Abstract
Like many other RNA viruses, severe acute respiratory syndrome coronavirus (SARS‐CoV) produces polyproteins containing several non‐structural proteins, which are then processed by the viral proteases. These proteases often exist within the polyproteins, and are excised by their own proteolytic activity (‘autoprocessing’). It is important to investigate the autoprocessing mechanism of these proteases from the point of view of anti‐SARS‐CoV drug design. In this paper, we describe a new method for investigating the autoprocessing mechanism of the main protease (M pro), which is also called the 3C‐like protease (3CL pro). Using our method, we measured the activities, under the same conditions, of the mature form and pro‐forms with the N‐terminal pro‐sequence, the C‐terminal pro‐sequence or both pro‐sequences, toward the pro‐form with both N‐ and C‐terminal pro‐sequences. The data indicate that the pro‐forms of the enzyme have proteolytic activity, and are stimulated by the same proteolytic activity. The stimulation occurs in two steps, with approximately eightfold stimulation by N‐terminal cleavage, approximately fourfold stimulation by C‐terminal cleavage, and 23‐fold stimulation by the cleavage of both termini, compared to the pro‐form with both the N‐ and C‐terminal pro‐sequences. Such cleavage mainly occurs in a trans manner; i.e. the pro‐form dimer cleaves the monomeric form. The stimulation by N‐terminal pro‐sequence removal is due to the cis (intra‐dimer and inter‐protomer) effect of formation of the new N‐terminus, whereas that by C‐terminal cleavage is due to removal of its trans (inter‐dimer) inhibitory effect. A numerical simulation of the maturation pathway is presented.
Keywords: 3CL protease, autoprocessing, cell‐free protein synthesis, SARS‐CoV
The use of a newly developed method for measuring the activities of the mature and pro‐forms of the main protease (Mpro; or 3C‐like protease, 3CLpro) of severe acute respiratory syndrome coronavirus (SARS‐CoV) revealed important aspects of its autoprocessing mechanism, allowing a numerical simulation of the maturation pathway.
Abbreviations
- SARS‐CoV
severe acute respiratory syndrome coronavirus
Introduction
Severe acute respiratory syndrome (SARS) is a highly lethal infectious disease. It was first discovered in Guangdong Province, China, in November 2002, and spread to 26 countries with 8098 cases, including 774 deaths, over 8 months until 5 July 2003, when the World Health Organization reported that the last human chain of transmission in that epidemic had been broken 1. SARS coronavirus (SARS‐CoV) is the etiological agent of this disease. The 3C‐like protease (3CLpro, also referred to as the main protease, Mpro) of the virus is a key enzyme, as it cleaves several sites to produce non‐structural proteins that are essential for genome replication and virion production, such as an RNA‐dependent RNA polymerase, a helicase, ribonucleases and 3CLpro itself, from two types of polyproteins (pp1a and pp1ab) 2.
RNA viruses encode polyproteins containing several non‐structural proteins, which are then processed by the viral proteases. However, these proteases exist within the polyproteins, and are excised by their own proteolytic activity (‘autoprocessing’). These proteases often function as homo‐dimers. SARS‐CoV 3CLpro exists as a homo‐dimer, in which each protomer has an active site 3. Precise processing of its N‐terminus is important for its proteolytic activity 4, 5, 6. Indeed, expression of enzymes with two to five additional vector‐derived amino acid residues or without the first amino acid residue displayed reduced activity (1–40%) 5, 6. This is because the processed N‐terminus of one protomer is required for formation of the oxyanion hole in the active site of the other protomer in the homo‐dimer 3. Although the mature protease cleaves both the N‐ and C‐terminus of the 3CLpro precursor from the replicase polyproteins, it is not clear how the first 3CLpro molecule is excised and becomes mature after SARS‐CoV infection of human cells.
The difficulty in investigating the autoprocessing mechanism lies in the ability to measure the activity of each pro‐form of the enzyme, because (a) a chain reaction exists, in which the processed and activated enzyme (product) reacts with the precursor molecule (substrate), and (b) the enzyme protomer dimerizes with a substrate protomer, even if the catalytically inactive mutant pro‐form of the enzyme (such as C145A or H41A 3) is used as the substrate. We developed a new method to investigate autoprocessing of SARS‐CoV 3CL protease, which completely eliminated these problems. In this paper, we describe the activities, measured under the same conditions, of the mature form and pro‐forms with the N‐terminal pro‐sequence, C‐terminal pro‐sequence and both pro‐sequences toward the pro‐form with both N‐ and C‐terminal pro‐sequences. Furthermore, by use of two types of substrates, we determined that the first N‐terminal processing from the polyprotein occurs in a trans manner, i.e. the pro‐form dimer cleaves the monomeric form of the pro‐form, and determined the inhibitory modes of the N‐ and C‐terminal pro‐sequences, i.e., the N‐terminal pro‐sequence has a cis (intra‐dimer and inter‐protomer) effect, and the C‐terminal pro‐sequence has a trans (inter‐dimer) effect.
Results
In vitro 3CLpro autoprocessing system
First we developed a SARS 3CL protease autoprocessing system by use of the Escherichia coli cell‐free protein synthesis system, with the N‐ and C‐terminal 10 amino acid pro‐sequences accompanied by an S tag and a His tag, respectively (Fig. 1A). The S tag is a 15 amino acid residue peptide tag derived from nuclease S, which can be detected by the S protein, derived from the other part of nuclease S. Although, stretches of more than several hundred amino acid residues are associated with the 3CLpro region on both sides of the polyprotein in infected cells, we assumed that the effects of elements upstream and downstream of the 10 amino acid regions tested are minimal.
Figure 1.
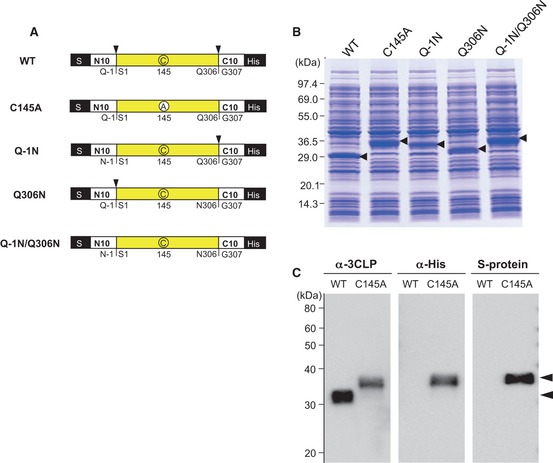
Cell‐free synthesis of the mature and pro‐forms of SARS‐CoV 3CL protease. (A) Expressed proteins. Sites cleaved by autoprocessing are indicated by arrowheads. The active cysteine residue (C) or inactivated residue (A), by a cysteine to alanine mutation in each construct, is circled. (B) SDS/PAGE gel stained using Coomassie Brilliant Blue G‐250. Estimated migration positions for each species after expected cleavage are indicated by arrowheads. (C) SDS/PAGE followed by western blotting analysis. Estimated migration positions for each species after expected cleavage are indicated by arrowheads.
Over the course of the cell‐free protein synthesis reaction (30 °C, 4 h), the N‐ and C‐terminal processing sites of the catalytically active construct (wild‐type, WT in Fig. 1A) were completely cleaved by the enzyme's endogenous proteolytic activity, as shown in Fig. 1B,C (WT). The 34 kDa band of the catalytically active construct (WT) in Fig. 1B was stained using an antibody against SARS‐CoV 3CLpro, but not by those against either the His or S tag. This mature form was excised from the gel, and its N‐terminal amino acid sequence was determined to be SGFRKMAFP. Its molecular mass was determined by MALDI‐TOF mass spectrometry as 33 792 Da, which agreed well with its calculated molecular mass (33 845 Da). However, the catalytically inactive form (C145A), in which the active residue Cys145 was converted to Ala, was unable to process either the N‐terminal pro‐sequence or the C‐terminal pro‐sequence, and resulted in the 41 kDa band in Fig. 1B (calculated molecular mass 40 525 Da), which reacted with antibodies against the His and S tags (Fig. 1C). These observations are consistent with those previously reported 7.
As 3CLpro requires a glutamine residue at the P1 position of the substrate 8, mutation of Q–1 to N and Q306 to N resulted in loss of cleavage at these sites (Fig. 1A,B). However, interestingly, the N‐terminal uncleavable mutant Q–1N was able to cleave its own C‐terminal processing site, and the C‐terminal uncleavable mutant, Q306N, was able to cleave its own N‐terminal processing site. The processed site for Q–1N was assumed to be the same as the C‐terminal processing site for WT, and that for Q306N was assumed to be the same as the N‐terminal processing site for WT, based on their molecular weights estimated by SDS/PAGE (Fig. 1B).
Measuring the activity of 3CLpro by the trans‐processing system
To investigate the activities of the pro‐forms of SARS‐3CLpro in detail, we developed a ‘trans‐cleaving assay’, using a substrate in which the core region of the 3CL protease was exchanged with GFP (green fluorescent protein) (Fig. 2A). This substrate comprises the S tag, the N‐terminal pro‐sequence of 10 amino acid residues, the N‐terminal sequence of 10 amino acid residues, GFP, the C‐terminal sequence of 10 amino acid residues, the C‐terminal pro‐sequence of 10 amino acid residues, and the 6xHis tag. The substrate molecule (GFP substrate) and the enzyme molecule (3CLpro, mature or pro‐form) were each synthesized using the E. coli cell‐free protein synthesis system at 30 °C for 4 h, as described above. The protein solutions were then mixed together and incubated for 1 h at 30 °C. The mature and pro‐enzymes (Q–1N, Q306N and Q–1N/Q306N) cleaved the N‐ and C‐terminal processing sites in the substrate molecule, and thus we were able to estimate the activities of the pro‐enzymes toward these processing sites. The enzyme solution synthesized by the cell‐free protein synthesis reaction was serially diluted (2‐, 4‐, 8‐, 16‐, 32‐fold, etc), mixed with the substrate molecule (1 : 1), and incubated for 1 h (Fig. 2B). We estimated the activity of the enzyme (or pro‐enzyme) toward the N‐ and C‐terminal processing sites in the GFP substrate on the basis of the dilution ratio at which 50% cleavage was achieved (Fig. 2C).
Figure 2.
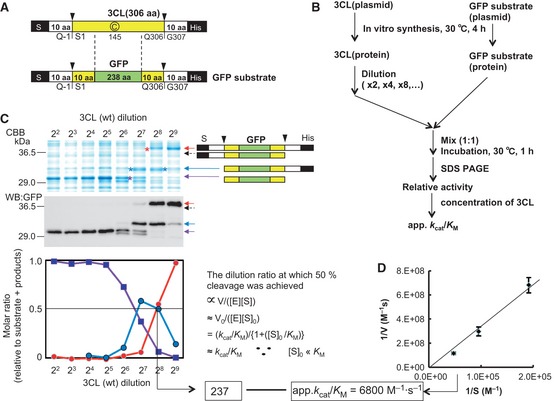
Trans‐processing assay. (A) The enzyme molecule (3CL) and the GFP substrate. Arrowheads indicate sites cleaved by autoprocessing during cell‐free protein synthesis. (B, C) Method for the trans‐cleavage assay. The substrate molecule [GFP or pro‐3CL(C145A)] and the enzyme molecule (3CL pro, mature or pro‐form), synthesized by the E. coli cell‐free protein synthesis system, were mixed together, incubated for 1 h at 30 °C, and analyzed by SDS/PAGE. In (C), the red arrows and asterisk indicate the GFP substrate, the blue arrows and asterisk indicate the N‐terminally processed substrate, and the purple arrows and asterisk indicate the substrate processed at both termini, as confirmed by western blotting analysis using an antibody against GFP. The C‐terminal processed substrate did not appear, but its expected positions are indicated by black dashed arrows. The activity of the enzyme for a processing site was estimated on the basis of the dilution ratio at which 50% cleavage was achieved. This value was then converted to the k cat/K M value using the concentration of the enzyme species. The standard k cat/K M value was obtained by the slope of the plot shown in (D). (D) Lineweaver–Burk plot for the wild‐type (mature form of 3CLpro) toward the N‐terminal processing site of the GFP substrate. The enzyme, the mature (wild‐type) 3CL pro and the GFP substrate were prepared by the E. coli cell‐free protein synthesis system. The substrate solution was diluted, mixed with 256‐fold diluted mature 3CL pro enzyme solution, and incubated at 30 °C for 15 min. The enzyme, substrate and product concentrations were measured by quantification of the respective bands on an SDS/PAGE gel stained with Coomassie Brilliant Blue G‐250.
The calculated molecular masses of the GFP substrate and the pro‐form of 3CL with both the N‐ and C‐terminal pro‐sequences (Q‐1N/Q306N) are 35 723 and 40 526 Da, respectively. However, under some conditions, the mobility of GFP‐fused proteins in SDS/PAGE is smaller than the calculated values, i.e. the estimated molecular masses are higher 9, presumably as a result of the covalently formed chromophore and partially formed secondary/tertiary structures. In our experiments, the migration of the GFP substrate and its cleaved products represented masses approximately 10 kDa greater than their actual molecular masses, and approximately 5 kDa greater than those of the corresponding pro‐3CLpro species, as confirmed by western blotting (Fig. 2C).
Kinetic parameters for the mature 3CLpro toward the GFP substrate
We attempted to measure the k cat and K M values using a gel‐based assay. The GFP substrate and the mature 3CLpro enzyme were prepared as described above, and their concentrations were estimated by Coomassie Brilliant Blue G‐250 staining of the gel, using BSA as a standard. The substrate (41.8 μm) was diluted 1‐, 2‐ or 4‐fold, and was mixed with 256‐fold diluted mature 3CLpro enzyme solution (0.088 μm) (1 : 1). After 0, 15, 30 and 60 min incubations at 30 °C, the N‐terminal processing rates were estimated by SDS/PAGE, and the data from the 15 min incubation were used for estimation of the initial velocities (data not shown). Although the K M value could not be estimated, because it is much larger than the concentrations of the substrates used, the k cat/K M per protomer was estimated as 6800 m −1·s−1 from the Lineweaver–Burk plot (Fig. 2D). Strictly speaking, kinetic parameters must be measured under conditions where the employed substrate concentration is comparable to the K M value. However, in this case, the substrate concentrations were quite low compared to the K M value. Therefore, the k cat/K M value obtained is not precise, but may be used as an apparent value. The mature form of SARS‐CoV 3CLpro exists as a homo‐dimer, and the k cat/K M value of the dimer is 13 600 m −1·s−1. Considering the differences in the substrates, this value is comparable to that reported by Xue et al. 5 (26 500 m −1·s−1). They used a synthetic peptide as a substrate, whereas we used proteins synthesized by the cell‐free translation system. To our knowledge, this is the first report of the k cat/K M value for SARS‐CoV 3CLpro toward a protein substrate rather than a peptide substrate.
Activities of the mature and pro‐forms of the 3CLpro enzyme toward the GFP substrate and the inactive 3CLpro pro‐form (C145A)
The mature enzyme, its pro‐forms, the GFP substrate and the catalytically inactive pro‐form of 3CLpro(C145A) [pro‐3CL(C145A)] were produced by the E. coli cell‐free protein synthesis system, as described above. In each experiment, the reactions were fractionated by SDS/PAGE (Figs 1B and 3, and data not shown), and then the concentration of each species was analyzed using the band stained by Coomassie Brilliant Blue G‐250 (Fig. 3A), with BSA as a standard, as described above and in Experimental procedures. Typically, the molar concentration of each species determined by its calculated molecular mass is 22.6 μm for the mature enzyme, 14.4 μm for the pro‐form with the N‐terminal pro‐sequence (Q–1N), 17.3 μm for the pro‐form with the C‐terminal pro‐sequence (Q306N), 18.7 μm for the pro‐form with both N‐ and C‐terminal pro‐sequences (Q–1N/Q306N), and 41.8 μm for the GFP substrate. These values were used for calculation of the k cat/K M values, as described below.
Figure 3.
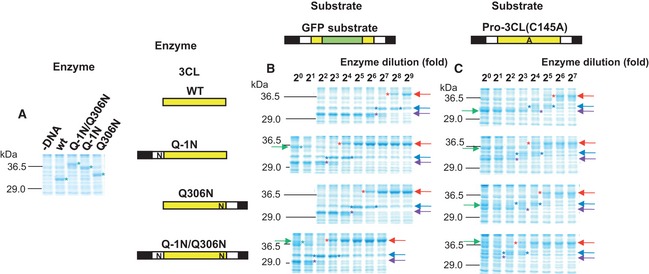
Trans‐processing assay using the GFP substrate and the pro‐3CL(C145A) substrate. Red arrows and asterisks indicate unprocessed molecules. Blue arrows and asterisks indicate N‐terminally processed molecules. Purple arrows and asterisks indicate molecules processed at both termini. Green arrows and asterisks indicate enzymes added. (A) Enzyme species produced by the E. coli cell‐free protein synthesis system. The lane ‘–DNA’ insicates the control cell‐free protein synthesis reaction without any plasmid DNAs. (B, C) Processing reactions using the GFP substrate (B) and the catalytically inactive pro‐form of 3CL pro [pro‐3CL(C145A)] (C).
The activities of the enzyme and its pro‐forms were estimated on the basis of the dilution ratios at which 50% cleavage of the substrates was achieved at 30 °C and 1 h (Fig. 3B). Under these conditions, the velocities of the cleavage reactions were the same, and thus the dilution ratios are proportional to the velocities of the cleavage reactions catalyzed by a certain amount of the enzyme. In turn, they are proportional to the V max/K M values, because the K M is much higher than the concentration of the substrate used (Fig. 2C). These V max/K M values may be converted to k cat/K M values using the concentrations of the enzyme species estimated by the SDS/PAGE analyses described above, and the activities are indicated by their k cat/K M values in Table 1 and Fig. 4. All molecules had processing activities towards both the N‐ and C‐terminal processing sites of the GFP substrate (Fig. 4): the pro‐form with the N‐terminal pro‐sequence (Q–1N) had approximately 19% activity, the pro‐form with the C‐terminal pro‐sequence (Q306N) had approximately 33% activity, and the pro‐form with both pro‐sequences had approximately 4% activity as compared to the mature form (WT). The cleavability of the C‐terminal processing site (VTFQ↓GKF) of the GFP substrate was approximately half of that of the N‐terminal processing site (AVLQ↓SGF) (0.56 by the mature enzyme, 0.60 by Q–1N, 0.50 by Q306N, and 0.49 by Q–1N/Q306N). These values are in good agreement with that obtained in a previous study of the mature enzyme (0.41) toward synthetic peptides 10. These values also explain the order of cleavage of the two processing sites in the catalytically inactive pro‐3CLpro mutant (C145A) with the mature enzyme 9: first at the N‐terminal site, and then at the C‐terminal site. The values may also be explained by the difference at the P2 position (Leu for the N‐terminal site, Phe for the C‐terminal site), according to recent detailed analyses by Chuck et al. 11. They estimated the substrate specificity by mutating each of the residues, one by one, at the P5–P3′ positions of the N‐terminal processing site. Therefore, there is no difference in accessibility between the N‐ and C‐terminal processing sites in the GFP substrate.
Table 1.
Enzymatic activities (k cat/K M per protomer). The values indicated by the superscripts a–h were used for estimation of the k cat/K M of dimers (Table 2)
| Enzyme | (k cat/K M)protomer × 10−3 (m −1·s−1) | |||
|---|---|---|---|---|
| Toward N‐terminal site | Toward C‐terminal site | |||
| GFP substrate | 3CL(C145A) substrate | GFP substrate | 3CL(C145A) substrate | |
| Wild‐type (mature form) | 6.78a ± 0.75 | 2.35 ± 0.81 | 3.82e ± 0.20 | 0.73 ± 0.07 |
| Q–1N (with N‐terminal pro‐sequence) | 1.23b ± 0.15 | 0.93 ± 0.10 | 0.75f ± 0.02 | 0.43 ± 0.16 |
| Q306N (with C‐terminal pro‐sequence) | 2.39c ± 0.05 | 1.15 ± 0.03 | 1.19g ± 0.12 | 0.44 ± 0.06 |
| Q–1N/Q306N (with both pro‐sequences) | 0.30d ± 0.02 | 0.41 ± 0.03 | 0.15h ± 0.01 | 0.19 ± 0.03 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 4.
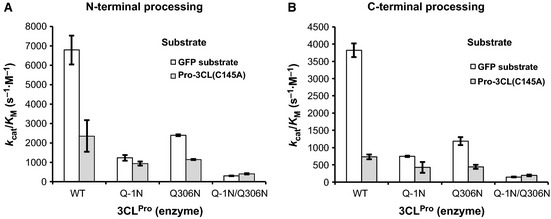
Activities of the mature and pro‐forms of SARS‐CoV 3CL pro (Table 1). The dilution ratios at which 50% cleavage was achieved were converted to k cat/K M values, based on that for the mature enzyme (WT) toward the GFP substrate (6800 m −1·s−1). Values are means ± standard errors (n = 3). (A) Activities toward the N‐terminal processing site. (B) Activities toward the C‐terminal processing site.
Trans or cis reaction?
Exact processing of the N‐terminus of SARS 3CLpro is required for efficient cleavage activity 5. However, as shown in Fig. 1, the pro‐forms of 3CLpro synthesized by the E. coli cell‐free protein synthesis system had sufficient proteolytic activity to process their own autoprocessing sites. Moreover, they were able to process a substrate in which the core region was replaced with the GFP protein. Although SARS‐CoV 3CLpro forms a homo‐dimer 3, this GFP‐replaced substrate molecule does not dimerize with 3CLpro. Therefore, these pro‐forms efficiently process the autoprocessing sites in a trans manner on the GFP substrate (Figs 2, 3B, and 4). These pro‐forms also efficiently processed the autoprocessing sites on the pro‐form of a catalytically inactive 3CLpro(C145A) mutant [pro‐3CL(C145A)] (Figs 3C and 4). The fact that 3CLpro species with the N‐terminal pro‐sequence (Q–1N and Q–1N/Q306N) cleaved the autoprocessing site of 3CLpro(C145A) as efficiently as the GFP‐replaced substrate suggests an autoprocessing mechanism in which the pro‐3CLpro dimer (enzyme) cleaves the site in the monomeric form of the pro‐form (trans reaction). Although an X‐ray crystallographic analysis revealed that the N‐terminus of the mature form in the dimeric structure of 3CLpro is buried inside the molecule 3, this protease exists in equilibrium between the dimer and monomer in solution 7, 12. The pro‐sequences enhance dissociation of the dimer 7, 12, and both the N‐ and C‐terminus of the monomer molecule are exposed 13, 14, 15. Therefore, it is plausible that the pro‐form also exists in a dimer and monomer equilibrium, and that the dimer molecule easily processes the monomer molecule.
Inhibitory effects of the N‐ and C‐terminal pro‐sequences
The presence of either the N‐terminal pro‐sequence (Q–1N) or the C‐terminal pro‐sequence (Q306N) reduces the activity toward the GFP substrate (Fig. 4). However, there are some differences in the processability of the substrate molecules. We used two substrates: pro‐3CL(C145A), the model substrate of the pro‐form, and the GFP substrate, in which the core region is replaced by the GFP protein (Fig. 2A). The difference between these two substrates is the ability to dimerize with the enzyme protomer (the mature and pro‐forms of 3CLpro). The GFP substrate acts only as a substrate, whereas pro‐3CL(C145A) acts as both substrate and protomer in a dimer. As each protomer of the SARS 3CLpro dimer has an active site, the concentration of the active site in the reaction solution does not change when using the pro‐3CL(C145A) substrate, even if the active protomer dimerized with the pro‐3CL(C145A) substrate. Therefore, the difference in reactivity between the GFP substrate and the pro‐3CL(C145A) substrate reflects the influence of the N‐ or C‐terminal pro‐sequence of 3CLpro(C145A) on the active site of the catalytically active protomer with which it dimerizes.
The fact that the activity of the pro‐form with the C‐terminal pro‐sequence (Q306N) toward the pro‐3CL (C145A) substrate was lower than that toward the GFP substrate (Fig. 4) indicates that the pro‐3CL(C145A) substrate dimerized with the pro‐form (Q306N), and its N‐terminal pro‐sequence inhibited the activity. In contrast, there was no difference in the activities of the pro‐form with both the N‐ and C‐terminal pro‐sequences (Q–1N/Q306N) toward the pro‐3CL(C145A) substrate and the GFP substrate (Fig. 4). As this pro‐form (Q–1N/Q306N) has the N‐terminal pro‐sequence, dimerization with the pro‐3CL(C145A) substrate did not affect its activity further through the N‐terminal pro‐sequence.
It is worth mentioning the minimal difference in the activities of the pro‐form with the N‐terminal pro‐sequence (and without the C‐terminal pro‐sequence) (Q–1N) between the two substrates (Fig. 4). As there is an activity difference between the mature form (WT) and the pro‐form with the C‐terminal pro‐sequence (Q306N) toward the GFP substrate, the C‐terminal pro‐sequence has an inhibitory effect. However, this inhibitory effect is not achieved through either dimerization or an inter‐protomer interaction in the dimer. Two possibilities may be considered. One is that the C‐terminal pro‐sequence of the protomer reduces its own activity (‘intra‐protomer’). The other is that the C‐terminal pro‐sequence inhibits another enzyme dimer, e.g. through competitive inhibition (‘inter‐dimer’). The latter proposal is more plausible, as judged from the physical relationship between the C‐terminus and the active site of a protomer in the crystal structure of the mature enzyme [3].
Discussion
Dimerization of pro‐forms
Hsu et al. 7 reported dissociation constants (Kd values) for dimerization of the wild‐type (mature form) and mutants (C145A) with extra N‐ or C‐terminal amino acids (10aa–C145A and C145A–10aa) as 0.35, 17.2 and 5.6 nm, respectively. The catalytically active pro‐forms Q–1N and Q306N, for which the activities were measured in our study, had almost the same structures as 10aa–C145A and C145A–10aa. The differences are that our preparations have enzymatic activity (i.e., they are not the C145A mutants), Q‐1N (Gln to Asn) and/or Q306N (Gln to Asn) mutations, and S and/or His tag moieties. In our study, the activity of each pro‐form was based on the concentration of the pro‐form enzyme at which 50% cleavage of the substrate was achieved. These concentrations were 47.7, 263.9 and 135.3 nm for the mature form, Q–1N and Q306N, respectively. Using the K d values (mentioned above) from Hsu et al. 7, the populations of the dimer of each species were estimated as 94%, 84%, and 87%, respectively. Therefore, we assumed that the major part of each of protomer species exists as a homo‐ or hetero‐dimer under our experimental conditions.
Maturation pathway
Although the pro‐form of 3CLpro with both the N‐ and C‐terminal 10 amino acid residue pro‐sequences processed both processing sites itself (Fig. 1B,C, WT), the paths from the pro‐form to the mature form are complicated. There may be four types of protomer molecules (pro‐form with N‐ and C‐terminal pro‐sequences, pro‐form with the N‐terminal pro‐sequence, pro‐form with the C‐terminal pro‐sequence, and the mature form), and they form several types of homo/hetero‐dimers with each other. Thus, during the course of maturation, several types of dimer species co‐exist and cleave each other's pro‐sequences, thus changing the types of dimeric forms and finally generating the mature form. However, we can simulate this pathway, as described below.
During maturation, 10 types of dimeric forms exist, and their activities toward the N‐ and C‐terminal processing sites were estimated (Table 2) from the data in Table 1. Based on the assumption that, between any dimeric protomer species, the monomer/dimer exchange rate is so fast that the population of each dimer at each time is proportional to the population of the protomers forming the dimer, the maturation process was simulated as shown in Fig. 5A. This assumption is supported by the difference in the inhibitory effects of the N‐terminal pro‐sequences of the two substrates, the pro‐3CL(C145A) and GFP substrate, on the N‐terminally processed enzyme/pro‐enzyme (WT or Q306N; Fig. 4). As described in Results, this difference indicates that dimerization of the enzyme protomer (WT or Q306N) and the substrate pro‐3CL(C145A), i.e. inter‐exchange of a protomer between the enzyme dimer and the substrate pro‐3CL(C145A) dimer, occurs under our experimental conditions. This conversion must occur through the monomeric form.
Table 2.
Estimated k cat/K M values of the enzyme dimers. These values were calculated from the (k cat /K M) values per protomer, indicated by superscripts a–h in Table 1
| Enzyme dimer species | (k cat/K M)dimer × 10−3 (m −1·s−1) | |
|---|---|---|
| Toward the N‐terminal processing site | Toward the C‐terminal processing site | |
Dimer 1 
|
0.60 (d + d) | 0.29 (h + h) |
Dimer 2 
|
2.69 (c + d) | 1.32 (g + h) |
Dimer 3 
|
1.53 (b + d) | 0.89 (f + h) |
Dimer 4 
|
4.79 (c + c) | 2.37 (g + g) |
Dimer 5 
|
3.62 (b + c) | 1.93 (f + g) |
Dimer 6 
|
7.08 (a + d) | 3.97 (e + h) |
Dimer 7 
|
2.45 (b + b) | 1.49 (f + f) |
Dimer 8 
|
9.18 (a + c) | 5.00 (e + g) |
Dimer 9 
|
8.01 (a + b) | 4.56 (e + f) |
Dimer 10 
|
13.57 (a + a) | 7.64 (e + e) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 5.
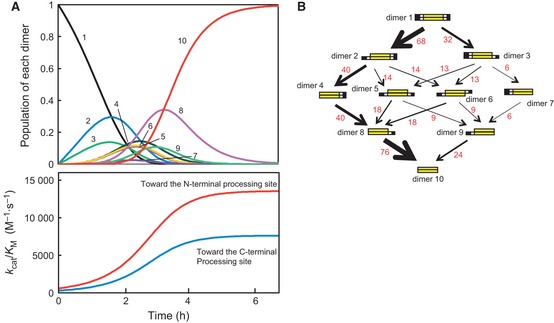
Simulation of the maturation path of SARS‐CoV 3CL pro (Table 2). (A) Four types of protomers were assumed to be present (the mature type, the protomer with the N‐terminal pro‐sequence, the protomer with the C‐terminal pro‐sequence, and the protomer with both pro‐sequences). At time t, the population of each dimer was calculated from the population of protomers forming the dimer. The N‐terminal processing activity [k cat /K M (N)] and the C‐terminal processing activity [k cat /K M (C)] were then calculated by using the estimated k cat /K M value of each dimer (Table 2). Using these values, the population of protomers at time t + dt was calculated. At time 0, all protomers were the protomer with both pro‐sequences, i.e. all dimers were dimer 1. The numbers 1, 2, 3, etc, indicate the populations of dimer 1, dimer 2, dimer 3, etc. Although the time scale changes according to the initial concentration of dimer 1 at time 0, the shape and size of each curve are independent of the initial concentration. The time scale indicated is for the initial condition of 100 nm dimer 1. Parameters: at time = 0 s, the concentration of dimer 1 = 100 nm, the concentration of dimer 2 = the concentration of dimer 3 … = the concentration of dimer 10 = 0 m, dt = 100 s. (B) Maturation path of SARS‐CoV 3CL pro. The red numbers indicate rough estimates of the percentage of each reaction path on the basis of the simulated data (A).
At time t, the population of each dimer was calculated from the population of protomers forming the dimer. The N‐terminal processing activity [k cat/K M (N)] and the C‐terminal processing activity [k cat/K M(C)] were then calculated using the estimated k cat/K M value for each dimer (Table 2). Using these values, the population of protomers at time t + dt was calculated.
At time 0, only one dimer species, dimer 1, is assumed to exist (indicated by ‘1’; Fig. 5A). The other species (dimers 2–10; indicated by ‘2’– ‘10’) emerge as the processing reaction continues, and finally all of the molecules become the mature form, dimer 10. The total activities toward the N‐ and C‐terminal processing sites are also indicated. Although the time scale changes according to the initial concentration of dimer 1 at time 0, the shape and size of each curve are independent of the initial concentration. Based on these simulated data, the conversion paths of the dimers are indicated in Fig. 5B.
The first cleavage
Hsu et al. 7 investigated the processing of a model molecule (Trx‐10aa‐3CLpro(C145A)‐10aa‐GST), by the mature 3CL protease, and found that N‐terminal processing occurred before C‐terminal processing. This is consistent with the higher cleavability of the N‐terminal processing site than the C‐terminal processing site, as reported by Fan et al. 10 and found in the present study. They proposed a mechanism for autoprocessing of polyproteins in which the first mature 3CLpro molecule is formed: first, cleavage of the N‐terminal processing site occurs, the dimeric structure forms as the mature 3CLpro, and C‐terminal processing then occurs in an inter‐molecular manner 7. They also solved the crystal structure of an inactive mutant (C145A) of the mature form of 3CLpro, in which the C‐terminal moiety of a protomer in the dimer was bound to the active site of a protomer in the adjacent asymmetric unit, and proposed that this structure represented that just after C‐terminal processing had occurred 7.
Thus, the most important step in the maturation process of 3CLpro appears to be N‐terminal autoprocessing of the 3CLpro moiety in the polyprotein 7, 12. As protomer dimerization had been assumed to be essential for the proteolytic activity of 3CLpro, Hsu et al. 7 proposed that the 3CLpro moiety of the polyprotein dimerized in a different manner to dimerization of the mature form, in which the N‐terminal processing site of a protomer was cleaved by the active site of the other protomer (cis cleavage). After cleavage of both N‐termini of the protomers, rearrangement of the protomers occurs to form the same dimeric structure as that of the mature enzyme. Subsequently, cleavage of the C‐terminal processing site of the other molecule of the N‐terminally processed dimer, as well as cleavage of the N‐ and C‐terminal processing sites in other polyprotein molecules, occurs. However, the possibility of a first cis cleavage of the N‐terminus of the polyprotein dimer is based only on the fact that the N‐terminus of a protomer is near the active site of the other protomer in the crystal structure of the mature enzyme 3. This possibility was supported by Chen et al. 16, who demonstrated that mutants of the 3CLpro pro‐form that exist in the monomeric form, as determined by a gel‐filtration assay, had N‐terminal processing activities. In contrast, Li et al. 12 proposed a trans mechanism for the first N‐terminal processing of the polyprotein, in which the 3CLpro moiety forms the dimeric structure of the mature form, and cleaves the N‐terminus of another polyprotein molecule. They used two types of pro‐3CLpro molecules with an N‐terminal pro‐sequence of six amino acids, accompanied by CFP and YFP at the N‐ and C‐termini, respectively: one contains an active (wild‐type) protease site and and inactive N‐terminal processing site (Q–1E) (enzyme), and the other contains an inactive protease site (C145A) and an active (wild‐type) N‐terminal processing site (substrate). They demonstrated cleavage of the processing site in the substrate molecule (C145A). However, this approach did not eliminate the cis reaction by which the enzyme molecule and the substrate molecule dimerize. Although our results also did not completely eliminate the possibility of the cis reaction, they indicated that the trans reaction is sufficiently robust for the autoprocessing, and is the main path for the first N‐terminal processing of the 3CLpro moiety in the polyprotein.
Liberation of 3CLpro from the polyprotein at the ER membrane
SARS‐CoV contains RNA(+) as its genome in the virion particle 8. After infection of cells, this RNA(+) is introduced into the cytosol and translated to form two large polyproteins, pp1a (4382 amino acids) and pp1ab (7073 amino acids), the latter of which is produced by a −1 translational frameshift occurring just upstream of the stop codon of pp1a. These polyproteins are processed by PL2pro and 3CLpro to form proteins nsp1–nsp11 from pp1a, and proteins nsp1–nsp10 and nsp12–nsp16 from pp1ab (nsp5 is 3CLpro). These proteins form a multi‐subunit protein complex called the ‘viral replicase complex’, which generates a nested set of viral sub‐genomic segments by complicated reactions 8. Only three cleavages (between nsp1 and nsp2, nsp2 and nsp3, and nsp3 and nsp4) are performed by PL2pro, and the other 11 sites are cleaved by 3CLpro. However, the cleavages by PL2pro are suggested to occur before those by 3CLpro 17, and proteolysis between nsp3 and nsp4 by PL2pro may occur before 3CLpro activation. At this stage, 3CLpro (nsp5) is located between nsp4 and nsp6. Both nsp4 and nsp6 are membrane proteins. Oostra et al. reported that the N‐ and C‐termini of both the nsp4 and nsp6 proteins are on the cytoplasmic side of the ER 17, 18, so the 3CLpro moiety is on the cytoplasmic side. Moreover, they proposed the membrane topology for polyprotein pp1a, in which all of the cleavage sites cleaved by 3CLpro, including its own N‐ and C‐terminal processing, are on the cytoplasmic side, and are accessible by mature 3CLpro and the 3CLpro moiety in the polyprotein 18.
Using our new method, we were able to analyze the enzymatic activities of the mature form and three pro‐forms (with the N‐terminal pro‐sequence, the C‐terminal pro‐sequence, or both pro‐sequences) of the 3CL protease towards the N‐ and C‐terminal processing sites, under the same conditions. We identified the inhibitory effect of the C‐terminal pro‐sequence, by which the C‐terminal pro‐sequence of a dimer molecule inhibits the activity of another enzyme dimer molecule (inter‐dimer manner).
The assay system developed here is a convenient method to investigate the autoprocessing mechanisms of other viral proteases.
Experimental procedures
Plasmids
The DNA fragment of part of the SARS‐CoV cDNA, encompassing the N‐terminal proximal 10 amino acids, the coding region of 3CLpro, and the C‐terminal proximal 10 amino acids, was prepared by PCR using oligonucleotides 5′‐CGTGGATCCCAGACATCAATCACTTCTGCTGTTCTGCAGAGTGGTTTTAGGAAAATGGCATTCCC‐3′ and 5′‐GGTGCTCGAGAGTGCCCTTAACAATTTTCTTGAACTTACCTTGGAAGGTACACCAGAGCATTGTC‐3′), and inserted into the BamHI and XhoI sites of the pET29a vector (Merck KGaA/Novagen, Darmstadt, Germany). The encoded protein ([S tag]‐GS‐QTSITSAVLQ‐[3CLpro]‐GKFKKIVKGT‐LE‐HHHHHH), in which GS and LE are derived from the BamHI (GGATCC) and XhoI (CTCAGA) sites of the pET29a vector, and QTSITSAVLQ and GKFKKIVKGT are from the adjacent pro‐sequences in the SARS‐CoV polyprotein, was expressed by the E. coli cell‐free protein synthesis system. The Gln–1 to Asn (Q–1N), Gln306 to Asn (Q306N) and Cys145 to Ala (C145A) mutations were introduced by Quik‐Change mutagenesis (Agilent Technologies/Stratagene, Santa Clara, CA, USA) using with the oligonucleotides 5′‐CAATCACTTCTGCTGTTCTGAACAGTGGTTTTAGGAAAATGGC‐3′ and 5′‐GCCATTTTCCTAAAACCACTGTTCAGAACAGCAGAAGTGATTG‐3′ for Q–1N, 5′‐GCTCTGGTGTTACCTTCAACGGTAAGTTCAAGAAAATTG‐3′ and 5′‐CAATTTTCTTGAACTTACCGTTGAAGGTAACACCAGAGC‐3′ for Q309N, and 5′‐GGTTCTTTCCTTAATGGATCAGCTGGTAGTGTTGGTTT TAAC‐3′ and 5′‐GTTAAAACCAACACTACCAGCTGATCCATTAAGGAAAGAACC‐3′ for C145A. The GFP substrate was created by replacing the region encoding amino acid residues 11–296 with that encoding the GFP protein, by PCR using four primers (5′‐TTCTCCTTTACTCATTGACGGGAATGCCATTTTCCTAAAACCACTC‐3′, 5′‐GATGAGCTCTACAAAGTTAGACAATGCTCTGGTGTTACCTTCCAAG‐3′, 5′‐GCATTCCCGTCAATGAGTAAAGGAGAAGAACTTTTC‐3′, and 5′‐GCATTGTCTAACTTTGTAGAGCTCATCCATGCCATG‐3′). The protein expressed from this plasmid is [S tag]‐GS‐QTSITSAVLQSGFRKMAFPS‐[GFP]‐VRQCSGVTFQGKFKKIVKGT‐LE‐HHHHHH. The plasmids were amplified in the E. coli DH5α strain, purified using a PureLink HiPure Plasmid Filter Midiprep Kit (Life Technologies/Invitrogen, Carlsbad, CA, USA), and used in the E. coli cell‐free protein synthesis system.
Cell‐free protein synthesis
The E. coli cell‐free protein synthesis system has been described previously 19, 20. A 60 μL reaction solution, containing 55 mm HEPES/KOH buffer (pH 7.5), 16 μg·mL−1 plasmid DNA, 1.7 mm dithiothreitol, 1.2 mm ATP, 0.8 mm each of CTP, GTP and UTP, 80 mm creatine phosphate, 250 μg·mL−1 creatine kinase, 0.64 mm 3′,5′‐cyclic AMP, 68 μm l(–)‐5‐formyl‐5,6,7,8‐tetrahydrofolic acid, 175 μg·mL−1 E. coli total tRNA, 210 mm potassium glutamate, 27.5 mm ammonium acetate, 10.7 mm magnesium acetate, 1.0 mm each of standard 20 amino acids, 93 μg·mL−1 T7 RNA polymerase, and E. coli S30 extract at a final attenuance at 260 nm of standard approximately 60, was placed in a dialysis cup, Slide‐A‐Lyzer MINI dialysis unit (10K molecular weight cut‐off; Thermo Scientific, Waltham, MA, USA). Dialysis was performed at 30 °C for 4 h with moderate shaking against 1.0 mL of the same reaction solution, but lacking DNA, tRNA, creatine kinase, T7 RNA polymerase and S30 extract (dialysis buffer).
Trans‐processing assay
The enzyme molecule (3CLpro, mature or pro‐form) was synthesized by the E. coli cell‐free protein synthesis system and diluted using the dialysis buffer described above. The substrate molecule [3CLpro(C145A) or the GFP substrate] was also synthesized by the E. coli cell‐free protein synthesis system. The two solutions were mixed together at a 1 : 1 ratio, incubated for 1 h at 30 °C, and analyzed by SDS/PAGE.
SDS/PAGE
SDS/PAGE analyses of proteins were performed using 12.5% or 15% polyacrylamide mini‐gels (10 cm × 6 cm) in Laemmli's system 21. After electrophoresis, the gels were stained with Coomassie Brilliant Blue G‐250 using Quick‐CBB Plus (Wako Pure Chemical Industries, Osaka, Japan), and each band on the gel was quantified using imagej (http://rsb.info.nih.gov/ij/) software. Background subtractions were performed for each of the bands using the control value (–DNA in Fig. 3A) of the corresponding region of the same area.
Western blotting
Mouse monoclonal antibody against His‐tag (Novagen), mouse anti‐SARS‐CoV 3CLpro monoclonal antibody (Genesis Biotech Inc., Taipei, Taiwan), and mouse monoclonal antibody against GFP (Medical & Biological Laboratories Co. Ltd, Nagoya, Japan) were used as the primary antibodies for His‐tag, 3CLpro and GFP detection, respectively, and the S‐protein horseradish peroxidase conjugate (Merck KGaA/Novagen, Darmstadt, Germany) was used for S tag detection.
MALDI‐TOF mass spectrometry and N‐terminal amino acid sequence determination
The protein sample was excised from the SDS/PAGE gel and analyzed by MALDI‐TOF mass spectrometry using sinapinic acid (3,5‐dimethoxy‐4‐hydroxy cinnamic acid) as the MALDI matrix and a Life Technologies/Applied Biosystems (Carlsbad, CA, USA) Voyager biospectrometry workstation, according to the manufacturer's instructions. The N‐terminal amino acid sequence of the protein was analyzed using Life Technologies/Applied Biosystems Procise 494‐ht protein sequencer, according to the manufacturer's instructions.
Acknowledgements
We thank Hideaki Tanaka and Mariko Ikeda (RIKEN Systems and Structural Biology Center) for technical support. This work was supported by a Grant‐in‐Aid for Scientific Research (C) (20570115), by the RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses, and by the Targeted Proteins Research Program, Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1. World Health Organization (2004) WHO Guidelines for the Global Surveillance of Severe Acute Respiratory Syndrome (SARS). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2. Ziebuhr J (2004) Molecular biology of severe acute respiratory syndrome coronavirus. Curr Opin Microbiol 7, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H et al (2003) The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100, 13190–13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuo C‐J, Chi Y‐H, Hsu JT‐A & Liang P‐H (2004) Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem Biophys Res Commun 318, 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xue X, Yang H, Shen W, Zhao Q, Li J, Yang K, Chen C, Jin Y, Bartlam M & Rao Z (2007) Production of authentic SARS‐CoV MPro with enhanced activity: application as a novel tag‐cleavage endopeptidase for protein overproduction. J Mol Biol 366, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verschueren KHG, Pumpor K, Anemüller S, Chen S, Mesters JR & Hilgenfeld R (2008) A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem Biol 15, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu M‐F, Kuo C‐J, Chang K‐T, Chang H‐C, Chou C‐C, Ko T‐P, Shr H‐L, Chang G‐G & Wang AH‐J (2005) Mechanism of the maturation process of SARS‐CoV 3CL protease. J Biol Chem 280, 31257–31266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiel V, Ivanov KA, Putic A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW et al (2003) Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84, 2305–2315. [DOI] [PubMed] [Google Scholar]
- 9. Inoue S & Tsuji FI (1994) Aequorea green fluorescent protein. Expression of the gene and fluorescent characteristics of the recombinant protein. FEBS Lett 341, 277–280. [DOI] [PubMed] [Google Scholar]
- 10. Fan K, Wei P, Feng Q, Chen C, Hung C, Ma L, Lai B, Pei J, Liu Y, Chen J et al (2004) Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C‐like proteinase. J Biol Chem 279, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chuck C‐P, Chong L‐T, Chen C, Chow H‐F, Wan DC‐C & Wong K‐B (2010) Profiling of substrate specificity of SARS‐CoV 3CLpro . PLoS One 5, e13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li C, Qi Y, Teng X, Yang Z, Wei P, Zhang C, Tan L, Zhou L, Liu Y & Lai L (2010) Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C‐like protease. J Biol Chem 285, 28134–28140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen S, Hu T, Zhang J, Chen J, Chen K, Ding J, Jiang H & Shen X (2008) Mutation of Gly‐11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C‐like protease. Crystal structure with molecular dynamics simulations. J Biol Chem 283, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi J, Sivaraman J & Song J (2008) Mechanism for controlling the dimer–monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C‐like protease. J Virol 82, 4620–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu T, Zhang Y, Li L, Wang K, Chen S, Chen J, Ding J, Jiang H & Shen X (2009) Two adjacent mutations on the dimer interface of SARS coronavirus 3C‐like protease cause different conformational changes in crystal structure. Virology 388, 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen S, Jonas F, Shen C & Hilgenfeld R (2010) Liberation of SARS‐CoV main protease from the viral polyprotein: N‐terminal autocleavage does not depend on the mature dimerization mode. Protein Cell 1, 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJM & de Haan CAM (2007) Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol 81, 12323–12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oostra M, Hagemeijer MC, van Gent M, Bekker CPJ, te Lintelo EG, Rottier PJM & de Haan CAM (2008) Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol 82, 12392–12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A & Yokoyama S (2004) Preparation of Escherichia coli cell extract for highly productive cell‐free protein expression. J Struct Funct Genomics 5, 63–68. [DOI] [PubMed] [Google Scholar]
- 20. Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T & Yokoyama S (1999) Cell‐free production and stable‐isotope labeling of milligram quantities of proteins. FEBS Lett 442, 15–19. [DOI] [PubMed] [Google Scholar]
- 21. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]


