Abstract
After fusion with the cellular plasma membrane or endosomal membranes, viral particles are generally too large to diffuse freely within the crowded cytoplasm environment. Thus, they will never reach the cell nucleus or the perinuclear areas where replication or reverse transcription usually takes place. It has been proposed that many unrelated viruses are transported along microtubules in a retrograde manner using the cellular dynein machinery or, at least, some dynein components. A putative employment of the dynein motor in a dynein‐mediated transport has been suggested from experiments in which viral capsid proteins were used as bait in yeast two‐hybrid screens using libraries composed of cellular proteins and dynein‐associated chains were retrieved as virus‐interacting proteins. In most cases DYNLL1, DYNLT1 or DYNLRB1 were identified as the dynein chains that interact with viral proteins. The importance of these dynein–virus interactions has been supported, in principle, by the observation that in some cases the dynein‐interacting motifs of viral proteins altered by site‐directed mutagenesis result in non‐infective virions. Furthermore, overexpression of p50 dynamitin, which blocks the dynein–dynactin interaction, or incubation of infected cells with peptides that compete with viral polypeptides for dynein binding have been shown to alter the viral retrograde transport. Still, it remains to be proved that dynein light chains can bind simultaneously to incoming virions and to the dynein motor for retrograde transport to take place. In this review, we will analyse the association of viral proteins with dynein polypeptides and its implications for viral infection.
Keywords: dynein, DYNLL1, DYNLT1, infection, retrograde transport, virus
After fusion with the cellular plasma membrane or endosomal membranes, viral particles are generally too large to diffuse freely within the crowded cytoplasm environment. It has been proposed that many unrelated viruses are transported along microtubules in a retrograde manner using the cellular dynein machinery or, at least, some dynein components, in most cases the light chains DYNLL1, DYNLT1 or DYNLRB1.
Abbreviations
- ASFV
African swine fever virus
- DYNC1H
dynein heavy chain
- DYNC1I
intermediate chain
- DYNC1LI
light intermediate chain
- DYNLL1
dynein light chain LC8
- DYNLRB1
dynein light chain roadblock
- DYNLT1
dynein light chain Tctex
- GFP
green fluorescent protein
- HHV
human herpes virus
- HIV
human immunodeficiency virus
- HSV
herpes simplex virus
- MTOC
microtubule organizing centre
- PV
papillomavirus
- RV
rabies virus
- siRNA
small interfering RNA
Introduction
The function of eukaryotic cells relies on the transport of macromolecules and small organelles throughout the cytoplasm. Microtubules are polar cytoskeletal filaments assembled from thousands of α/β tubulin heterodimers which are nucleated and organized by the perinuclear microtubule organizing centre (MTOC). Whereas kinesin and dynein motors use microtubules to move cargo throughout the cytoplasm, myosin motors interact with actin filaments [1, 2, 3]. Cytoplasmic dynein, frequently in cooperation with its cofactor dynactin, is a minus‐end‐directed microtubule associated motor responsible for retrograde transport (towards the nucleus) in eukaryotic cells [4, 5, 6]. This molecular motor is a large multiprotein complex of approximately 1.2 MDa that contains heavy chains, intermediate chains, light intermediate chains and light chains. Cytoplasmic dynein plays critical roles in a variety of eukaryotic cellular functions, including Golgi maintenance, nuclear migration, retrograde axonal transport and organelle positioning. In addition, cytoplasmic dynein is involved in numerous aspects of mitosis, such as spindle formation and organization, spindle orientation and mitotic checkpoint regulation [7, 8].
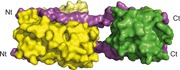
Cargoes transported by dynein are linked to the motor via the tail, which consists of an N‐terminal section of one or more heavy chains and a number of associated polypeptides [4, 5, 9, 10]. In general, the cytoplasmic dynein complex is resolved on SDS/polyacrylamide gels into subunit polypeptides of ∼ 530 (dynein heavy chains, DYNC1H), ∼ 74 (intermediate chains, DYNC1I), ∼ 53–59 (light intermediate chains, DYNC1LI) and ∼ 10–14 kD (light chains). Cytoplasmic dynein heavy chain, with ∼ 4650 amino acids in humans, is among the largest polypeptides found in mammalian cells. Dimeric dyneins have a conserved tail structure in which the heavy chains dimerize through protein–protein interactions mediated by amino acids 300–1140 in the tail region (Fig. 1) [11]. In addition, dynein heavy chain residues 446–701 and 649–800 are involved in intermediate chain and light intermediate chain binding respectively [11]. In the case of cytoplasmic dynein, intermediate chain is also able to form homodimers through interactions mediated by residues 151–250 [12]. Furthermore, in mammals, the 74 kDa cytoplasmic intermediate chain is homologous to the 67 kDa axonemal intermediate chain [13]. Three highly conserved light chains, also shared by axonemal dyneins, Tctex1 (DYNLT1), LC8 (DYNLL1) and roadblock (LC7 or DYNLRB1), bind to distinct regions of the intermediate chains, always as homodimers [14, 15]. DYNLL, a ubiquitous molecule, is the most highly conserved among light chains and, interestingly, in spite of lacking sequence homology displays a three‐dimensional fold almost identical to that of DYNLT [16, 17, 18]. Both DYNLT and DYNLL form homodimers with α‐helices flanking a shared central β‐sheet, with the peptides from interacting partners lengthening the preformed β‐strand. Indeed, according to the recently published crystal structures [19, 20] the two short, consecutive protein stretches of dynein intermediate chain that bind to DYNLT and DYNLL adopt an extended conformation (Fig. 1). However, DYNLRB, also a ubiquitous component of cytoplasmic dyneins, is structurally different from Tctex1 and LC8, with a dimer interface that includes the coiled‐coiled pairing of two α‐helices [21]. Unlike when binding to DYNLT or DYNLL, two sequential helical segments of dynein intermediate chain associate to DYNLRB (Fig. 1). In addition to these three light chain homodimers, there is also a dimer of DYNC1LI bound to the tail of cytoplasmic dyneins. Consistent with a role in cargo binding, DYNC1LI has been shown to bind to the core centrosomal protein pericentrin [22]. Thus, since dynein light and light intermediate chains appear to associate with cargo, it has been suggested that viruses might target these cellular components during infection processes.
Figure 1.
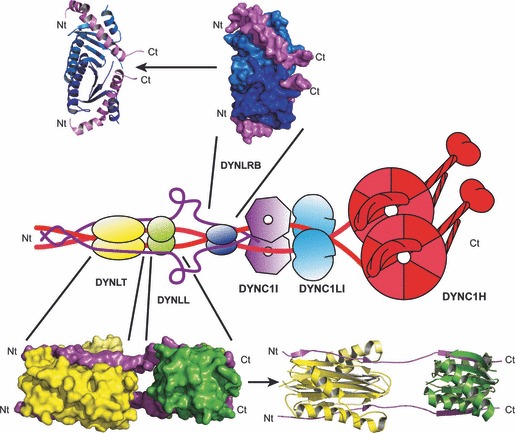
General architecture of the cytoplasmic dynein motor complex. The positions of DYNLL (dynein light chain LC8, green), DYNLT (dynein light chain Tctex, yellow), DYNLRB (dynein light chain roadblock, dark blue), DYNC1I (dynein intermediate chain, violet; the carboxy‐terminal WD40 repeat (β‐propeller) is depicted as a heptagon), DYNC1L (dynein light intermediate chain, light blue) and DYNC1H (dynein heavy chain, red) are shown. The crystallographic structures of the DYNLT1 homodimer (yellow) and DYNLL1 homodimer (green) binding to adjacent sequences from the dynein intermediate chain (violet) are shown at the bottom (PDB accession number http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3FM7). In addition, the crystallographic structure of the DYNLRB homodimer (PDB accession number http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3L9K) depicted in blue is shown on top. Note that the crystal structure of the motor domain of the dynein heavy chain has also been recently reported [119].
In human cells, the DYNLT1 and DYNLL1 homodimers bind to LGMAKITQVDF and KETQTP motifs respectively, both contiguously positioned in the intermediate chain sequence in which they adopt an extended conformation (Fig. 1) [19, 20, 23, 24]. It is generally assumed that most of the cellular and viral polypeptides that also bind to DYNLT1 or DYNLL1 have protein stretches with similarities to these intermediate chain sequences and bind in a similar fashion [25, 26, 27, 28].
Following cell entry, several viruses exploit the cellular cytoplasmic transport mechanisms to allow them to travel long distances through the cytoplasm and reach their site of replication [29, 30, 31, 32] (Fig. 2). Viruses require active transport along microtubules since diffusion of particles larger than 50 nm in diameter is restricted by the structural organization of the cytoplasm [33]. Experiments in which microtubule‐depolymerizing agents such as colchicine, nocodazole or vinblastine were used have shown that the integrity of the microtubules is essential for virus infection. Many pathogens causing widespread illness including herpes simplex virus (HSV) [34, 35], adenovirus [35, 36], hepatitis B virus [37], human cytomegalovirus [38], human immunodeficiency virus (HIV) [39], African swine fever virus (ASFV) [40], parvovirus [41], influenza virus [42], papillomavirus (PV) [43] and rabies virus (RV) [44] rely on microtubules for efficient nuclear targeting and successful infection.
Figure 2.
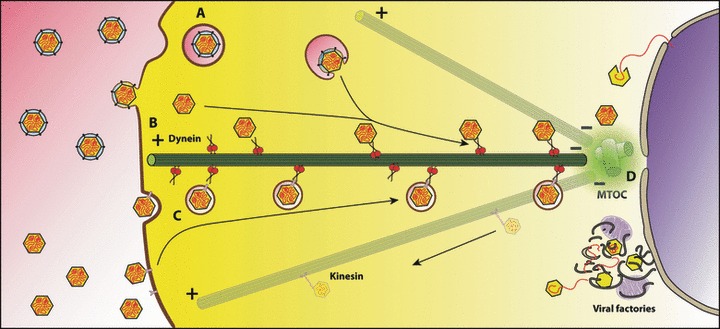
Viral retrograde transport model. Both the entry of the viral particle through the endosome pathway (A, C) and the direct fusion of the viral envelope to the plasma membrane (B) lead to the retrograde transport along microtubules using the cytoplasmic the dynein motor. Viral capsid proteins might associate to dynein directly (A) or a cellular receptor might bind simultaneously to a viral protein and the dynein motor (C). After reaching the MTOC at the microtubular minus end viruses are uncoated and directed to the sites of replication, production and assembly of the new viral proteins (D) such as the nucleus or the viral factories. The newly assembled particles might become transported to the cell periphery by the anterograde transport machinery.
Labelling of unenveloped viruses, such as SV40 [45] or adenoviruses [46], with fluorescent dyes has proved extremely useful to show their retrograde transport. Likewise, to initiate infection, herpes virus must attach to cell surface receptors, fuse its envelope to the plasma membrane and allow the de‐enveloped capsid to be transported to the nuclear pores [47, 48, 49]. More recently, video microscopy using green fluorescent protein (GFP) tagged viral proteins has also demonstrated the retrograde advance of viruses towards the cell nucleus. In particular, retrograde transport of HSV in axons has been visualized using time‐lapse fluorescent microscopy [50]. Similarly, GFP‐tagged poliovirus receptor, which associates to dynein light chain DYNLT1, colocalizes with Alexa fluor 555‐labelled poliovirus and both undergo retrograde transport along microtubules of cultured motor neurons [51]. Likewise, GFP‐tagged ASFV protein p54 was found associated to microtubules during infection and nocodazole treatment abrogated this association [52].
Remarkably, when virus proteins were used as baits and screened against a library of cellular proteins, several dynein polypeptides, mostly dynein light chains DYNLL1 (LC8), DYNLT1 (Tctex) and DYNLRB1 (roadblock), were retrieved as interacting partners.
On the basis of these results, the hijack of the dynein motor by numerous viruses has been proposed as a common mechanism for virus delivery near the cell nucleus replication site [29, 30, 31, 32]. In this review we will analyse in detail this association and its biological significance.
Herpesviruses
Alphaherpesviruses such as HSV‐1 are unique parasites of the vertebrate peripheral nervous system. Primary infection usually occurs at an epithelial surface, after which the virus invades the termini of sensory and autonomic neurons that innerve the infected tissue. HSV‐1 binds to cell surface receptors, then loses its envelope after cell fusion and subsequently virion components, including the tegument and capsid layers together with the double‐stranded DNA, are transported in a retrograde manner along axons towards the cell bodies of these neurons [50, 53, 54]. Since simple diffusion does not allow the viral components to travel long distances, cellular microtubule‐based motors must be involved in alphaherpesvirus transport [31, 55]. In fact, the dynein motor is known to co‐localize with inbound cytosolic capsids of HSV‐1 [34, 56], virus infection can be blocked by over‐expression of the dynactin subunit p50 (dynamitin) [56] and proteins from both the capsid and the tegument of HSV‐1 are known to associate with dynein protein members. Likewise, immunofluorescence studies of pig monocytes infected with pseudorabies virus, also a member of the Alphaherpesvirinae family, showed a clear co‐localization with dynein [57].
Protein UL34 of HSV‐1 is an integral membrane protein that is targeted to the inner nuclear membrane and only transiently associated with viral particles during the passage through the nuclear envelope during egress of newly assembled capsids from the nucleus into the cytosol [58]. Surprisingly, it has been reported that UL34 is associated with the microtubular network and binds to the dynein intermediate chain (DYNC1I1) [59]. The importance of this interaction is hard to evaluate in vivo, since UL34 from HSV‐1 and the homologous genes in all other herpesviruses encode for an essential protein [60].
Using a yeast two‐hybrid screen, fifteen HSV‐1 proteins were confronted with dynein light chains DYNLL1, DYNLT1 and DYNLT3, and a strong interaction could be detected between both UL35(VP26) and UL46(VP11/12) and the dynein light chain DYNLT1 and its homolog DYNLT3 [61]. In fact recombinant capsids of HSV‐1 were microinjected into the cytoplasm, and those decorated with VP26 showed a stronger tendency to accumulate at the nuclear envelope [61]. Besides a role of VP26 in recruiting the dynein motor, these data may also suggest that VP26 is somehow involved in binding to the nuclear pores. Nevertheless, additional HSV‐1 proteins must associate with the dynein motor, since capsids from HSV‐1 mutants that lack VP26 can still bind to purified dynein [62, 63]. In addition, studies with HSV‐1 lacking VP26 have shown no significant effect on dynein‐dependent retrograde viral transport in cell cultures [64]. Recent results seem to indicate that both dynein (retrograde movement motor) and kinesin (anterograde movement motor) can bind isolated capsids of HSV‐1 in vitro [62], hence raising the possibility that this association might be important for certain steps of the viral life cycle or that these two cellular motors must somehow be coordinated for viral infection to succeed [62]. Likewise, it has been reported that VP26 of pseudorabies virus is also not required for intracellular transport [65]. Finally, HSV‐1 mutants lacking VP26 are not impaired in animal experiments that rely on axonal transport [66].
Moreover, HSV‐1 UL9, a protein that binds to the viral origin of replication, displays a consensus DYNLL1 binding motif (746‐KSTQT‐750) that is functional when tested as an isolated dodecapeptide [28].
The Betaherpesvirus human herpes virus 7 (HHV‐7) is known to infect CD4+ T lymphocytes and epithelial cells of salivary glands. Its protein U19, probably a transactivator according to its similarity to human cytomegalovirus UL38, displays a consensus sequence for DYNLL1 binding (RSTQT) repeated in tandem at its carboxy terminus. Using a pepscan technique, these sequences were found to efficiently associate with DYNLL1 [28]. Interestingly, no similarity exists between HHV‐7 and HHV‐6 at this region, thus indicating that the association to this dynein light chain might be restricted to the HHV‐7 isolate.
Rabies virus
RV belongs to the lyssavirus genus of the Rhabdoviridae family, with rabies and Mokola virus as reference strains. They are small, enveloped, single‐stranded negative‐sense RNA viruses, whose genome is tightly encapsidated into a ribonucleoprotein complex with the viral proteins of the nucleocapsid, the RNA polymerase and the phosphoprotein P as non‐catalytic cofactor. RV is a highly neurotrophic virus that enters the organism by bites or injuries in the skin and muscle, where it replicates. It then enters the neuronal endings of peripheral nerves, such as neuromuscular junctions, to reach the central nervous system and causes lethal encephalitis in animals and humans. After receptor binding, RV enters its host cells through the endosomal pathway via a low‐pH‐induced membrane fusion process catalysed by the glycoprotein G, a major determinant for RV neuropathogenicity [67]. The retrograde transport along microtubules has been shown recently by using fluorescently labelled RV glycoprotein. Incubation of in vitro differentiated NS20Y neuroblastoma cells with fluorescently labelled virus clearly showed the transport in the retrograde direction over long distances in neurites [68]. Subsequently, all transcription and replication events take place in the cytoplasm, inside a specialized virus factory referred to as the Negri body [69].
Experiments performed simultaneously by two independent groups used the rabies and Mokola virus phosphoproteins as baits in yeast two‐hybrid screens and retrieved DYNLL1 as an interacting partner using PC12 cells and human brain libraries [70, 71]. Fine mapping of the DYNLL1 binding site within the P phosphoprotein of these two lyssaviruses revealed the presence of a KSTQT motif in rabies and KSIQI motif in Mokola that constituted the interacting polypeptide stretch. The P phosphoprotein of rhabdovirus is a cofactor of the RNA polymerase complex and, in fact, facilitates the binding of the polymerase to the N‐RNA complex [67]. Using the SAD‐D29 low‐virulent strain of RV, deletion of the DYNLL1 binding motif in the P phosphoprotein resulted in a remarkable viral attenuation after intramuscular but not after intracranial inoculation [72]. Unfortunately, no difference could be observed between wild‐type and recombinant viruses in which the DYNLL1 binding sequence had been deleted in the P phosphoprotein when non‐attenuated viral strains were used. Other authors have concluded that the deletion of the DYNLL1 binding site in phosphoprotein P did not produce a biologically important impairment of viral transport in the nervous system [73].
Some recent data seem to indicate that mutations in the DYNLL1 binding site within the P phosphoprotein of RV significantly attenuated viral transcription and replication in the central nervous system, hence showing that DYNLL1 binding to the viral protein has a more crucial role in viral polymerase activity than in the intracellular transport of the virus [74]. This is in agreement with the nuclear staining of the P phosphoprotein where it co‐localizes with promyelocytic leukaemia protein [75]. In this context, it is interesting to note that the DYNLL1 binding motif of the phosphoprotein of RV when fused to a reporter protein is not able, by itself, to promote active import into the cell nucleus although it can facilitate nuclear protein import when appended to proteins with nuclear localization sequences [76].
African swine fever virus
ASFV, the only member of the family Asfarviridae, is a large double‐stranded DNA virus that codes for approximately 150 proteins. ASFV enters the cell by dynamin‐ and clathrin‐dependent endocytosis, and its infectivity depends on the acidification of the endosome [77]. Elegant studies by Alonso and co‐workers have shown that ASFV p54, a major protein of virion membranes, associates with DYNLL1, which allows the transport of the virus to the MTOC in the cell perinuclear area [78]. Fine mapping in yeast two‐hybrid assays, site‐directed mutagenesis and the presence of a polypeptide stretch in p54 with the sequence TASQT that closely resembles the DYNLL1 consensus binding motifs [25, 27] led to the identification of the DYNLL1 binding region in p54 [78, 79]. In fact, small peptide inhibitors that display this binding sequence together with an internalization sequence can disrupt the interaction between p54 and DYNLL1 altering both infectivity and the viral egress [79]. Likewise, the inhibition of the dynein–dynactin complex formation by the overexpression of p50 dynamitin blocks ASFV transport in infected cells [78]. Thus, the interaction of ASFV p54 with DYNLL1 is required for efficient infectivity, virus replication and viral production yields.
Papillomavirus
PVs are small non‐enveloped double‐stranded DNA viruses that infect the stratified epithelia of skin and mucous membranes. The icosaedric capsid contains 360 copies of the major capsid protein L1 and up to 72 molecules of the minor capsid protein L2 [80]. Whereas the L2 protein is required for egress of the viral genome from endosomes, L1 does not appear to exit the endosomal compartment [81]. Based on immunofluorescence and co‐immunoprecipitation experiments, the L2 protein was found attached to microtubules after uncoating of incoming human PV pseudovirions [82]. Then, the minor capsid protein L2 accompanies the viral DNA to the nucleus and subsequently to the subnuclear promyelocytic leukemia protein bodies [83]. Since L2 and the viral genome co‐localize in the nucleus at promyelocytic leukemia protein bodies, it has been suggested that they are associated in the nucleus forming a complex [83]. Site‐directed mutagenesis and deletion studies showed that the carboxy‐terminal region of L2 somehow associates with the dynein motor [82]. This observation is in agreement with previous experiments that had shown that bovine PV binds to microtubules and becomes transported along them, and suggested the possibility that dynein is involved in this process [84].
Recently, a yeast two‐hybrid screen using PV L2 as bait against a human cDNA library retrieved dynein light chain DYNLT1 as a tight binder. In addition, in vitro binding studies and cotransfection experiments in HeLa cells proved that L2 was also able to bind to its homologue DYNLT3 [43]. Subsequent studies have shown that depletion of DYNLT1 or DYNLT3 using small interfering RNA (siRNA) treatment inhibited human PV‐16 infection, whereas infection was increased after overexpression of these dynein light chains [43].
It must also be noted that human PV has another polypeptide, termed E4 (also known as E1^E4), that is expressed from an E1^E4 spliced mRNA prior to the assembly of infectious virions and accumulates to very high levels in cells supporting productive infection [85]. Several PV types, such as 08, 47 or 21, display a polypeptide stretch with the sequence KQTQT that conforms a consensus binding sequence for DYNLL1. In vitro binding assays have shown that this sequence does, indeed, bind to DYNLL1 tightly [28], although no studies have been performed yet to demonstrate that this interaction occurs during viral infection.
Poliovirus
Poliovirus is an enteric virus that rarely causes disease in humans. Nevertheless, in the pre‐vaccine era ∼ 1% of infected individuals developed paralytic poliomyelitis due to viral invasion of the central nervous system and destruction of motor neurons. To gain access and sustain infection in neurons, a neurotropic virus such as poliovirus must be able to efficiently traffic in axons, which can be up to 1 m long. CD155, the human poliovirus receptor, is a member of the immunoglobulin superfamily, with three linked extracellular Ig‐like domains followed by a membrane‐spanning domain and a short cytoplasmic domain. Intramuscularly inoculated poliovirus is known to become incorporated into neural cells after binding to the first Ig‐like domain of CD155 followed by endocytosis [86, 87]. Then, the cytoplasmic domain of CD155 is known to associate to the dynein light chain DYNLT1 [86, 88] and subsequently the endosomes, together with the CD155‐bound poliovirus, undergo retrograde transport along microtubules through the axon to the neural‐cell body, where the uncoating and replication of poliovirus occur [51, 87].
Alternative splicing generates two membrane‐bound CD155 isoforms: CD155α and CD155δ. Yeast two‐hybrid analyses have identified the 50‐residue cytoplasmic domain of CD155α and the 25‐residue cytoplasmic domain of CD155δ as DYNLT1 binding partners [87, 88]. Subsequent studies have revealed that a basic motif adjacent to the transmembrane domain is required for efficient binding. In addition, purified recombinant DYNLT1 binds to the cytoplasmic domain when fused to glutathione S‐transferase in vitro [87].
Retrovirus
Spumaviruses, also known as foamy viruses, target the microtubule organizing centre prior to nuclear translocation. Hence, centrosomal targeting of incoming viral proteins and subsequent viral replication can be inhibited by nocodazole treatment [89]. The efficiency of MTOC targeting was analysed by using various GFP‐tagged Gag mutant constructs of human foamy virus transfected in cultured cells, and a region located around amino acids 150–180 was found necessary for this subcellular localization. In this regard, a Leu171Gly Gag mutant displayed drastically reduced infectivity of the proviral clone [90]. Interestingly, when COS6 cells were transfected with wild‐type Gag, but not with its Leu171Gly mutant, dynein light chain DYNLL1 could be co‐immunoprecipitated. However, the direct interaction between DYNLL1 and human foamy virus Gag protein has not been unequivocally proved [90].
HIV enters the cells following virus binding to CD4 and co‐receptors and the fusion of the viral membrane with the plasma membrane of the cell. During passage through the cytosol, the viral RNA genome is reverse transcribed into DNA within a structure named the reverse transcription complex that, eventually, must be imported into the nucleus, where the HIV genome is integrated into a chromosome. Initial reports concluded that depolymerization of cell microtubules with nocodazole had little effect on virus infection whereas actin depolymerization had a profound effect on infection [91]. Subsequent studies used a GFP‐tagged Vpr incorporated into virions and in vivo fluorescence to show a microtubule‐dependent transport towards the MTOC positioned in perinuclear areas [39]. Interestingly, the individual treatment of infected cells with either nocodazole or the F‐actin inhibitor latranculin B did not impede the movement of GFP‐labelled particles, whereas the simultaneous treatment with both compounds led to a cessation of movement. This suggests that HIV movement inside the cell depends on both actin and the microtubule network. Furthermore, the direct implication of dynein in HIV movement was further shown when infected cells were injected with anti‐dynein antibodies and viral migration along microtubule networks decreased significantly [39]. Nonetheless, in the case of HIV and its binding to the dynein motor the exact viral protein and its dynein partner remain to be identified.
By means of broad yeast two‐hybrid screens, HIV integrase was found to bind to the yeast dynein light chain Dyn2p, the orthologue of mammalian DYNLL1. When analysed inside yeast, HIV integrase associates to the microtubular network and accumulates at the spindle pole body, the yeast equivalent of mammalian perinuclear MTOC [92]. In fact, nocodazole treatment of transfected yeast or transfection of the GFP–integrin construct in a Δdyn2 mutant strain resulted in the aberrant localization of HIV integrase. Nevertheless, it is not known if HIV integrase binds to DYNLL1 in mammalian cells or if this association is required for efficient virus infection.
Interestingly, the matrix protein of Mason–Pfizer monkey virus, the archetypal D‐type retrovirus, binds directly to dynein light chain DYNLT1, according to yeast two‐hybrid assays, in vitro association of recombinant proteins and in cell immunoprecipitation assays [93]. Indeed, this association might be responsible for the retrograde transport of Gag‐synthesizing polysomes alongside microtubules or perhaps for other steps of the viral cycle. It is not known, nonetheless, if other retroviral matrix proteins associate to DYNLT1 as well.
Finally, the dynein motor has also been involved in the regulation of viral Gag and viral genomic RNA egress on endosomal membranes. In this regard, following transcription and nuclear export, the viral genomic RNA might transit towards the MTOC where it interacts with Gag proteins in a dynein‐mediated process [94].
Adenovirus
Adenoviruses are 90–100 nm diameter non‐enveloped dsDNA viruses that exit to the cytosol soon after receptor‐mediated endocytosis. Early studies revealed that adenoviruses associate to the dynein motor [36, 95, 96] and microinjection of function‐blocking anti‐dynein but not anti‐kinesin antibodies abolished the viral nuclear localization, consistent with a net minus‐end‐directed motility [97]. It was then suggested that the subcellular transport of adenoviruses is the result of the equilibrium between dynein (retrograde movement) and kinesin (anterograde movement) forces [36, 46, 98].
Type 2 adenovirus E3 protein, a polypeptide involved in the downregulation of the host’s immune response, binds to a small GTPase (RRAG) that, in turn, is associated with the dynein light chain DYNLT1 [99]. Since this viral polypeptide is not a structural component of the virion, the biological significance of the interaction remains unclear.
Dynein has been implicated in the transport of naked viral capsids from endosomes to the nuclear periphery after virus uncoating in the endosomes [97]. However, incubation of HeLa cells with recombinant adenovirus penton base protein clearly shows that a significant population of the viral protein traffics in a retrograde manner towards the cell nucleus. Cell treatment with nocodazole or transfection with p50/dynamitin abrogates retrograde transport by at least ∼ 50% [100].
Recent analysis has revealed that the viral capsid hexon subunit interacts directly with the dynein intermediate chain [101]. Using immunoprecipitation studies as well as antibody microinjection experiments the adenovirus hexon binding site was selectively localized to a single site within the intermediate chain and no significant interactions were observed with any of the three dynein light chains DYNLL1, DYNLT1 or DYNLRB1 [101].
Other viruses
Several other virus proteins have been reported to interact with the dynein motor. For instance, the E protein of severe acute respiratory syndrome coronavirus, a small integral membrane protein of 76 amino acids, associates, directly or indirectly, with dynein heavy chain when overexpressed in Vero cells [102]. In addition, its non‐structural protein 3 is also known to bind to multiple cellular proteins in infected Vero cells, including the dynein heavy chain, although it is not known if this binding is direct or mediated by other dynein light chains [102].
Yeast two‐hybrid screening has also revealed that virion protein 35 of ebolavirus binds to DYNLL1 through the consensus binding sequence SQTQT [103]. Interestingly, VP35 inhibits type I interferon production, thereby suppressing host innate immunity, an activity analogous to that of the P protein from RV, which also binds to DYNLL1.
In the case of canine parvovirus the role of dynein in viral infection has been inferred not only from the observation that intact microtubules are required for the traffic of viral particles towards the nucleus but also by the fact that microinjection of anti‐dynein antibodies reduced the nuclear accumulation of viral capsids and immunoprecipitation of dynein in infected cells co‐immunoprecipitated viral capsid proteins [41, 104].
A very recent paper described a silencing screen using siRNAs targeted against 5516 different cellular genes, with each gene being covered by three independent siRNAs. When Borna disease virus was used to infect an oligodendroglial cell line, silencing of dynein light chain DYNLRB1 significantly blocked virus infection. Although the exact viral protein that associated to DYNLRB1 has yet to be identified, this elegant approach has revealed the implication of this dynein light chain in viral infection [105].
The dynein motor is also involved in influenza virus infection. Surprisingly, endosomal acidification of this pathogen occurs in perinuclear areas after a dynein‐mediated retrograde transport has taken place, as demonstrated by experiments using anti‐dynein antibody injection [42].
Finally, using a pepscan technique and after screening multiple viral polypeptides with putative DYNLL1 binding sequences, polypeptides from Amsacta moorei entomopoxvirus, the polymerase from Vaccinia virus or Yam mosaic potyvirus polyprotein were shown to bind to this dynein light chain Nevertheless, additional studies are clearly needed in order to ascertain if these interactions do, indeed, take place during the virus infective cycle.
Selected interactions between viral polypeptides and dynein chain proteins are summarized in Table 1.
Table 1.
Selected viral proteins involved in a direct interaction with dynein polypeptides. The most recent dynein nomenclature [10] is used: DYNLL (dynein light chain LC8), DYNLT (dynein light chain Tctex), DYNLRB (dynein light chain roadblock), DYNC1LI (dynein light intermediate chain) and DYNC1I (dynein intermediate chain).
| Virus | Family | Protein that binds to a dynein polypeptide | Dynein protein | Reference |
|---|---|---|---|---|
| Herpes simplex | Alphaherpesvirinae | Viral UL34 | DYNC1I1a | [59] |
| Viral UL9 (helicase) | DYNLL1 | [28] | ||
| Viral UL35 (VP26) | DYNLT1 and DYNLT3 | [61] | ||
| Herpesvirus 7 | Betaherpesvirinae | Viral U19 | DYNLL1 | [28] |
| African swine fever | Asfarviridae | Viral p54 | DYNLL1 | [78] |
| Mokola | Rhabdoviridae | Viral phosphoprotein (P) | DYNLL1 | [71] |
| Rabies | Rhabdoviridae | Viral phosphoprotein (P) | DYNLL1 | [70, 71] |
| Papillomavirus | Papillomaviridae | Viral minor capsid protein L2 | DYNLT1 and DYNLT3 | [43, 82] |
| Viral probable protein E4 | DYNLL1 | [28] | ||
| Borna disease | Bornaviridae | Probably viral G surface glycoprotein | DYNLRB1 | [105] |
| Poliovirus | Picornaviridae | Cellular CD155 receptor | DYNLT1 | [86, 88] |
| Human immunodeficiency | Retroviridae | Viral integrase | Dyn2p (yeast orthologue of DYNLL1) | [92] |
| Mason–Pfizer monkey | Retroviridae | Viral matrix | DYNLT1 | [93] |
| Adenovirus | Adenoviridae | Viral capsid hexon | DYNC1LI2 DYNC1LI1 | [101] |
| Ebolavirus | Filoviridae | Viral phosphoprotein (VP35) | DYNLL1 | [103] |
The dimer–dimer hypothesis
Both DYNLL1 and DYNLT1 are protein members of the dynein motor in which they bind contiguously to the dynein intermediate chain [19, 20, 24]. However, only about 40% of total DYNLL1 associates to the dynein intermediate chain in a microtubule pellet of rat brain [106]. Likewise, a significant fraction of DYNLT1 is not associated to microtubules in fibroblasts, as shown by sequential immunoprecipitation [107].
Although they share no significant sequence similarity, dynein light chains DYNLL1 and DYNLT1 display a very similar three‐dimensional structure and adopt identical ‘geometric specificity’ upon binding to protein ligands [19, 20]. Both of these small proteins are homodimers and structurally consist of two α‐helices followed by five β‐strands, with the second β‐strand being swapped between protomers. The proteins that bind to either DYNLL1 or DYNLT1 do so through polypeptide stretches that adopt an extended β‐strand conformation that inserts into the ligand binding grooves. The consensus protein sequence necessary for binding to dynein light chain DYNLT1 is not well known. However, numerous atomic coordinates are available for dynein light chain DYNLL1 in association with protein partners, and in all cases GIQVD or KXTQT motifs, or variations of them, are inserted into the DYNLL1 binding grove [24, 25, 27, 108]. Using yeast two‐hybrid and mutagenesis experiments, the binding region to DYNLL1 has been narrowed to TASQT for ASFV p54 [78] and KSTQT and KSIQI in the case of the P protein of rabies and Mokola viruses respectively [70, 71] and SQTQT in the case of protein VP35 of ebolavirus [103]. The presence of these sequences that closely resemble the KXTQT motif in various viral proteins [28] suggests that virus association also occurs with the viral protein adopting an extended antiparallel β strand that fits into the DYNLL1 groove and extends the pre‐existing β‐sheet. The atomic coordinates of the modeled ASFV p54–DYNLL1 complex clearly indicate that this is indeed the case [26]. In fact, RV P protein and the pro‐apoptotic Bcl‐2 family member Bim display an identical DYNLL1 binding sequence (DKSTQT) and the published NMR structure of the DYNLL1–Bim complex also shows the β‐sheet augmentation mode of binding [18].
Dynein light chains have been proposed to mediate cargo binding for their cellular transport. DYNLL1 is a bivalent molecule and many of its interacting partners are dimeric (or oligomeric) proteins [20]. However, linking cargo molecules to dynein is not easily reconciled with binding data [109]. In fact, DYNLL1 binding affinity for a dimeric partner is significantly higher compared with the same partner as a monomer [19, 20]. This observation has led to the hypothesis that two identical polypeptide segments from a dimeric partner occupy both of the binding grooves of DYNLL1 and DYNLT1. If this is the case, it is hard to accept that one DYNLL1 binding site is occupied by a viral protein whereas the other is occupied by the dynein intermediate chain as would be required for retrograde transport. Although this is theoretically possible, binding of a viral polypeptide to either DYNLL1 or DYNLT1 would require the displacement of the dynein intermediate chain from one of the binding sites, which is a thermodynamically unfavourable process. In this regard, it must be mentioned that, for instance, in the case of DYNLT1, peptides from the intermediate chain compete with the G protein β subunit [110], an indication that both peptides bind at the same location and therefore DYNLT1 cannot be simultaneously binding to both proteins. Therefore, if the DYNLL1 dimer (and by analogy the DYNLT1 dimer) binds to either two chains of the dynein intermediate chain or two chains of putative cargo proteins at the same location, how can viruses be transported in a retrograde manner towards the minus end of microtubules associated to the dynein motor?
It is conceivable that viral polypeptides, either as part of the virion or detached after uncoating, associate to dynein light chains using both binding sites simultaneously when these light chains are not part of the dynein motor [Fig. 3A, binding modes (a) and (b)]. Hence, binding to dynein light chains might promote dimerization of viral polypeptides through the binding to intrinsically disordered regions, a function that has recently been assigned to DYNLL1 [111, 112]. This would mean that the interaction of viral polypeptides with dynein light chains is not responsible for the association with microtubules and might be responsible for other processes during the infective cycle. Moreover, this would rationalize the fact of why several viral proteins that are neither envelope glycoproteins nor belong to the virion capsid associate to dynein light chains.
Figure 3.
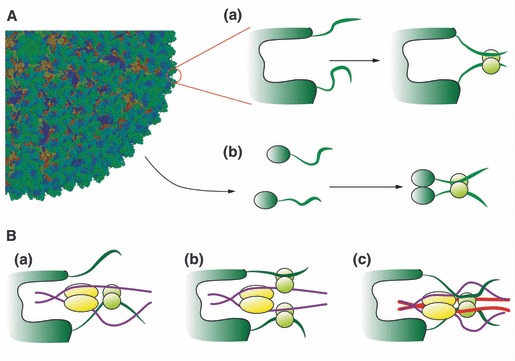
(A) Model for the interaction of a generic viral capsid with cytoplasmic, non‐microtubule‐associated DYNLL. It is then conceivable that the DYNLL homodimer might bind simultaneously to two viral polypeptides when part of the viral capsid (a) or when soluble after viral disassembly (b). This is in agreement with the modeled solution structure of the complex of DYNLL1 with p54 of ASFV [26]. (B) Three hypotheses for the association of viral proteins to the dynein molecular motor. One viral polypeptide displaces a dynein intermediate chain from one binding side of the DYNLL homodimer (a). Two DYNLL homodimers associate to one dynein intermediate chain and to one viral polypeptide simultaneously (b). Binding of the viral polypeptides displaces the dynein intermediate chains from the DYNLL binding grooves but DYNLL remains part of the dynein motor through the binding to the dynein heavy chain (red) (c).
If we focus on DYNLL1 only (a similar case could be put forward for viral polypeptides that associate to DYNLT1) three different situations might explain the proposed dynein motor–virus association responsible for the aforementioned retrograde transport.
-
1
It is conceivable that viral proteins could interact with DYNLL1 with one binding site occupied by the dynein intermediate chain and the opposite site of the same homodimer occupied by the viral protein (Fig. 3B‐a).
-
2
However, since binding of DYNLL1 to dimeric partners is energetically favourable over monomeric partners, it is then conceivable that viral proteins might adopt a conformation that facilitates the binding of two dynein light chain dimers with viral polypeptides alternating with dynein intermediate chains in each groove of the homodimer (Fig. 3B‐b).
-
3
Alternatively, virus polypeptides might bind simultaneously to the two equivalent sites within DYNLL1, hence displacing the dynein intermediate chain, but with the light chains still being part of the dynein motor through interactions with other dynein proteins (Fig. 3B‐c). This might occur if the dynein heavy chain (coloured in red) could associate to DYNLL1 through a different surface, such as its α‐helices.
Virus, dynein light chains and apoptosis
Bim and Bmf are two pro‐apoptotic BH3‐only proteins that signal to the cell death machinery by sensing cellular damage. In healthy cells both Bim and Bmf are sequestered away from the sites where pro‐survival Bcl‐2 family members reside (fundamentally the endoplasmic reticulum and mitochondria membranes) through interaction with dynein light chain proteins. In these cells the light chain component of the myosin V motor complex, DYNLL2, binds to the polypeptide stretch DKATQTL present in Bmf [113], whereas the equivalent component of cytoplasmic dynein, DYNLL1, binds to the polypeptide stretch DKSTQTP present in the Bim isoforms BimL and BimEL [114]. In response to apoptotic stimuli that impact upon the motor complexes, Bim or Bmf in complex with their respective light chains are released into the cytoplasm where they can interact with pro‐survival Bcl‐2 proteins via their BH3 domains.
Since virus infection is frequently associated with cell apoptosis [115, 116] it has been suggested that viral polypeptides and Bim or Bmf compete for the binding of dynein light chains DYNLL1 and DYNLL2. Thus, binding of a viral polypeptide to DYNLL1 or DYNLL2 might release Bim or Bmf which, in turn, would translocate to the mitochondria and initiate apoptosis. In this regard, transfection of Vero cells with ASFV p54 but not with a mutant protein that cannot bind to DYNLL1 triggers the release of microtubule‐associated Bim and the concomitant caspase‐9 and caspase‐3 activation [117]. Hence, apoptosis induced by p54 results from the direct competition between Bim and p54 for their binding to DYNLL1, which suggests that virus–dynein interactions might be important not only in retrograde transport. Likewise, HIV Tat, which binds to microtubules through residues 35–50, induces apoptosis in a Bim‐dependent manner [118].
Conclusions
In the past few years, numerous studies of virus–host interactions have revealed the role of the dynein motor and the integrity of microtubules in virus infection. The development of microscopy techniques has also enabled the retrograde movement of viruses in the cellular cytoplasm to be tracked. In addition, the in vitro binding and transport assays using complete viral capsids and intact microtubule motors may be instrumental in further characterizing potential functions of the interactions between dynein light chains and viral proteins. Moreover, several viral polypeptides are known to associate to dynein light chains, although many of them do not belong to capsid proteins. However, dynein light chains appear also as homodimers in the cellular cytoplasm, without being part of the dynein motor. Therefore, it remains to be unambiguously established if the interaction of viral polypeptides with dynein light chains links the virion to the dynein motor when bound to microtubules and if this interaction is responsible for the observed viral retrograde transport. Hence, co‐immunoprecipitation of dynein intermediate or heavy chains bound to incoming virions would be an adequate experiment to prove that the interaction of certain viral polypeptides with dynein light chains does indeed link virions to microtubules via the dynein motor. Since DYNLL1 (and probably DYNLT1) work as dimerization clamps that bind to intrinsically disordered regions of proteins it is also conceivable that the dimerization of viral polypeptides might be important for certain viral infective processes distinct from retrograde transport. This might be the case of the interaction of DYNLL1 with the phosphoproteins of RV, Mokola virus and ebolavirus. On the other hand, a conundrum therefore exists in that certain envelope or capsid viral proteins bind to dynein light chains at the same site used by dynein intermediate chain. Thus, whether the binding of viral proteins to dynein light chains displaces dynein intermediate chain and how it is performed remains to be established.
Acknowledgements
This work was supported by grants from the Ministerio de Ciencia e Innovación BFU2009‐10442 and BQU2008‐0080, and by the Consolider‐Ingenio Centrosome 3D CSD2006‐00023. We are especially grateful to Dr Douglas Laurents (Intituto de Química‐Física Rocasolano) for extensive English editing and helpful suggestions.
References
- 1. Schliwa M & Woehlke G (2003) Molecular motors. Nature 422, 759–765. [DOI] [PubMed] [Google Scholar]
- 2. Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- 3. Sakakibara H & Oiwa K (2011) Molecular organization and force‐generating mechanism of dynein. FEBS J 278, 2964–2979. [DOI] [PubMed] [Google Scholar]
- 4. Kardon JR & Vale RD (2009) Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oiwa K & Sakakibara H (2005) Recent progress in dynein structure and mechanism. Curr Opin Cell Biol 17, 98–103. [DOI] [PubMed] [Google Scholar]
- 6. King SM (2000) The dynein microtubule motor. Biochim Biophys Acta 1496, 60–75. [DOI] [PubMed] [Google Scholar]
- 7. Bader JR & Vaughan KT (2010) Dynein at the kinetochore: timing, interactions and functions. Semin Cell Dev Biol 21, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vallee RB, Varma D & Dujardin DL (2006) ZW10 function in mitotic checkpoint control, dynein targeting and membrane trafficking: is dynein the unifying theme? Cell Cycle 5, 2447–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hook P & Vallee RB (2006) The dynein family at a glance. J Cell Sci 119, 4369–4371. [DOI] [PubMed] [Google Scholar]
- 10. Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, McIntosh JR, Porter ME, Schroer TA, Vaughan KT, Witman GB et al. (2005) Cytoplasmic dynein nomenclature. J Cell Biol 171, 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tynan SH, Gee MA & Vallee RB (2000) Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem 275, 32769–32774. [DOI] [PubMed] [Google Scholar]
- 12. Lo KW, Kan HM & Pfister KK (2006) Identification of a novel region of the cytoplasmic dynein intermediate chain important for dimerization in the absence of the light chains. J Biol Chem 281, 9552–9559. [DOI] [PubMed] [Google Scholar]
- 13. Paschal BM, Mikami A, Pfister KK & Vallee RB (1992) Homology of the 74‐kD cytoplasmic dynein subunit with a flagellar dynein polypeptide suggests an intracellular targeting function. J Cell Biol 118, 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makokha M, Hare M, Li M, Hays T & Barbar E (2002) Interactions of cytoplasmic dynein light chains Tctex‐1 and LC8 with the intermediate chain IC74. Biochemistry 41, 4302–4311. [DOI] [PubMed] [Google Scholar]
- 15. Susalka SJ & Pfister KK (2000) Cytoplasmic dynein subunit heterogeneity: implications for axonal transport. J Neurocytol 29, 819–829. [DOI] [PubMed] [Google Scholar]
- 16. Wu H, Maciejewski MW, Marintchev A, Benashski SE, Mullen GP & King SM (2000) Solution structure of a dynein motor domain associated light chain. Nat Struct Biol 7, 575–579. [DOI] [PubMed] [Google Scholar]
- 17. Wu H, Maciejewski MW, Takebe S & King SM (2005) Solution structure of the Tctex1 dimer reveals a mechanism for dynein–cargo interactions. Structure 13, 213–223. [DOI] [PubMed] [Google Scholar]
- 18. Fan J, Zhang Q, Tochio H, Li M & Zhang M (2001) Structural basis of diverse sequence‐dependent target recognition by the 8 kDa dynein light chain. J Mol Biol 306, 97–108. [DOI] [PubMed] [Google Scholar]
- 19. Hall J, Karplus PA & Barbar E (2009) Multivalency in the assembly of intrinsically disordered dynein intermediate chain. J Biol Chem 284, 33115–33121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC & Hendrickson WA (2007) Structural and thermodynamic characterization of a cytoplasmic dynein light chain–intermediate chain complex. Proc Natl Acad Sci USA 104, 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hall J, Song Y, Karplus PA & Barbar E (2010) The crystal structure of dynein intermediate chain–light chain roadblock complex gives new insights into dynein assembly. J Biol Chem 285, 22566–22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purohit A, Tynan SH, Vallee R & Doxsey SJ (1999) Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol 147, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vaughan PS, Leszyk JD & Vaughan KT (2001) Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J Biol Chem 276, 26171–26179. [DOI] [PubMed] [Google Scholar]
- 24. Rapali P, Szenes A, Radnai L, Bakos A, Pál G & Nyitray L (2011) DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J 278, 2980–2996. [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez‐Crespo I, Yelamos B, Roncal F, Albar JP, Ortiz de Montellano PR & Gavilanes F (2001) Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett 503, 135–141. [DOI] [PubMed] [Google Scholar]
- 26. Garcia‐Mayoral MF, Rodriguez‐Crespo I & Bruix M (2011) Structural models of DYNLL1 with interacting partners: African swine fever virus protein p54 and postsynaptic scaffolding protein gephyrin. FEBS Lett 585, 53–57. [DOI] [PubMed] [Google Scholar]
- 27. Lo KW, Naisbitt S, Fan JS, Sheng M & Zhang M (2001) The 8‐kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem 276, 14059–14066. [DOI] [PubMed] [Google Scholar]
- 28. Martinez‐Moreno M, Navarro‐Lerida I, Roncal F, Albar JP, Alonso C, Gavilanes F & Rodriguez‐Crespo I (2003) Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett 544, 262–267. [DOI] [PubMed] [Google Scholar]
- 29. Sodeik B (2002) Unchain my heart, baby let me go – the entry and intracellular transport of HIV. J Cell Biol 159, 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greber UF & Way M (2006) A superhighway to virus infection. Cell 124, 741–754. [DOI] [PubMed] [Google Scholar]
- 31. Dohner K, Nagel CH & Sodeik B (2005) Viral stop‐and‐go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol 13, 320–327. [DOI] [PubMed] [Google Scholar]
- 32. Henry T, Gorvel JP & Meresse S (2006) Molecular motors hijacking by intracellular pathogens. Cell Microbiol 8, 23–32. [DOI] [PubMed] [Google Scholar]
- 33. Luby‐Phelps K (2000) Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol 192, 189–221. [DOI] [PubMed] [Google Scholar]
- 34. Sodeik B, Ebersold MW & Helenius A (1997) Microtubule‐mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol 136, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mabit H, Nakano MY, Prank U, Saam B, Dohner K, Sodeik B & Greber UF (2002) Intact microtubules support adenovirus and herpes simplex virus infections. J Virol 76, 9962–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP & Greber UF (1999) Microtubule‐dependent plus‐ and minus end‐directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol 144, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Funk A, Mhamdi M, Lin L, Will H & Sirma H (2004) Itinerary of hepatitis B viruses: delineation of restriction points critical for infectious entry. J Virol 78, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ogawa‐Goto K, Tanaka K, Gibson W, Moriishi E, Miura Y, Kurata T, Irie S & Sata T (2003) Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J Virol 77, 8541–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M & Hope TJ (2002) Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jouvenet N, Monaghan P, Way M & Wileman T (2004) Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J Virol 78, 7990–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suikkanen S, Aaltonen T, Nevalainen M, Valilehto O, Lindholm L, Vuento M & Vihinen‐Ranta M (2003) Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J Virol 77, 10270–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lakadamyali M, Rust MJ, Babcock HP & Zhuang X (2003) Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider MA, Spoden GA, Florin L & Lambert C (2011) Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13, 32–46. [DOI] [PubMed] [Google Scholar]
- 44. Ceccaldi PE, Gillet JP & Tsiang H (1989) Inhibition of the transport of rabies virus in the central nervous system. J Neuropathol Exp Neurol 48, 620–630. [DOI] [PubMed] [Google Scholar]
- 45. Pelkmans L, Kartenbeck J & Helenius A (2001) Caveolar endocytosis of simian virus 40 reveals a new two‐step vesicular‐transport pathway to the ER. Nat Cell Biol 3, 473–483. [DOI] [PubMed] [Google Scholar]
- 46. Leopold PL & Crystal RG (2007) Intracellular trafficking of adenovirus: many means to many ends. Adv Drug Deliv Rev 59, 810–821. [DOI] [PubMed] [Google Scholar]
- 47. Marozin S, Prank U & Sodeik B (2004) Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell‐cell contact sites. J Gen Virol 85, 775–786. [DOI] [PubMed] [Google Scholar]
- 48. Cunningham AL, Diefenbach RJ, Miranda‐Saksena M, Bosnjak L, Kim M, Jones C & Douglas MW (2006) The cycle of human herpes simplex virus infection: virus transport and immune control. J Infect Dis 194(Suppl 1), S11–S18. [DOI] [PubMed] [Google Scholar]
- 49. Tomishima MJ, Smith GA & Enquist LW (2001) Sorting and transport of alpha herpesviruses in axons. Traffic 2, 429–436. [DOI] [PubMed] [Google Scholar]
- 50. Bearer EL, Breakefield XO, Schuback D, Reese TS & LaVail JH (2000) Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc Natl Acad Sci USA 97, 8146–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ohka S, Sakai M, Bohnert S, Igarashi H, Deinhardt K, Schiavo G & Nomoto A (2009) Receptor‐dependent and ‐independent axonal retrograde transport of poliovirus in motor neurons. J Virol 83, 4995–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hernaez B, Escribano JM & Alonso C (2006) Visualization of the African swine fever virus infection in living cells by incorporation into the virus particle of green fluorescent protein‐p54 membrane protein chimera. Virology 350, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ojala PM, Sodeik B, Ebersold MW, Kutay U & Helenius A (2000) Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro . Mol Cell Biol 20, 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee GE, Murray JW, Wolkoff AW & Wilson DW (2006) Reconstitution of herpes simplex virus microtubule‐dependent trafficking in vitro . J Virol 80, 4264–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Enquist LW, Husak PJ, Banfield BW & Smith GA (1998) Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res 51, 237–347. [DOI] [PubMed] [Google Scholar]
- 56. Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R & Sodeik B (2002) Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13, 2795–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Van de Walle GR, Favoreel HW, Nauwynck HJ, Van Oostveldt P & Pensaert MB (2002) Antibody‐induced internalization of viral glycoproteins in pseudorabies virus‐infected monocytes and role of the cytoskeleton: a confocal study. Vet Microbiol 86, 51–57. [DOI] [PubMed] [Google Scholar]
- 58. Baines JD (2007) Envelopment of herpes simplex virus nucleocapsids at the inner nuclear membrane In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, Chapter 11 (Arvin A, Campadelli‐Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R. & Yamanishi K, eds), Cambridge University Press, Cambridge. [PubMed] [Google Scholar]
- 59. Ye GJ, Vaughan KT, Vallee RB & Roizman B (2000) The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J Virol 74, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roller RJ, Zhou Y, Schnetzer R, Ferguson J & DeSalvo D (2000) Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol 74, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Douglas MW, Diefenbach RJ, Homa FL, Miranda‐Saksena M, Rixon FJ, Vittone V, Byth K & Cunningham AL (2004) Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J Biol Chem 279, 28522–28530. [DOI] [PubMed] [Google Scholar]
- 62. Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A & Sodeik B (2010) Plus‐ and minus‐end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6, e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ & Sodeik B (2006) The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro . Traffic 7, 227–237. [DOI] [PubMed] [Google Scholar]
- 64. Dohner K, Radtke K, Schmidt S & Sodeik B (2006) Eclipse phase of herpes simplex virus type 1 infection: efficient dynein‐mediated capsid transport without the small capsid protein VP26. J Virol 80, 8211–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Antinone SE, Shubeita GT, Coller KE, Lee JI, Haverlock‐Moyns S, Gross SP & Smith GA (2006) The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J Virol 80, 5494–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Desai P, DeLuca NA & Person S (1998) Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247, 115–124. [DOI] [PubMed] [Google Scholar]
- 67. Albertini AA, Schoehn G, Weissenhorn W & Ruigrok RW (2008) Structural aspects of rabies virus replication. Cell Mol Life Sci 65, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klingen Y, Conzelmann KK & Finke S (2008) Double‐labeled rabies virus: live tracking of enveloped virus transport. J Virol 82, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y & Blondel D (2009) Functional characterization of Negri bodies (NBs) in rabies virus‐infected cells: evidence that NBs are sites of viral transcription and replication. J Virol 83, 7948–7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raux H, Flamand A & Blondel D (2000) Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74, 10212–10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jacob Y, Badrane H, Ceccaldi PE & Tordo N (2000) Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol 74, 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mebatsion T (2001) Extensive attenuation of rabies virus by simultaneously modifying the dynein light chain binding site in the P protein and replacing Arg333 in the G protein. J Virol 75, 11496–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rasalingam P, Rossiter JP, Mebatsion T & Jackson AC (2005) Comparative pathogenesis of the SAD‐L16 strain of rabies virus and a mutant modifying the dynein light chain binding site of the rabies virus phosphoprotein in young mice. Virus Res 111, 55–60. [DOI] [PubMed] [Google Scholar]
- 74. Tan GS, Preuss MA, Williams JC & Schnell MJ (2007) The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc Natl Acad Sci USA 104, 7229–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, De The H & Chelbi‐Alix MK (2002) Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21, 7957–7970. [DOI] [PubMed] [Google Scholar]
- 76. Moseley GW, Roth DM, DeJesus MA, Leyton DL, Filmer RP, Pouton CW & Jans DA (2007) Dynein light chain association sequences can facilitate nuclear protein import. Mol Biol Cell 18, 3204–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hernaez B & Alonso C (2010) Dynamin‐ and clathrin‐dependent endocytosis in African swine fever virus entry. J Virol 84, 2100–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alonso C, Miskin J, Hernaez B, Fernandez‐Zapatero P, Soto L, Canto C, Rodriguez‐Crespo I, Dixon L & Escribano JM (2001) African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light‐chain dynein. J Virol 75, 9819–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hernaez B, Tarrago T, Giralt E, Escribano JM & Alonso C (2010) Small peptide inhibitors disrupt a high‐affinity interaction between cytoplasmic dynein and a viral cargo protein. J Virol 84, 10792–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT & Trus BL (2008) Arrangement of L2 within the papillomavirus capsid. J Virol 82, 5190–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kamper N, Day PM, Nowak T, Selinka HC, Florin L, Bolscher J, Hilbig L, Schiller JT & Sapp M (2006) A membrane‐destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol 80, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Florin L, Becker KA, Lambert C, Nowak T, Sapp C, Strand D, Streeck RE & Sapp M (2006) Identification of a dynein interacting domain in the papillomavirus minor capsid protein l2. J Virol 80, 6691–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Day PM, Baker CC, Lowy DR & Schiller JT (2004) Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci USA 101, 14252–14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu WJ, Qi YM, Zhao KN, Liu YH, Liu XS & Frazer IH (2001) Association of bovine papillomavirus type 1 with microtubules. Virology 282, 237–244. [DOI] [PubMed] [Google Scholar]
- 85. Doorbar J, Elston RC, Napthine S, Raj K, Medcalf E, Jackson D, Coleman N, Griffin HM, Masterson P, Stacey S et al. (2000) The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J Virol 74, 10081–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ohka S & Nomoto A (2001) Recent insights into poliovirus pathogenesis. Trends Microbiol 9, 501–506. [DOI] [PubMed] [Google Scholar]
- 87. Ohka S, Matsuda N, Tohyama K, Oda T, Morikawa M, Kuge S & Nomoto A (2004) Receptor (CD155)‐dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. J Virol 78, 7186–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mueller S, Cao X, Welker R & Wimmer E (2002) Interaction of the poliovirus receptor CD155 with the dynein light chain Tctex‐1 and its implication for poliovirus pathogenesis. J Biol Chem 277, 7897–7904. [DOI] [PubMed] [Google Scholar]
- 89. Saib A, Puvion‐Dutilleul F, Schmid M, Peries J & de The H (1997) Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J Virol 71, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Petit C, Giron ML, Tobaly‐Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H & Saib A (2003) Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J Cell Sci 116, 3433–3442. [DOI] [PubMed] [Google Scholar]
- 91. Bukrinskaya A, Brichacek B, Mann A & Stevenson M (1998) Establishment of a functional human immunodeficiency virus type 1 (HIV‐1) reverse transcription complex involves the cytoskeleton. J Exp Med 188, 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Desfarges S, Salin B, Calmels C, Andreola ML, Parissi V & Fournier M (2009) HIV‐1 integrase trafficking in S. cerevisiae: a useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast 26, 39–54. [DOI] [PubMed] [Google Scholar]
- 93. Vlach J, Lipov J, Rumlova M, Veverka V, Lang J, Srb P, Knejzlik Z, Pichova I, Hunter E, Hrabal R et al. (2008) D‐retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex‐1 of dynein. Proc Natl Acad Sci USA 105, 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lehmann M, Milev MP, Abrahamyan L, Yao XJ, Pante N & Mouland AJ (2009) Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J Biol Chem 284, 14572–14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kelkar SA, Pfister KK, Crystal RG & Leopold PL (2004) Cytoplasmic dynein mediates adenovirus binding to microtubules. J Virol 78, 10122–10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Suomalainen M, Nakano MY, Boucke K, Keller S & Greber UF (2001) Adenovirus‐activated PKA and p38/MAPK pathways boost microtubule‐mediated nuclear targeting of virus. EMBO J 20, 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez‐Boulan E & Crystal RG (2000) Dynein‐ and microtubule‐mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther 11, 151–165. [DOI] [PubMed] [Google Scholar]
- 98. Dohner K & Sodeik B (2005) The role of the cytoskeleton during viral infection. Curr Top Microbiol Immunol 285, 67–108. [DOI] [PubMed] [Google Scholar]
- 99. Lukashok SA, Tarassishin L, Li Y & Horwitz MS (2000) An adenovirus inhibitor of tumor necrosis factor alpha‐induced apoptosis complexes with dynein and a small GTPase. J Virol 74, 4705–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rentsendorj A, Xie J, MacVeigh M, Agadjanian H, Bass S, Kim DH, Rossi J, Hamm‐Alvarez SF & Medina‐Kauwe LK (2006) Typical and atypical trafficking pathways of Ad5 penton base recombinant protein: implications for gene transfer. Gene Ther 13, 821–836. [DOI] [PubMed] [Google Scholar]
- 101. Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP & Vallee RB (2009) Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Alvarez E, DeDiego ML, Nieto‐Torres JL, Jimenez‐Guardeno JM, Marcos‐Villar L & Enjuanes L (2010) The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non‐structural protein 3 and is ubiquitinated. Virology 402, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kubota T, Matsuoka M, Chang TH, Bray M, Jones S, Tashiro M, Kato A & Ozato K (2009) Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J Virol 83, 6952–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Suikkanen S, Saajarvi K, Hirsimaki J, Valilehto O, Reunanen H, Vihinen‐Ranta M & Vuento M (2002) Role of recycling endosomes and lysosomes in dynein‐dependent entry of canine parvovirus. J Virol 76, 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Clemente R, Sisman E, Aza‐Blanc P & de la Torre JC (2010) Identification of host factors involved in borna disease virus cell entry through a small interfering RNA functional genetic screen. J Virol 84, 3562–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. King SM, Barbarese E, Dillman JF III, Patel‐King RS, Carson JH & Pfister KK (1996) Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem 271, 19358–19366. [DOI] [PubMed] [Google Scholar]
- 107. Tai AW, Chuang JZ & Sung CH (1998) Localization of Tctex‐1, a cytoplasmic dynein light chain, to the Golgi apparatus and evidence for dynein complex heterogeneity. J Biol Chem 273, 19639–19649. [DOI] [PubMed] [Google Scholar]
- 108. Radnai L, Rapali P, Hodi Z, Suveges D, Molnar T, Kiss B, Becsi B, Erdodi F, Buday L, Kardos J et al. (2010) Affinity, avidity, and kinetics of target sequence binding to LC8 dynein light chain isoforms. J Biol Chem 285, 38649–38657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bergen JM & Pun SH (2007) Evaluation of an LC8‐binding peptide for the attachment of artificial cargo to dynein. Mol Pharm 4, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sachdev P, Menon S, Kastner DB, Chuang JZ, Yeh TY, Conde C, Caceres A, Sung CH & Sakmar TP (2007) G protein beta gamma subunit interaction with the dynein light‐chain component Tctex‐1 regulates neurite outgrowth. EMBO J 26, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Barbar E (2008) Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47, 503–508. [DOI] [PubMed] [Google Scholar]
- 112. Hodi Z, Nemeth AL, Radnai L, Hetenyi C, Schlett K, Bodor A, Perczel A & Nyitray L (2006) Alternatively spliced exon B of myosin Va is essential for binding the tail‐associated light chain shared by dynein. Biochemistry 45, 12582–12595. [DOI] [PubMed] [Google Scholar]
- 113. Puthalakath H, Villunger A, O’Reilly LA, Beaumont JG, Coultas L, Cheney RE, Huang DC & Strasser A (2001) Bmf: a proapoptotic BH3‐only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science 293, 1829–1832. [DOI] [PubMed] [Google Scholar]
- 114. Puthalakath H, Huang DC, O’Reilly LA, King SM & Strasser A (1999) The proapoptotic activity of the Bcl‐2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3, 287–296. [DOI] [PubMed] [Google Scholar]
- 115. Perkins D (2005) Virus signaling and apoptosis in the central nervous system infection. Front Biosci 10, 2804–2819. [DOI] [PubMed] [Google Scholar]
- 116. Nguyen ML & Blaho JA (2007) Apoptosis during herpes simplex virus infection. Adv Virus Res 69, 67–97. [DOI] [PubMed] [Google Scholar]
- 117. Hernaez B, Diaz‐Gil G, Garcia‐Gallo M, Ignacio Quetglas J, Rodriguez‐Crespo I, Dixon L, Escribano JM & Alonso C (2004) The African swine fever virus dynein‐binding protein p54 induces infected cell apoptosis. FEBS Lett 569, 224–228. [DOI] [PubMed] [Google Scholar]
- 118. Chen D, Wang M, Zhou S & Zhou Q (2002) HIV‐1 Tat targets microtubules to induce apoptosis, a process promoted by the pro‐apoptotic Bcl‐2 relative Bim. EMBO J 21, 6801–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Carter AP, Cho C, Jin L & Vale RD (2011) Crystal structure of the dynein motor domain. Science 331, 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]


