Abstract
PURPOSE
Patients with acute myeloid leukemia (AML) in remission remain at risk for relapse even after allogeneic hematopoietic cell transplantation (alloHCT). AML measurable residual disease (MRD) status before alloHCT has been shown to be prognostic. Whether modulation of the intensity of the alloHCT conditioning regimen in patients with AML who test positive for MRD can prevent relapse and improve survival is unknown.
METHODS
Ultra-deep, error-corrected sequencing for 13 commonly mutated genes in AML was performed on preconditioning blood from patients treated in a phase III clinical trial that randomly assigned adult patients with myeloid malignancy in morphologic complete remission to myeloablative conditioning (MAC) or reduced-intensity conditioning (RIC).
RESULTS
No mutations were detected in 32% of MAC and 37% of RIC recipients; these groups had similar survival (3-year overall survival [OS], 56% v 63%; P = .96). In patients with a detectable mutation (next-generation sequencing [NGS] positive), relapse (3-year cumulative incidence, 19% v 67%; P < .001) and survival (3-year OS, 61% v 43%; P = .02) was significantly different between the MAC and RIC arms, respectively. In multivariable analysis for NGS-positive patients, adjusting for disease risk and donor group, RIC was significantly associated with increased relapse (hazard ratio [HR], 6.38; 95% CI, 3.37 to 12.10; P < .001), decreased relapse-free survival (HR, 2.94; 95% CI, 1.84 to 4.69; P < .001), and decreased OS (HR, 1.97; 95% CI, 1.17 to 3.30; P = .01) compared with MAC. Models of AML MRD also showed benefit for MAC over RIC for those who tested positive.
CONCLUSION
This study provides evidence that MAC rather than RIC in patients with AML with genomic evidence of MRD before alloHCT can result in improved survival.
INTRODUCTION
Acute myeloid leukemia (AML) is a cancer that is typically widely disseminated at diagnosis; however, unlike many other metastatic cancers, cytotoxic chemotherapy is often effective at achieving morphologic complete remission (CR). Allogeneic hematopoietic cell transplantation (alloHCT) is a standard postremission strategy to prevent relapse in patients with AML.1 This approach is suboptimally effective, however, with death as a result of AML representing the most common form of transplant treatment failure.2
Transplant-related morbidity and mortality is also problematic and results in underuse of alloHCT in patients with AML who may benefit. The toxicity from alloHCT is decreasing though,3 in part because of the adoption of preparative conditioning regimens of reduced intensity. Large retrospective analyses4 have suggested that reduced transplant-related mortality (TRM) in reduced-intensity conditioning (RIC) is counterbalanced by increased relapse rates, but 2 randomized phase III trials5-7 that compared RIC with myeloablative conditioning (MAC) failed to clearly establish which approach is preferable in patients with AML eligible for either form of conditioning.
Morphologic CR in AML represents a highly heterogenous state with a wide range of residual leukemia burden that ranges from truly cured to relapse imminent.8-14 It has been shown that measurable residual disease (MRD) testing, using either flow cytometric or molecular methods, for patients with AML in CR at the pretransplantation landmark time point can predict post-transplantation relapse and survival.9,11,15 Given the genetic etiology of AML16-18 and the success of monitoring by quantitative polymerase chain reaction (qPCR) in acute promyelocytic and NPM1-mutated AMLs,10,19 there is interest in the detection and quantification of AML-associated mutations to estimate residual disease burden using next-generation sequencing (NGS).13,20-24 In the current study, we performed unbiased ultra-deep DNA-based NGS on preconditioning blood samples taken before random assignment to either MAC or RIC to determine the impact of alloHCT conditioning intensity on patients with AML in morphologic CR but with genomic evidence of residual disease.
METHODS
Clinical Cohort
We performed ultra-deep DNA sequencing of blood samples from patients treated in the Blood and Marrow Transplant Clinical Trials Network (BMT CTN) 0901 (ClinicalTrials.gov identifier: NCT01339910), a phase III randomized clinical trial that compared outcomes by conditioning intensity in adult patients with myeloid malignancy undergoing an alloHCT while in morphologic CR (ie, < 5% marrow myeloblasts at the time of pretransplantation assessment).5 Frozen whole blood collected during remission before the conditioning regimen was available from 190 of the 218 patients with AML and came equally from those randomly assigned to either MAC or RIC. Patients were well matched for baseline characteristics, including age, sex, comorbidity, disease risk, disease duration, cytogenetics, donor type and match, graft type, and antithymocyte globulin (ATG) use (Table 1). Extended follow-up of patients enrolled in this protocol was extracted from the Center for International Blood and Marrow Transplant Research (CIBMTR) research database. Median follow-up in survivors was > 49 months. Patient characteristics and clinical outcomes were aligned with those previously reported for patients with AML in this trial (Data Supplement, online only). All patients provided written informed consent to participate in both the BMT CTN 0901 trial and the CIBMTR research database. This study was approved by the BMT CTN and CIBMTR and conducted with approval of the National Marrow Donor Program institutional review board.
TABLE 1.
Patient Clinical Characteristics

Sequencing
Targeted DNA sequencing using a custom 51-Kb–anchored multiplex PCR-based panel designed to incorporate molecular barcode/unique molecular identifiers (UMIs) and cover regions of 13 commonly mutated genes in AML (ASXL1, DNMT3A, FLT3, IDH1, IDH2, JAK2, KIT, NPM1, NRAS, RUNX1, SF3B1, TET2, and TP53) was performed on 400 ng of genomic DNA isolated from each blood sample. A dual pre- and post-PCR–separated automated liquid-handling robot workflow was used for library preparation. Paired-end 150-bp sequencing was performed with unique dual-sample indices on a HiSeq 2500 system (rapid run mode; Illumina, San Diego, CA), with an average of 41 million paired-end reads per sample. Details are provided in the Data Supplement.
Bioinformatics
Consensus sequences based on UMI read families were mapped to human genome version hg19 (build GRCh37). De novo variant calls using a minimum allele frequency of 0.001 were filtered using information with regard to UMI read families, background error rate models, unique start sites, strand-specific priming, and homopolymer runs to determine high-confidence, error-corrected mutations. Alternative approaches were used for insertional mutations in NPM1 and FLT3 internal tandem duplication (ITD). Confirmation of error-corrected NGS specificity was achieved with orthogonal validation of variants by digital droplet PCR on the RainDrop platform (RainDance Technologies, Lexington, MA). Details are provided in the Data Supplement.
Statistical Analysis
Kaplan-Meier estimation and log-rank test were used for the analyses of overall survival (OS) and relapse-free survival (RFS) end points. Cumulative incidence of relapse end points with TRM as a competing risk were evaluated using the method of Gray. Cox proportional hazards regression models were fit by forcing conditioning intensity, disease risk, and donor type into all models and using stepwise selection to identify additional variables (from among recipient sex, age, comorbidity index, and use of ATG) associated with all different end points. Details are provided in the Data Supplement.
RESULTS
Mutations Detected
In patients in morphologic CR, 275 mutations were detected, with a median variant allele frequency (VAF) of 0.5% (Fig 1A; Data Supplement). In samples with mutations detected, the median number of variants was 2 (range, 1-9 variants; Fig 1B). Mutations were well distributed between patients who subsequently received MAC or RIC (n = 139 v 136). Patients with mutations were older than those without mutations (median age, 57 v 50 years, respectively; P < .001) but did not differ for other measured baseline factors (Data Supplement). No mutations were detected in 32% of MAC and 37% of RIC samples.
FIG 1.
Detection of mutations in the blood of patients with acute myeloid leukemia (AML) during complete remission (CR). (A) The allele frequency of each mutation detected in each gene in the blood of patients with AML during morphologic CR before transplantation is shown. Allele frequencies for FLT3 internal tandem duplication (ITD) and NPM1 variants detected at < 0.1% are plotted at 0.1%. The majority of variants (88%) had an allele frequency < 10%, with exceptions primarily occurring in genes known to be associated with clonal hematopoiesis. (B) The total number of mutations detected per patient and the distribution across patients are shown, including patients with at least 1 mutation detectable in DNMT3A, TET2, or ASXL1 (DTA) genes and those with no detectable DTA mutation. (C) A network analysis of the co-occurrence of mutations on the gene level within patient samples is shown. Each gene is depicted as a node, and mutational co-occurrence is depicted as an edge, where node color is representative of the total number of genes found to be concurrently mutated and the distance between nodes is proportional to the rate of co-occurrence across all patient samples.
Mutations were found in all genes evaluated. Within patient samples, patterns of mutational co-occurrence between genes were observed (Figs 1B and C). DNMT3A, TET2, and ASXL1 (DTA) mutations, associated with age-related clonal hematopoiesis,13,25,26 were frequently observed, with at least 1 DTA mutation detected in 45% of patients. The median detected VAF for DTA mutations (1.0%; range, 0.1%-46.8%) was higher than for non-DTA mutations (0.4%; range, 0.03%-41.3%; P < .001). Patients with DTA mutations were older than those without DTA mutations (median age, 58 v 52 years, respectively; P < .001). Forty-five percent of patients with DTA mutations also had at least 1 non-DTA mutation detected. As would be expected with previously treated patients with cancer, detection of TP53 mutations was also common.27 TP53 mutations were typically found at lower VAF than DTA mutations (median, 0.4%; range, 0.01%-31.0%; P = .01).
Given that the original mutational profile present at diagnosis was not available for comparison, orthogonal validation using digital droplet PCR was performed for 15% of the mutations detected by ultra-deep sequencing in preconditioning remission samples. All were confirmed, with excellent correlation in allele frequency between the 2 methodologies (Data Supplement).
Clinical End Points
TRM was higher in those who underwent MAC (27% at 3 years; 95% CI, 18% to 36%) compared with those who received RIC (9% at 3 years; 95% CI, 4% to 15%; P = .001). No differences in TRM were observed between groups on the basis of mutation detection status (Fig 2A).
FIG 2.
Impact of conditional intensity and mutational status on clinical outcomes. (A) Differences in rates of transplant-related mortality (TRM) were identified between subgroups defined by conditioning intensity and mutational status (P = .02). TRM was significantly higher in patients who underwent treatment with myeloablative conditioning (MAC) v reduced-intensity conditioning (RIC; P = .001), but there was no difference on the basis of mutational status (P = .8). Rates of relapse were different between subgroups (P < .001), with RIC having a higher relapse rate than MAC (P < .001) and the highest rate occurring in next-generation sequencing (NGS) positive patients who received RIC (P < .001). (B) In patients with no mutations detected (NGS negative), overall survival (OS) did not differ on the basis of conditioning intensity (3-year OS, 63% RIC v 56% MAC; P = .96). However, in those with detectable mutations, survival was significantly worse in those who received RIC (3-year OS, 43% RIC v 61% MAC; P = .02).
Overall, 41% of patients experienced relapse after transplantation. The majority of relapses occurred within the first year (77%), with a median time to relapse of 3.3 months in those randomly assigned to RIC v 8.1 months in those randomly assigned to MAC. Most patients who experienced relapse (71%) had a mutation detectable in blood before conditioning. The relapse rate was higher in RIC than in MAC (1-year cumulative incidence, 47% v 15%; P < .001). Those with a mutation detected (NGS positive) who were randomly assigned to RIC experienced a higher relapse rate than those randomly assigned to MAC (1-year cumulative incidence, 58% v 14%; P < .001; Fig 2A).
There were significant survival differences between groups defined by NGS and conditioning regimen (Fig 2B). For patients with no mutations detected, survival was not significantly different based on conditioning intensity (3-year OS, 63% RIC v 56% MAC; P = .96). However, for patients with mutations detected, those randomly assigned to RIC experienced inferior survival compared with those randomly assigned to MAC (3-year OS, 43% v 61%, respectively; P = .02).
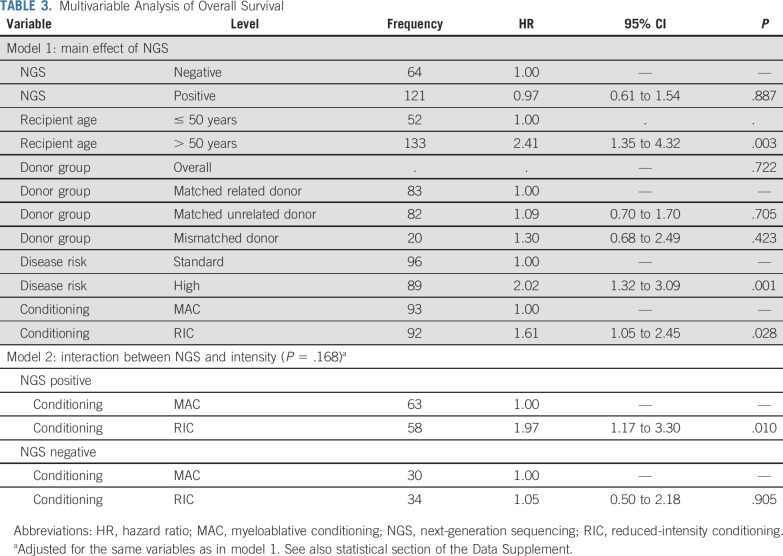
In multivariable analysis for those patients who were NGS positive pretransplantation, adjusting for disease risk and donor group, RIC was significantly associated with increased relapse (hazard ratio [HR], 6.38; 95% CI, 3.37 to 12.10; P < .001), decreased RFS (HR, 2.94; 95% CI, 1.84 to 4.69; P < .001), and decreased OS (HR, 1.97; 95% CI, 1.17 to 3.30; P = .01) compared with MAC (Tables 2 and 3).
TABLE 2.
Multivariable Analysis of Relapse
TABLE 3.
Multivariable Analysis of Overall Survival
Risk by Mutation Type
Mutations detected before treatment give an insight into leukemia biology and have utility in AML risk stratification.1,17,18,28 Whether mutations have different prognostic implications when detected in clinical remission at an extremely low frequency is unknown. In addition, while all 13 genes sequenced here have been found to be recurrently mutated in patients with AML,17,18,28 several of these mutations have also been found in aging adults without myeloid malignancy25,26 and in patients with non-AML cancer after chemotherapy.27 Without diagnostic samples available to test, it is not possible to determine whether mutations detected are found within the leukemic clone. We therefore evaluated the risk of relapse associated with detection of a mutation for each gene sequenced.
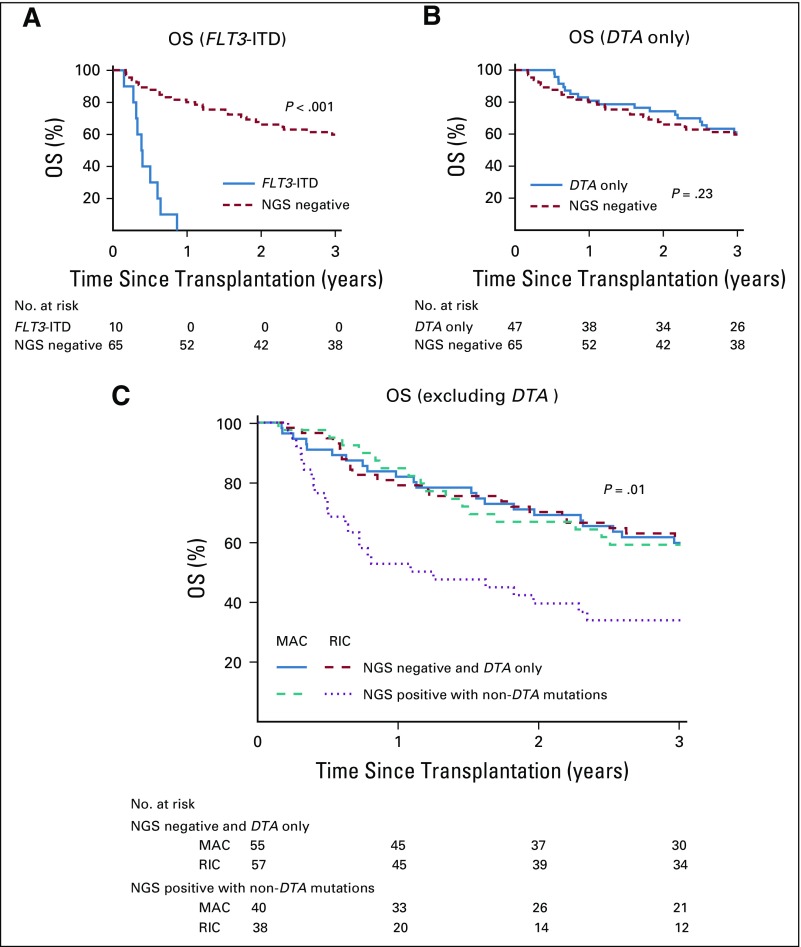
The prognostic implications of detecting a mutation differed according to subsequent conditioning intensity (Fig 3; Data Supplement). For example, excluding those who died as a result of TRM, all patients with a detectable IDH1, IDH2, SF3B1, or non-ITD FLT3 mutation who received RIC experienced relapse when compared with no relapses in those with such mutations who received MAC. Detection of mutated NPM1 (NPM1mut) and/or FLT3-ITD was found to be highly associated with relapse. NPM1mut is well validated as a marker of AML MRD10,14 and was detected in 11 patients (VAF, 0.03%-2.19%), of whom 9 experienced relapse. Four patients with detectable NPM1mut were randomly assigned to MAC, including 2 who did not experience relapse. FLT3-ITD was detected in 10 patients (VAF, 0.03%-3.97%), 5 of whom also had detectable NPM1mut. All 10 patients died after transplantation, 1 as a result of TRM and 9 as a result of relapse (Fig 4A). Of the 10 patients reported as having an FLT3-ITD mutation present at initial AML diagnosis, 7 had no detectable FLT3-ITD pretransplantation, of whom only 1 experienced relapse.
FIG 3.
Relapse outcomes of patients with specific gene mutations detected. The percentage of patients with mutations in each gene separated by conditioning intensity and excluding those with transplant-related mortality (TRM) is shown. For multiple genes (IDH1, IDH2, SF3B1, non–internal tandem duplication [ITD] FLT3, NPM1), differences in whether a patient experienced relapse v no relapse were observed on the basis of treatment with reduced-intensity conditioning (RIC) v myeloablative conditioning (MAC).
FIG 4.
Prognostic implications of detected gene mutations. (A) Patients with a detectable FLT3 internal tandem duplication (ITD) mutation had a significantly decreased rate of overall survival (OS) compared with patients with no detectable mutation (next-generation sequencing [NGS] negative; P < .001), while (B) patients with mutations detected only in ≥ 1 of DNMT3A, TET2, or ASXL1 (DTA) showed no difference in OS (P = .23). (C) A model was generated by considering those patients with mutations only in DTA as negative. Using this definition, OS for patients with detectable non-DTA mutations treated with reduced-intensity conditioning (RIC) was significantly worse than for other subgroups (P = .01). MAC, myeloablative conditioning.
DTA mutations13,23 did not seem to have prognostic implications for survival (Fig 4B). Exclusion of DTA mutations previously described as nonprognostic in AML post-treatment was evaluated. This new definition resulted in 41% of patients classified as positive (40% RIC v 42% MAC). Consistent with classification using the broader definition, positive patients who received RIC experienced higher rates of relapse (3-year cumulative incidence, 72% v 15%; P < .001) and lower survival rates (3-year OS, 34% v 59%; P = .01) compared with those randomly assigned to MAC (Fig 4C). Additional exploratory analyses that exclude TP53 mutations or limit the range of allele frequencies considered were also strongly supportive of the findings of this study (Data Supplement).
DISCUSSION
The purpose of alloHCT in patients with AML in CR is to prevent relapse. Our central hypothesis was that the benefit of MAC in preventing relapse in patients with AML in morphologic CR would be greatest for those with genomic evidence of residual leukemia. We confirm here that the detection of an AML-associated mutation in patients with AML in morphologic CR before RIC alloHCT is associated with a higher relapse rate after transplantation.23 We also show that such determination can be made without reference to those mutations previously present at initial diagnosis. We demonstrate that the prognostic impact of detectable mutations at CR pre-alloHCT depends on conditioning intensity, with higher relapse and worse survival in those who were randomly assigned to receive RIC rather than MAC.
Samples from initial diagnosis and post-transplantation time points were not available for assessment to determine an association with those variants detected in remission before transplantation. Because of the nature of the sample type available, we were unable to perform other AML MRD assessments, such as qPCR, flow cytometry, or single-cell sequencing; overlap between these approaches and NGS is not addressed by this study. Similarly, DNA from blood at a single time point did not allow assessment of kinetics of mutational clearance,20,29 clonal architecture,30-32 or immune factors.33 It is possible that mutation detection rates would have differed in bone marrow aspirate; however, such samples were not available for testing.
Consistent with prior reports,13,23 DTA mutations were among the most commonly detected in this study but had limited prognostic value. In addition, we found a higher rate of TP53 mutations than previously described in newly diagnosed adult patients with AML.17,18,28 TP53 mutations in AML are believed to be associated with resistance to conventional cytotoxic chemotherapy (but possibly not to decitabine21) and are also detectable in the blood after chemotherapy in older adults with nonhematologic malignancies.27 Single-cell sequencing may have allowed determination if DTA, TP53, and co-occurring mutations were within a single leukemic clone, which would have helped to refine prognostic implications. The proportion of patients assigned to positive test status by diagnostic mutation agnostic sequencing (66%) is within the range of studies using NGS at remission to detect mutations present at diagnosis (45%-79%)22,23 but is higher than the MRD-positive fraction reported in analogous cohorts at the same treatment time point using qPCR (21%)15 or flow cytometry (24%).9 Detectable mutations may not always represent AML MRD. Models that exclude DTA (41%) or DTA and TP53 (23%) have characteristics consistent with AML MRD tests previously reported9,15,23 and continue to strongly demonstrate superiority of MAC over RIC for patients who test positive. The rate of relapse in the RIC arm of BMT CTN 09015 was higher than that observed in another randomized phase III trial6,7 but is likely due to the different RIC regimens used. The relapse rate in the NGS-negative patients who received RIC in this study was similar to that reported for RIC regimens in a large retrospective analysis from CIBMTR.34 We considered only patients with AML here, and the impact of conditioning intensity on residual disease in patients with myelodysplasia may be different.29,35,36
This study provides evidence that MAC rather than RIC in patients with AML and genomic evidence of MRD before alloHCT results in lower relapse and improved survival. It is conceivable that previously observed lower relapse rates in MRD-positive patients with AML who receive cord blood transplants compared with other allogeneic transplant types may also be related to conditioning intensity.37 Given that MAC is not a viable conditioning option for many patients with AML because of age or comorbidity, this study helps to focus efforts on those at highest risk of post-transplantation relapse. While MRD-based treatment approaches after transplantation have been reported,38 targeted AML therapies are now available,39,40 which may have particular utility in those demonstrated in the current study to be at highest relapse risk.
In patients with no mutations detected in blood before alloHCT, the increased relapse in the RIC arm was balanced by increased TRM in the MAC arm, which resulted in equivalent post-transplantation survival for these approaches. Future studies should confirm the hypothesis that RIC remains a reasonable approach in AML, even in those eligible for MAC, when no genomic evidence of residual disease is detected. The NGS-negative group in this cohort was small (34%) but included false negatives likely because of the limited number of genes sequenced in this study. It is conceivable that future work with broader sequencing, at the same depth, could better identify those for whom RIC is the preferable approach. Larger data sets may also allow determination of what type and level of residual leukemic clones are susceptible to which conditioning intensity and lead to a better understanding of the impact of allogeneic immune effects in relapse prevention.
ACKNOWLEDGMENT
This work used the National Heart, Lung, and Blood Institute Sequencing and Genomics Core and the computational resources of the National Institutes of Health high-performance computing Biowulf cluster (http://hpc.nih.gov).
PRIOR PRESENTATION
Presented at the 24th European Hematology Association Congress, Amsterdam, the Netherlands, June 13-16, 2019.
SUPPORT
Supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) and by Grant No. U10HL069294 to the Blood and Marrow Transplant Clinical Trials Network from NHLBI and the National Cancer Institute. This study used BMT CTN 0901 research materials, biospecimens, and clinical trial data provided by the BMT CTN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
See accompanying Editorial on page 1249
AUTHOR CONTRIBUTIONS
Conception and design: Christopher S. Hourigan, Laura W. Dillon, Brent R. Logan, Edwin P. Alyea, H. Joachim Deeg, Mehdi Hamadani, Bart L. Scott
Provision of study material or patients: Christopher S. Hourigan, Edwin P. Alyea, H. Joachim Deeg, Sergio Giralt, Alan Howard, Richard T. Maziarz, David L. Porter, Bart L. Scott, Mitchell E. Horwitz
Collection and assembly of data: Christopher S. Hourigan, Laura W. Dillon, Gege Gui, Jack Ghannam, Yuesheng Li, Edwin P. Alyea, Asad Bashey, H. Joachim Deeg, Sergio Giralt, Alan Howard, Bart L. Scott, Marcelo C. Pasquini, Mitchell E. Horwitz
Data analysis and interpretation: Christopher S. Hourigan, Laura W. Dillon, Gege Gui, Brent R. Logan, Mingwei Fei, Jack Ghannam, Abel Licon, Edwin P. Alyea, Asad Bashey, H. Joachim Deeg, Steven M. Devine, Hugo F. Fernandez, Sergio Giralt, Mehdi Hamadani, Alan Howard, Richard T. Maziarz, David L. Porter, Bart L. Scott, Erica D. Warlick, Marcelo C. Pasquini, Mitchell E. Horwitz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia With Genomic Evidence of Residual Disease
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/journal/jco/site/ifc..
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Christopher S. Hourigan
Research Funding: Merck Sharp & Dohme (Inst), Sellas Life Sciences (Inst)
Other Relationship: QIAGEN, ArcherDx
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1021742/summary
Brent R. Logan
Consulting or Advisory Role: Eisai, Daiichi Sankyo, Enlivex Therapeutics, Gamida Cell
H. Joachim Deeg
Honoraria: Incyte
Steven M. Devine
Honoraria: Kiadis
Consulting or Advisory Role: Bristol-Myers Squibb
Research Funding: Orca Biosystems (Inst), Kiadis (Inst)
Travel, Accommodations, Expenses: Orca Biosystems
Hugo F. Fernandez
Honoraria: Incyte, Jazz Pharmaceuticals
Speakers’ Bureau: Sanofi
Sergio Giralt
Honoraria: Celgene, Takeda Pharmaceuticals, Amgen, Jazz Pharmaceuticals, Sanofi
Consulting or Advisory Role: Celgene, Takeda Pharmaceuticals, Sanofi, Jazz Pharmaceuticals, Amgen, Janssen Pharmaceuticals, Actinuum, Bristol-Myers Squibb, Johnson & Johnson, Pfizer
Research Funding: Celgene (Inst), Takeda Pharmaceuticals, Miltenyi Biotec (Inst), Johnson & Johnson, Amgen, Actinuum, Sanofi
Travel, Accommodations, Expenses: Celgene, Sanofi, Amgen, Jazz Pharmaceuticals
Mehdi Hamadani
Honoraria: Celgene
Consulting or Advisory Role: MedImmune, Cellerant Therapeutics, Janssen Research & Development, Incyte, Pharmacyclics, ADC Therapeutics, Puma Biotechnology (I), Verastem
Speakers’ Bureau: Genzyme, Celgene, AstraZeneca
Research Funding: Takeda Pharmaceuticals, Spectrum Pharmaceuticals, Otsuka US, Astellas Pharma, Genzyme
Alan Howard
Travel, Accommodations, Expenses: Jazz Pharmaceuticals (Inst)
Richard T. Maziarz
Honoraria: Juno Therapeutics
Consulting or Advisory Role: Novartis, Incyte, Kite Pharma
Patents, Royalties, Other Intellectual Property: Athersys shared patent on the use of mesenchymal stromal cells for treatment of graft-versus-host disease
Travel, Accommodations, Expenses: Novartis, Juno Therapeutics, Incyte, Kite Pharma
David L. Porter
Employment: Genentech (I), Roche (I)
Stock and Other Ownership Interests: Genentech (I), Roche (I)
Consulting or Advisory Role: Novartis, Kite Pharma, Incyte, Gerson Lehrman Group, Bellicum Pharmaceuticals, Glenmark
Research Funding: Novartis
Patents, Royalties, Other Intellectual Property: Patent/royalty rights
Travel, Accommodations, Expenses: Kite Pharma, Novartis, Janssen Pharmaceuticals
Other Relationship: National Marrow Donor Program, American Board of Internal Medicine
Bart L. Scott
Consulting or Advisory Role: Celgene, Acceleron Pharma, Astex Pharmaceuticals
Speakers’ Bureau: Alexion Pharmaceuticals, Celgene, Jazz Pharmaceuticals, Novartis
Research Funding: Celgene (Inst)
Marcelo C. Pasquini
Honoraria: Celgene
Consulting or Advisory Role: Pfizer, Medigene, Celgene, Amgen
Research Funding: Kite Pharma, Gilead, Novartis, Celgene, Bristol-Myers Squibb
Mitchell E. Horwitz
Honoraria: Molmed
Consulting or Advisory Role: AbbVie
Research Funding: Gamida Cell (Inst), CSL Behring (Inst), Incyte (Inst), Astellas Pharma (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2. D’Souza A, Fretham C: Current use and outcome of hematopoietic stem cell transplantation: CIBMTR summary slides, 2018. https://www.cibmtr.org.
- 3.Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luger SM, Ringdén O, Zhang MJ, et al. Similar outcomes using myeloablative vs reduced-intensity allogeneic transplant preparative regimens for AML or MDS. Bone Marrow Transplant. 2012;47:203–211. doi: 10.1038/bmt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott BL, Pasquini MC, Logan BR, et al. Myeloablative versus reduced-intensity hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol. 2017;35:1154–1161. doi: 10.1200/JCO.2016.70.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornhäuser M, Kienast J, Trenschel R, et al. Reduced-intensity conditioning versus standard conditioning before allogeneic haemopoietic cell transplantation in patients with acute myeloid leukaemia in first complete remission: A prospective, open-label randomised phase 3 trial. Lancet Oncol. 2012;13:1035–1044. doi: 10.1016/S1470-2045(12)70349-2. [DOI] [PubMed] [Google Scholar]
- 7.Fasslrinner F, Schetelig J, Burchert A, et al. Long-term efficacy of reduced-intensity versus myeloablative conditioning before allogeneic haemopoietic cell transplantation in patients with acute myeloid leukaemia in first complete remission: Retrospective follow-up of an open-label, randomised phase 3 trial. Lancet Haematol. 2018;5:e161–e169. doi: 10.1016/S2352-3026(18)30022-X. [DOI] [PubMed] [Google Scholar]
- 8.Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013;10:460–471. doi: 10.1038/nrclinonc.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araki D, Wood BL, Othus M, et al. Allogeneic hematopoietic cell transplantation for acute myeloid leukemia: Time to move toward a minimal residual disease-based definition of complete remission? J Clin Oncol. 2016;34:329–336. doi: 10.1200/JCO.2015.63.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 11.Buckley SA, Wood BL, Othus M, et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: A meta-analysis. Haematologica. 2017;102:865–873. doi: 10.3324/haematol.2016.159343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hourigan CS, Gale RP, Gormley NJ, et al. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31:1482–1490. doi: 10.1038/leu.2017.113. [DOI] [PubMed] [Google Scholar]
- 13.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378:1189–1199. doi: 10.1056/NEJMoa1716863. [DOI] [PubMed] [Google Scholar]
- 14.Schuurhuis GJ, Heuser M, Freeman S, et al. Minimal/measurable residual disease in AML: A consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131:1275–1291. doi: 10.1182/blood-2017-09-801498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hourigan CS, Goswami M, Battiwalla M, et al. When the minimal becomes measurable. J Clin Oncol. 2016;34:2557–2558. doi: 10.1200/JCO.2016.67.6395. [DOI] [PubMed] [Google Scholar]
- 16.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimwade D, Jovanovic JV, Hills RK, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27:3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 20.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314:811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch JS, Petti AA, Miller CA, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375:2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T, Moon JH, Ahn JS, et al. Next-generation sequencing-based posttransplant monitoring of acute myeloid leukemia identifies patients at high risk of relapse. Blood. 2018;132:1604–1613. doi: 10.1182/blood-2018-04-848028. [DOI] [PubMed] [Google Scholar]
- 23.Thol F, Gabdoulline R, Liebich A, et al. Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood. 2018;132:1703–1713. doi: 10.1182/blood-2018-02-829911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillon LW, Hayati S, Roloff GW, et al. Targeted RNA-sequencing for the quantification of measurable residual disease in acute myeloid leukemia. Haematologica. 2019;104:297–304. doi: 10.3324/haematol.2018.203133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21:374–382.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duncavage EJ, Jacoby MA, Chang GS, et al. Mutation clearance after transplantation for myelodysplastic syndrome. N Engl J Med. 2018;379:1028–1041. doi: 10.1056/NEJMoa1804714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cocciardi S, Dolnik A, Kapp-Schwoerer S, et al. Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat Commun. 2019;10:2031. doi: 10.1038/s41467-019-09745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Galen P, Hovestadt V, Wadsworth Ii MH, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281.e24. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christopher MJ, Petti AA, Rettig MP, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med. 2018;379:2330–2341. doi: 10.1056/NEJMoa1808777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eapen M, Brazauskas R, Hemmer M, et al. Hematopoietic cell transplant for acute myeloid leukemia and myelodysplastic syndrome: Conditioning regimen intensity. Blood Adv. 2018;2:2095–2103. doi: 10.1182/bloodadvances.2018021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kröger N, Iacobelli S, Franke GN, et al. Dose-reduced versus standard conditioning followed by allogeneic stem-cell transplantation for patients with myelodysplastic syndrome: A prospective randomized phase III study of the EBMT (RICMAC Trial) J Clin Oncol. 2017;35:2157–2164. doi: 10.1200/JCO.2016.70.7349. [DOI] [PubMed] [Google Scholar]
- 36.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milano F, Gooley T, Wood B, et al. Cord-blood transplantation in patients with minimal residual disease. N Engl J Med. 2016;375:944–953. doi: 10.1056/NEJMoa1602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platzbecker U, Middeke JM, Sockel K, et al. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): An open-label, multicentre, phase 2 trial. Lancet Oncol. 2018;19:1668–1679. doi: 10.1016/S1470-2045(18)30580-1. [DOI] [PubMed] [Google Scholar]
- 39.DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 40.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]