Dev Biol 277, 92–101. [DOI] [PubMed] [Google Scholar]
- 26. Ishaq M, Hu J, Wu X, Fu Q, Yang Y, Liu Q & Guo D (2008) Knockdown of cellular RNA helicase DDX3 by short hairpin RNAs suppresses HIV‐1 viral replication without inducing apoptosis. Mol Biotechnol 39, 231–238. [DOI] [PubMed] [Google Scholar]
- 27. Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T & Kato N (2007) DDX3 DEAD‐box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81, 13922–13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao X, Yang ZL, Xiong L, Zou Q, Yuan Y, Li J, Liang L, Chen M & Chen S (2013) Nectin‐2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol 6, 179–190. [PMC free article] [PubMed] [Google Scholar]
- 29. Gu L, Fullam A, Brennan R & Schroder M (2013) Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol Cell Biol 33, 2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oshiumi H, Ikeda M, Matsumoto M, Watanabe A, Takeuchi O, Akira S, Kato N, Shimotohno K & Seya T (2010) Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS‐1‐mediated IFN‐beta induction. PLoS One 5, e14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schroder M, Baran M & Bowie AG (2008) Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon‐mediated IRF activation. EMBO J 27, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soulat D, Burckstummer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T & Superti‐Furga G (2008) The DEAD‐box helicase DDX3X is a critical component of the TANK‐binding kinase 1‐dependent innate immune response. EMBO J 27, 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fullam A & Schroder M (2013) DExD/H‐box RNA helicases as mediators of anti‐viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta 1829, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawai T & Akira S (2008) Toll‐like receptor and RIG‐I‐like receptor signaling. Ann N Y Acad Sci 1143, 1–20. [DOI] [PubMed] [Google Scholar]
- 35. Robbins MA & Rossi JJ (2005) Sensing the danger in RNA. Nat Med 11, 250–251. [DOI] [PubMed] [Google Scholar]
- 36. Saito T, Owen DM, Jiang F, Marcotrigiano J & Gale M Jr (2008) Innate immunity induced by composition‐dependent RIG‐I recognition of hepatitis C virus RNA. Nature 454, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horner SM & Gale M Jr (2013) Regulation of hepatic innate immunity by hepatitis C virus. Nat Med 19, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li XD, Sun L, Seth RB, Pineda G & Chen ZJ (2005) Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 102, 17717–17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ & Chung RT (2006) Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol 80, 9226–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H & Ryu WS (2010) Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PLoS Pathog 6, e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oshiumi H, Sakai K, Matsumoto M & Seya T (2010) DEAD/H BOX 3 (DDX3) helicase binds the RIG‐I adaptor IPS‐1 to up‐regulate IFN‐beta‐inducing potential. Eur J Immunol 40, 940–948. [DOI] [PubMed] [Google Scholar]
- 42. Kok KH, Lui PY, Ng MH, Siu KL, Au SW & Jin DY (2011) The double‐stranded RNA‐binding protein PACT functions as a cellular activator of RIG‐I to facilitate innate antiviral response. Cell Host Microbe 9, 299–309. [DOI] [PubMed] [Google Scholar]
- 43. Cardenas WB, Loo YM, Gale M Jr, Hartman AL, Kimberlin CR, Martinez‐Sobrido L, Saphire EO & Basler CF (2006) Ebola virus VP35 protein binds double‐stranded RNA and inhibits alpha/beta interferon production induced by RIG‐I signaling. J Virol 80, 5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li S, Min JY, Krug RM & Sen GC (2006) Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double‐stranded RNA. Virology 349, 13–21. [DOI] [PubMed] [Google Scholar]
- 45. Kew C, Lui PY, Chan CP, Liu X, Au SW, Mohr I, Jin DY & Kok KH (2013) Suppression of PACT‐induced type I interferon production by herpes simplex virus 1 Us11 protein. J Virol 87, 13141–13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H et al (2013) Mutual antagonism between the Ebola virus VP35 protein and the RIG‐I activator PACT determines infection outcome. Cell Host Microbe 14, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PC, Yuen KY et al (2014) Middle east respiratory syndrome coronavirus 4a protein is a double‐stranded RNA‐binding protein that suppresses PACT‐induced activation of RIG‐I and MDA5 in the innate antiviral response. J Virol 88, 4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Owsianka AM & Patel AH (1999) Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257, 330–340. [DOI] [PubMed] [Google Scholar]
- 49. You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH & Lee YH (1999) Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol 73, 2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mamiya N & Worman HJ (1999) Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274, 15751–15756. [DOI] [PubMed] [Google Scholar]
- 51. Lee JW, Liao PC, Young KC, Chang CL, Chen SS, Chang TT, Lai MD & Wang SW (2011) Identification of hnRNPH1, NF45, and C14orf166 as novel host interacting partners of the mature hepatitis C virus core protein. J Proteome Res 10, 4522–4534. [DOI] [PubMed] [Google Scholar]
- 52. Peters GA, Hartmann R, Qin J & Sen GC (2001) Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol 21, 1908–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patel CV, Handy I, Goldsmith T & Patel RC (2000) PACT, a stress‐modulated cellular activator of interferon‐induced double‐stranded RNA‐activated protein kinase, PKR. J Biol Chem 275, 37993–37998. [DOI] [PubMed] [Google Scholar]
- 54. Koscianska E, Starega‐Roslan J & Krzyzosiak WJ (2011) The role of Dicer protein partners in the processing of microRNA precursors. PLoS One 6, e28548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bass BL (2000) Double‐stranded RNA as a template for gene silencing. Cell 101, 235–238. [DOI] [PubMed] [Google Scholar]
- 56. Mello CC & Conte D Jr (2004) Revealing the world of RNA interference. Nature 431, 338–342. [DOI] [PubMed] [Google Scholar]
- 57. Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17, 449–459. [DOI] [PubMed] [Google Scholar]
- 58. Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P & Ding SW (2006) RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW & Voinnet O (2013) Antiviral RNA interference in mammalian cells. Science 342, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee Y, Hur I, Park SY, Kim YK, Suh MR & Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO J 25, 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen YC, Su WC, Huang JY, Chao TC, Jeng KS, Machida K & Lai MM (2010) Polo‐like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J Virol 84, 7983–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]


