Abstract
Entry of enveloped viruses into host cells is mediated by their surface envelope glycoproteins (Env). On the surface of the virus, Env is in a metastable, prefusion state, primed to catalyze the fusion of the viral and host membranes. An external trigger is needed to promote the drastic conformational changes necessary for the fusion subunit to fold into the low‐energy, 6‐helix bundle. These triggers typically facilitate pH‐independent entry at the plasma membrane or pH‐dependent entry in a low‐pH endosomal compartment. The α‐retrovirus avian sarcoma leukosis virus (ASLV) has a rare, 2‐step entry mechanism with both pH‐dependent and pH‐independent features. Here, we present the 2.0‐Å‐resolution crystal structure of the ASLV transmembrane (TM) fusion protein. Our structural and biophysical studies indicated that unlike other pH‐dependent or pH‐independent viral TMs, the ASLV fusion subunit is stable irrespective of pH. Two histidine residues (His490 and His492) in the chain reversal region confer stability at low pH. A structural comparison of class I viral fusion proteins suggests that the presence of a positive charge, either a histidine or arginine amino acid, stabilizes a helical dipole moment and is a signature of fusion proteins active at low pH. The structure now reveals key residues and features that explain its 2‐step mechanism, and we discuss the implications of the ASLV TM structure in the context of general mechanisms required for membrane fusion.—Aydin, H., Smrke, B.M., Lee, J. E. Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2‐step entry mechanism. FASEB J. 27, 5059–5071 (2013). http://www.fasebj.org
Keywords: ASLV, TM, helix dipole moment, HTLV‐1
Abbreviations
- ASLV
avian sarcoma leukosis virus
- CD
circular dichroism
- CR
chain reversal
- EBOV
Ebola virus
- Env
envelope glycoprotein
- HR
heptad repeat
- HTLV‐1
human T‐lymphotropic virus 1
- IAV
influenza A virus
- LCMV
lymphocytic choriomeningitis virus
- MARV
Marburg virus
- MoMLV
Moloney murine leukemia virus
- PDB
Protein Data Bank
- RSV
Rous sarcoma virus
- SU
surface [viral attachment subunit]
- TM
transmembrane [viral fusion subunit]
Invasion of a cell by enveloped viruses requires the attachment and fusion of the host and viral membranes (1). This process is mediated by one or more virionattached envelope glycoproteins (Envs; refs. 1, 2), classified into 3 groups (class I, II, and III) based on structural characteristics (3, 4). Many of the world's greatest viral threats, such as paramyxo‐, orthomyxo‐, filo‐, corona‐, arena‐ and retroviruses, display glycoproteins belonging to the class I family. Class I Envs are initially translated as a single polypeptide chain with a type I transmembrane anchor (5, 6). Proteolytic processing of Env forms separate surface (SU) attachment and transmembrane (TM) fusion subunits (5, 7) that protrude on the viral surface as a fusion‐competent, metastable, trimeric peplomer.
Class I viral glycoproteins are triggered through two general mechanisms: pH‐independent entry at the plasma membrane, or pH‐dependent entry at a low pH endosome (reviewed in ref. 8). In the pH‐independent mechanism, such as those employed by retro‐, corona‐, or paramyxoviruses, receptor binding by the attachment subunit triggers conformational changes in the Env to mediate the fusion event (9). In contrast, for pH‐dependent viral entry, such as those employed by flavi‐, arena‐, or influenza A viruses (IAVs), the acidic environment of the endosome triggers the Env conformational changes (10, 11). Following triggering, structural constraints on the hydrophobic fusion peptide or loop is released to allow its insertion into the target host membrane to form an extended prehairpin intermediate. Class I viral fusion proteins are thought to adopt a central trimeric α‐helical structure at the prehairpin intermediate state where its N‐terminal end is anchored to the target host lipid bilayer and the C‐terminal end embedded in the viral membrane. Multiple fusion proteins may be necessary to anchor and complete the membrane merger process. Subsequently, conformational changes to the fusion subunit allow the prehairpin intermediate to collapse. The C‐terminal hydrophobic helical region folds back and packs against the central trimeric α‐helical structure. As the extended prehairpin intermediate folds up, this brings both the viral and host membranes together to promote mixing of the two outer leaflets and the formation of the hemifusion intermediate. The formation of the final low‐energy, postfusion 6‐helix bundle then completes the formation of the fusion pore. The fusion of two lipid bilayers is a thermodynamically feasible process but comes with a high energetic price. The free energy released from the conformational changes of the prefusion glycoprotein to the low‐energy 6‐helix bundle drives the fusion reaction.
Interestingly, the α‐retrovirus avian sarcoma leukosis virus (ASLV) enters cells through a rare, 2‐step mechanism that has characteristics of the entry mechanisms of disparate viruses, such as HIV‐1 and IAV (12, 13). The unique mechanism of ASLV entry makes it an excellent model system to study the general viral entry processes (12). ASLV Env contains an N‐terminal SU subunit that is involved in receptor binding and a C‐terminal TM subunit that contains the fusion machinery (14–16). The receptor for the ASLV subgroup A is Tva, a protein that belongs to the low‐density lipoprotein receptor family (17). The ASLV Env hydrophobic internal fusion loop is sequestered in the prefusion, metastable state, but on receptor binding at neutral pH and at a temperature >22°C, conformational changes are triggered to expose and insert the fusion loop into the cell plasma membrane (18–20). This triggering of the fusion subunit by receptor binding is pH‐independent and is analogous to the effects of conformational changes on HIV‐1 gp160 on CD4 and chemokine receptor binding. However, previous studies show that in the presence of bafilomycin, an agent that raises the pH of endosomes, viral reverse transcripts were not detected in infected cells. This finding thus suggests a key role for low pH in entry (12). Additional studies on ASLV support the role of low pH being required to induce hemifusion and stabilization of the 6‐helix bundle (21, 22). The second step in ASLV entry is reminiscent of the low‐pH requirements of IAV and other pH‐dependent viruses.
To explore the rare, 2‐step entry mechanism of ASLV, we determined the crystal structure of the ASLV TM core. While a number of class I viral fusion proteins have been determined structurally (23–27), the ASLV TM is the first crystal structure of a viral fusion subunit that undergoes a hybrid entry mechanism. Interestingly, the ASLV TM is stable to all pH conditions, as opposed to other class I viral fusion proteins. Moreover, the ASLV TM has a strong negatively charged helix dipole at the chain‐reversal region that is stabilized by two histidine residues at low pH. A structural comparison of class I viral fusion proteins suggests that the presence of a positive charge from a histidine or arginine residue is a signature of fusion proteins that function at low pH. We also show that hydrophobic interactions between heptad repeat 1 (HR1) and HR2 and a single arginine residue (Arg473) located at the center of the molecule are important for the stability of the 6‐helix bundle. This study identified the residues that are important to the 2‐step entry mechanism and implications on our general understanding of viral fusion processes are discussed.
MATERIALS AND METHODS
Expression and purification of ASLV TM
The DNA sequence of the ASLV TM (residues 455–525) was codon‐optimized for expression in Escherichia coli, and the gene was synthesized and cloned into pET‐46 EK/LIC (EMD Millipore, Billerica, MA, USA) for expression of an N‐terminal 6‐His‐tagged protein with a TEV cleavage site. Cys506 was mutated to a serine to prevent nonspecific intersubunit disulfide bond formation. The expression vector was transformed into the E. coli SHuffle T7 expression cell line (New England Biolabs, Ipswich, MA, USA). A 20 ml overnight culture was used to inoculate 1 L of LB‐Miller medium, and cells were grown at 37°C to an OD600 of 0.6. Expression of the ASLV TM was induced with a final concentration of 0.5 mM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG). The temperature was lowered to 18°C, and cells were harvested 18 h postinduction by centrifugation at 4000 rpm (Sorvall SLC‐4000 rotor; Thermo Scientific, Waltham, MA, USA) for 20 min. Cells were resuspended in 25 ml 1 × Ni‐NTA binding buffer (50 mM Tris‐HCl, pH 7.5; 300 mM NaCl; and 20 mM imidazole) supplemented with 0.05% (w/v) CHAPS and 1 × EDTA‐free complete protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Cells were then lysed at 30 kpsi using a hydraulic cell disruption system (Constant Systems, Kenneshaw, GA, USA) and centrifuged at 17,000 rpm (Sorvall SS‐34 rotor) for 45 min to remove the cell debris. The supernatant was then applied to a 2‐ml Ni‐NTA column (Thermo Pierce, Rockford, IL, USA) by gravity flow at 1 ml/min. Protein was eluted in sequential steps using increasing concentrations of imidazole (125, 250, 375, or 500 mM in 1 × Ni‐NTA buffer). Samples containing ASLV TM were concentrated and further purified by size‐exclusion chromatography on a Superdex‐75 10/300 GL column (GE Life Sciences, Piscataway, NJ, USA) equilibrated with 10 mM Tris‐HCl (pH 7.5), 150 mM NaCl, and 0.05% (w/v) CHAPS. The concentration of purified protein was determined from the absorbance at 280 nm, and purity was confirmed by SDS‐PAGE and mass spectrometry.
Crystallization and structure determination of ASLV TM
The protein was concentrated to 10 mg/ml using an Amicon Ultra‐0.5 concentrator (Millipore, Billerica, MA, USA), and initial sparse matrix crystallization screens were performed in sitting‐drop 96‐well plates using an Art Robbins Phoenix liquid handling system (Art Robbins Instruments, Sunnyvale, CA, USA). Rod‐shaped crystals were obtained by hanging‐drop vapor diffusion in 20% (w/v) 2‐methyl‐2,4‐pentanediol, 5% (w/v) PEG 8000, and 0.1 M sodium cacodylate (pH 6.5). Crystals were soaked in a mother liquor solution containing 30% (v/v) glycerol for cryoprotection and then flash‐cooled in liquid nitrogen. ASLV TM diffraction data were collected on a Rigaku FR‐E Superbright X‐ray generator and Saturn A200 HD CCD detector (Rigaku Corp., Woodlands, TX, USA) at the Structural Genomics Consortium (Toronto, ON, Canada). Data were reduced and scaled using d∗trek (Rigaku; ref. 28). The structure of ASLV TM was determined by molecular replacement using the program Phaser (29), and a truncated, polyalanine search model of the Moloney murine leukemia virus (MoMLV) fusion glycoprotein [Protein Data Bank (PDB) code: 1MOF; ref. 30]. A clear solution was found with one monomer in the asymmetric unit. Alternating rounds of manual model rebuilding in Coot (31) followed by simulated annealing torsion angle refinement with TLS refinement using Phenix.refine (32) were performed until the R work/R free converged. Data collection and refinement statistics are presented in Table 1 . Analysis of protein dipole moments were performed using the Protein Dipole Moments Server (33). ASLV TM crystal structure and structure factors were deposited into the Protein Data Bank with accession code 4JPR.
Table 1.
Data collection and refinement statistics for ASLV TM
| Statistic | Value |
|---|---|
| Data collection | |
| Space group | H3 |
| Unit cell dimensions | |
| a, b, c (Å) | 42.5, 42.5, 120 |
| α, β, γ (deg) | 90, 90, 120 |
| Wavelength (Å) | 1.54 |
| Resolution range (Å) | 35.2–2.0 |
| Total reflections (n) | 28,380 |
| Unique reflections (n) | 5371 (444) |
| Completeness (%) | 97.9 (81.8) |
| R merge (%) | 2.6 (30.8) |
| Redundancy | 5.3 (2.5) |
| I/ σ (I) | 28.6 (2.7) |
| Refinement | |
| Molecules in asymmetric unit (n) | 1 |
| Residues (n) | 76 |
| Water molecules (n) | 7 |
| Chloride atoms (n) | 1 |
| R work/R free (%) | 20.7/23.7 |
| Average B factor (Å2) | |
| Overall | 38.0 |
| Protein | 38.1 |
| Solvent | 38.0 |
| RMSD bonds (Å) | 0.014 |
| RMSD angles (deg) | 1.36 |
| Ramachandran plot statistics (%) | |
| Most favored region | 97.3 |
| Additional allowed region | 2.7 |
| Disallowed region | 0.0 |
Values in parentheses refer to reflections in the outer resolution shell: 2.07–2.00 Å. R merge = ΣΣj |Ij – 〈I〉|/Σ |〈I〉|, where Ij and 〈I〉 represent the diffraction intensity values of the individual measurements and the corresponding mean values, and the summation is over all unique measurements. R work = (Σhkl ‖F obs| – k |F calc ‖)/(Σhkl |F obs|), where F obs and F calc are the observed and calculated structure factors, respectively. For R free, the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
Circular dichroism (CD) spectroscopy and thermal melts
Human T cell leukemia virus 1 (HTLV‐1) gp21 was expressed and purified as described previously (25, 34). ASLV TM and HTLV‐1 fusion proteins were buffer exchanged by size‐exclusion chromatography on a Superdex‐75 10/300 column into 10 mM potassium phosphate, 150 mM NaCl, and 0.05% (w/v) CHAPS at pH 6.5 and 7.5, respectively. CD spectra of wild‐type and mutant ASLV TM and HTLV‐1 gp21 (concentrations ranging from 0.2 to 2 mg/ml) were acquired on a Jasco J‐810 spectropolarimeter (Jasco, Inc., Easton, MD, USA) using 1 mm quartz cuvettes at 25°C. Data were collected between 190 and 250 nm and averaged over 5 accumulations. Raw data were then converted to molar ellipticity (deg cm2 dmol–1) for analysis. Thermal denaturation assays were carried out at a single wavelength (222 nm) by monitoring the change in ellipticity as a function of temperature (20–99°C) at a rate of 5°C/min. Thermal melts as a function of pH were performed under the following buffer conditions: pH 5.0–5.5, sodium acetate (NaOAc) buffer (10 mM NaOAc, 150 mM NaCl, and 0.05% w/v CHAPS); pH 6.0–7.5, potassium phosphate buffer (10 mM K2HPO4/KH2PO4, 150 mM NaCl, and 0.05% w/v CHAPS); pH 7.5–8.5, Tris‐HCl buffer (10 mM Tris‐HCl, 150 mM NaCl, and 0.05% w/v CHAPS). All thermal denaturation data were normalized between 0 (folded) and 1 (unfolded) and were fit to a nonlinear biphasic sigmoidal curve in GraphPad (GraphPad, San Diego, CA, USA).
RESULTS AND DISCUSSION
Overall structure of ASLV TM
The ASLV TM core is the first structure determined from the α‐retrovirus family. Each ASLV TM monomer consists of a long α‐helical N‐terminal segment (residues 455–489), a disulfide‐linked chain reversal (CR) region (residues 490–502), a tether (residues 503–508), and a short C‐terminal α‐helical segment (residues 509–525) (Fig. 1 A, B). Three ASLV TM monomers form a 24‐ × 24‐ × 60‐Å trimeric protomer that adopts the typical 6‐helix bundle conformation (Fig. 1 C) seen in the postfusion state of other class I viral fusion proteins (reviewed in refs. 3, 5, 35).
Figure 1.
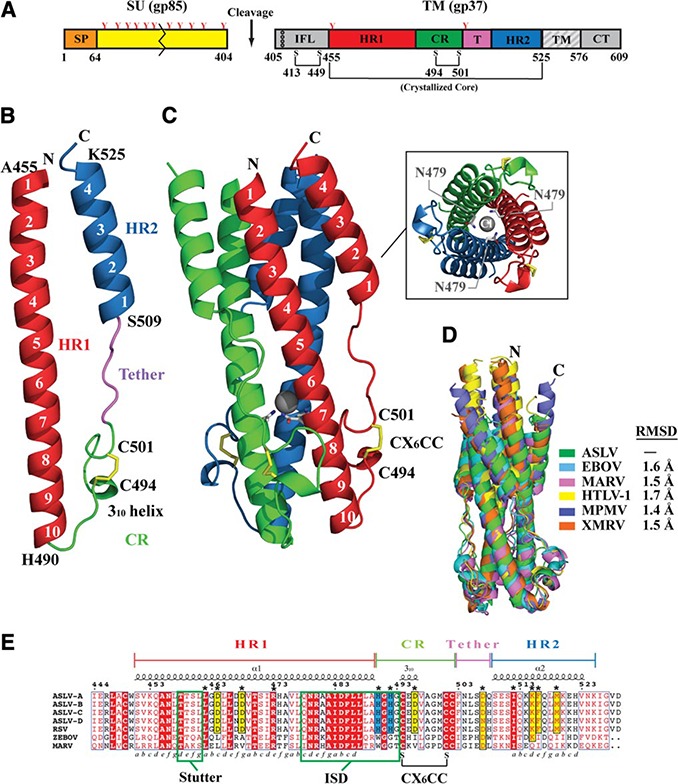
Structure of ASLV TM. A) Schematic of the ASLV Env. CR, chain reversal region; CT, cytoplasmic tail; HR1, heptad repeat 1 region; HR2, heptad repeat 2 region; IFL, internal fusion loop; SP, signal peptide; SU, surface attachment subunit; TM, transmembrane domain. Colored regions correspond to the ASLV TM core that was crystallized. Red Y‐shaped symbols denote N‐linked glycans. TM fusion subunit contains 3 disulfide linkages: one within the hydrophobic internal fusion loop, one in the chain‐reversal region, and an intermolecular covalent linkage between the SU and TM. B) Monomer of the ASLV TM. Features in the TM are color coded to the regions shown in panel A. C) Trimeric ASLV TM postfusion peplomer. The 3 ASLV TM monomers are shown in red, blue, and green. Inset: view of ASLV TM down the 3‐fold axis, showing the chloride ion bound by 3 aspargine residues within the inner HR1 core. D) Structural superimposition of ASLV TM and other retroviral and filoviral fusion subunits. PDB coordinate files used are as follows: ASLV, 4JPR; EBOV, 2EBO; MARV, 4G2K; HTLV‐1, 1MG1; MPMV, 4JF3; XMRV, 4JGS. E) Primary sequence alignment of ASLV TM subtypes A–D, Rous sarcoma virus (RSV) TM, EBOV GP2, and MARV GP2. The 3–4 periodicity of the HRs is shown below the alignment. Stutter region, immunosuppressive domain (ISD), and CX6CC motif are highlighted within labeled green boxes.
N‐terminal HR1 region
The N‐terminal segment of each ASLV TM monomer consists of a 10‐turn α‐helix; the polypeptide chain changes direction after this segment. The N‐terminal helix displays the typical HR motif (a, b, c d, e, f, g), where a and d positions are apolar residues that are located at the coiled coil interface, and e and g are solvent‐exposed polar residues (36, 37). The repeating 3‐4‐3‐4‐3‐4 periodicity of the HR allows the hydrophobic leucine, isoleucine, and valine residues to form the “knobs‐into‐holes” packing with a neighboring chain to form the trimeric core (36, 37). The hydrophobic knob‐into‐holes packing is interrupted by a single aspargine layer (Asn479) that interacts with a chloride ion within the inner core (Fig. 1 C). In addition, the ASLV TM N‐terminal HR1 region has a stutter‐phase shift that corresponds to a 4‐residue insertion, which results in core packing discontinuities and underwinding of the coiled‐coil core (residues 458–461). Similar stutters and chloride binding sites have also been noted in other members of the class I fusion proteins, such as those from lymphocytic choriomeningitis virus (LCMV), Ebola virus (EBOV), Marburg virus (MARV), and severe acute respiratory syndrome (SARS) virus (23, 26, 30, 38). Another feature of the ASLV TM is the similarity of its HR1 base region (residues 478–504) with a putative immunosuppressive domain found in EBOV and MARV (~50% sequence identity), and β‐, δ‐, and γ‐retroviruses (~36% sequence identity) (39). Synthetic peptides of the immunosuppressive motif from EBOV or MARV modulated the expression of interferon‐γ, IL‐2 and IL‐10 cytokines, lower CD4+ and CD8+ T‐cell activation, and increase immune cell apoptosis (40).
CR region
The CR region (residues 490–502) allows the protein to change direction to complete a 180° turn that places the polypeptide chain antiparallel to the N‐terminal HR1 helix. The CR region contains a random coil His‐Gly‐His‐Gly loop, a 310 helix, and a highly conserved CX6CC motif (residues 494–502). The CX6CC motif forms an internal disulfide bond (C494–C501) that stabilizes the loop containing the 310 helix. In the δ and γ‐retroviruses, such as HTLV‐1 and MoMLV, the SU subunit carries a CXXC thiol‐disulfide exchange motif that is involved in disulfide bond formation to the third cysteine in the CX6CC motif (41, 42). A thiol‐disulfide exchange induces isomerization of the SU‐TM disulfide bond to initiate fusion. In ASLV Env and filovirus glycoproteins, the SU subunit lacks the CXXC disulfide exchange motif, but the attachment subunit remains covalently linked to the TM. In ASLV, in lieu of the CXXC thiol–disulfide exchange motif (43), it was proposed that the low pH might trigger the structural rearrangements observed during viral fusion (5, 12, 43, 44).
Tether and C‐terminal HR2 region
Six residues (residues 503–508) in a random coil conformation act as a tether between the CR and the C‐terminal HR2 region. The C‐terminal HR2 region forms 4 helical turns that pack between two HR1 helices. The HR1‐HR2 interface is stabilized by hydrophobic interactions with residues Ile512, Phe516, Met519, and Val523. Moreover, Lys515 forms an intersubunit electrostatic ion‐pair with Asp464 in the N‐terminal HR1.
Structural similarity of ASLV TM with other retroviral and filoviral fusion subunits
Postfusion class I viral fusion subunits all adopt a 6‐helix bundle conformation. Side‐by‐side analysis of these structures reveals significant variation in the lengths of the HR1 and HR2 helices, presence of stutter regions, and surface electrostatic potential. Structural analysis previously demonstrated that the fusion machineries of β‐, δ‐, and γ‐retroviruses are similar to EBOV and MARV (refs. 25, 26, 45, and unpublished results). This finding led to the hypothesis that the retroviral and filoviral fusion subunits diverged from a common ancestral fusion protein. Not surprisingly, the ASLV TM also shares a strong overall structural similarity with transmembrane subunits of the β‐, δ‐, and γ‐retroviruses and the glycoproteins of EBOV and MARV (Fig. 1 D). A closer inspection, however, reveals differences in the CR region. In all characterized β‐, δ‐, and γ‐retroviral and filoviral fusion subunits, a 310 helix is formed directly after the central HR1 helix, which is followed by a critical glycine‐glycine (Gly‐Gly) linker and a disulfide bond‐stabilized single‐turn α‐helix in the CX6CC motif. Mutations to the Gly‐Gly motif in HTLV‐1 Env render the virus defective in fusion and prevent the formation of the 6‐helix bundle in the maltose‐binding protein‐gp21 chimera (25, 34). In the ASLV TM, no 310 helix is directly after the HR1 core, and the Gly‐Gly pair is interrupted by His490 and His492 to give a His‐Gly‐His‐Gly motif. Moreover, the short α‐helix observed in other retroviruses is replaced by a 310 helix. These features of the ASLV CR region, in particular the His‐Gly‐His‐Gly motif, are important for viral fusion (46) and are conserved in all ASLV subtypes (Fig. 1 E).
ASLV TM is stable over a wide pH spectrum, unlike other retroviral TMs
The 2‐step entry mechanism of ASLV requires that the envelope glycoprotein function at both neutral and low pH. We performed CD thermal denaturation assays to investigate the stability of the ASLV fusion subunit as a function of pH (pH 5.0 to 8.5). At neutral pH, the denaturation of ASLV TM was irreversible with a melting temperature (T m) of 67°C. This finding is consistent with the melting temperatures of other class I viral fusion proteins (47–49). Although thermodynamic parameters (i.e., ΔG° values) cannot be calculated from an irreversible thermal denaturation study, the apparent Tm provides a simple measure of protein stability. The ASLV TM was stable over a wide pH range (Fig. 2 A), with a slightly increased apparent melting temperature (ΔTm=2–5°C) at pH values corresponding to early (pH 6.0–6.5) and late (pH 5.0–6.0) endosomal stages of the viral life cycle. Considering the nature of the ASLV 2‐step entry mechanism, our results are in alignment with the biology of the virus. Initially, receptor binding of ASLV SU triggers conformational changes at the cell surface, where the ASLV TM structure is rearranged to form the extended prehairpin intermediate at neutral pH. Then, low pH conditions trigger additional conformational changes to form the hemifusion intermediate and postfusion 6‐helix bundle conformation. Given that ASLV TM must interact at both the plasma and endocytic membranes, the TM must be stable over a wide range of pH.
Figure 2.
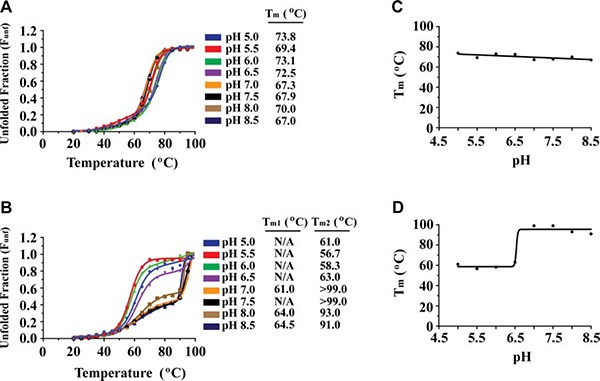
ASLV TM stability as a function of pH. A, B) CD thermal denaturation profiles of ASLV TM (A) and HTLV‐1 gp21 (B) at pH values between 5.0 and 8.5. CD signal was normalized between 0 (folded) and 1 (unfolded). C, D) Plot of Tm vs. pH for ASLV TM (C) and HTLV‐1 gp21 (D). ASLV TM is stable from pH 5.0 to pH 8.5, whereas HTLV‐1 gp21 is highly stable at pH > 7.0.
The stability of ASLV TM over a wide range of pH values is unique to this viral fusion protein. In contrast, fusion proteins of viruses that traffic to the endosome, such as influenza A, EBOV, and MARV, are most stable only at low pH (48–50). Previous studies of EBOV GP2 revealed a dramatic decrease in melting temperature (ΔTm=37°C) on a pH change from 5.3 to 6.1 (48). For viruses that fuse at the plasma membrane, such as the β‐, δ‐, and γ retroviruses, we have shown that the fusion subunits are most stable at neutral pH (Fig. 2 B). Thermal denaturation analysis of HTLV‐1 TM revealed that at values of pH < 7.0, the Tm decreased by >40°C relative to that at neutral pH, suggesting a dramatic destabilization of the fusion protein (Fig. 2 B). At values of pH > 7.0, HTLV‐1 gp21 was very stable and did not fully denature even at 99°C. In addition, the thermal melts above pH 7.0 had a biphasic nature, which was not seen in ASLV TM. The first transition (Tm1) likely corresponds to the denaturation of the HR2 helix and the second and larger transition (Tm2) to denaturation of the HR1 trimeric core. At values of pH < 7.0, a cooperative single transition was observed, suggesting that the HR2 region is not as tightly associated to the HR1 core. One explanation for the neutral‐pH‐dependent stability of HTLV‐1 gp21 is electrostatic interactions. Previous structural analysis of retroviral fusion subunits revealed that HTLV‐1 gp21 contains an extensive series of inter‐ and intrachain salt bridges between HR1 and HR2 regions (25, 51). The relative contribution of electrostatic interactions to protein stability predominates at neutral pH as both side chains are fully ionized (reviewed in ref. 52). However, as the virus is taken up into the low‐pH endosomes, the strength of the ion pair is thus reduced. Moreover, HTLV‐1 gp21 has 3 histidine residues (His348, His365, and His409) positioned in a hydrophobic cavity at the HR1‐HR2 interface. At endosomal pH, these histidine residues are protonated and the placement of an unpaired positive charge in a hydrophobic pocket may explain why HTLV‐1 gp21 is unstable at low pH. Taken together, our results are in excellent agreement with a neutral pH environment being required to mediate the host cell fusion, as low pH entry pathways are not a viable route for HTLV‐1 entry.
Electrostatic interactions are not critical for ASLV TM postfusion stability
The ASLV fusion protein contains 3 sets of salt bridges (Fig. 3 A): between the HR1 and HR2 helices, between neighboring HR1‐HR1 interfaces at the trimeric core, and in the CR region. Specifically, at the HR1‐HR2 interface, a single interchain electrostatic interaction is present between Lys515 on the HR2 helix and Asp464 on the HR1 trimeric core. In the HR1 trimeric core, an interchain salt bridge is located between two neighboring HR1‐HR1 helices. Arg473 is located centrally on HR1 and makes an electrostatic interaction with Asp468 in a neighboring HR1 helix. A third intrachain salt bridge is situated in the CR region between His492 and Asp496.
Figure 3.
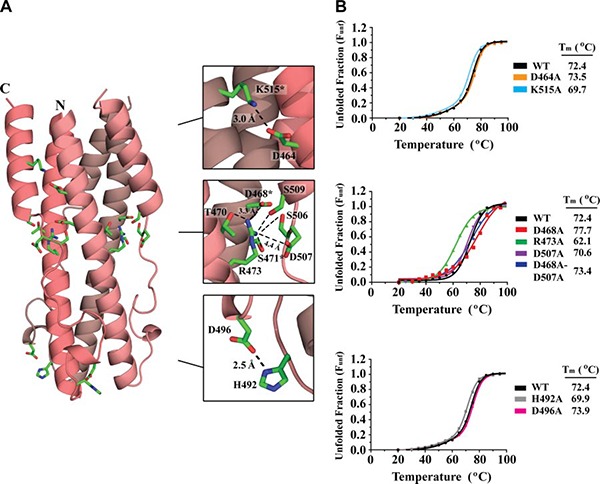
ASLV TM electrostatic interactions. A) Ribbon diagram of ASLV TM with ion‐pair interactions shown as green sticks. Each TM monomer is shown in a different shade of pink. Inset: zoomed view of electrostatic interactions between the HR1‐HR2 and HR1‐HR1 interfaces and the CR region. Distances between residues are shown in angstroms; asterisks indicate residues contributed by a neighboring molecule. B) CD thermal denaturation profiles of wild‐type and salt bridge ASLV TM mutants. CD signals were normalized between 0 (folded) and 1 (unfolded); melting temperatures are shown at right.
To understand the role of the ASLV TM salt bridges on protein stability, the positively and negatively charged residues were mutated to alanine residues, and the effects on protein stability were monitored by CD spectroscopy as a function of temperature. The CD wavelength scan indicated that the alanine mutations did not change the overall spectral characteristics of the protein, suggesting that the secondary structural elements remained intact. The Tm of the wild‐type ASLV TM was 72.4°C (Fig. 3 B). Interestingly, alanine mutations to the salt bridge pair that link the HR1 and HR2 helices (Asp464Ala and Lys515Ala) or the ion pair in the CR region (His492Ala and Asp496Ala) had little to no effect on the stability of the protein (ΔTm=3.0°C) (Fig. 3 B). This finding is in stark contrast to glycoproteins from other retroviruses, such as HTLV‐1 gp21, XMRV TM, and MPMV TM that have intricate networks of inter‐ and intrachain electrostatic interactions in the fusion subunit. Mutations to residues involved in electrostatic interactions between the HR1 and HR2 regions have profound effects (ΔTm>30°C) on fusion protein stability and viral entry (34, unpublished data).
An Asp468‐Arg473 ion pair located between HR1 helices within the ASLV TM trimeric core is unique. To our knowledge, no other class I fusion protein has an ion pair at the HR1 trimer interface. Typically, the interactions between HR1‐HR1 helices are largely mediated by hydrophobic forces. An alanine mutation to Arg473 resulted in a 10°C decrease in the apparent Tm compared to wild‐type protein, suggesting Arg473 plays a strong role in ASLV TM stability. However, an alanine mutation to Asp468, the ion‐pair partner to Arg473, stabilized the fusion protein with a 5°C increase in melting temperature (Fig. 3 B). We believe that the increase in the melting temperature is due to the formation of a new salt bridge to Asp507 located on the outer HR2 helix. This new ion pair will stabilize the inner core (HR1) to the outer layer (HR2) of the 6‐helix bundle, thus leading to the observed increased Tm. An alanine mutation to Asp507, as expected, did not affect the Tm, indicating that this residue is not involved in stabilizing the fusion protein. We decided to make a double mutation encompassing both Asp468Ala and Asp507Ala. The removal of all the negatively charged partners to Arg473 should decrease the stability of the fusion subunit. However, the Asp468Ala‐Asp507Ala double mutant revealed no major changes in apparent Tm compared to wild‐type (Fig. 3 B). This strongly suggests that the HR1‐HR1 ion‐pair does not play a role in stabilizing the inner trimeric core. Instead, Arg473 is likely involved in stabilizing the HR1‐HR2 interface through a network of hydrogen bonds made between Arg473 to Ser471, Ser506, and Ser509 (Fig. 3 A). Overall, our data suggest that electrostatic interactions between HR1‐HR1 and HR1‐HR2 regions of the ASLV TM fusion subunit are not critical for the stability of the postfusion 6‐helix bundle structure.
Hydrophobic interactions between the HR1 and HR2 regions stabilize the postfusion conformation
Considering the minimal contribution of ASLV TM electrostatic interactions to the stability of the fusion subunit, we wanted to investigate the role of hydrophobic residues that line the HR1‐HR2 interface (Fig. 4 A). We mutated Leu461, Ile512, Phe516, and Met519 residues at the HR1‐HR2 interface to an alanine. Mutations on 3 hydrophobic residues within the HR2 helical region (Ile512, Phe516, and Met519) decreased the stability by 5–7°C compared to wild‐type, whereas the Leu461Ala mutation on HR1 destabilized ASLV TM substantially by reducing the melting temperature by >10°C (Fig. 4 B). Leu461 is located on HR1 and makes van der Waals interactions with Met519 and Phe516 from HR2. Previous viral entry assays identified that a single alanine mutation to Leu461 significantly hinders the infectivity of the virus (53). The importance of hydrophobic residues in stabilizing the fusion protein is consistent with the nature of the ASLV lifecycle. Hydrophobic interactions are independent of pH, and thus are able to stabilize the postfusion state in a broad range of pH environments.
Figure 4.
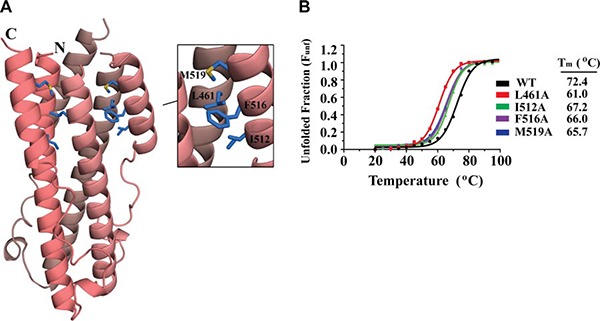
ASLV TM hydrophobic interactions. A) Ribbon diagram of ASLV TM with hydrophobic interactions between the HR1‐HR2 regions. Each TM monomer is shown in a different shade of pink. Inset: zoomed view of the hydrophobic interactions (blue sticks). B) CD thermal denaturation profiles of wild‐type and hydrophobic ASLV TM mutants. CD signals were all normalized between 0 (folded) and 1 (unfolded); melting temperatures are shown at right.
Histidine residues in the CR region stabilize a negative helix dipole at low pH
Two conserved histidine residues (His490 and His492) are at the CR region of the ASLV TM. His490 is positioned immediately after the HR1 region following a short stretch of hydrophobic residues (FLLLA). The side chain of the His490 faces into the center of the hydrophobic inner trimeric TM core (Fig. 5 A). In contrast, His492 faces out into the solvent and forms an intrachain ion pair with Asp496. To investigate the role of these histidine residues for the postfusion conformation of the protein, we mutated His490 and His492 to alanine residues, and monitored the melting temperatures of the ASLV TM mutants as a function of pH (pH 5.0–8.0). Neither His490Ala nor His492Ala had significantly different thermal stabilities from that of the wild‐type protein at pH 6.5, 7.5, or 8.0 (Fig. 5 B). However, at pH 5.0, mutation of either His490 or His492 to alanine significantly destabilized the ASLV TM by ~20°C compared to the wild‐type TM. Mutation of His490 had the larger effect on the stability of the protein, decreasing the Tm below 50°C at pH 5.0, whereas mutation of His492 reduced the melting temperature to 56°C. Delos et al. (46) previously showed that alanine substitutions to His490 or to His492 did not hinder the membrane association of the TM during the pH‐independent, receptor‐mediated first step of viral fusion, but inhibited the entry of infectious ASLV particles for complete fusion. Subsequently, their pH profiling studies on CR histidines demonstrated that although His490Ala was not significantly different from wild‐type, but His492Ala shifted the pH required for activation by 0.6 pH units (46). Delos et al. (46) suggested that His492 acts as a trigger to release structural constraints on the fusion subunit on encountering a low‐pH environment.
Figure 5.

ASLV TM CR region histidine residues are important for stability at low pH. A) Left panels: ribbon diagrams of ASLV TM, HTLV‐1 gp21, IAV HA2, and EBOV GP2. Molecules are viewed down the 3‐fold axis with the CR region facing toward the viewer. Right panels: electrostatic potential mapped onto the molecular surface. Red and blue regions denote negative and positive charges, respectively. Positively charged residues that stabilize the negative helix dipole moment are highlighted as green sticks. B) CD thermal denaturation profiles of ASLV TM His490Ala, His490Arg, His492Ala, His492Glu, and HTLV‐1 gp21 Lys394His at pH 8.0, 7.5, 6.5, and 5.0. CD signal was normalized between 0 (folded) and 1 (unfolded).
Our results show that both His490 and His492 play a major role for the stability of the ASLV TM postfusion conformation. We hypothesize that His490 and His492 are important in stabilizing a helix dipole moment on the fusion subunit. From prefusion and postfusion crystal structures of the EBOV and IAV envelope glycoproteins (24, 26, 54–56), it is clear that, on triggering, the HR1 region remodels into a single long α‐helix. As the HR1 α‐helix extends, the alignment of individual dipoles from the carbonyl groups along the helical axis place increasing net positive and negative charges at the N‐ and C‐terminal poles of the helix, respectively (57, 58). The negatively and positively charged helix dipole moments are stabilized by oppositely charged end‐capping residues (57, 59, 60). Unbalanced helix dipole moments have been shown to destabilize helical bundles (57, 58, 61). Calculation of the electrostatic potential on the surface of the ASLV TM reveals negatively and positively charged regions at the poles of the 6‐helix bundle. At low pH, the ASLV TM histidine residues will have a positive charge and, due to their positions at the base of the central HR1 trimeric helix, are in excellent position to stabilize the negative dipole moment (Fig. 5 A). It is not clear whether any residues are present that cap the positively charged dipole moment, as most crystal structures lack the fusion peptide and transmembrane anchor regions of the fusion subunit.
Helix dipole moments are strongest in pH‐dependent viral fusion proteins
All class I viral fusion proteins adopt an α‐helical postfusion structure to overcome the large energy barrier of the viral and host cell membrane merger (5). To our knowledge, the role of a helix dipole moment on postfusion protein stability has not been investigated for viral class I fusion proteins. Currently, representative models exist in the PDB of viral fusion proteins from viruses that undergo either a pH‐dependent or pH‐independent entry mechanism. ASLV, LCMV, and IAV are well‐characterized viruses that require low pH for activation, whereas retroviruses are prototypical viruses for pH‐independent entry (12, 50, 62–66). Assessment of these structures revealed that the helix dipole moments of the viral fusion subunits from three different viral families (ASLV, LCMV, and IAV) appear to have a three‐ to four‐fold larger dipole moment compared to other retroviral (HTLV‐1, BLV, MoMLV, and HERV‐FRD) and filoviral (EBOV and MARV) fusion proteins (Table 2 ). From our analysis, a trend exists where viruses undergoing a pH‐dependent activation (ASLV, LCMV, and IAV) have a stronger helix dipole moment than those that enter through a pH‐independent mechanism (MoMLV, HERV‐FRD, HTLV‐1, and BLV). The filovirus (EBOV and MARV) GP2 structures are an exception to this helix dipole trend. However, Ebola virus is a rather unique pH‐dependent virus. Whereas other pH‐dependent viruses utilize the low‐pH environment of the endosome to activate its viral glycoprotein for fusion, the low‐pH requirement for EBOV entry is linked to the activation of a cellular protease to cleave EBOV GP for fusion (67–69). Moreover, the EBOV and MARV GP2 structures are highly similar to the retroviral fusion subunits (Fig. 1 D). Therefore, while EBOV GP2 is classified as a pH‐dependent virus, its viral fusion protein may have characteristics, including its helix dipole properties, more similar to the pH‐independent viruses. It has been shown that electrostatic interactions between protonated histidine and a negative helix dipole are more effective at low pH (59).
Table 2.
Calculated dipole moments of various trimeric viral fusion TM proteins
| PDB code | Virus | Viral family | Atoms (n)a | Residue length of HR1/HR2 | Dipole moment (Debye)a |
|---|---|---|---|---|---|
| pH‐dependent fusion | |||||
| 4JPR | ASLV | α‐Retroviridae | 687 | 39/16 | 621 |
| 3MK0 | LCMV | Arenaviridae | 875 | 48/14 | 850 |
| 1HTM | IAV | Orthomyxoviridae | 989 | 65/17 | 802 |
| 2EBO | EBOV | Filoviridae | 601 | 36/14 | 261 |
| 4G2K | MARV | Filoviridae | 662 | 41/15 | 262 |
| pH‐independent fusion | |||||
| 1MG1 | HTLV‐1 | δ‐Retroviridae | 663 | 46/8 | 176 |
| 2XZ3 | BLV | δ‐Retroviridae | 746 | 52/8 | 232 |
| 1Y4M | HERV‐FRD | γ‐Retroviridae | 399 | 31/0b | 275 |
| 1MOF | MoMLV | γ‐Retroviridae | 418 | 33/0b | 210 |
aCalculated for a single‐fusion TM monomer. bHR2 helix is disordered in the crystal structure.
Stabilization of a significant helix dipole moment may provide a contribution to the energetics for host and viral membrane fusion.
Insights into the general mechanisms of viral fusion
While significant diversity exists among viral fusion proteins in terms of architecture, size, and physiological properties, the mechanisms of viral fusion are generally conserved. The two‐step nature of ASLV allows us to reevaluate and compare the existing crystal structures of viral fusion proteins to identify general trends and features required for fusion (Fig. 6 ).
Figure 6.
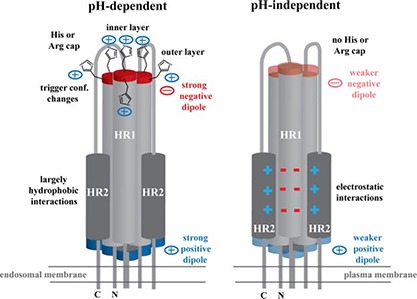
Summary of pH‐dependent and pH‐independent fusion protein properties. Cartoon shows the viral fusion subunit in the postfusion conformation. Viruses that require low pH for entry, such as ASLV, IAV, and LCMV, fuse at the endosomal membrane and have strong positive and negative charges at their ends due to its helix dipole moment. Negatively charged helix dipole is stabilized by two layers of positively charged histidine or arginine capping residues (only histidine is shown). In addition, the outer layer of histidine residues may be involved in triggering conformational changes in the prefusion viral glycoprotein on entering a low‐pH environment. HR1‐HR2 interface is in general mediated by largely hydrophobic interactions. Viruses that undergo a pH‐independent entry process, such as retroviruses, fuse at the plasma membrane. The helix dipole moment is decreased, and no histidine or arginine residues are located in the CR region to stabilize the negative helix dipole. Moreover, the HR1‐HR2 interface is stabilized with a larger number of electrostatic interactions rather than hydrophobic forces.
Fusion proteins are most stable at the pH of the site of fusion
In general, the stability of the fusion proteins tends to mimic the environment where they fuse. For example, it was recently shown that EBOV fusion occurs at an endolysosome, where the pH is lower than 5.0 (70). Interestingly, the EBOV and MARV GP2 fusion subunits are most stable at values of pH < 5.5 (48, 49). This trend is also seen in viruses that fuse at the plasma membrane, such as HTLV‐1, where its fusion protein (gp21) is most stable at neutral pH. In ASLV TM, where the virus undergoes a 2‐step entry mechanism at the plasma membrane followed by endosomal activation, the TM is equally stable at low and neutral pHs.
Electrostatic charges are important to the stability of pH‐independent viral TMs
The β‐, δ‐, and δ‐retroviruses are well known to undergo a fusion process at the plasma membrane at neutral pH (62, 63). Structures of the β‐, δ‐, and γ‐retroviral fusion subunits reveal a clustering of electrostatic interactions at the HR and CR regions (ref. 34 and unpublished results). The presence of salt bridges on pH‐independent viral TMs aligns with the life cycle of retroviruses, as the electrostatic interactions will provide the utmost stability at neutral pH. Biophysical and mutagenic studies revealed that the HR1‐HR2 interface and CR regions of these retroviral TMs are strongly stabilized by its electrostatic interactions (unpublished results). Moreover, previous reports showed that the alanine mutations to these residues resulted in melting temperature changes of >30°C and rendered the virus incapable of entry (34). For instance, Maerz et al. (34) identified a CR region salt bridge (Arg380 and Glu398) on HTLV‐1 gp21, and an asparagine substitution on Glu398 residue reduced the fusion activity by more than 5‐fold compared to wild‐type. Similar results were obtained from a previously identified HIV‐1 salt bridge (Lys574 and Asp632) and inhibitors binding to a hydrophobic pocket that interrupts this salt bridge were shown to hinder the cellular entry of infectious HIV‐1 particles (71–74). As mentioned previously, the ASLV fusion subunit also displays a series of salt bridges between its HR1 and HR2 helices. Despite ASLV being a retrovirus, its fusion protein has evolved to downplay the requirement of these electrostatic interactions on the stability of the postfusion 6‐helix bundle. The ASLV HR1‐HR2 interface is instead stabilized through hydrophobic interactions, in line with viruses that undergo a low pH‐dependent entry. Therefore, viral fusion proteins that mediate entry at the plasma membrane tend to display salt bridges in its HR and CR regions, whereas viruses entering through an endosomal pathway tend to utilize nonionizable hydrophobic residues for stability.
Positive charge clusters in the CR region of pH‐dependent viral TMs
Structural analysis of class I fusion proteins from viruses that require low pH for entry, such as influenza A virus HA2, EBOV GP2, MARV GP2, and LCMV GP2, reveals a layering of outer and inner positive charges at the HR1 base or CR region (Fig. 5 A). In ASLV TM, IAV HA2, EBOV GP2 MARV GP2, and LCMV GP2, a histidine residue is present in an outer layer on the fusion protein. In EBOV GP2 and MARV GP2, the outer histidine is conserved structurally with ASLV TM His492, suggesting an important role for the outer histidine. In IAV HA2, the outer histidine (His142) is suggested to act as a pH sensor to trigger conformational changes at low pH (75). Based on the prefusion EBOV GP structure (55), His602 is at the GP1‐GP2 interface, thus supporting a role in also activating the glycoprotein for fusion. Histidine residues are ideal low‐pH sensors, as the pKa of its imidazole side chain is 6.0. Thus, at the pH of an early or late endosome, histidine residues become protonated to trigger conformational changes. However, in addition to its potential role in conformational changes, the outer layer positive charge may be involved in stabilizing the negative helix dipole moment. Mutation of ASLV TM His492Ala and the addition of a negative charge (His492Glu) decreased the apparent Tm by >20°C at low pH (Fig. 5 B), suggesting that the presence of a positive charge on the histidine residue is important for stabilization of the fusion subunit. In contrast, the incorporation of a histidine residue on the outer layer of HTLV‐1 gp21 (Lys394His), as expected, destabilized the fusion protein at all pH values (Fig. 5 B). Due to the weak dipole moment of HTLV‐1 gp21, a protonated histidine residue did not increase the stability at low pH. At pH values >7.0, a neutral histidine residue destabilized the fusion subunit, suggesting that a positive charge in the outer layer is important for postfusion HTLV‐1 gp21 stability. In summary, the outer layer positive charge may play two roles: triggering the conformational changes for fusion, and stabilizing the negatively charged helix dipole in the postfusion 6‐helix bundle.
Inner layer of positive charge is important in stabilizing the negative dipole moment
A positive charge is also commonly found on an inner layer of the fusion subunit. In ASLV TM, IAV HA2, EBOV GP2 MARV GP2, and LCMV GP2, a positive charge is found on an inner layer of the HR1 core. In IAV HA2 and ASLV TM, His106 and His490 reside at the base of the HR1 core, respectively. Histidine residues located at the C‐terminus of a helix have been commonly shown to interact with a helix dipole to increase protein stability through charge‐dipole interactions (57, 59, 60). In EBOV GP2, MARV GP2, and LCMV GP2, an arginine residue is located at the base of HR1 in a position similar to His490 in ASLV TM. The substitution of His490Arg in ASLV TM resulted in maintained thermal stability (Tm>70°C) over a wide pH range (Fig. 5 B). Our results show that a positively charged arginine residue can also stabilize the postfusion conformation of viral fusion proteins at low pH. Regardless of the virus and type of amino acid, the positive charge at the base of the helical core can counterbalance the negative helix dipole moment. For the fusion proteins of pH‐independent viruses, such as HTLV‐1 and MoMLV, no inner layer histidine or positively charged residues exist. Given that the pH‐independent viral fusion subunits have a smaller negative helix dipole moment, end capping of the helix dipole moment may not be necessary for this class of fusion proteins. In summary, a basic residue is likely required on an inner layer of the HR1 core to counterbalance the negative helix dipole moment and stabilize the postfusion 6‐helix bundle for viral fusion proteins that encounter a low pH environment.
The research was supported in part by University of Toronto startup funds, a Canada Research Chair in Structural Virology, a Canadian Institutes of Health Research (CIHR) open operating grant (MOP‐115066), and a New Investigator award (MSH‐113554) to J.E.L. H.A. was supported by a University of Toronto graduate fellowship, and B.M.S. was funded by an Ontario Genomics Institute Summer Research Fellowship and Canadian Society for Immunology Summer Internship Award. The authors thank the Molecular Structure and Function program at the Hospital for Sick Children Research Institute (Toronto, ON, Canada) for generous access to a circular dichroism spectropolarimeter, and Aiping Dong, Dr. Aled Edwards, and Dr. Cheryl Arrowsmith (Structural Genomics Consortium, Toronto, ON, Canada) for access to the X‐ray diffraction facility. ASLV TM crystal structure and structure factors have been deposited into the Protein Data Bank (accession code 4JPR).
Aydin, H. , Smrke, B.M., Lee, J. E. Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2‐step entry mechanism. FASEB J. 27, 5059–5071 (2013). http://www.fasebj.org
REFERENCES
- 1. Harrison, S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weissenhorn, W. , Dessen, A. , Calder, L. J. , Harrison, S. C. , Skehel, J. J. , and Wiley, D. C. (1999) Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 3. Plemper, R. K. (2011) Cell entry of enveloped viruses. Curr. Opin. Virol. 1, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dimitrov, D. S. (2004) Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. White, J. M. , Delos, S. E. , Brecher, M. , and Schornberg, K. (2008) Structures and mechanisms of viral membrane fusion proteins. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Markovic, I. , Leikina, E. , Zhukovsky, M. , Zimmerberg, J. , and Chernomordik, L. V. (2001) Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 155, 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bentz, J. (2000) Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 78, 886–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernandez, L. D. , Hoffman, L. R. , Wolfsberg, T. G. , and White, J. M. (1996) Virus‐cell and cell‐cell fusion1. Ann. Rev. Cell. Dev. Biol. 12, 627–661 [DOI] [PubMed] [Google Scholar]
- 9. Morizono, K. , and Chen, I. S. (2011) Receptors and tropisms of envelope viruses. Curr. Opin. Virol. 1, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skehel, J. J. , and Wiley, D. C. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Ann. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 11. Lee, K. K. (2010) Architecture of a nascent viral fusion pore. EMBO J. 29, 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mothes, W. , Boerger, A. L. , Narayan, S. , Cunningham, J. M. , and Young, J. A. T. (2000) Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103, 679–689 [DOI] [PubMed] [Google Scholar]
- 13. Barnard, R. J. O. , Narayan, S. , Dornadula, G. , Miller, M. D. , and Young, J. A. T. (2004) Low pH is required for avian sarcoma and leukosis virus env‐dependent viral penetration into the cytosol and not for viral uncoating. J. Virol. 78, 10433–10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barnard, R. J. O. , Elleder, D. , and Young, J. A. T. (2006) Avian sarcoma and leukosis virus‐receptor interactions: From classical genetics to novel insights into virus–cell membrane fusion. Virology 344, 25–29 [DOI] [PubMed] [Google Scholar]
- 15. Damico, R. , Rong, L. , and Bates, P. (1999) Substitutions in the receptor‐binding domain of the avian sarcoma and leukosis virus envelope uncouple receptor‐triggered structural rearrangements in the surface and transmembrane subunits. J. Virol. 73, 3087–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delos, S. E. , Godby, J. A. , and White, J. M. (2005) Receptor‐induced conformational changes in the SU subunit of the avian sarcoma/leukosis virus A envelope protein: implications for fusion activation. J. Virol. 79, 3488–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bates, P. , Young, J. A. , and Varmus, H. E. (1993) A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 18. Earp, L. J. , Delos, S. E. , Netter, R. C. , Bates, P. , and White, J. M. (2003) The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77, 3058–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith, J. G. , Mothes, W. , Blacklow, S. C. , and Cunningham, J. M. (2004) The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 78, 1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilbert, J. M. , Hernandez, L. D. , Balliet, J. W. , Bates, P. , and White, J. M. (1995) Receptor‐induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 69, 7410–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuyama, S. , Delos, S. E. , and White, J. M. (2004) Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78, 8201–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melikyan, G. B. , Barnard, R. J. O. , Markosyan, R. M. , Young, J. A. T , and Cohen, F. S. (2004) Low pH is required for avian sarcoma and leukosis virus env‐induced hemifusion and fusion pore formation but not for pore growth. J. Virol 78, 3753–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Igonet, S. , Vaney, M.‐C. , Vonhrein, C. , Bricogne, G. , Stura, E. A. , Hengartner, H. , Eschli, B. , and Rey, F. A. (2011) X‐ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl Acad. Sci. U. S. A. 108, 19967–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bullough, P. A. , Hughson, F. M. , Skehel, J. J. , and Wiley, D. C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 [DOI] [PubMed] [Google Scholar]
- 25. Kobe, B. , Center, R. J. , Kemp, B. E. , and Poumbourios, P. (1999) Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose‐binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. U. S. A. 96, 4319–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weissenhorn, W. , Carfí, A , Lee, K‐H , Skehel, J. J. , and Wiley, D. C. (1998) Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 2, 605–616 [DOI] [PubMed] [Google Scholar]
- 27. Weissenhorn, W. , Dessen, A. , Harrison, S. C. , Skehel, J. J. , and Wiley, D. C. (1997) Atomic structure of the ectodomain from HIV‐1 gp41. Nature 387, 426–430 [DOI] [PubMed] [Google Scholar]
- 28. Pflugrath, J. W. (1999) The finer things in X‐ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 29. McCoy, A. J. , Grosse‐Kunstleve, R. W. , Adams, P. D. , Winn, M. D. , Storoni, L. C. , and Read, R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fass, D. , Harrison, S. C. , and Kim, P. S. (1996) Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3, 465–469 [DOI] [PubMed] [Google Scholar]
- 31. Emsley, P. , and Cowtan, K. (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Adams, P. D. , Afonine, P. V. , Bunkóczi, G. , Chen, V. B. , Davis, I. W. , Echols, N. , Headd, J. J. , Hung, L.‐W. , Kapral, G. J. , Grosse‐Kunstleve, R. W. , McCoy, A. J. , Moriarty, N. W. , Oeffner, R. , Read, R. J. , Richardson, D. C. , Richardson, J. S. , Terwilliger, T. C. , and Zwart, P. H. (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felder, C. E. , Prilusky, J. , Silman, I. , and Sussman, J. L. (2007) A server and database for dipole moments of proteins. Nucleic Acids Res. 35, W512–W521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maerz, A. L. , Center, R. J. , Kemp, B. E. , Kobe, B. , and Poumbourios, P. (2000) Functional implications of the human T‐lymphotropic virus type 1 transmembrane glycoprotein helical hairpin structure. J. Virol. 74, 6614–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kielian, M. , and Rey, F. A. (2006) Virus membrane‐fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown, J. H. , Cohen, C. , and Parry, D. A. D. (1996) Heptad breaks in α‐helical coiled coils: Stutters and stammers. Proteins Struct. Funct. Bioinfo. 26, 134–145 [DOI] [PubMed] [Google Scholar]
- 37. Mason, J. M. , and Arndt, K. M. (2004) Coiled coil domains: stability, specificity, and biological implications. ChemBioChem 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 38. Duquerroy, S. , Vigouroux, A. , Rottier, P. J. M. , Rey, F. A. , and Bosch, B. J. (2005) Central ions and lateral asparagine/glutamine zippers stabilize the post‐fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology 335, 276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gallaher, W. R. (1996) Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85, 477–478 [DOI] [PubMed] [Google Scholar]
- 40. Yaddanapudi, K. , Palacios, G. , Towner, J. S. , Chen, I. , Sariol, C. A. , Nichol, S. T. , and Lipkin, W. I. (2006) Implication of a retrovirus‐like glycoprotein peptide in the immunopathogenesis of Ebola and Marburg viruses. FASEB J. 20, 2519–2530 [DOI] [PubMed] [Google Scholar]
- 41. Wallin, M. , Ekström, M. , and Garoff, H. (2004) Isomerization of the intersubunit disulphide‐bond in Env controls retrovirus fusion. EMBO J. 23, 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li, K. , Zhang, S. , Kronqvist, M. , Wallin, M. , Ekström, M. , Derse, D. , and Garoff, H. (2008) Intersubunit disulfide isomerization controls membrane fusion of human T‐cell leukemia virus. Env. J. Virol. 82, 7135–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li, K. , Zhang, S. , Kronqvist, M. , Ekström, M. , Wallin, M. , and Garoff, H. (2007) The conserved His8 of the Moloney murine leukemia virus Env SU subunit directs the activity of the SU–TM disulphide bond isomerase. Virology 361, 149–160 [DOI] [PubMed] [Google Scholar]
- 44. Babel, A. R. , Bruce, J. , and Young, J. A. T. (2007) The hr1 and fusion peptide regions of the subgroup B avian sarcoma and leukosis virus envelope glycoprotein influence low pH‐dependent membrane fusion. PLoS ONE 2, e171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koellhoffer, J. F. , Malashkevich, V. N. , Harrison, J. S. , Toro, R. , Bhosle, R. C. , Chandran, K. , Almo, S. C. , and Lai, J. R. (2012) Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry 51, 7665–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delos, S. E. , La, B. , Gilmartin, A. , and White, J. M. (2010) Studies of the “chain reversal regions” of the avian sarcoma/leukosis virus (ASLV) and Ebola virus fusion proteins: analogous residues are important, and a His residue unique to EnvA affects the pH dependence of ASLV entry. J. Virol. 84, 5687–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weissenhorn, W. , Wharton, S. A. , Calder, L. J. , Earl, P. L. , Moss, B. , Aliprandis, E. , Skehel, J. J. , and Wiley, D. C. (1996) The ectodomain of HIV‐1 env subunit gp41 forms a soluble, alphahelical, rod‐like oligomer in the absence of gp120 and the N‐terminal fusion peptide. EMBO J. 15, 1507–1514 [PMC free article] [PubMed] [Google Scholar]
- 48. Harrison, J. S. , Higgins, C. D. , Chandran, K. , and Lai, J. R. (2011) Designed protein mimics of the Ebola virus glycoprotein GP2 α‐helical bundle: stability and pH effects. Prot. Sci. 20, 1587–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harrison, J. S. , Koellhoffer, J. F. , Chandran, K. , and Lai, J. R. (2012) Marburg virus glycoprotein GP2: pH‐dependent stability of the ectodomain α‐helical bundle. Biochemistry 51, 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wiley, D. C. , and Skehel, J. J. (1987) The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann. Rev. Biochem. 56, 365–394 [DOI] [PubMed] [Google Scholar]
- 51. Lamb, D. , Schüttelkopf, A. W. , van Aalten, D. M. F. , and Brighty, D. W. (2011) Charge‐surrounded pockets and electrostatic interactions with small ions modulate the activity of retroviral fusion proteins. PLoS Pathog. 7, e1001268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bosshard, H. R. , Marti, D. N. , and Jelesarov, I. (2004) Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recog. 17, 1–16 [DOI] [PubMed] [Google Scholar]
- 53. Netter, R. C. , Amberg, S. M. , Balliet, J. W. , Biscone, M. J. , Vermeulen, A. , Earp, L. J. , White, J. M. , and Bates, P. (2004) Heptad repeat 2‐based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J. Virol. 78, 13430–13439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malashkevich, V. N. , Schneider, B. J. , McNally, M. L. , Milhollen, M. A. , Pang, J. X. , and Kim, P. S. (1999) Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9‐Å resolution. Proc. Natl. Acad. Sci. U. S. A. 96, 2662–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee, J. E. , Fusco, M. L. , Hessell, A. J. , Oswald, W. B. , Burton, D. R. , and Saphire, E. O. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson, I. A. , Skehel, J. J. , and Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. 289, 366–373, 1981. [DOI] [PubMed] [Google Scholar]
- 57. Sǎli, D. , Bycroft, M. , and Fersht, A. R. (1988) Stabilization of protein structure by interaction of α‐helix dipole with a charged side chain. Nature 335, 740–743 [DOI] [PubMed] [Google Scholar]
- 58. Sengupta, D. , Behera, R. N. , Smith, J. C. , and Ullmann, G. M. (2005) The α helix dipole: screened out? Structure 13, 849–855 [DOI] [PubMed] [Google Scholar]
- 59. Sancho, J. , Serrano, L. , and Fersht, A. R. (1992) Histidine residues at the N‐ and C‐termini of alpha‐helixes: perturbed pKas and protein stability. Biochemistry 31, 2253–2258 [DOI] [PubMed] [Google Scholar]
- 60. Prévost, M. (1996) Concurrent interactions contribute to the raised pKaof His18 in barnase. J. Mol. Biol. 260, 99–110 [DOI] [PubMed] [Google Scholar]
- 61. Ben‐Tal, N. , and Honig, B. (1996) Helix‐helix interactions in lipid bilayers. Biophys. J. 71, 3046–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McClure, M. O. , Sommerfelt, M. A. , Marsh, M. , and Weiss, R. A. (1990) The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71, 767–773 [DOI] [PubMed] [Google Scholar]
- 63. Côté, M. , Zheng, Y.‐M. , Liu, S.‐L. (2012) Membrane fusion and cell entry of XMRV are pH‐independent and modulated by the envelope glycoprotein's cytoplasmic tail. PLoS ONE 7, e33734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rojek, J. M. , Perez, M. , and Kunz, S. (2008) Cellular entry of lymphocytic choriomeningitis virus. J. Virol. 82, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quirin, K. , Eschli, B. , Scheu, I. , Poort, L. , Kartenbeck, J. , and Helenius, A. (2008) Lymphocytic choriomeningitis virus uses a novel endocytic pathway for infectious entry via late endosomes. Virology 378, 21–33 [DOI] [PubMed] [Google Scholar]
- 66. Doms, R. W. , and Helenius, A. (1986) Quaternary structure of influenza virus hemagglutinin after acid treatment. J. Virol. 60, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaletsky, R. L. , Simmons, G. , and Bates, P. (2007) Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81, 13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schornberg, K. , Matsuyama, S. , Kabsch, K. , Delos, S. , Bouton, A. , and White, J. (2006) Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80, 4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chandran, K. , Sullivan, N. J. , Felbor, U. , Whelan, S. P. , and Cunningham, J. M. (2005) Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Carette, J. E. , Raaben, M. , Wong, A. C. , Herbert, A. S. , Obernosterer, G. , Mulherkar, N. , Kuehne, A. I. , Kranzusch, P. J. , Griffin, A. M. , Ruthel, G. , Cin, P. D. , Dye, J. M. , Whelan, S. P. , Chandran, K. , and Brummelkamp, T. R. (2011) Ebola virus entry requires the cholesterol transporter Niemann‐Pick C1. Nature 477, 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Eckert, D. M. , Malashkevich, V. N. , Hong, L. H. , Carr, P. A. , and Kim, P. S. (1999) Inhibiting HIV‐1 entry: discovery of D‐peptide inhibitors that target the gp41 coiled‐coil pocket. Cell 99, 103–115 [DOI] [PubMed] [Google Scholar]
- 72. Liu, K. , Lu, H. , Hou, L. , Qi, Z. , Teixeira, C. , Barbault, F. , Fan, B.‐T. , Liu, S. , Jiang, S. , and Xie, L. (2008) Design, synthesis, and biological evaluation of N‐carboxyphenylpyrrole derivatives as potent HIV fusion inhibitors targeting gp41. J. Med. Chem. 51, 7843–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Katritzky, A. R. , Tala, S. R. , Lu, H. , Vakulenko, A. V. , Chen, Q‐Y. , Sivapackiam, J. , Pandya, K. , Jiang, S. , and Debnath, A. K. (2009) Design, synthesis, and structure‐activity relationship of a novel series of 2‐Aryl 5‐(4‐oxo‐3‐phenethyl‐2‐thioxothiazolidinylidenemethyl)furans as HIV‐1 entry inhibitors. J. Med. Chem. 52, 7631–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang, H. , Qi, Z. , Guo, A. , Mao, Q. , Lu, H. , An, X. , Xia, C. , Li, X. , Debnath, A. K. , Wu, S. , Liu, S. , and Jiang, S. (2009) ADS‐J1 inhibits human immunodeficiency virus type 1 entry by interacting with the gp41 pocket region and blocking fusion‐active gp41 core formation. Antimicrob. Agents Chemother. 53, 4987–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kampmann, T. , Mueller, D. S. , Mark, A. E. , Young, P. R. , and Kobe, B. (2006) The role of histidine residues in low‐pH‐mediated viral membrane fusion. Structure 14, 1481–1487 [DOI] [PubMed] [Google Scholar]


