Abstract
Investigating 5-methylcytosine (5mC) has led to many hypotheses regarding molecular mechanism underlying human diseases and disorders. Many of these studies, however, utilize bisulfite conversion alone, which cannot distinguish 5mC from its recently discovered oxidative product, 5-hydroxymethylcytosine (5hmC). Furthermore, previous array-based technologies do not have the necessary probes to adequately investigate both modifications simultaneously. In this manuscript, we used technical replicates of DNA from human brain, human blood, and human saliva, in combination with oxidative bisulfite conversion and Illumina’s Infinium MethylationEPIC array, to analyze 5mC and 5hmC at more than 650 000 and 450 000 relevant loci, respectively, in the human genome. We show the presence of loci with detectable 5mC and 5hmC to be equally distributed across chromosomes and genomic features, while also being present in genomic regions with transcriptional regulatory properties. We also describe 2528 5hmC sites common across tissue types that show a strong association with immune-related functions. Lastly, in human brain, we show that 5hmC accounts for one-third of the total signal from bisulfite-converted data. As such, not only do our results confirm the efficacy and sensitivity of pairing oxidative bisulfite conversion and the EPIC array to detect 5mC and 5hmC in all three tissue types, but they also highlight the importance of dissociating 5hmC from 5mC in future studies related to cytosine modifications.
Keywords: 5-methylcytosine, 5-hydroxymethylcytosine, InfiniumEPIC, oxBS
Introduction
Studying epigenetic modifications to elucidate molecular mechanisms underlying disease has been in the forefront of research over the last decade, especially with respect to neurodevelopmental and neuropsychiatric disorders. The most common epigenetic modification is 5-methylcytosine (5mC), which is involved in transcriptional silencing, X-chromosome inactivation, and genomic imprinting, to name a few [1]. Although regarded as a genomically stable mark, recent advances have shed light on 5mC’s oxidative products, namely 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), which arise through successive oxidation reactions catalyzed by the ten-eleven translocation (TET) enzymes [2–5]. Given the growing interest in the study of these modifications in the mammalian genome, a pressing issue is the development of low-cost, high-throughput technologies to analyze them [6].
For many years, the ‘gold standard’ to methylation analyses has revolved around traditional sodium bisulfite conversion. Using this approach, cytosine, 5fC, and 5caC are converted to uracil following sodium bisulfite conversion and appear as thymine after sequencing, whereas 5mC and 5hmC are protected from this conversion and are read as cytosines after sequencing [7, 8]. The addition of a chemical (oxBS-Seq) [8] or enzymatic (TAB-Seq) [9] oxidation reaction, where 5hmC is converted to 5fC or 5mC is converted to 5caC, respectively, prior to sodium bisulfite conversion, has allowed for the discrimination of 5mC from 5hmC. However, since sequencing technologies to study genome-wide 5hmC at base pair resolution remain expensive when used genome wide with sufficient resolution, alternative methods are required to increase accessibility. Both TAB- and oxBS-Seq can be combined with reduced-representation bisulfite sequencing (RRBS) to interrogate CG-rich regions, such as CpG islands and promoters [10, 11]. Although this can dramatically decrease sequencing costs, low levels of 5hmC in these regions [8] make it unlikely to yield relevant results. Similarly, combining TAB or oxBS with the Illumina Infinium 450K Methylation array (450K array) [12–14] is not ideal because the probes on this array were specifically designed to investigate differential 5mC in tissues with low levels of 5hmC. This latter point is apparent in previous reports using oxBS combined with the 450K array, where a maximum of 80 000 loci on this array were found to contain 5hmC in two human brain regions [13, 15].
Most recently, an update to the 450K array was presented. The Infinium MethylationEPIC array (EPIC array) contains probes covering more than 850 000 CpGs in the human genome, including more than 90% of the probes on the 450K array. Of particular interest, the additional probes on the EPIC array have been designed to cover ENCODE enhancer and open chromatin regions, FANTOM5 enhancers, DNase hypersensitivity sites, non-CpG methylation sites identified in stem cells, and miRNA promoters, each of which has the potential to yield functionally relevant data in a variety of phenotypes.
In this report, using technical replicates of DNA from human brain, blood, and saliva, we combine oxBS with Illumina’s EPIC array (oxBS-EPIC) to evaluate whether the EPIC array is suitable for studying 5hmC in these tissues. With information on more than 400 000 relevant loci, we show this technology to be suitable for large-scale 5mC and 5hmC analyses.
Materials and methods
Subjects
Brain tissue from the cerebellum of an adult male, psychiatrically healthy control was obtained from the Douglas–Bell Canada Brain Bank (DBCBB), whereas human saliva and blood samples were obtained from two separate adult male, psychiatrically healthy individuals, respectively. Tissue from the DBCBB was dissected at 4°C, snap-frozen in liquid nitrogen, and stored at −80°C following standard procedures. The Quebec Coroner’s office assessed the cause of death, and subsequently, the DBCBB obtained information on the subject’s mental health through psychological autopsies using the Structured Clinical Interviews for DSM-IV Axis 1 [16]. In addition, the brain tissue sample was assessed for absence of pathological processes by a neuropathologist. Written informed consent was obtained from next-of-kin (postmortem sample) or the individual himself (blood and saliva samples), and the Douglas Institute Research Ethics Board approved this study.
DNA extraction
Genomic DNA was extracted from brain tissue and blood using QIAGEN’s QIAmp DNA Mini Kit (QIAGEN, cat #: 51304) and FlexiGene DNA kit (QIAGEN, cat. #: 51206), respectively. Saliva was collected in DNA Genotek’s Oragene OG-250 containers (DNA Genotek, cat. #: OG-250) and genomic DNA was extracted using DNA Genotek’s prepIT-L2P reagents (DNA Genotek, cat. #: PT-L2P). NanoDrop 2000 spectrophotometer and Quant-IT PicoGreen (Thermo Scientific, cat. #: P7589) were used to assess the DNA quality and concentration.
Oxidative bisulfite conversion
The oxidative bisulfite conversion reaction was done using the CEGX True Methyl kit (Cambridge Epigenetix, Cat. #: CEGXTMS). Briefly, 1 μg of DNA from all samples was purified and denatured. DNA from each subject was then split in two equal reactions, one of which underwent chemical oxidation followed by bisulfite conversion, the other underwent mock oxidation (oxidant replaced by water) followed by bisulfite conversion. All bisulfite reactions were cleaned up using a bead-based purification and final elution was in 12 μl of elution buffer.
Infinium MethylationEPIC array
Immediately following elution, all reactions were processed through the EPIC array protocol. Briefly, 7 μl of converted DNA was denatured with 1 μl of 0.4N sodium hydroxide prior to whole genome amplification on the MSA4 plate. All other steps were followed as per manufacturer’s guidelines. Array bead chips were scanned on Illumina HiScan.
Data processing
Illumina’s Infinium assays utilize methylated and unmethylated probes that bind to specific loci to determine the (hydroxy)methylation values at a given CpG. Beta values represent the measured (hydroxy)methylation values, based on the intensities of these two probes. Beta values range from 0 to 1, and can be thought of as a methylation percentage. For example, a beta value of 0.2 can be thought of as 20% (hydroxy)methylation. Processing and statistical analysis of the EPIC array data was performed using the R statistical language version 3.3.0 and various packages of the Bioconductor project [17]. Specifically, minfi version 1.18.2 [18] was used to load, apply SWAN normalization [19], annotate probes, and filter probes with high detection P-values (>0.01) across multiple samples (30%), probes mapping to known SNPs, and non-CpG methylation probes.
The threshold for calling 5hmC or 5mC was set to 0.0799, which corresponds to the 95th percentile of all negative ΔβBS-oxBS values observed across all samples and sites [15]. Detection of 5hmC or 5mC with the same tissue was called if the threshold value was exceeded in both replicates.
Representative gene analysis
Gene models as defined in the Bioconductor package TxDb.Hsapiens.UCSC.hg19.knownGene version 3.2.2 were used to define TSS5000 (5 kb upstream), gene body, and TES (5 kb downstream) regions. Genes for which one region overlapped with any region of a different gene were excluded. The remaining 13 695 TSS5000, gene bodies, and TES5000 were each split in 100 bins of equal relative size. Site measurements (βBS, βox, ΔβBS-oxBS) were averaged within each bin and then averaged across genes.
Results
Quality control measures of the oxBS assay and EPIC array
As global levels of 5hmC differ between tissue types [20–22], here, in technical replicates from three human biological tissues, we sought to compare whether the additional probes on the EPIC array yield valuable data for each modification. To do so, DNA from human cerebellum, blood, and saliva was extracted and split in two. Each aliquot was processed through the oxBS protocol independently and was loaded into separate wells on the EPIC array bead chip. Interrogation of the oxBS digestion and sequencing controls (Supplementary Fig. S1) and comparison of the bisulfite conversion efficiencies from the oxBS kit with that of the commonly used Zymo EZ DNA Methylation bisulfite conversion kit (Supplementary Fig. S2) confirm the efficiency of both the oxidative and the bisulfite conversion reagents in the oxBS kit. Furthermore, quality control measures from the array, which show comparable total signal between samples, appropriate hierarchical clustering, and expected multidimensional scaling, can be found in Supplementary Fig. S3.
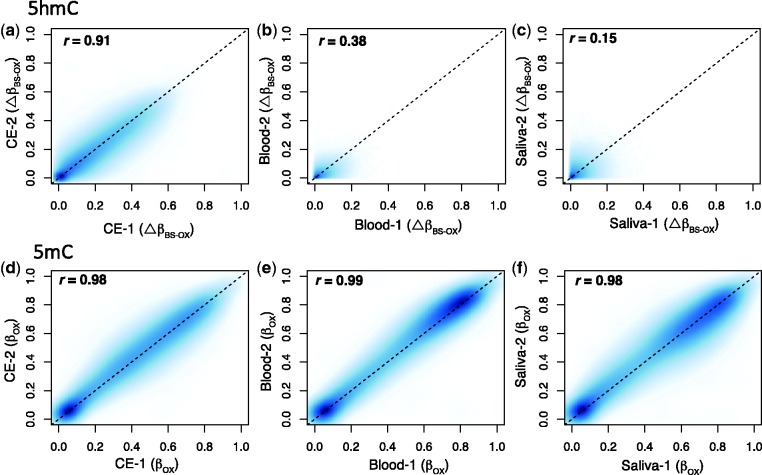
Using oxBS, 5mC values are represented by the beta values in the oxBS fraction (βoxBS), whereas 5hmC values (ΔβBS-oxBS) are determined by subtracting βoxBS from the beta values in the BS fraction (βBS). Technical replicates in all three specimens showed significant linear correlations for both ΔβBS-oxBS (cerebellum: r = 0.91; blood: r = 0.38; saliva: r = 0.15; P < 0.05 for all) and βoxBS (cerebellum: r = 0.98; blood: r = 0.99; saliva: r = 0.98; P < 0.05 for all) (Fig. 1a–f). The variability in the ΔβBS-oxBSr-coefficients across the three tissue types appears to be a consequence of the lower levels of 5hmC in blood and saliva. As such, these results suggest this method to be highly replicable.
Figure 1.
oxBS-EPIC shows strong replicability in cerebellum, blood, and saliva. Scatter plots confirm the replicability of 5hmC in (a) brain (r = 0.91), (b) blood (r = 0.38), and (c) saliva (r = 0.15) and 5mC in (d) brain (r = 0.98), (e) blood (r = 0.99), and (f) saliva (r = 0.98) across two technical replicates. P-values are < 0.05 for all comparisons.
Differing levels of 5mC and 5hmC in human tissues
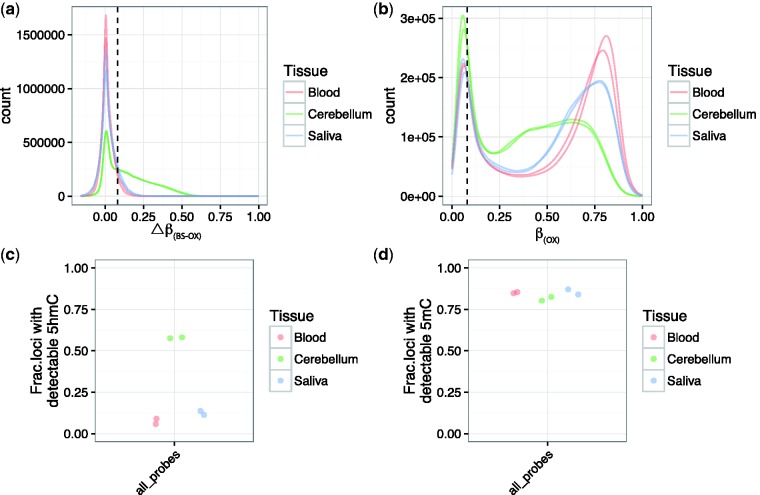
After filtering based on quality control parameters, we were left with 831 828 probes. ΔβBS-oxBS and βoxBS values were mostly positive in all three tissue types, as expected (Fig. 2a and b). There were, however, negative ΔβBS-oxBS values, which likely represent technical variations resulting from the oxBS and EPIC array protocols. To account for these areas of potential error, and in line with previous work using oxBS on the 450K array [15], we considered probes to contain a detectable level of 5hmC if the probe’s ΔβBS-oxBS > 0.0799 in both replicates, which represents the 95th percentile of negative ΔβBS-oxBS values (see “Materials and Methods” section for additional information). Common to both replicates, there were 432 418 and 660 880 probes containing detectable 5hmC and 5mC in human cerebellum, 18 999 and 697 291 probes containing detectable 5hmC and 5mC in blood, and 27 478 and 694 234 probes containing detectable 5hmC and 5mC in saliva (Fig. 2c and d). Of these detectable probes, we found 2528 5hmC-containing and 642 032 5mC-containing loci to be common across all three tissues types (Supplementary Fig. S4). Probes containing detectable 5hmC common across the three tissue types showed a similar distribution of ΔβBS-oxBS compared to the genome-wide probe set. Similarly, these probes were found across all chromosomes and genomic features (Supplementary Fig. S4). Interestingly, gene ontology analyses showed these common probes with detectable 5hmC to be related primarily to immune system and immune response (Supplementary Table S1). The low number of overlapping sites for 5hmC is explained by the low abundance of this modification in tissues other than the brain [13, 22, 23], and in this case in blood and saliva. In addition, this is consistent with the expectation that 5hmC shows a very strong tissue-specific variation [20–22,24–26]. The overlap of 5mC-containing probes across three tissue types suggests a higher stability of 5mC across tissues, although this may be a function of the loci specifically chosen for this array. Together, these results show the ability of the EPIC array to detect and differentiate 5mC and 5hmC, especially in human brain tissue, when combined with oxBS.
Figure 2.
Both 5hmC and 5mC are present at detectable levels in all three tissue types. (a) 5hmC and (b) 5mC detectability was determined using a Δβ threshold of 0.0799 (dashed line). (c) An average of 57.8 ± 0.3%, 7.4 ± 1.6%, and 12.5 ± 1.2% of loci contained detectable 5hmC in cerebellum, blood, and saliva, respectively. (d) Of all loci, >80% showed detectable 5mC in all three tissue types, with an average of 81.3 ± 1.1%, 85.1 ± 0.3%, and 85.5 ± 1.5% in cerebellum, blood, and saliva, respectively. Data from (c) and (d) include ‘all probes’ regardless of annotation or genomic location.
Characterizing 5mC and 5hmC across chromosomes and genomic features
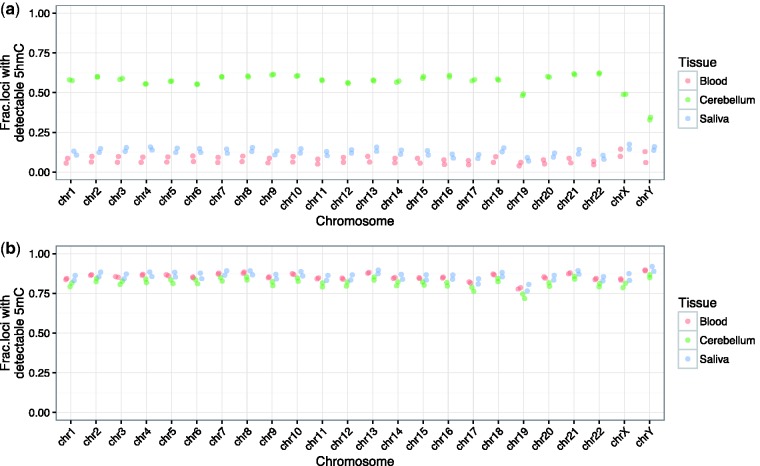
Within each tissue, we aimed to see whether the number of probes with detectable 5mC and 5hmC varied across chromosomes. To do so, we plotted the number of loci with detectable 5hmC or 5mC per given chromosome, corrected for the total number of probes mapping to that particular chromosome on the array. For blood and saliva, although the absolute number of probes is low, we did observe an even distribution of detectable 5hmC loci across chromosomes. Interestingly, in cerebellum, >50% of probes on each autosomal chromosome contain detectable 5hmC levels, with chromosome 19 being the only exception. Also in this tissue, chromosome X shows a similar fraction of detectable 5hmC compared to the autosomes, whereas chromosome Y shows a reduction (Fig. 3a). Unlike 5hmC, and as expected given the purpose of the array, >75% of probes contain detectable 5mC across autosomes and sex chromosomes, regardless of tissue, with chromosome 19 again being the sole exception with the fraction being just short of this percentage (Fig. 3b). The decrease in loci containing both 5mC and 5hmC on chromosome 19 could be a result of it being the chromosome with the greatest gene density (Supplementary Fig. S5) [27]. Nevertheless, the consistency of 5hmC and 5mC across chromosomes reinforces the value of this approach in genome-wide cytosine modification analyses and provides insight into the stability of 5hmC in mammalian brain DNA.
Figure 3.
The EPIC array contains an equal distribution of probes with detectable 5hmC and 5mC across chromosomes. (a) Although the fraction of probes containing 5hmC in blood and saliva is greatly reduced, all three tissue types show an equal distribution across autosomes. Of particular importance, in cerebellum, >50% of all probes on autosomes show detectable 5hmC, with the exception of chromosome 19, at>48%. Detectability is reduced on sex chromosomes, primarily chromosome Y. (b) Also with the exception of chromosome 19, 5mC is detectable at >75% of probes on all chromosomes in all three tissue types, including chrX and chrY.
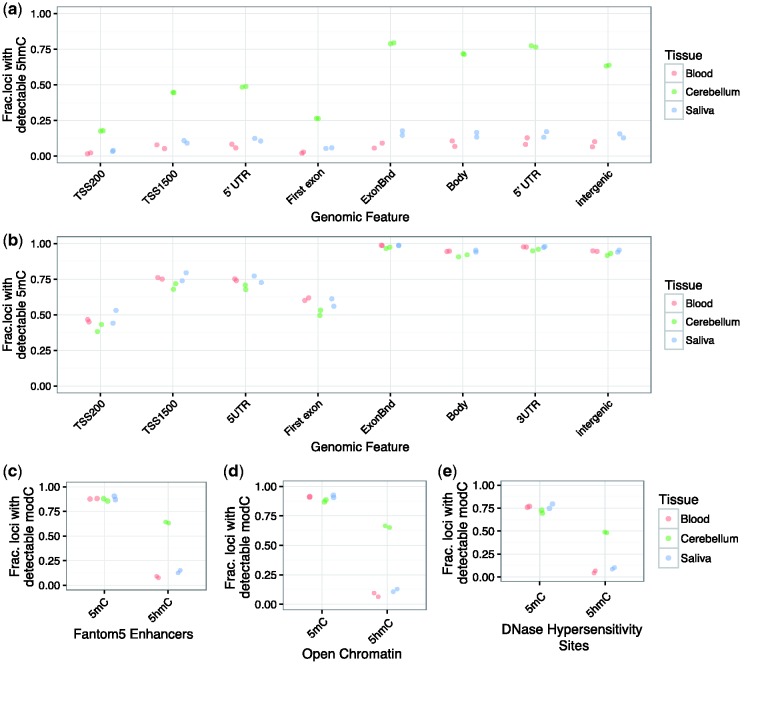
We next looked at patterns of loci with detectable 5hmC and 5mC probes across genomic features annotated on the array, which include regions downstream of the transcription start sites (TSS200 and TSS1500), the 5′ untranslated region (5′ UTR), the first exon, exon boundaries, gene bodies, 3′ UTR, and intergenic regions. Similar to the analyses across chromosomes, we again plotted the number of loci with detectable 5hmC or 5mC per genomic feature, corrected for the total number of probes on the array mapping to that particular genomic feature. We show an increase in the number of probes with detectable 5hmC in gene bodies, exon boundaries, and intergenic regions, whereas the regions within 200 bp of the TSS and the annotated first exon showed the lowest levels of 5hmC-detecting probes (Fig. 4a). Although the fraction of probes containing detectable 5hmC is not an indication of the absolute levels of this modification in the particular region, these findings are in line with previous reports highlighting the abundance of 5hmC in gene bodies and enhancer regions [28, 29]. Following the same pattern of abundance as 5hmC from one feature to the next, nearly 100% of probes contained within the annotated exon boundaries, gene body, 3′ UTR, and intergenic regions showed detectable 5mC, while this value was reduced to <75% for upstream regions of the gene (TSS200, TSS1500, 5′ UTR, and first exon) (Fig. 4b). Certainly, the large number of probes containing 5mC is not surprising since these probes were chosen specifically for this methylation array. Nevertheless, this profile was consistent across all tissue types, which lends to the notion that 5mC shows stable patterns across tissue types.
Figure 4.
The additional probes added to the EPIC array provide substantial 5hmC data in regions likely to be phenotypically relevant. (a) The fraction of probes containing detectable 5hmC in cerebellum is highest in the gene body compared to proximal promoter regions and the first exon. Detectable 5hmC probes in blood and saliva follow a similar pattern, but, as expected, are greatly reduced in number. (b) The fraction of probes containing detectable 5mC is similar in all three tissue types and also shows the highest levels in intragenic and intergenic regions, apart from regions proximal to the TSS. The additional probes on the EPIC array in the (c) Fantom5 enhancers, (d) open chromatin regions, and (e) DNase hypersensitivity sites show a strong abundance of probes containing both 5mC and 5hmC, suggesting that this array is more suitable for discovery of differentially modified regions in case–control analyses.
We further analyzed annotated regions with putative functional properties, such as Fantom5 enhancer regions, DNase hypersensitivity sites, and regions of open chromatin. The fraction of loci with detectable 5mC was always >75%, regardless of tissue (Fig. 4c–e, left half of graphs), whereas, for human brain in particular, 50% of the probes within these regions contained detectable 5hmC (Fig. 4c–e, right half of graphs). As expected due to the lower global levels of 5hmC, around 10% of probes associated with Fantom5 enhancers, DNase hypersensitivity sites, and regions of open chromatin contained detectable 5hmC in blood and saliva. Although gene ontology analyses did not identify specific differences in functional enrichment of 5hmC between DNase hypersensitivity sites, regions of open chromatin, and Fantom5 enhancer regions (Supplementary Tables S2–S4, respectively), in their entirety, these results confirm the utility of the tested methodology to identify 5hmC in different tissues, especially human brain.
Variance of 5mC and 5hmC across a representative gene
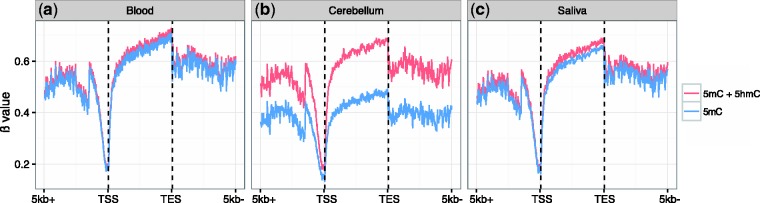
The abundance of 5hmC in human brain and the inability of standard methods commonly used to study 5mC to differentiate between 5mC and 5hmC raises the question of how much of the variance in cytosine modifications across a gene may be accounted for by 5mC alone. To do so, we created an archetype gene comprising a 5 kb region upstream of the TSS, a region between the TSS and the TES, and a 5 kb region downstream of the TES. This archetype included βoxBS (5mC) and βBS (5mC + 5hmC) values representative of >13 695 genes on this array. Given the low levels of 5hmC in blood and saliva, nearly all of the signal from the array was accounted for by 5mC (Fig. 5a and c). However, our results showed that the 5mC signal in human cerebellum accounted for approximately two-third of the total signal from cytosine modifications (Fig. 5b), at least in the genomic regions covered by the EPIC array. This finding supports the idea that studies in mammalian brain using bisulfite conversion alone may not properly measure total 5mC content, and that part of the published results may be explained by differences in 5hmC levels.
Figure 5.
5mC accounts for only two-third of the variance in cytosine modifications in human cerebellum. For blood (a), cerebellum (b), and saliva (c), βoxBS (blue lines, 5mC) and βBS (red lines, 5mC + 5hmC) were plotted from 5 kb upstream of the TSS (5 kb+) to the TSS, across the intragenic region to the TES, and 5 kb downstream of the TES (5 kb−) for >13 695 genes containing detectable 5mC. For blood and saliva, the vast majority of cytosine modification signal from bisulfite-converted DNA is that of 5mC, with the gene body region in saliva samples showing slightly more contribution from 5hmC. For cerebellum, however, 5mC contributes to only two-third of the cytosine modification signals, not only in the gene body but also across the entire representative genome.
Discussion
Enrichment-based approaches, such as 5hmC immunoprecipitation (hMeDIP) or selective chemical labeling (hMe-SEAL), have been combined with next-generation sequencing to allow for an effective, low-cost whole-genome analysis of 5hmC. To achieve base pair resolution, methodologies utilize modified bisulfite conversion protocols, such as oxBS- and TAB-Seq. However, with the high cost associated with whole-genome studies, alternative approaches became necessary. In this manuscript, we combine oxBS with the EPIC array to study both 5mC and 5hmC. Our results demonstrate not only the replicability of this methodology in all three tissue types, but also its ability to effectively study 5hmC in >400 000 loci in human brain.
The increasing number of publications showing 5hmC in different tissue types brings to light several important caveats to consider when looking at previously published bisulfite-based results. Data generated using a bisulfite alone, whether combined with whole- or reduced-genome sequencing arrays, prevent researchers from dissociating 5mC from 5hmC. Certainly, this issue will be more prominent in studies using DNA from mammalian brain, regardless of platform, but it should not be overlooked in studies using tissues with lower levels of 5hmC or in studies using previous array technologies. As we move forward, researchers who generate data using bisulfite conversion alone should consider using the term cytosine modifications rather than methylation. Alternatively, the success of the oxBS-EPIC methodology presented in this manuscript provides researchers with an effective approach to simultaneously investigate both modifications, thereby adding validity to their findings.
The oxBS-EPIC methodology presented here may also provide insight into the debate on the function of cytosine modifications. On the one hand, it is proposed that 5mC’s oxidative products are destined for demethylation. Specifically, 5fC and 5caC are targeted and excised by thymine DNA glycosylase and the base excision repair pathway [30, 31]. However, given the relative abundance of 5hmC in DNA from mammalian brain tissue we show here and shown elsewhere [2, 32], it is thought that this modification may also be stable [33] and act as a transcriptional regulator [28, 29, 32, 34]. Of particular interest, compared to the 450K array, the EPIC array includes an additional 400 000 probes, most of which are located in regions such as Fantom5 enhancers, ENCODE enhancers and open chromatin regions, and DNase 1 hypersensitivity sites. The majority of the probes with detectable 5hmC discovered in this manuscript are located within these added probes, each of which has proposed transcriptional regulatory properties. Taken together, although the oxidation of 5mC to 5hmC can technically be deemed as demethylation, thereby making 5hmC an intermediate molecule in the demethylation pathway, its presence in mammalian DNA certainly has biological relevance in the realm of gene activation and repression.
Supplementary Material
Acknowledgements
We would like to thank Cambridge Epigenetix and Illumina for providing the reagents for the oxidative bisulfite conversion and the Infinium MethylationEPIC arrays, respectively.
Conflict of interest statement
None declared.
Funding
This work is supported by grants to G.T. from the Canadian Institute of Health Research [MOP93775, MOP11260, MOP119429, and MOP119430], from the National Institutes of Health [1R01DA033684], and an investigator-initiated grant from Pfizer Canada, as well as by the Fonds de recherche du Québec - Santé through a Chercheur National salary award and through the Quebec Network on Suicide, Mood Disorders and Related Disorders; and by a grant to G.B. from Genome Canada. J.A.G. is supported by a CIHR Frederick Banting and Charles Best Doctoral fellowship. P.E.L. is supported by the Fondation Fyssen, the Canadian Institutes of Health Research, the American Foundation for Suicide Prevention, the Fondation Deniker, and the Fondation pour la Recherche Médicale.
References
- 1. Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010;11:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009;324:929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324:930–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ito S, D'Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010;466:1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011;333:1300–03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet 2014;15:647–61. [DOI] [PubMed] [Google Scholar]
- 7. Huang Y, Pastor WA, Shen Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 2010;5:e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Booth MJ, Branco MR, Ficz G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012;336:934–37. [DOI] [PubMed] [Google Scholar]
- 9. Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012;149:1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gu H, Smith ZD, Bock C, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011;6:468–81. [DOI] [PubMed] [Google Scholar]
- 11. Lee YK, Jin S, Duan S, et al. Improved reduced representation bisulfite sequencing for epigenomic profiling of clinical samples. Biol Proced Online 2014;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nazor KL, Boland MJ, Bibikova M, et al. Application of a low cost array-based technique - TAB-Array - for quantifying and mapping both 5mC and 5hmC at single base resolution in human pluripotent stem cells. Genomics 2014;104:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stewart SK, Morris TJ, Guilhamon P, et al. oxBS-450K: a method for analysing hydroxymethylation using 450K BeadChips. Methods 2015;72:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Field SF, Beraldi D, Bachman M, et al. Accurate measurement of 5-methylcytosine and 5-hydroxymethylcytosine in human cerebellum DNA by oxidative bisulfite on an array (OxBS-array). PLoS One 2015;10:e0118202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lunnon K, Hannon E, Smith RG, et al. Variation in 5-hydroxymethylcytosine across human cortex and cerebellum. Genome Biol 2016;17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dumais A, Lesage AD, Lalovic A, et al. Is violent method of suicide a behavioral marker of lifetime aggression? Am J Psychiatry 2005;162:1375–78. [DOI] [PubMed] [Google Scholar]
- 17. Huber W Carey VJ Gentleman R. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol 2012;13:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Globisch D, Munzel M, Muller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010;5:e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011;2011:870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nestor CE, Ottaviano R, Reddington J, et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res 2012;22:467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Godderis L, Schouteden C, Tabish A, et al. Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res Int 2015;2015:845041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kinney SM, Chin HG, Vaisvila R, et al. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem 2011;286:24685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munzel M, Globisch D, Bruckl T, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angewandte Chemie 2010;49:5375–77. [DOI] [PubMed] [Google Scholar]
- 26. Terragni J, Bitinaite J, Zheng Y, et al. Biochemical characterization of recombinant beta-glucosyltransferase and analysis of global 5-hydroxymethylcytosine in unique genomes. Biochemistry 2012;51:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Surralles J, Sebastian S, Natarajan AT. Chromosomes with high gene density are preferentially repaired in human cells. Mutagenesis 1997;12:437–42. [DOI] [PubMed] [Google Scholar]
- 28. Gross JA, Pacis A, Chen GG, et al. Characterizing 5-hydroxymethylcytosine in human prefrontal cortex at single base resolution. BMC Genomics 2015;16:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wen L, Li X, Yan L, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol 2014;15:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011;333:1303–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen L, Wu H, Diep D, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 2013;153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bachman M, Uribe-Lewis S, Yang X, et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature Chem 2014;6:1049–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mellen M, Ayata P, Dewell S, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012;151:1417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.