Abstract
The family M1 of Zn‐dependent aminopeptidases comprises members of closely related enzymes which are known to be involved in a variety of physiologically important processes. On the basis of two highly conserved peptide motifs, we have identified a new member of this family by PCR amplification and cDNA‐library screening. The longest ORF encodes a protein of 930 residues. It contains the HEXXH(X)18E Zn‐binding motif and displays high homology to the other M1 family members except for its N‐terminus for which a signal sequence of 20 residues can be predicted. This interpretation was supported by expressing fusion proteins formed with green fluorescent protein which localized to intracellular vesicles in COS‐7 and BHK cells. Northern‐blot analysis revealed ubiquitous expression of a major 3.1‐kb transcript. For enzymatic studies, the complete protein was expressed in Sf 9 insect cells. When aminoacyl β‐naphthylamides were used as substrates, efficient hydrolysis was only observed for Leu (and to a lesser extent Met). The activity was inhibited by chelators of bivalent cations and by other known aminopeptidase inhibitors, but surprisingly puromycin was without effect. This newly identified puromycin‐insensitive leucyl‐specific aminopeptidase is a signal‐sequence‐bearing member of family M1 and may be another example of the small subset of substrate‐specific peptidases.
Keywords: family‐specific PCR, metallopeptidase, puromycin, signal peptide, Zn‐dependent aminopeptidase
Abbreviations
- PILS‐AP
puromycin‐insensitive leucyl‐specific aminopeptidase
- PSA
puromycin‐sensitive aminopeptidase
- TRH‐DE
thyrotropin‐releasing hormone‐degrading ectoenzyme
- EGFP
enhanced green fluorescent protein
Aminopeptidases are exopeptidases that catalyze the hydrolysis of amino‐acid residues from the N‐terminus of peptide or protein substrates. They are widely distributed throughout the animal and plant kingdoms and are found in subcellular organelles, in the cytoplasm, and as membrane components. These enzymes are involved in a variety of physiologically important processes such as protein processing and turnover, regulation of peptide hormone action, viral infection, tissue invasion, and cell cycle control [1]. Many of the aminopeptidases known are metalloenzymes containing a central Zn2+ ion, which is crucial for enzymatic activity. According to the structurally based classification of proteolytic enzymes suggested by Rawlings and Barrett [2], the majority of these enzymes belong to clan MA, family M1 of Zn‐dependent exopeptidases (reviewed in [3]).
The most prominent member within the group of mammalian enzymes of family M1 is aminopeptidase N, one of the major proteins of the microvillar membrane of the small intestinal and kidney epithelial cells (reviewed in [4]). Aminopeptidase N displays a broad substrate spectrum and hydrolyzes neutral N‐termini. In brain, it may participate in the inactivation of enkephalins and other neuropeptides. On hematopoietic cells, it has been identified as the cell‐surface differentiation antigen CD13. Interestingly, certain viruses (e.g. coronavirus 229E) misuse aminopeptidase N as a receptor enabling host cell infection. Aminopeptidase A is highly homologous to aminopeptidase N but prefers acidic N‐terminal residues. Like aminopeptidase N, it is concentrated on brush‐border membranes and on certain hematopoietic cells. In brain, aminopeptidase A has been implicated in the metabolism of angiotensin II to angiotensin III (reviewed in [5]). Two more membrane‐anchored mammalian M1 family members have been described; the thyrotropin‐releasing hormone (TRH)‐degrading ectoenzyme (TRH‐DE) seems to represent the rare case of a substrate‐specific exopeptidase. It is the only ectoenzyme known that is capable of degrading TRH (pyroGlu‐His‐Pro‐NH2) by hydrolyzing the pyroGlu‐His bond. Its hormonal regulation in the anterior pituitary indicates that it may function as a regulatory element for the control of TRH‐stimulated thyrotropin and prolactin release, whereas in brain it may act as a terminator for neuronal TRH signals (reviewed in [6]). The insulin‐regulated membrane aminopeptidase from rat adipocytes is also known as human placental leucine aminopeptidase/oxytocinase. Although its physiological function is unclear, its insulin‐induced redistribution from intracellular vesicles to the plasma membrane argues for a specific role in peptide hormone processing (reviewed in [7]).
Besides these four type‐II ectoenzymes, three cytoplasmic M1 members have been characterized. Aminopeptidase B specifically removes basic residues from the N‐terminus of substrate peptides [8] and is found in a variety of tissues. It is closely related to leukotriene‐A4 hydrolase, an enzyme originally identified by its specific epoxide hydrolase activity [9]. It was later shown to be an aminopeptidase of broad specificity [10]. A soluble puromycin‐sensitive aminopeptidase (PSA) has been described in a variety of tissues. PSA inhibition by low puromycin concentrations that do not yet interfere with protein synthesis may directly lead to cell cycle arrest at the G2/M phase and induction of apoptosis [11].
In this paper we describe the molecular cloning and initial characterization of the eighth mammalian member of family M1, a puromycin‐insensitive leucyl‐specific aminopeptidase (PILS‐AP). This enzyme displays a high degree of structural homology to the other M1 members within the catalytically important regions but differs with respect to its inhibition profile, the presence of a signal peptide, and its unusual substrate specificity.
Materials and methods
Materials
All chemicals were of analytical grade and obtained from Sigma or Bachem Biochemica, Heidelberg, Germany. Radiochemicals were from Hartmann Analytic, Braunschweig, Germany. Oligonucleotides and G418 were from Gibco‐BRL, and restriction endonucleases were from Gibco‐BRL or New England Biolabs.
Family‐specific PCR
mRNA was prepared from whole rat (Sprague–Dawley) pituitaries by affinity chromatography on paramagnetic oligo‐dT polystyrene beads as described previously [12]. Poly(A)‐rich RNA (5 µg) was reverse‐transcribed using 500 ng oligo‐dT18 as primer and 200 units of a modified Moloney murine leukemia virus reverse transcriptase (Stratascript; Stratagene) at 44 °C for 90 min. Completely degenerated oligonucleotides were constructed on peptide motifs that are conserved among the members of family M1. Additional restriction sites were incorporated at the 5′ ends to facilitate subcloning. The N‐terminal peptide motif chosen was FPCFDEP (F‐primer: GAATTYCCNTGYTTYGAYGARCC), and the C‐terminal motif was HQWFGN/D (R‐primer: ACGAGCTCNCCRAACCAYTGRTG). Amplifications were achieved by 40 cycles (30 s at 95 °C, 10 s at 55 °C, 60 s at 75 °C) with Pfu‐DNA polymerase (Stratagene) under standard conditions. The PCR products were analyzed by agarose gel electrophoresis, subcloned into pBluescript KSII + and sequenced by dideoxy chain‐termination reactions with [35S]dATP[αS] and T7 DNA polymerase (United States Biochemicals).
Isolation of cDNA clones
cDNA was synthesized from rat pineal‐gland mRNA, size‐fractionated on Sephacryl S‐500 columns, ligated to λ‐ZAP‐Express vector arms and packaged with the help of Gigapack III Gold packaging extract as recommended by the supplier (Stratagene). From the high‐molecular‐mass cDNA fraction (average size range 3–8 kb), 1.5 million independent phages were obtained. The complete library was amplified once. The isolated PCR product was labeled with [α‐32P]dCTP by random priming using Magenta polymerase (Stratagene), and library screening was performed by standard methods [13]. Positive phages were plaque purified, and the inserts were prepared as pBK‐CMV plasmids by in vivo excision via superinfection with ExAssist helper phages (Stratagene). Overlapping sequences of both DNA strands were obtained by dideoxy chain‐termination reactions with an Applied Biosystems 371 automated sequencer employing fluorescent dNTPs (ABI) and a battery of sequence‐specific oligonucleotides. Sequence analyses were performed using the program macmolly tetra (SoftGene GmbH, Berlin, Germany). Sequence comparisons were performed with the blast and prosite algorithms using the default parameters to search the National Center for Biotechnology Information non‐redundant protein and DNA databases [14, 15].
Expression as fusion protein with enhanced green fluorescent protein (EGFP)
The codon of the last residue (Leu930) and the stop codon (CTGTGA) were converted to an XhoI site (CTCGAG) by PCR‐based mutagenesis. The complete ORF and the 5′ UTR were fused in frame N‐terminal to a red‐shifted variant of EGFP [16], taking advantage of the introduced XhoI site and the unique SpeI site of the multiple cloning region of clone 11 and restriction sites NheI and XhoI of pEGFP‐N1 (Clontech). COS‐7 and BHK cells were transfected with plasmid pPILS‐AP‐EGFP using lipofectamine PLUS as suggested by the supplier (Gibco‐BRL). Stable clones expressing recombinant EGFP‐tagged PILS‐AP were selected by propagation in the presence of G418 (475 µg·mL−1). Cells were harvested, washed in NaCl/Pi, fixed with 3% formaldehyde in NaCl/Pi for 20 min at room temperature and washed in NaCl/Pi again. Cells expressing recombinant PILS‐AP‐EGFP were viewed by an Olympus BH‐2 fluorescence microscope equipped with the appropriate filter set, and pictures were recorded with an Olympus camera.
Expression of recombinant protein in Sf 9 insect cells
A 32‐bp NotI–PstI fragment encoding the Flag epitope (DYKDDDDK) was introduced into the baculovirus transfer expression plasmid pBacPak9 (Clontech). A 1‐kb fragment of PILS‐AP including the start ATG codon (bold) was amplified by PCR with primers Fmut1 GGGGTACCGAGCTCAAATATGCCCTCTCTTCTTTCCCTAGTA and R960 (reverse complementary to nucleotides 940–980) introducing a SacI restriction site (underlined) in front of the start ATG codon (bold) and used to replace the corresponding sequences of the cDNA. The complete ORF of the resulting clone was excised by SacI–XhoI digestion and fused in frame N‐terminal to the Flag epitope bearing pBacPak9 creating pBac‐PILS‐AP‐Flag. The encoded protein thus differs only by the presence of 17 additional residues at the C‐terminus that allowed immunocytochemical detection. Recombinant baculovirus was produced with the BaculoGold System as recommended by the supplier (Pharmingen, San Diego, CA, USA) using empty pBacPAK9 as control and pBac‐PILS‐AP‐Flag together with linearized BaculoGold AvNPV DNA. Single viruses were isolated by plaque assay, amplified and used to infect cultures of 50 mL Sf 9 cells grown in Grace's Insect Medium (Gibco‐BRL). Four days after infection, cells were harvested by centrifugation and washed three times with NaCl/Pi. The resulting cell pellet was homogenized on ice in 30 mL 20 mm sodium phosphate buffer, pH 7.3, with the aid of a glass/Teflon homogenizer. Crude cell fractionation was achieved by successive centrifugation at 3000 g for 10 min and 75 000 g for 2 h. The pellets were washed twice with NaCl/Pi and resuspended in 10 mL 20 mm sodium phosphate buffer, pH 7.3. Protein concentrations were determined using the Bio‐Rad protein assay with BSA as standard.
Northern‐blot and Southern‐blot analysis
Total RNA was prepared from various rat tissues by use of RNeasy columns as suggested by the supplier (Qiagen). Samples (30 µg per lane) were size‐fractionated in a denaturing formaldehyde/agarose gel, capillary‐transferred to a nylon membrane (Nytran NY 12 N; Schleicher & Schuell, Dassel, Germany) and analyzed under high‐stringency conditions. Rat and mouse genomic DNA was isolated by standard methods and completely digested by the indicated restriction endonucleases. The fragments were size‐fractionated in 1.0% agarose gels, denaturated and capillary‐transferred to the membrane. Hybridizations were carried out at 42 °C (Northern blot) or 38 °C (Southern blot) in 50% formamide, 6 × NaCl/Pi/EDTA (1 × NaCl/Pi/EDTA = 0.15 m NaCl, 10 mm sodium phosphate, 1 mm EDTA, pH 7.4), 0.5% SDS and 100 µg·mL−1 salmon sperm DNA with randomly labeled cDNA probes of high specific activity (> 109 c.p.m.·µg−1). The PCR fragment was used as a probe for the Northern‐blot analysis, and an N‐terminal fragment including the 5′ UTR and the first 260 bp of rat PILS‐AP coding sequence was used as a probe for Southern‐blot analysis. After extensive washing to final stringencies of 0.1 × NaCl/Pi/EDTA/0.3% SDS for 30 min at 60 °C (Northern blot) or 0.2 × NaCl/Pi/EDTA/0.3% SDS for 30 min at 55 °C (Southern blot), the signals were detected by exposure to X‐ray films (XOMat; Kodak) and quantified by analysis with a phosphorimager (Fujix BAS 1000; Fuji). The signals obtained for glyceraldehyde‐3‐phosphate dehydrogenase mRNA were used to calculate relative expression levels.
Western‐blot analysis
SDS/PAGE was carried out as described by Laemmli [17]. In brief, 10 µg protein was separated in 7.5% acrylamide gels and transferred by use of a semidry electrophoretic transfer cell (Bio‐Rad) on to nitrocellulose membranes (Immobilon; Millipore). Blots were blocked with 1% BSA in 50 mm Tris/HCl, pH 7.5, containing 150 mm NaCl and 0.05% Tween 20, at room temperature for 3 h. Monoclonal primary antibodies to FLAG (anti‐FLAG M2; Integra Biosciences, Fernwald, Germany) were applied for 5 h at room temperature in the same buffer and detected by the enzymatic activity of secondary anti‐mouse alkaline phosphatase‐conjugated IgG (Bio‐Rad). The molecular masses were assessed by comparison with a high‐range molecular‐mass marker (Boehringer‐Mannheim).
Assay for aminopeptidase activity
Recombinant PILS‐AP was produced in Sf 9 cells as described above. Aminopeptidase activity was measured in the crude homogenates from the control virus‐infected and PILS‐AP‐infected Sf 9 cells. Standard assays were performed with 5 µg protein diluted in 0.5 mL 20 mm sodium phosphate buffer, pH 7.3. To suppress endogenous aminopeptidase activity of Sf 9 cells, all reaction buffers were supplemented with 5 µm puromycin. The reaction was started by adding 500 µL 0.2 mm substrate (aminoacyl β‐naphthylamide derivatives in 20 mm sodium phosphate buffer, pH 7.3), and the incubation was allowed to proceed at 30 °C for 30 min. Liberated β‐naphthylamide was determined by fluorescence spectrometry (excitation at 340 nm, emission at 410 nm) using a Perkin–Elmer LS‐3B fluorescence spectrometer [18]. For the determination of substrate specificity, all 20 proteinogenic aminoacyl β‐naphthylamide derivatives were assayed as described above; for Cys‐β‐naphthylamide, the reaction buffer was supplemented with 1 mm dithiothreitol.
For inhibitor studies, the indicated substances were preincubated with the enzyme on ice for 5 min unless stated otherwise. Kinetic constants were determined for both Leu‐β‐naphthylamide and Met‐β‐naphthylamide hydrolysis by Lineweaver–Burk analysis. The two substrates were prepared separately at concentrations ranging from 5 to 1000 µm, and initial velocities were calculated. The reaction was started by adding 5 µg protein in 10 µL 20 mm sodium phosphate buffer, pH 7.3, to the prewarmed substrates. For analysis of the pH‐dependence, Leu‐β‐naphthylamide hydrolysis was determined in the pH range 5.5–9 with 20 mm sodium phosphate buffer. Temperature dependence was determined by analyzing Leu‐β‐naphthylamide hydrolysis at 25 −60 °C under standard assay conditions.
Results
cDNA cloning of rat PILS‐AP
All members of family M1 of Zn‐dependent aminopeptidases share two well‐conserved peptide motifs of six to seven residues. To characterize additional members (e.g. aminopeptidase W) that might belong to this family, completely degenerated oligonucleotides were deduced and used to amplify the respective cDNA fragments from rat pituitary. Among the products obtained was one fragment that was highly homologous but did not match perfectly to any of the known aminopeptidases. Using this PCR product as a probe, we isolated three independent clones from a rat pineal‐gland cDNA library. The largest (no. 20) contained a cDNA insert of 5 kb; the others (nos 6 and 11) had inserts of 2.9 and 2.7 kb, respectively (Fig. 1A). Despite the differences in total size, clones 11 and 20 had the same 5′‐terminal sequences (except for three more nucleotides in clone 20) and identical 3′ ends [except for a longer poly(A) tail in clone 11]. Clone 6 was identical with clone 11 except for the first 257 nucleotides, which were missing. Clones 20 and 11 were sequenced completely. The sequences were identical except for an inserted fragment of 2.1 kb in clone 20 starting at nucleotide 2697. Alignment with the other members of family M1 revealed that this position maps to an exon/intron boundary in human aminopeptidase N [19], mouse aminopeptidase A [20], and human TRH‐DE [21]. The putative retained intron in clone 20 displays the invariant splice site consensus residues (AGgt–agGT), and the splice acceptor site is preceded by a pyrimidine‐rich sequence (75% pyrimidines within the 20 nucleotides in front of the GT‐acceptor site). Interestingly, the sequences of both the 5′ end of this intron and the 5′ end of the following exon are very similar, and the first 10 nucleotides display 80% identity (consensus: GTAAARGGRT). This may explain a high rate of alternative usage as the acceptor site (see Fig. 2B for details).
Figure 1.
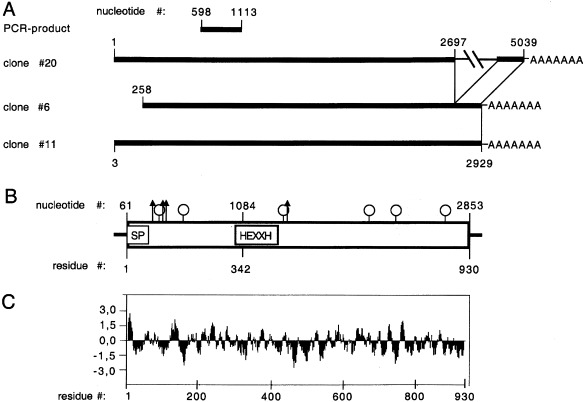
Schematic illustration of the cDNA cloning strategy, the major transcript and the deduced protein. (A) Summary of the relative positions of the PCR product and the cDNA clones. Numbers correspond to the composite cDNA. The inserted sequence of clone 20 and the poly(A) tail are indicated. (B) Schematic structure of the major transcript and the deduced protein. Horizontal lines indicate the 5′ UTR and the 3′ UTR. The open box represents the ORF; SP indicates the potential signal peptide, and HEXXH the Zn‐binding motif. Arrows and lollipops indicate the positions of predicted O‐glycosylation and N‐glycosylation sites, respectively. (C) Hydrophobicity profile of the deduced protein.
Figure 2.

Deduced primary structure of PILS‐AP. (A) Protein sequence deduced from the major 3.1‐kb transcript. The amino‐acid residues are numbered, the potential N‐terminal signal peptide is indicated by italic letters, and the consensus sequence of Zn‐dependent aminopeptidases is bold. Arrows indicate the positions of the family‐specific primers used; potential O‐glycosylation and N‐glycosylation sites are underlined. (B) Comparison of the region where the cDNA clones diverge. The consensus sites for intron/exon boundaries are underlined; the potential intronic sequences of cDNA clone 20 are in lower case. Note that the 10 bp following the boundary are highly homologous (indicated by bold letters).
cDNA sequence analysis
The first Met codon (nucleotides 61–63) is located within a sequence (GCAAGCATGC) resembling the Kozak consensus sequence for translational initiation (GCCRCCATGG), although it is not optimal [22]. It is preceded by a short 5′ UTR that contains three in‐frame stop codons (at nucleotides 10, 16 and 46). Alternative ATG initiation codons further downstream (nucleotides 152, 178, 212) do not match the consensus sequence to a higher degree. The longest ORF starts at the aforementioned ATG (nucleotide 61) and encodes a protein of 930 amino acids with a calculated molecular mass of 106 kDa (Fig. 1B). Hydropathy analysis using the Kyte and Doolittle algorithm [23] predicts a single hydrophobic region at the N‐terminus (Fig. 1C). A network‐based analysis employing the signal prediction server SignalP V1.1 (http://genome.cbs.dtu.dk/services/SignalP/index.html) predicts this stretch to represent an N‐terminal signal sequence (Fig. 2A). The size of the predicted signal peptide agrees well with the average length of 22.6 residues, as determined from over 1000 individual eukaryotic signal sequences [24]. The consensus sequence for Zn‐dependent metallopetidases His‐Glu‐Xaa‐Xaa‐His is found at residues 342–346 and followed by a second Glu separated by 18 residues, which has been shown to participate in Zn binding [25]. A third Glu (residue 309) is found at a conserved position in the context of the invariant triad Met‐Glu‐Asn shared by all M1 members. Site‐directed mutagenesis studies with aminopeptidase N characterized this residue to be involved in the binding of the free N‐terminus of the substrate [26]. Another residue (Tyr427) is found at a conserved distance from the Zn‐binding motif in a homologous environment and has been shown to participate in stabilizing the transition‐state complex in aminopeptidase A [27]. The sequence predicts six potential N‐glycosylation sites and four O‐glycosylations sites. The 3′ UTR extends for only 60 nucleotides and contains a polyadenylation signal like sequence AGTAAA (nucleotides 2890–2896) at a distance of 15 nucleotides from the poly(A) tail. The finding that all three independent clones share the same site of polyadenylation indicates that it should represent the authentic 3′ end of the transcript. The same applies for the 5′ end, as it is shared by two of the isolated clones and the 3‐nucleotide longer version of clone 20 aligns perfectly with a recently released mouse expressed sequence tag (accession no. AT119040, GI:3519364), which shows 92% identity over its total length of 605 nucleotides, indicating that it partially represents the mouse ortholog of PILS‐AP.
Homology to other peptidases
A comparison of the deduced amino‐acid sequence with the NCBI protein database revealed strong homology to other mammalian Zn‐dependent aminopeptidases. Greatest similarity is found for KIAA0525, a 6.3‐kb cDNA isolated from a human brain cDNA library [28]. This sequence seems to represent the major part of the human ortholog without the initiation codon, the potential signal peptide, and the following 42 N‐terminal residues. Among the functionally characterized M1 family members, rat insulin‐regulated aminopeptidase and its human ortholog placental leucine aminopeptidase/oxytocinase display greatest similarity (46% amino‐acid identity). With respect to the other mammalian Zn‐dependent aminopeptidases, overall amino‐acid identity ranges from 32% for mouse PSA, 32% for rat TRH‐DE, 32% for mouse aminopeptidase A, 31% for rat aminopeptidase N to 23% for rat aminopeptidase B and 22% for rat leukotriene‐A4 hydrolase. The similarities are highest around the Zn‐binding motif in the region that was used for the PCR‐based cloning (Fig. 3).
Figure 3.

Comparison of PILS‐AP with the other M1 family members. Alignment of the primary sequences in the most conserved region surrounding the known catalytic residues, Zn ligands and the substrate binding Glu (marked by asterisks). Identical residues conserved in all M1 members are shaded. For clarity, the less homologous sequences of aminopeptidase B and leukotriene‐A4 hydrolase have been omitted. IRAP, insulin‐regulated aminopeptidase; AP‐N, AP‐A, aminopeptidases N and A, respectively.
Functional expression
To analyze the enzymatic properties, PILS‐AP was expressed by baculovirus‐mediated transfection of Sf 9 insect cells. Immunoreactive FLAG‐tagged fusion proteins were detected by Western‐blot analysis as a doublet migrating at 110 kDa and 100 kDa (Fig. 4). The recombinant enzyme displayed aminopeptidase activity. Cell fractionation yielded the majority of active PILS‐AP in the high‐speed supernatant as a soluble enzyme. This preparation was used for characterization of the enzymatic properties of PILS‐AP. Of the 20 aminoacyl‐β‐naphthylamide derivatives tested, only Leu‐β‐naphthylamide and Met‐β‐naphthylamide were effectively cleaved (Fig. 5). Comparing the relative activities under the conditions described, Leu‐β‐naphthylamide turned out to be a four times better substrate than Met‐β‐naphthylamide. Hydrolysis of the remaining aminoacyl‐β‐naphthylamide derivatives was minimal (< 2% of Leu‐β‐naphthylamide hydrolysis). Aminopeptidase activity obeyed Michaelis–Menten kinetics. Analysis by a Lineweaver–Burk plot yielded a graph of first order leading to a K m value of 65.4 µm for Leu‐β‐naphthylamide hydrolysis (Fig. 6A) and 77.4 µm for Met‐β‐naphthylamide hydrolysis. Analysis of the temperature‐dependence and pH‐dependence for Leu‐β‐naphthylamide hydrolysis showed optimal enzymatic activities at 40 °C and at pH 8.0 (Fig. 6B,C).
Figure 4.
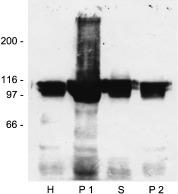
Recombinant expression of PILS‐AP in insect cells. Aliquots (10 µg protein per lane) of the Sf9 cell fractions were separated by SDS/PAGE, and recombinant PILS‐AP was detected using antibodies directed against the FLAG‐epitope. The molecular mass (kDa) of standard proteins is indicated on the left. H, homogenate; P1, 3000 g pellet; S, high‐speed supernatant; P2, high‐speed pellet.
Figure 5.
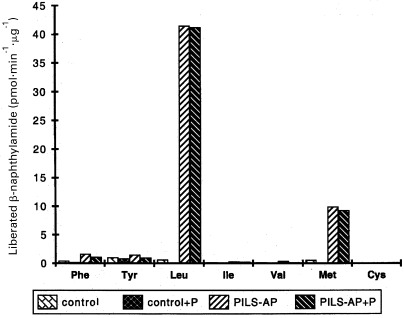
Substrate specificity. The aminopeptidase activities towards the indicated aminoacyl‐β‐naphthylamide derivatives are given. Control and PILS‐AP‐baculovirus infected Sf9 cells were homogenized, and aliquots of the high‐speed supernatants (5 µg protein) were incubated at 30 °C for 30 min with the aminoacyl‐β‐naphthylamide derivatives (100 µm final concentration) in the presence (+ P) or absence of puromycin (5 µm). Liberated β‐naphthylamide was determined fluorimetrically. The activities towards Ala‐, Arg‐, Asn‐, Asp‐, Gln‐, Glu‐, Gly‐, His‐, Lys‐, Pro‐, Ser‐ and Trp‐β‐naphthylamide were below 0.1 pmol liberated β‐naphthylamide·min−1·µg−1.
Figure 6.
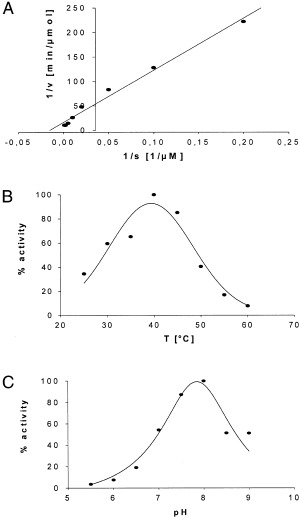
Determination of Km and temperature‐dependence and pH‐dependence for Leu‐β‐naphthylamide hydrolysis by recombinant PILS‐AP. Sf9 cells were infected with PILS‐AP encoding virus, homogenized and a high‐speed supernatant was prepared. The rate of Leu‐β‐naphthylamide hydrolysis was measured under the conditions described; all data were recorded in triplicate. (A) K m was determined by Lineweaver–Burk analysis. (B) Temperature‐dependence was determined over the range 25–60 °C. (C) pH‐dependence was analyzed from pH 5.5 to pH 9.0.
Effects of proteinase inhibitors
Various chemicals were tested for their effects on PILS‐AP activity (Table 1). Puromycin turned out not to inhibit the activity of the recombinant enzyme but effectively reduced background aminopeptidase activity from Sf 9 cells (Fig. 5). Thus, all reaction buffers for the other substances were supplemented with 5 µm puromycin. The serine protease inhibitor di‐isopropyl fluorophosphate and the SH‐group‐reactive agent 2‐iodoacetamide were without effect. Strong inhibition was achieved with l‐leucinethiol, a very effective inhibitor of porcine aminopeptidase N [29]. Dithiothreitol inhibited PILS‐AP only at exceedingly high concentrations. The cation chelator 1,10‐phenanthroline effectively inhibited PILS‐AP activity, but not the isomers 1,7‐phenanthroline and 4,7‐phenanthroline. The effects of EDTA were only moderately dependent on concentration, but showed a strong dependence on preincubation time; almost complete inactivation of PILS‐AP activity was observed after a preincubation period of 10 days.
Table 1.
Effect of various protease inhibitors on PILS‐AP activity. The indicated substances were preincubated with the enzyme on ice for 5 min and the reaction was started by adding Leu‐β‐naphthylamide to 0.1 mm.
| Chemical (preincubation) | Concentration (µm) | Inhibition (%) |
|---|---|---|
| None | 0 | |
| Di‐isopropyl fluorophosphate | 2000 | 7 |
| 2‐Iodoacetamide | 2000 | 5 |
| Dithiothreitol | 100 1000 10 000 | 2 11 61 |
| 1,10‐Phenanthroline | 100 200 | 74 88 |
| 4,7‐Phenanthroline | 100 200 | 23 37 |
| 1,7‐Phenanthroline | 100 200 | 11 21 |
| EDTA (5 min, 4 °C) EDTA (20 h, 4 °C) EDTA (48 h, 4 °C) EDTA (240 h, 4 °C) | 1000 1000 1000 1000 | 0 19 49 96 |
| Phosphoramidon | 10 | 0 |
| l‐Leucinethiol | 0.1 1.0 | 85 97 |
| Leucinol | 100 | 0 |
| Puromycin | 1.0 5.0 | 0 0 |
| Actinonin | 1.0 | 23 |
| Amastatin | 1.0 | 44 |
| Bestatin | 1.0 | 11 |
Distribution of PILS‐AP mRNA in rat tissues
The expression of PILS‐AP in rat tissues was assessed by Northern‐blot analysis employing the PCR product corresponding to the Zn‐binding region as a radioactive probe. Transcripts were easily detected by overnight exposure, indicating relatively high expression levels (Fig. 7). A major band of 3.1 kb was detected in the total RNA preparations from a wide range of rat tissues, with higher staining intensities in the normalized samples from adrenals, spleen, lung, and thymus. An additional less intense band running around the position of the 28S rRNA (i.e. at 5 kb) was observed in all RNA preparations analyzed and especially from cultured cell lines (rat somatomammotropic GH3 cells or fibroblast‐like rat‐1 cells).
Figure 7.
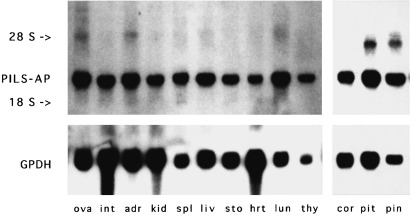
Northern‐blot analysis. PILS‐AP cDNA fragments were radioactively labeled and used to hybridize a nylon membrane with size‐separated preparations of RNA from rat tissues (ova, ovaries; int, intestine; adr, adrenals; kid, kidney; spl, spleen; liv, liver; sto, stomach; hrt, heart; lun, lung; thy, thymus; cor, cortex cerebrum; pit, pituitary gland; pin, pineal gland). The 28S rRNA and 18S rRNA bands are indicated. GPDH, glyceraldehyde‐3‐phosphate dehydrogenase.
Southern‐blot analysis
Genomic DNA from rat kidney and mouse brain was digested to completion by a variety of restriction endonucleases, and the fragments were separated according to size and blotted. The N‐terminal fragment including the 5′ UTR and the first 260 bp of rat PILS‐AP coding sequence was used as a probe. The simple pattern of bands observed indicates that PILS‐AP is encoded by a single‐copy gene in mouse and rat and that the rodent sequences are highly homologous (not shown).
Subcellular distribution
To investigate the subcellular distribution of PILS‐AP, crude cell fractionation was performed with Sf 9 cells expressing the recombinant enzyme bearing a C‐terminal FLAG‐tag (Fig. 4). The high‐speed supernatant contained > 80% of PILS‐AP activity; some activity was also found in the 3000 g pellet and in the 75 000 g pellet (not shown). To analyze its distribution in mammalian cells, BHK and COS‐7 cells were transfected with expression plasmids encoding EGFP (pEGFP) or a fusion protein of PILS‐AP with EGFP at the C‐terminus (pPILS‐AP‐EGFP). Stable transformants were selected by G418 treatment, grown on glass slides, fixed and examined with a fluorescence microscope. Cells transfected with pEGFP showed bright fluorescence distributed over the whole cell body (not shown). The fluorescence signals from pPILS‐AP‐EGFP‐transfected cells were considerably lower in intensity but focused on and specifically localized to vesicular structures (Fig. 8).
Figure 8.
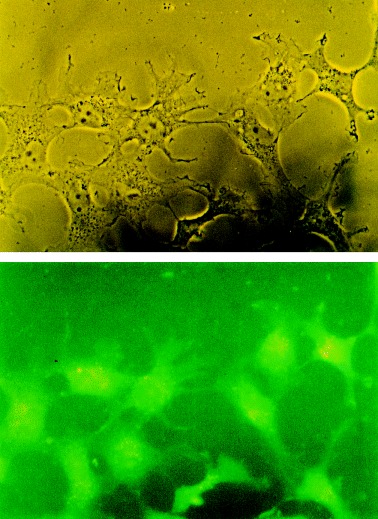
Subcellular distribution. Localization of fluorescent PILS‐AP fusion proteins in mammalian cells. COS‐7 cells were stably transfected with pPILS‐AP‐EGFP, fixed with formaldehyde and examined by fluorescent microscopy. Original magnification: 400 ×. Top, phase contrast; bottom, fluorescence.
Discussion
The brain and pituitary possess a large array of proteolytic enzymes engaged in the metabolism of biologically active neuropeptides. While working on the highly specific metallopeptidase TRH‐DE, we realized that there were similarities to the published data on the X‐Trp‐specific hydrolase (aminopeptidase W, EC 3.4.11.16) with respect to size, Zn‐dependence, topology and neuronal localization [30, 31]. Thus we rationalized that it should belong to the same family of metallopeptidases, and therefore started a PCR‐based screening approach. The cDNA clones isolated seem to represent the major and a minor incompletely/alternatively spliced transcript of a new member of family M1 of Zn‐dependent metallopeptidases (PILS‐AP). Its mRNA is easily detected in all rat tissues and cell lines analyzed so far, indicating ubiquitous expression. Moreover, expressed sequence tags from mouse and man can be assigned to represent PILS‐AP orthologs by displaying a high degree of sequence conservation. In man, these sequences have been derived from various cDNA sources (heart, pancreas, fat, ovary, testis, etc.), thus corroborating its widespread distribution.
The primary sequence of PILS‐AP, as predicted from the major transcript, contains all of the known functionally important residues shared by the members of family M1 of Zn‐dependent aminopeptidases. Besides these homologies, important differences in substrate specificity, the subcellular localization and enzyme chemical characteristics are evident, indicating that PILS‐AP is not aminopeptidase W, but an as yet uncharacterized enzyme. PILS‐AP does not accept basic or acidic substrates like aminopeptidase A, aminopeptidase B or leukotriene‐A4 hydrolase. The other M1 members, i.e. insulin‐regulated aminopeptidase (placental leucine aminopeptidase/oxytocinase), aminopeptidase N and PSA, prefer neutral and/or hydrophobic residues. This preference partially overlaps with the spectrum of PILS‐AP, but its limited substrate range, i.e. Leu‐β‐naphthylamide and Met‐β‐naphthylamide as the sole aminoacyl‐β‐naphthylamide derivatives accepted, clearly distinguishes it from these aminopeptidases, which also hydrolyze chemically similar residues such as Val‐β‐naphthylamide or Ile‐β‐naphthylamide to a comparable degree. Another striking difference from PSA is the insensitivity of PILS‐AP to puromycin, a substance that in our expression system, i.e. Sf 9 cells, was able to inhibit the endogenous insect Leu‐cleaving activities almost completely. Recently the purification of a neuron‐specific Leu‐cleaving aminopeptidase from rat brain has been described [32]. Although the substrate specificity is very similar (Leu and Met derivatives as the preferred substrates), the clear difference in sensitivity towards puromycin again allows unequivocal discrimination. Another well‐characterized Leu‐preferring aminopeptidase is the ubiquitously expressed cytosolic leucine aminopeptidase, which has been best characterized from bovine lens [33]. It is a soluble hexameric Zn‐dependent enzyme composed of identical subunits of 54 kDa each in its native form. Several leucine aminopeptidases from various organisms have been characterized and collectively constitute clan MF, family M17 of cocatalytic leucyl‐metallopeptidases [34]. Again, these enzymes differ from PILS‐AP by sequence and by the relaxed substrate specificity hydrolyzing Leu‐β‐naphthylamide and most N‐terminal amino acids with comparable rates [35]. Moreover, the identity of mammalian leucine aminopeptidase (EC 3.4.11.1) with proline aminopeptidase (EC 3.4.11.5) has been demonstrated [36]. In addition, the K m for Leu‐β‐naphthylamide hydrolysis by leucine aminopeptidase is substantially higher (250 µm vs. 65 µm for PILS‐AP) and leucine aminopeptidase is clearly more sensitive to l‐leucinethiol and to the naturally occurring inhibitors bestatin and amastatin. With respect to the metal‐chelating inhibitors, PILS‐AP could be effectively inhibited by 1,10 phenanthroline and EDTA. This was expected in analogy with the other M1 members, but interestingly the effect of EDTA became maximal only after prolonged preincubation periods, which is generally not the case for the other metallopeptidases. The observed kinetics of inhibition of PILS‐AP with EDTA is very reminiscent of the EDTA effects on TRH‐DE, the only M1 family member with high substrate specificity that is known to have a highly conserved structure [21] and to bind Zn2+ very tightly [37]. An even more pronounced time‐dependence was observed for astacin, a small metallopeptidase from crayfish, for which a 200‐h preincubation period was necessary to obtain 50% inhibition [38]. Taken together, the substrate spectrum of PILS‐AP is strikingly limited, and the sensitivity towards various inhibitors is consistent with a Zn‐dependent enzyme which binds the metal ion tightly. The deduced primary structure predicts a cleavable signal peptide for the N‐terminus. This feature is unique among the currently known mammalian M1 family members. Aminopeptidase A, aminopeptidase N, insulin‐regulated aminopeptidase and TRH‐DE contain N‐terminal domains of 23–25 consecutive hydrophobic residues which anchor these peptidases as type‐II ectoenzymes. In contrast, PSA is a cytosolic enzyme which may functionally interact with proteasomes [11]. Although it has been found partially associated with membrane, this interaction is clearly different from integral membrane proteins, and the active site was localized within the cytosol [39]. Aminopeptidase B and the closely related leukotriene‐A4 hydrolase are soluble enzymes which can be found as secreted aminopeptidases, although no classical signal peptide can be predicted in the primary sequences. With respect to PILS‐AP, our analysis of the subcellular localization of the EGFP‐fusion proteins in mammalian cells argues in favor of the functionality of its signal peptide. The nature of the vesicles to which the enzyme was targeted and the possibility of regulated secretion remain crucial questions to be addressed in the future. Given the high homology to the other M1 members and the widespread expression of PILS‐AP, the narrow substrate specificity is a rather atypical feature among the known peptidases [40], and it is increasingly puzzling that nature developed such a multitude of peptidases which are all capable of cleaving N‐terminal Leu residues.
Acknowledgements
We thank Birgit Kühlein, Anke Rosebrock and Uwe Grunenberg for expert technical assistance, Ralf Sigmund for valuable help with computer‐based analysis, Anne Pötter for help with the insect cell culture, Rudi Bauerfeind and Beate Sodeik for experienced support with fluorescence microscopy, Stefan Turwitt, Theofilos Papadopoulos, Roland Rabeler, Daniel Brauer and Silke Tillmanns for stimulating discussions, and Valerie Ashe for linguistic help. This work was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Enzymes: leukotriene‐A4 hydrolase (EC 3.3.2.6); leucine aminopeptidase (EC 3.4.11.1); aminopeptidase N (EC 3.4.11.2); insulin‐regulatedmembrane aminopeptidase (EC 3.4.11.3); placental leucine aminopeptidase/oxytocinase (EC 3.4.11.3); aminopeptidase B (EC 3.4.11.6); aminopeptidase A (EC 3.4.11.7); puromycin‐sensitive aminopeptidase (EC 3.4.11.14); puromycin‐insensitive leucyl‐specific aminopeptidase (EC 3.4.11.‐); thyrotropin‐releasing hormone‐degrading ectoenzyme (EC 3.4.19.6).
Note: the nucleotide sequences reported in this paper have been submitted to the GenBank with accession numbers AF148323and AF148324.
References
- 1. Brownlees, J. & Williams, C.H. (1993) Peptidases, peptides, and the mammalian blood–brain barrier. J. Neurochem. 60, 793 780. [DOI] [PubMed] [Google Scholar]
- 2. Rawlings, N.D. & Barrett, A.J. (1993) Evolutionary families of peptidases. Biochem. J. 290, 205 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrett, A.J. , Rawlings, N.D. , Woessner, J.F. (1998). Handbook of Proteolytic Enzymes(Barrett A.J. Rawlings N.D. & Woessner J.F., eds), pp. 994 1032. Academic Press, London. [Google Scholar]
- 4. Noren, O. , Sjöström, H. , Olson, J. (1997) Cell‐Surface Peptidases in Health and Disease(Kenny A.J. & Boustead C.M., eds), pp. 175 192. BIOS Scientific Publishers Ltd., Oxford. [Google Scholar]
- 5. Wang, J. & Cooper, M.D. (1997) Cell‐Surface Peptidases in Health and Disease(Kenny A.J. & Boustead C.M., eds), pp. 193 202. BIOS Scientific Publishers Ltd., Oxford. [Google Scholar]
- 6. Bauer, K. (1995) Inactivation of thyrotropin‐releasing hormone (TRH) by the hormonally regulated TRH‐degrading ectoenzyme: a potential regulator of TRH‐signals? Trends Endocrinol. Metab. 6, 101 105.DOI: 10.1016/1043-2760(94)00216-q [DOI] [PubMed] [Google Scholar]
- 7. Keller, S.R. , Scott, H.M. , Mastick, C.C. , Aebersold, R. , Lienhard, G.E. (1995) Cloning and characterization of a novel insulin‐regulated membrane aminopeptidase from Glut4 vesicles. J. Biol. Chem. 270, 23612 23618. [DOI] [PubMed] [Google Scholar]
- 8. Cadel, S. , Pierotti, A.R. , Foulon, T. , Creminon, C. , Barre, N. , Segretain, D. , Cohen, P. (1995) Aminopeptidase‐B in the rat testes: isolation, functional properties and cellular localization in the seminiferous tubules. Mol. Cell. Endocrinol. 110, 149 160.DOI: 10.1016/0303-7207(95)03529-g [DOI] [PubMed] [Google Scholar]
- 9. Ohishi, N. , Izumi, T. , Minami, M. , Kitamura, S. , Seyama, Y. , Ohkawa, S. , Terao, S. , Yotsumoto, H. , Takaku, F. , Shimizu, T. (1987) Leukotriene A4 hydrolase in the human lung. Inactivation of the enzyme with leukotriene A4 isomers. J. Biol. Chem. 262, 10200 10205. [PubMed] [Google Scholar]
- 10. Haeggström, J.Z. , Wetterholm, A. , Vallee, B.L. , Samuelsson, B. (1990) Leukotriene A4 hydrolase: an epoxide hydrolase with peptidase activity. Biochem. Biophys. Res. Commun. 173, 431 437. [DOI] [PubMed] [Google Scholar]
- 11. Constam, D.B. , Tobler, A.R. , Rensing‐Ehl, A. , Kemler, I. , Hersh, L.B. , Fontana, A. (1995) Puromycin‐sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J. Biol. Chem. 270, 26931 26939. [DOI] [PubMed] [Google Scholar]
- 12. Schomburg, L. & Bauer, K. (1995) Thyroid hormones rapidly and stringently regulate the messenger RNA levels of the thyrotropin‐releasing hormone (TRH) receptor and the TRH‐degrading ectoenzyme. Endocrinology 136, 3480 3485. [DOI] [PubMed] [Google Scholar]
- 13. Sambrook, J. , Fritsch, E.F. , Maniatis, T. (1989) Molecular Cloning: a Laboratory Manual, pp. 2.114 2.117. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 14. Bairoch, A. (1991) PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 19, 2241 2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altschul, S.F. & Lipman, D.J. (1990) Protein database searches for multiple alignments. Proc. Natl Acad. Sci. USA 87, 5509 5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cormack, B.P. , Valdivia, R.H. , Falkow, S. (1996) FACS‐optimized mutants of the green fluorescent protein (GFP). Gene 173, 33 38.DOI: 10.1016/0378-1119(95)00685-0 [DOI] [PubMed] [Google Scholar]
- 17. Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227, 680 685. [DOI] [PubMed] [Google Scholar]
- 18. Greenberg, L.J. (1962) Fluorometric measurement of alkaline phosphatase and aminopeptidase activities in the order of 10(‐14) mole. Biochem. Biophys. Res. Commun. 9, 430 435. [DOI] [PubMed] [Google Scholar]
- 19. Lerche, C. , Vogel, L.K. , Shapiro, L.H. , Norén, O. , Sjöström, H. (1996) Human aminopeptidase N is encoded by 20 exons. Mammal. Genome 7, 712 713. [DOI] [PubMed] [Google Scholar]
- 20. Wang, J. , Walker, H. , Lin, Q. , Jenkins, N. , Copeland, N.G. , Watanabe, T. , Burrows, P.D. , Cooper, M.D. (1996) The mouse BP‐1 gene: structure, chromosomal localization, and regulation of expression by type I interferons and interleukin‐7. Genomics 33, 167 176.DOI: 10.1006/geno.1996.0180 [DOI] [PubMed] [Google Scholar]
- 21. Schomburg, L. , Turwitt, S. , Prescher, G. , Lohmann, D. , Horsthemke, B. , Bauer, K. (1999) Human TRH‐degrading ectoenzyme cDNA cloning, functional expression, genomic structure and chromosomal assignment. Eur. J. Biochem. 265, 415 422.DOI: 10.1046/j.1432-1327.1999.00753.x [DOI] [PubMed] [Google Scholar]
- 22. Kozak, M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867 19870. [PubMed] [Google Scholar]
- 23. Kyte, J. & Doolittle, R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105 132. [DOI] [PubMed] [Google Scholar]
- 24. Nielsen, H. , Engelbrecht, J. , Brunak, S. , von Heijne, G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1 6. [DOI] [PubMed] [Google Scholar]
- 25. Jongeneel, C.V. , Bouvier, J. , Bairoch, A. (1989) A unique signature identifies a family of zinc‐dependent metallopeptidases. FEBS Lett. 242, 211 214. [DOI] [PubMed] [Google Scholar]
- 26. Luciani, N. , Marie‐Claire, C. , Ruffet, E. , Beaumont, A. , Roques, B.P. , Fournie‐Zaluski, M.C. (1998) Characterization of Glu350 as a critical residue involved in the N‐terminal amine binding site of aminopeptidase N (EC 3.4.11.2): insights into its mechanism of action. Biochemistry 37, 686 692. [DOI] [PubMed] [Google Scholar]
- 27. Vazeux, G. , Iturrioz, X. , Corvol, P. , Llorens‐Cortès, C. (1997) A tyrosine residue essential for catalytic activity in aminopeptidase A. Biochem. J. 327, 883 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagase, T. , Ishikawa, K. , Miyajima, N. , Tanaka, A. , Kotani, H. , Nomura, N. , Ohara, O. (1998) Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro . DNA Res. 5, 31 39. [DOI] [PubMed] [Google Scholar]
- 29. Chan, W.W. (1983) l‐leucinethiol: a potent inhibitor of leucine aminopeptidase. Biochem. Biophys. Res. Commun. 116, 297 302. [DOI] [PubMed] [Google Scholar]
- 30. Gee, N.S. & Kenny, A.J. (1987) Proteins of the kidney microvillar membrane. Enzymic and molecular properties of aminopeptidase W. Biochem. J. 246, 97 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kenny, A.J. & Bourne, A. (1991) Cellular reorganisation of membrane peptidases in Wallerian degeneration of pig peripheral nerve. J. Neurocytol. 20, 875 885. [DOI] [PubMed] [Google Scholar]
- 32. Hui, K.S. , Saito, M. , Hui, M. (1998) A novel neuron‐specific aminopeptidase in rat brain synaptosomes. Its identification, purification, and characterization. J. Biol. Chem. 273, 31053 31060. [DOI] [PubMed] [Google Scholar]
- 33. Kim, H. & Lipscomb, W.N. (1993) X‐ray crystallographic determination of the structure of bovine lens leucine aminopeptidase complexed with amastatin: formulation of a catalytic mechanism featuring a gem‐diolate transition state. Biochemistry 32, 8465 8478. [DOI] [PubMed] [Google Scholar]
- 34. Straeter, N. & Lipscomb, W.N. (1998) Handbook of Proteolytic Enzymes(Barrett A.J., Rawlings N.D. & Woessner J.F., eds), pp. 1382 1389. Academic Press, London. [Google Scholar]
- 35. Henson, H. & Frohne, M. (1976) Crystalline leucine aminopeptidase from lens (alpha‐aminoacyl‐peptide hydrolase; EC 3.4.11.1). Methods Enzymol. 45, 504 520. [DOI] [PubMed] [Google Scholar]
- 36. Matsushima, M. , Takahashi, T. , Ichinose, M. , Miki, K. , Kurokawa, K. , Takahashi, K. (1991) Structural and immunological evidence for the identity of prolyl aminopeptidase with leucyl aminopeptidase. Biochem. Biophys. Res. Commun. 178, 1459 1464. [DOI] [PubMed] [Google Scholar]
- 37. Czekay, G. & Bauer, K. (1993) Identification of the thyrotropin‐releasing‐hormone‐degrading ectoenzyme as a metallopeptidase. Biochem. J. 290, 921 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stöcker, W. , Wolz, R.L. , Zwilling, R. , Strydom, D.A. , Auld, D.S. (1988) Astacus protease: a zinc metalloenzyme. Biochemistry 27, 5026 5032. [Google Scholar]
- 39. Dyer, S.H. , Slaughter, C.A. , Orth, K. , Moomaw, C.R. , Hersh, L.B. (1990) Comparison of the soluble and membrane‐bound forms of the puromycin‐sensitive enkephalin‐degrading aminopeptidases from rat. J. Neurochem. 54, 547 554. [DOI] [PubMed] [Google Scholar]
- 40. Turner, A.J. , Matsas, R. , Kenny, A.J. (1985) Are there neuropeptide‐specific peptidases? Biochem. Pharmacol. 34, 1347 1356. [DOI] [PubMed] [Google Scholar]


