Abstract
Bone loss in aging is linked with chronic low-grade inflammation and the accumulation of marrowfat in animals and humans. Peroxisome proliferator-activated receptor gamma (PPARγ), an adipogenic regulator, plays key roles in these biological processes. However, studies of the roles of PPARγ in age-related bone loss and inflammation are lacking. We hypothesized that deletion of PPARγ in bone marrow mesenchymal lineage cells would reduce bone loss with aging, potentially through a reduction in fat-generated inflammatory responses and an increase in osteoblastic activity. In the present study, we show that mice deficient of PPARγ in Dermo1-expressing mesenchymal lineage cells (Dermo1-Cre:PPARγ fl/fl) have reduced fat mass and increased cortical bone thickness but that deficiency of PPARγ had limited effect on protection of trabecular bone with aging as demonstrated by dual-energy X-ray absorptiometry, µCT, and histomorphometric analyses. Conditional knockout of PPARγ reduced serum concentrations of adipokines, including adiponectin, resistin, and leptin, and reduced marrow stromal cell expression levels of inflammation-related genes. Inflammation genes involved in the interferon signaling pathway were reduced the most. These results demonstrate that disruption of the master adipogenic regulator, PPARγ, has a certain protective effect on aging-induced bone loss, suggesting that regulation of adipose function and modulation of interferon signaling are among the key mechanisms by which PPARγ regulates bone homeostasis during aging process.
Keywords: PPARγ, Adipocyte, Aging, Inflammation, Bone loss
In humans, aging is associated with the loss of bone and the accumulation of marrowfat (1–3), and the process is coupled with chronic low-grade inflammation and decline of immune function (4–7). Peroxisome proliferator-activated receptor gamma (PPARγ) is a key transcription factor regulating adipocyte differentiation (8). Because the bone marrow mesenchymal stem/progenitor cell (BMSC) is a common precursor cell for osteoblasts and adipocytes (9,10), and the two differentiation pathways have a reciprocal relationship (11,12), targeting PPARγ has been thought a potential strategy in preventing bone loss in aging (13,14). Adipose tissue is an endocrine organ and produces many factors including adipokines, chemokines, and inflammatory cytokines that may contribute significantly to aging-induced bone loss (15,16) and elevated chronic inflammation status (17,18). We reported previously that mice deficient of PPARγ gene in type I collagen-expressing osteoprogenitor cells (PPARγ fl/fl:Col3.6Cre) shows a preferential bone mass increase at vertebra in adult mice (>3 months of age) (19), suggesting that PPARγ may play a more important role in bone homeostasis rather than in bone development and skeletal buildup at the young age.
In this study, we tested the hypothesis that deletion of PPARγ in BMSC would reduce bone loss with aging, potentially through a reduction in fat-generated inflammatory responses and an increase in osteoblastic activity. The hypothesis was tested using a Dermo-1-Cre:PPARγ fl/fl conditional knockout (cKO) mouse model. Unlike the Col3.6-Cre mouse line, in which the Cre is expressed in preosteoblasts/mature osteoblasts (20,21), the Dermo-1 promoter used in this study directs Cre expression in mesenchymal condensations, from which the multilineage mesenchymal stem cells originate (22,23). Thus, deletion of PPARγ with this Cre line should have broader impacts and provide new insights into how fat and fat-generated factors may contribute to chronic inflammation and bone loss in aging.
Materials and Methods
Experimental Animal
PPARγ cKO mice (Dermo1-Cre:PPARγ fl/fl) were generated by breeding a Dermo1-Cre transgenic mouse with a PPARγ-floxed (PPARγ fl/fl) mouse, both of which are on a mixture of CD1 and C57BL/6 background. The original Dermo1-Cre and PPARγ fl/fl mice were developed on C57BL/6 background (22,24). The original PPARγ fl/fl mouse was crossed with a Col3.6-Cre transgenic mouse that was developed on CD1 background (21) in our previous studies (19). The PPARγ fl/fl mice were since maintained on CD1 background. The Dermo1-Cre mouse was back crossed with CD1 mouse for more than 10 generations before breeding with PPARγ fl/fl mouse in this study. The coat color of all mice is white. The knockout mice and control mice have no distinguishable difference in general appearance or body weight. The Dermo1-Cre and PPARγ fl/fl mice were kindly provided by Dr. David M. Ornitz at the Washington University Medical School and by Dr. Frank Gonzalez at the National Cancer Institute, respectively. The Col3.6-Cre mouse used in our previous study was provided by Dr. Barbara E. Kream at the University of Connecticut Health Center.
Mice were gang housed (four to five mice per cage) in the Augusta University Laboratory Animal Service facility under a 12-hour dark–light cycle and fed with standard rodent chow (Teklad, cat no. 2918) and water ad libitum. All animal procedures were performed in accordance with a protocol (no. 2008-0302) approved by the Augusta University Institutional Animal Care and Use Committee (IACUC).
Genotype was analyzed using primers and PCR conditions described previously (21). In brief, tail DNA was extracted and amplified with one forward primer (F1: 5′-CTCCAATGTTCTCAAACTTAC-3′) and two reverse primers (R1: 5′-GATGAGTCATGTAAGTTGACC-3′; R2: 5′-GTATTCTATGGCTTCCAGTGC-3′). The expected sizes of the PCR products are wild-type allele ~250 bp, floxed allele ~285 bp, and null allele ~450 bp. PCR analysis of Cre was performed using primers 5′-GCATTTCTGGGGATTGCTTA-3′ (forward) and 5′-GTCATCCTTAGCGCCGTAAA-3′ (reverse). The expected size of PCR product for Cre is ~350 bp. The deletion strategy, primer location, gel image of genotyping PCR products, and the validation of PPARγ deletion in bone by qRT-PCR analysis of BMSCs and bone chip RNA are shown in Supplementary Figure 1.
Isolation of Bone Marrow Mesenchymal Stem/Progenitor Cells and Cell Culture
The BMSCs were isolated from long bones of PPARγ cKO and PPARγ fl/fl mice using a negative immunodepletion (magnetic beads conjugated with anti-mouse CD11b, CD45R/B220, and Pan DC) and positive immunoselection (anti-Sca-1 beads) procedure as previously described (25). The BMSCs isolated using this procedure can form mineralized bone nodules, adipocytes, and myotubes. Importantly, they are able to form osteoblast-like lining cells in vivo when injected into C57BL mice (25). The cells were cultured under standard culture conditions (37°C, 5% CO2; DMEM with high glucose supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin).
Serum Assays
Sera were collected after centrifugation of blood samples at 2,000 rpm for 10 minutes at 4°C and stored at −80°C in aliquots. Serum concentrations of resistin were measured using a fluorescent bead-based Luminex Assay Kit (cat no. LXSAMSM-05, R&D Systems) and a Luminex 200 instrument (Luminex Corporation, Austin, TX). Adiponectin and leptin were detected using Quantikine ELISA Kits (cat nos. MRP300 and MOB00, respectively, R&D Systems). All assays were performed according to the manufacturer’s instructions, and all samples were assayed in duplicates.
Nanostring Analysis of Inflammation Genes
Bone marrow was flushed from 7- and 18-month-old male mice and cultured for 7–10 days under standard culture conditions (37°C, 5% CO2; high glucose DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin). Total cellular RNAs were isolated using TRIzol reagent (cat no. 15596026, ThermoFisher Scientific) according to the manufacturer’s instructions. RNA samples were then sent to the Augusta University Cytogenetics Laboratory for quality control and analysis using a mouse inflammation panel (cat no. MmV2_Inflammation) and nCounter system (NanoString Technologies, Inc., Seattle, WA). Data were processed and evaluated against six positive and eight negative controls and normalized to four housekeeping genes using nSolver version 4.0.66 software.
Real-Time qRT-PCR Analysis
For qRT-PCR analysis, bone marrow was flushed from 15-month-old male mice and cultured for 7–10 days under standard culture conditions with medium change every 2–3 days. RNA isolation and quantitative analysis of levels of mRNA expression were performed as described previously (26) using TaqMan Reverse Transcription Reagents (Applied Biosystems) and a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). The mRNA levels were normalized to β-actin (internal control), and gene expression was presented as fold change. The sequences of primers used in real-time qRT-PCR are listed in Supplementary Table 1.
Dual-Energy X-ray Absorptiometry, μCT, and Histology and Histomorphometric Analyses
Bone mineral density (BMD) and bone mineral content (BMC) were measured by dual-energy X-ray absorptiometry (GE Lunar PIXImus System, software version 1.4x) as previously described (27). The stability of the measurement was controlled by performing a quality control procedure before acquiring images. Bone structural parameters were measured by μCT scan (μCT-40; Scanco Medical AG, Bassersdorf, Switzerland) as previously described (28,29). Bone tissues for histological and histomorphometric analyses were collected, processed, and analyzed following our previously described procedures (28). Standard bone histomorphometrical nomenclatures, symbols, and units were used as recommended by Parfitt and colleagues (30).
Bone Marrow Cell Culture and Differentiation
Bone marrow cells were flushed from long bones of mice and induced for osteoblast or adipocyte differentiation as previously described (25,31). Briefly, for osteoblast differentiation, cells were plated in 96-well plates in triplicate at a density of 1 × 104 cells/cm2 and next day treated with osteogenic medium consisting of DMEM supplemented with 10% FBS, 5 mM β-glycerolphosphate, 50 µM l-ascorbic acid-2-phosphate, and 100 nM dexamethasone (Dex). Mineralized bone nodules were visualized by Alizarin Red S staining 3 weeks after induction.
For adipocyte differentiation, cells were treated with adipogenic induction medium (DMEM containing 10% FBS, 1 µM Dex, 60 µM indomethacin, 10 µg/mL insulin, and 0.5 mM 3-isobutyl-1-methylxanthine) for 3 days and then switched to adipogenic maintenance medium (growth medium plus 10 µg/mL insulin) with media replaced every other day. Adipocytes were visualized by oil red O staining.
Statistical Analysis
Results are expressed as mean ± SD. All in vitro experiments (qRT-PCR, cell differentiation, etc.) were performed in triplicates with samples from the indicated number of mice. Data were analyzed using either unpaired t-test with Welch’s correction or two-way analysis of variance (age: 7 and 20 months and gender: male and female) followed by Tukey’s post hoc test (serum cytokines) with GraphPad Prism version 7 software. A p value less than .05 was considered significant.
Results
Characterization of Dermo1-Cre:PPARγ fl/fl cKO Mice
PPARγ cKO mice were generated by breeding a Dermo1-Cre transgenic mouse (22) with a PPARγ-floxed (PPARγ fl/fl) mouse (24). Genotypes of the resulting pups were detected by PCR analysis of tail genomic DNA using indicated primer pairs (Supplementary Figure 1A and B). Deletion of PPARγ was confirmed at mRNA level by qRT-PCR analysis of RNA samples isolated from purified BMSCs (Supplementary Figure 1C) and bone chips (Supplementary Figure 1D), and by the ability of bone marrow cells undergo adipocyte differentiation (Supplementary Figure 1E). Hematoxylin and eosin histology study of the epididymal fat tissue (from 3-month male mice) showed that the number of adipocytes (per square millimeter) was significantly reduced, but the size of adipocytes was significantly increased in PPARγ cKO mice, ~2.5× bigger than that in the control mice (Supplementary Figure 1G and H). The knockout mice and control mice had no distinguishable difference in body size or body weight (Supplementary Figure 1I and J). These results demonstrated that Dermo-1 promoter drives Cre expression and ablates PPARγ gene in bone tissue and adipose tissue. This is in contrast to Col3.6-Cre:PPARγ fl/fl mice, which showed no difference in the amount of body fat compared with their age- and gender-matched floxed controls (19).
Bone Phenotype of PPARγ cKO Mice
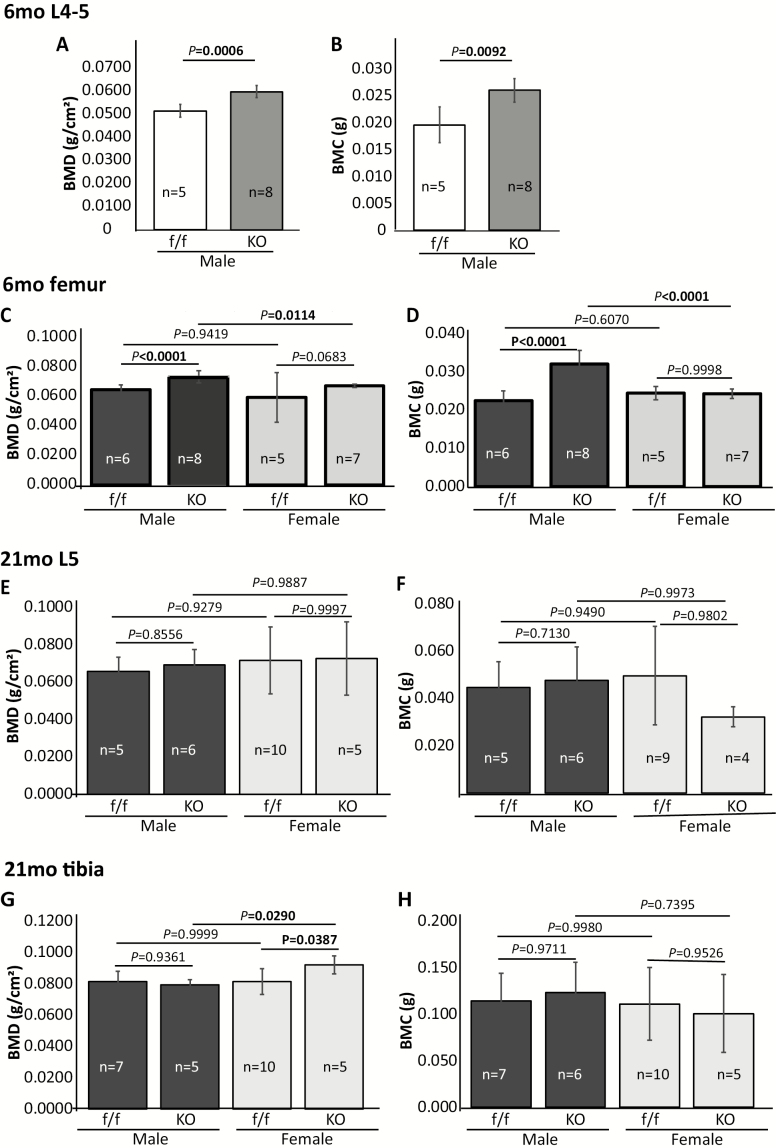
Dual-energy X-ray absorptiometry analysis showed that PPARγ cKO significantly increased BMD and BMC in vertebrae (Figure 1A and B) and femurs (Figure 1C and D) of mature adult male mice (6 months). A mild BMD, and not BMC, increase was detected in adult female cKO mice (p = .0683), but this increase did not reach to a level traditionally defined as statistically significant (p < .05). Male cKO mice had significantly higher BMD and BMC than female cKO mice, but these differences were not detected in control mice (Figure 1C and D). No significant differences in BMD and BMC were detected between cKO and controls in old (21 months) mice (Figure 1E–H), except in the tibiae of female cKO mice, which showed a significant BMD increase (p = .0387). As a result, the female cKO mice showed a significantly higher BMD (p = .0290) in tibiae than that in male cKO mice (Figure 1G).
Figure 1.
Dual-energy X-ray absorptiometry analysis showing BMD and BMC of 6-mo-old (A–D) and 21-mo-old (E–H) PPARγ cKO and floxed control mice. Values are given as mean ± SD. Sex, sample size, and p values are indicated. Vertebrae from 6-mo-old mice were analyzed only in males. BMD, bone mineral density; BMC, bone mineral content.
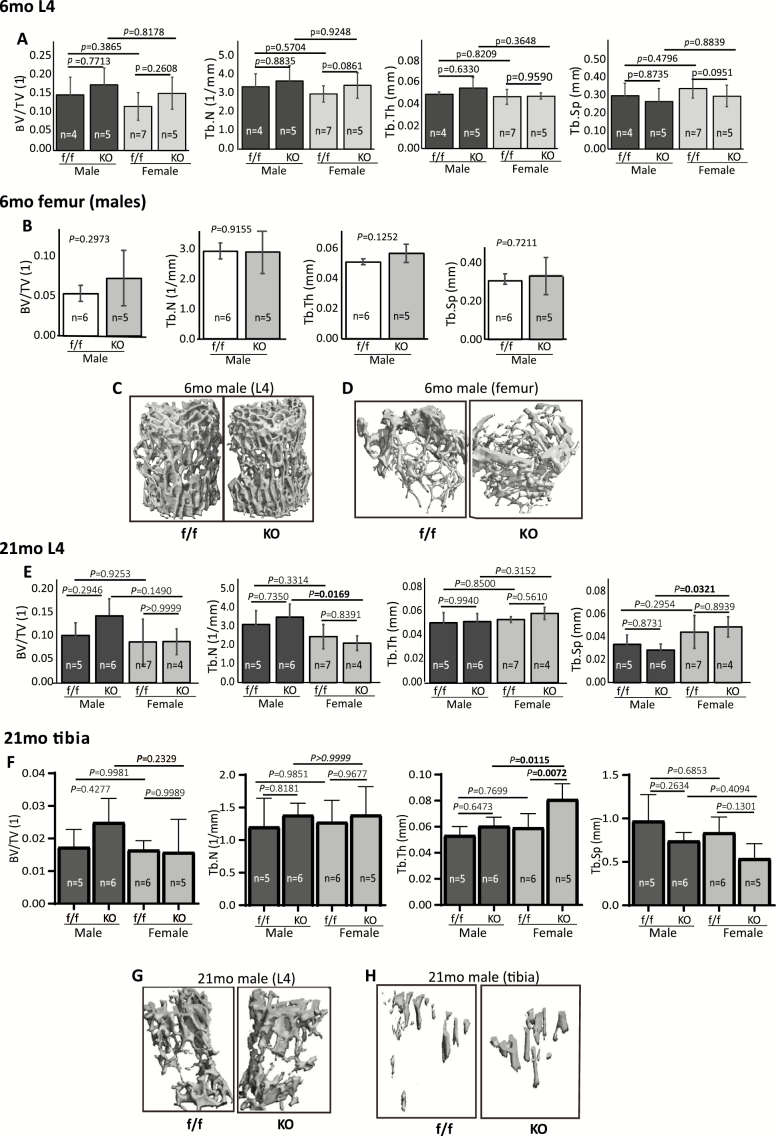
µCT analysis data showed that, separately, there were no significant differences in trabecular bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), or trabecular separation (Tb.Sp) between cKO and control mice in either male or female mice (Figure 2A and B, femurs were from males only). However, when male and female mice were combined together, we observed a significant increase in BV/TV (p = .0369), a mild increase in Tb.N (p = .0847), and a mild decrease in Tb.Sp (p = .0845) in vertebrae of the mature adult mice (Supplementary Figure 2A). Representative reconstructed 3D images of 6 months vertebra and femur (all males) are shown in Figure 2C and D, respectively. In old mice (21 months), µCT analysis of vertebrae (L4) showed no significant difference between cKO and floxed controls in either male or female mice (Figure 2E). However, the male cKO mice showed a significantly higher Tb.N (p = .0169) and lower Tb.Sp (p = .0321) compared with their female cKO counterpart. In old tibiae, PPARγ cKO significantly increased Tb.Th (p = .0072) in female mice, but not in male cKO mice (Figure 2F). When males and females were combined, we observed a significant decrease in Tb.Sp (p = .0379) and mild increases in Tb.N (p = .0929) and Tb.Th (p = .0715) (Supplementary Figure 2B). These results suggested that overall, PPARγ cKO had certain, but not striking, bone protective effect. Representative reconstructed 3D images of 21-month vertebra and femur (all males) are shown in Figure 2G and H, respectively.
Figure 2.
μCT analysis of trabecular bone. (A–D) μCT analysis of vertebra (A) and femur (B, males only) from 6-mo-old cKO and floxed control mice. (C, D) Representative 3D reconstructed images of vertebra (C) and femur (D) from 6-mo-old male mice. (E–H) μCT analysis of vertebra (E) and tibia (F) from 21-mo-old cKO and floxed control mice. (G, H) Representative 3D reconstructed images of vertebra (G) and tibia (H) from 21-mo-old cKO and floxed control mice. Values are given as mean ± SD. Sex, sample size, and p values are indicated. BV/TV, bone volume/tissue volume; cKO, conditional knockout; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation.
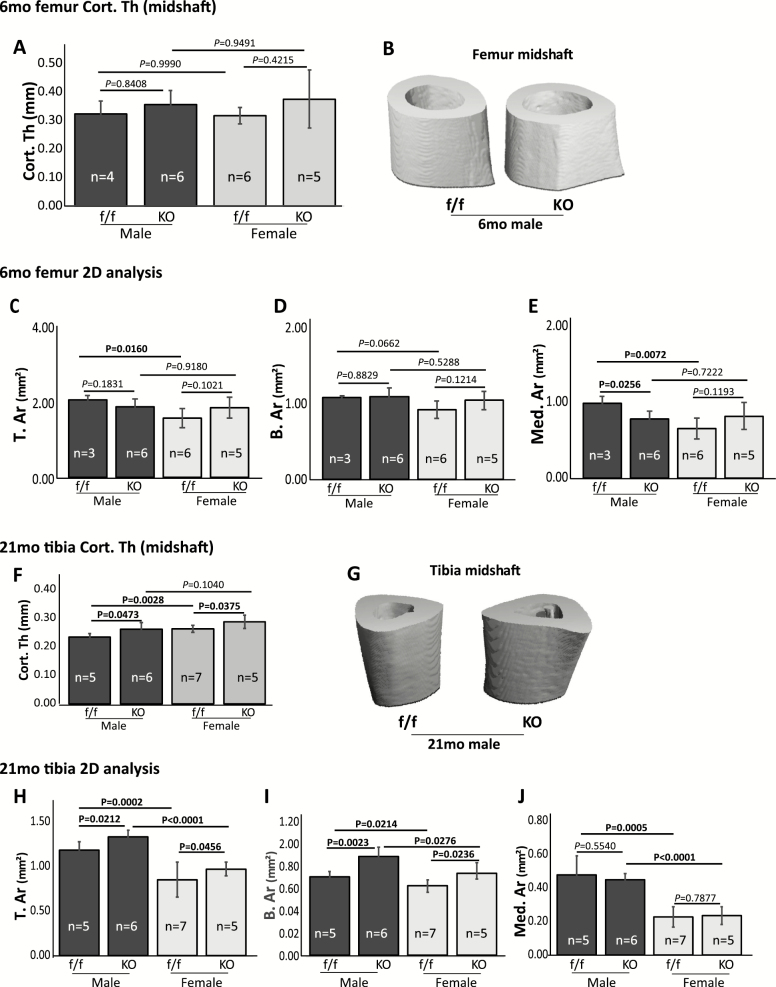
µCT analysis of cortical bone from mature adult mice (6 months) showed no difference between cKO and floxed control mice in cortical thickness (Cort. Th, femur midshaft) (Figure 3A and B), total cross-sectional tissue area (T. Ar), or total cross-sectional bone area (B. Ar) (Figure 3C and D). A significant decrease (p = .0256) in medullary area (Med. Ar) was detected in male cKO mice (Figure 3E). T. Ar and Med. Ar were significantly lower (p = .0160 and .0072, respectively) in floxed female mice than in their male counterpart (Figure 3C and E). The B. Ar of floxed female mice was lower than their male counterpart, but this difference was not statistically significant (p = .0662; Figure 3D). No significant differences were detected in these parameters between male cKO and female cKO mice. Interestingly, in old mice, PPARγ cKO significantly increased tibia Cort. Th (Figure 3F and G), T. Ar, and B. Ar (Figure 3H and I) in both male and female cKO mice. The Med. Ar showed no difference between cKO and control mice (Figure 3J). The T. Ar, B. Ar, and Med. Ar were significantly higher in male than that in female mice. Together, these results indicated that PPARγ cKO increased periosteal bone apposition in the tibia of old mice. Representative reconstructed 3D images of 6-month femur and 21-month tibia midshaft (all males) are shown (Figure 3B and G).
Figure 3.
μCT analysis of cortical bone. (A–E) μCT analysis of 6-mo-old femur midshaft Cort. Th (A) and cross-sectional area region of interest (C–E). (B) Representative 3D reconstructed images of femur midshaft from 6-mo male mice. (F–J) μCT analysis of 21-mo-old tibia midshaft Cort.Th (F) and cross-sectional area region of interest (H–J). (G) Representative 3D reconstructed images of tibia midshaft from 21-mo male mice. Values are given as mean ± SD. Sex, sample size, and p values are indicated. Cort.Th, cortical thickness; T.Ar, mean total cross-sectional tissue area; B.Ar, mean total cross-sectional bone area; Med. Ar, medullary area.
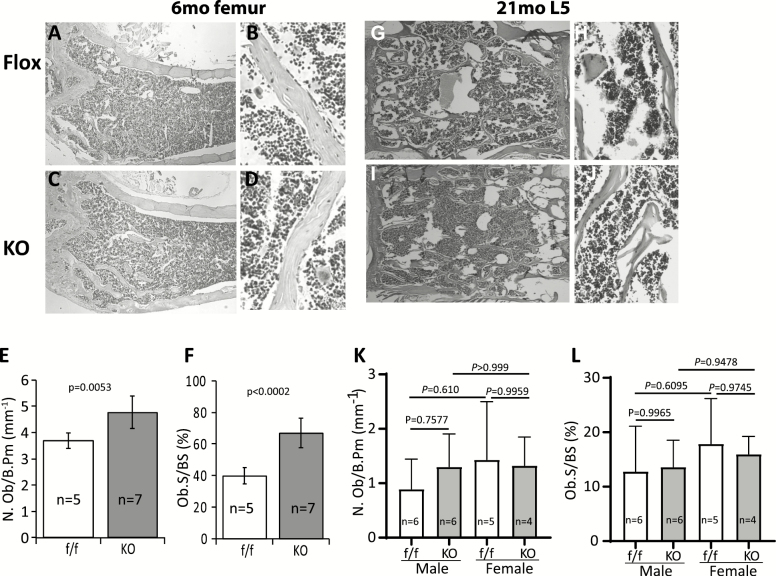
Histological and histomorphometric analyses showed that the osteoblast numbers and osteoblast covered bone surface areas were increased significantly in femurs of the mature adult cKO mice (Figure 4A–F), but these increases were not detected in vertebrae of the old cKO mice (Figure 4G–L, long bones not analyzed). B, D and H, J are the high-power images of femur and vertebra, respectively. In vitro cell culture and gene expression analysis showed that PPARγ cKO enhanced osteogenic differentiation as demonstrated by alizarin red S stain of the mineralized bone nodules (Supplementary Figure 3A and B), and increase in the expression of osteoblast lineage associated genes, including Runx2, alkaline phosphatase, and type I collagen as determined by real-time qRT-PCR analysis (Supplementary Figure 3C).
Figure 4.
Histology and histomorphometry analyses. (A–D) Representative hematoxylin and eosin (H&E) stained femur from 6-mo-old male mice. (B) and (D) are high-power images from selected areas. (E, F) Quantitative results showing osteoblast number and osteoblast covered bone surface area. (G–I) Representative H&E stained lumbar vertebra (L4) from 21-mo-old male mice. H and J are high-power images from selected areas. (K, L) Quantitative results showing osteoblast number and osteoblast covered bone surface area. Data are shown as mean ± SD. Sex, sample size, and p values are indicated. Results for 6-mo femur (E, F) are a mixture of four male and one female mice in control group and five male and two female mice in KO group.
Adipokines and Marrow Cell Inflammatory Genes Are Downregulated in PPARγ cKO Mice
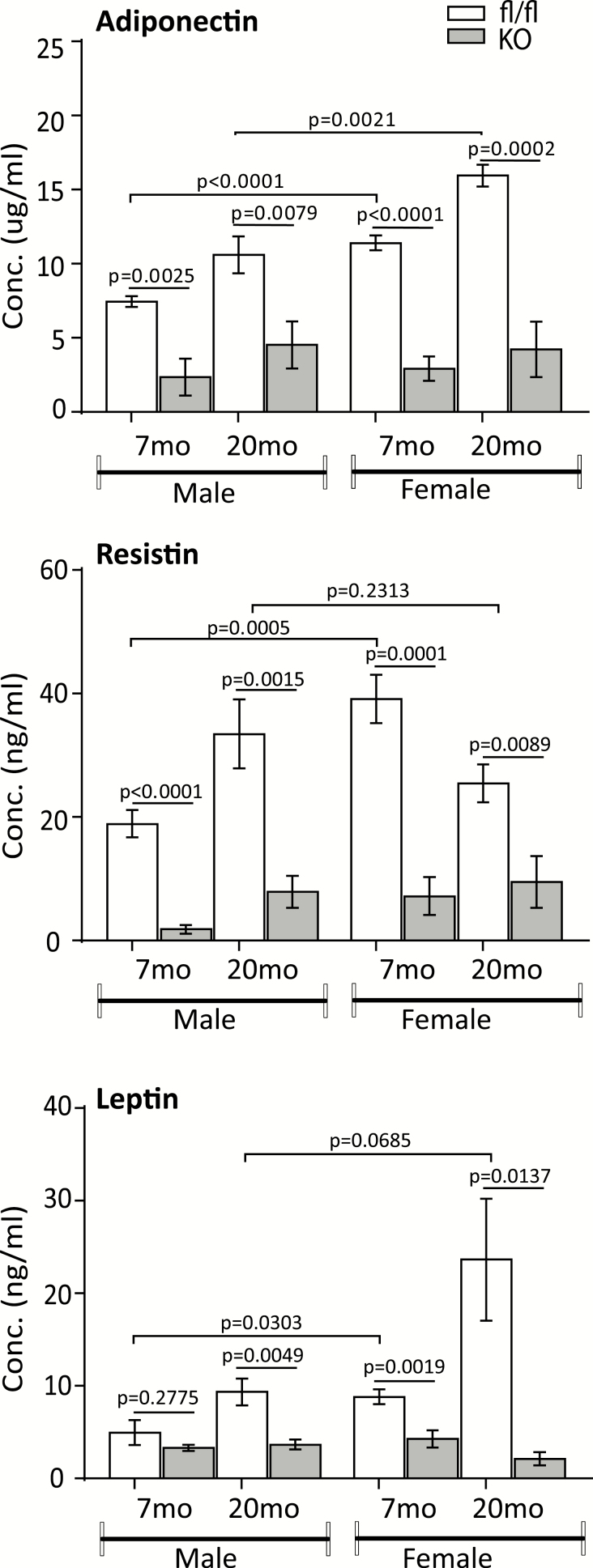
To assess the effect of PPARγ cKO on serum biochemistry, we measured levels of adipokines by Luminex or ELISA assays. Results showed that levels of adiponectin, resistin, and leptin were all decreased significantly in both adult (7 months) and old (20 months) cKO mice compared with their PPAR-floxed counterparts, except for leptin levels, which showed no significant change in adult male cKO mice (Figure 5). Levels of these adipokines were significantly higher in PPAR-floxed females than in males at younger age. In old mice, however, only the level of adiponectin, but not resistin and leptin, was significantly higher in females than in males. In contrast, levels of these adipokines showed no difference in PPAR cKO mice between males and females, except leptin, which is significantly higher in old male cKO mice than in female cKO mice.
Figure 5.
Serum levels of adipokines. Levels of adiponectin, resistin, and leptin from mature adult (7-mo) and old (20-mo) mice were measured by ELISA (adiponectin and leptin) or Luminex (resistin). Data are presented as means ± SD, n = 8–10.
We also measured basal serum concentrations of inflammatory cytokines and chemokines by Luminex multiplex immunoassays, but these were not dramatically changed because of the low concentrations of these factors (data not shown). These results may be due to the fact that mice were not challenged with inflammation inducers and the assay sensitivity. Using NanoString technology as an alternative approach, we isolated total cellular RNAs from bone marrow stromal cells and examined mRNA levels for a panel of inflammation-related genes. Results showed that PPAR cKO reduced expression levels of 11 genes by 40% or more in adult (7 months) and 44 genes in old (18 months) cKO mice, respectively, compared with their PPAR floxed control counterparts ("Table 1 and Supplementary Table 2 for raw data). Interestingly, we found that majority of these downregulated genes belong to a class of interferon (IFN)-stimulated genes. These include interferon regulatory factors (Irf1, -5, and -7), interferon-induced protein with tetratricopeptide repeats (Ifit2, -3), interferon-induced protein 44 (Ifi44), interferon inducible GTPase 1 (Iigp1), interferon-induced GTP-Binding protein Mx1 (Mx1), 2′-5′-oligoadenylate synthetase 2 (Oas2), and 2-5′-oligoadenylate synthetase-like 1 (Oasl1). The Mx1, Oas2, and Oasl1 all have potent antiviral activities. Secondary analysis by real-time qRT-PCR was hence performed to validate the NanoString data in RNA samples isolated from a different batch of mice (15-month-old male mice). Results showed that mRNA levels of Ifit2, -3, Ifi44, Ifi202b, Iigp1, Irf1, -5, -7, and IFNγ were all downregulated in PPAR cKO mice (Supplementary Figure 4). Together, these results confirmed that PPARγ is involved in the regulation of IFN-stimulated genes and provide new insight on the molecular mechanisms underlying inflammation and antiviral actions of PPARγ.
Table 1.
NanoString Analysis of Inflammation-Related Genes
| Gene | 18-mo f/f (n = 2) | 18-mo KO (n = 3) | 18-mo KO/fl | Gene | 7-mo f/f (n = 3) | 7-mo KO (n = 3) | 18-mo KO/fl |
|---|---|---|---|---|---|---|---|
| Ifit2 | 5,377.12 | 164.07 | 0.03 | Ifit2 | 92.45 | 46.32 | 0.68 |
| Ifit3 | 5,010.93 | 90.78 | 0.02 | Ifit3 | 45.40 | 22.71 | 0.15 |
| Ifi44 | 2,474.89 | 73.77 | 0.03 | Ifi44 | 25.90 | 12.99 | 0.36 |
| Ifi27l2a | 13,168.93 | 1,757.98 | 0.13 | Ifi27l2a | 879.06 | 439.60 | 0.54 |
| Oas2 | 701.86 | 51.72 | 0.07 | Oas2 | 20.40 | 10.32 | 0.69 |
| Iigp1 | 2,780.77 | 226.05 | 0.08 | Iigp1 | 82.05 | 41.04 | 0.34 |
| Oas1a | 4,901.52 | 795.96 | 0.16 | Oas1a | 237.44 | 118.81 | 0.58 |
| Mx1 | 947.32 | 184.71 | 0.19 | Mx1 | 72.63 | 36.39 | 0.57 |
| C3 | 992.97 | 145.11 | 0.15 | Irf7 | 113.07 | 56.57 | 0.44 |
| Stat1 | 8,184.33 | 1,157.42 | 0.14 | Cxcr4 | 61.13 | 30.68 | 0.68 |
| Stat2 | 2567.22 | 474.70 | 0.18 | H2-Eb1 | 32.72 | 16.40 | 0.43 |
| Ltb4r1 | 168.95 | 40.56 | 0.24 | Chi3l3 | 36.90 | 18.46 | 0.30 |
| Ptgs1 | 533.75 | 122.02 | 0.23 | Ptgs1 | 398.06 | 199.11 | 0.57 |
| Ccl5 | 813.31 | 65.37 | 0.08 | Ccl4 | 519.72 | 259.99 | 0.73 |
| Ccr1 | 4,056.88 | 1,039.19 | 0.26 | Cd4 | 578.78 | 289.46 | 0.56 |
Notes: RNA samples were isolated from marrow cell cultures of 7- and 18-mo-old male mice and analyzed. Listed are the top 15 most downregulated genes.
Discussion
In this study, we examined the role of PPARγ in aging-induced bone loss. Our data showed that mice carrying deletion of PPARγ gene in Dermo1-expressing cells (Dermo1-Cre:PPARfl/fl) have significantly increased cortical bone thickness in old age (21 months) and that loss of PPARγ in this mouse model had only a mild protective effect on aging-induced trabecular bone loss. It is worth mentioning that PPARγ cKO increased some (BMD, Tb.N), but not all, bone parameters more in the tibia of old female mice than in males (Figures 1G and 2F). This observation could be due to the relatively smaller sample size, and the conclusion needs to be validated further with more mice and at different ages. The other notable phenotype of these PPARγ cKO mice is a reduction of adiposity, which is characterized by a significant decrease in the number, but an increase in the size of the adipocytes. These phenotypical changes correlated with reduced levels of adipose-derived factors including adipokines and cytokines.
Contrary to the common hypothesis that deletion of PPARγ would dramatically increase bone mass and protect against aging-induced bone loss, our data showed that knockout PPARγ in dermo1-expressing mesenchymal lineage cells protected against aging-induced cortical bone loss, but the effect on preventing trabecular bone loss was mild, although PPARγ cKO increased bone acquisition in mature adult mice. Studies from other groups also failed to demonstrate a strong bone phenotype in similar PPARγ cKO mouse models, including Sox2-Cre:PPARγ f/f mice, which are lipodystrophic and exhibit a moderate BMD increase in vertebrae, but not in long bones (32), and Prx1-Cre:PPARγ f/f mice, which showed that PPARγ is dispensable for aging-induced bone loss (33), and Osx-Cre:PPARγ f/f mice, which showed a mild increase in BV with greater effect on Tb.N and Tb.Sp (34). The effect of global inhibition of PPARγ on bone is complex; while there is evidence that inhibition of PPARγ by its antagonists increases BV (35), there are also reports suggesting otherwise, although it is clear that inhibition of PPARγ suppresses adipocyte differentiation (36,37). These results indicated that alternative mechanisms such as regulation of adipose functions and PPARγ crosstalk with other pathways including mTOR (34), and IFN signaling pathways are also involved as mice deficient of PPARγ in adipose tissue, the lipodystrophic PPARγ hyp/hyp mice (38), and adipoq-Cre:PPARγ fl/fl mice (39) exhibit a high bone mass phenotype, and mice deficient of IFNγ signaling (Ifnγ −/− or IfnγR1−/−) have low bone mass and impaired MSC osteogenic differentiation ability (40,41), and administration of low-dose IFNγ increased bone mass in sham-operated animals and rescued ovariectomy-induced bone loss (40).
Our data showed that PPARγ knockout reduced the expression levels of interferon-stimulated genes, suggesting that PPARγ KO impaired IFNγ signaling. Studies of IFNγ on MSCs/osteoblasts are limited. Although not abundant, IFNγ is expressed by mouse MSCs (42), cultured human osteoblast-like cells (43), and stimulated primary human osteoblast cells and mouse calvarial osteoblast formation by upregulating osteogenic factors including Runx2, osterix, Alp, and osteocalcin (40). Though the literature on the roles of IFN-stimulated genes in osteoblast differentiation and bone formation is rare, Duque and colleagues showed that the expression levels of a set of these genes, including IFN-induced proteins (Ifit1, −15, −16), and IFN-induced transmembrane protein (Ifitm), among others, increased markedly during the early phase of hMSC osteogenic differentiation and the IFNγ produced by hMSC during this phase enhanced osteogenic differentiation in an autocrine manner (40). Although our NanoString analysis of inflammation genes was performed using RNA samples isolated from cultured bone marrow stromal cells that were heterogenic in nature, the effect of PPARγ deficiency on the expression of these genes should reflect the levels of these genes in mesenchymal lineage cells as our previous RNA-seq analysis data also showed significantly decreased expression of Ifi202b, Ifi44, Ifit1, Iigp1, and Mx2 in purified PPARγ KO MSCs (44). However, it is possible that the expression of these genes in hematopoietic lineage cells is also influenced, indirectly, by factors that are produced by the mesenchymal lineage cells, that is, adipocytes or PPARγ-deficient osteoprogenitor/bone cells.
The relationships between PPARγ and IFN or IFN-stimulated genes are complex and controversial. A study by Lin and colleagues showed that activation of PPARγ by its agonist ciglitazone time and dose dependently induces, and inhibition of PPARγ by its antagonist GW9962 suppresses the expression of Irf-1 in vascular smooth muscle cells (45). Lin and colleagues also showed that ciglitazone activates Irf-1 transcription in a PPARγ-dependent manner, as demonstrated by use of an adenovirus-mediated PPARγ overexpression and Irf-1 promoter-reporter assays (45). Contrary to our findings, Cunard and colleagues reported that IFNγ expression in T cells is inhibited by activation of PPARγ (46) and Welch and colleagues showed that induction of lipopolysaccharide and IFNγ target genes in macrophages is inhibited in a PPARγ-dependent manner with low concentrations of rosiglitazone or in a PPARγ-independent manner with high concentrations of rosiglitazone, possibly, through activation of PPARδ (47). Although in line with studies by Lin and colleagues, our data contradict with studies by Cunard and colleagues (46) and Welch and colleagues (47). The discrepancies reported from different groups could be due to (i) different experimental systems: Although the bone marrow stromal cells used in our study and the vascular smooth muscle cells used in Lin and colleagues’ study are mesenchymal lineage cells, the cells used in the studies by Cunard and colleagues (T cells) and Welch and colleagues (macrophages) are hematopoietic lineage cells; (ii) activation versus naive cells: although the cells used in the studies by Cunard and colleagues and Welch and colleagues were activated by different stimuli (Con A, PMA, ionomycin, LPS, thioglycollate, and IFN-γ) in different experiment settings (either individually or in combinations) before they were treated with PPARγ ligands, the bone marrow stromal cells used in our study were derived from PPARγ cKO mice with no additional manipulation. It is known that high doses and long durations are needed for PPARγ ligands to exert anti-inflammatory effects (48,49). Furthermore, there are evidence showing that PPARγ ligands can inhibit inflammatory response independent of PPARγ (50) and these ligands (agonists) can bind to, and activate the key anti-inflammatory mediator glucocorticoid receptor (51). In fact, the microarray data reported by Welch and colleagues showed a remarkable downregulation of IFN-stimulated genes (Ifit2, IFN-activated gene 203, MX1, IP10, and iNOS) in the untreated PPARγ −/− macrophages compared with untreated PPARγ +/+ macrophages (47), an experimental setting that is comparable with our experimental setting and results.
In summary, this study demonstrated that mice deficient of PPARγ in BMSC had decreased fat mass and increased cortical bone thickness in old mice, but the effect on trabecular bone mass enhancement was mild, compared with their age-matched control mice. Furthermore, the study showed that conditional knockout of PPARγ decreased expression of inflammation-related genes in marrow stromal cells of mice. These findings suggest that crosstalk between PPARγ and IFNγ signaling pathways may play an important role for bone homeostasis during aging.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Supplementary Figure 1. Characterization of Dermo1-Cre:PPARγ fl/fl mice. (A) Schematic diagram showing the knockout strategy for generation of PPARγ cKO mice. Approximate locations of primer pairs used for genotyping (F1, R1, R2) are indicated. (B) Agarose gel image showing the expected size of DNA fragments as determined by PCR using tail genomic DNA and indicated primers. (C, D) Real-time qRT-PCR analysis showing levels of PPARγ mRNA in BMSCs (C) and bone chips (D). (E) Oil red O stain of intracellular lipid droplets in differentiated bone marrow cell cultures. (F) Hematoxylin and eosin (H&E) stained sections of epididymal fat tissue from 3-mo-old male mice. Inserts show the images of enlarged boxed areas. (G, H) Quantitative results of F showing the number (G) and diameter (H) of the adipocytes, n = 5. (I) Body weight of male PPARγ-floxed and KO mice at 15–17 months of age. (J) Body weight of male and female mice combined at 15-17 months of age.
Supplementary Figure 2. μCT analysis showing bone structure parameters of 6-mo-old vertebrae (A) and 21-mo-old tibia (B) with data from male and female mice combined. Genotype, sample size, and p values are indicated.
Supplementary Figure 3. Osteogenic differentiation assay. (A) Whole bone marrow cells from 15-17-mo-old male mice were seeded in 96-well plates and subjected to standard osteogenic induction program. Cells were fixed and stained with alizarin red S for detection of mineralized bone nodules 21 days after induction. (B) Quantitative results from 3 independent experiments (n = 9 wells). (C) Real-time qRT-PCR analysis of Runx2, ALP and Col1a1 levels in marrow stromal cells. Data are shown as mean ± SD, n = 2 for control and 3 for cKO mice. Genotype and p values are indicated.
Supplementary Figure 4. Real-time qRT-PCR analysis. Bone marrow stromal cells from control and PPARγ cKO mice (15-mo-old males) were cultured for 7 days before harvesting for total cellular RNA isolation. Equal amounts of RNA were reverse transcribed and mRNA levels of the indicated genes analyzed. PCR reactions were performed in triplicates. Data are shown as mean ± SD and presented as fold changes. n = 3 and 4 for controls and cKO mice, respectively.
Supplementary Table 1. List of primers used in qRT-PCR analysis.
Supplemental Table 2. Normalized NanoString data exported from nSolver software (Excel file). Data were normalized to 4 housekeeping genes.
Acknowledgments
We thank Maribeth H. Johnson, MS, for assistance on statistical analysis.
Funding
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Numbers R01 AG046248 and P01 AG036675. Funding for J.C. was from the USDA Agricultural Research Service program “Food Factors to Prevent Obesity and Related Diseases,” Current Research Information System no. 5450-51000-048-00D.
Authors’ Contribution
The authors’ responsibilities were as follows: J.J.C. contributed to the study design, bone structure and serum assays, data interpretation, and manuscript preparation. X.S. contributed to the study design and implementation, data analysis and interpretation, and manuscript preparation. K.D., G.P., R.R., Y.S., Y.B., Z.H., and J.X. contributed to bone collection and analysis; M.M.L., M.W.H., and C.M.I. contributed to the study design and manuscript preparation.
Conflict of Interest
None reported.
References
- 1. Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol. 2006;2:35–43. doi: 10.1038/ncprheum0070 [DOI] [PubMed] [Google Scholar]
- 2. Demontiero O, Vidal C, Duque G. Aging and bone loss: new insights for the clinician. Ther Adv Musculoskelet Dis. 2012;4:61–76. doi: 10.1177/1759720X11430858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duque G Bone and fat connection in aging bone. Curr Opin Rheumatol. 2008;20:429–434. doi: 10.1097/BOR.0b013e3283025e9c [DOI] [PubMed] [Google Scholar]
- 4. Rossi DJ, Bryder D, Weissman IL. Hematopoietic stem cell aging: mechanism and consequence. Exp Gerontol. 2007;42:385–390. doi: 10.1016/j.exger.2006.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waterstrat A, Van Zant G. Effects of aging on hematopoietic stem and progenitor cells. Curr Opin Immunol. 2009;21:408–413. doi: 10.1016/j.coi.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 6. Vas V, Senger K, Dörr K, Niebel A, Geiger H. Aging of the microenvironment influences clonality in hematopoiesis. PLoS One. 2012;7:e42080. doi: 10.1371/journal.pone.0042080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosen ED, Sarraf P, Troy AE, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- 9. Ahdjoudj S, Lasmoles F, Oyajobi BO, Lomri A, Delannoy P, Marie PJ. Reciprocal control of osteoblast/chondroblast and osteoblast/adipocyte differentiation of multipotential clonal human marrow stromal F/STRO-1(+) cells. J Cell Biochem. 2001;81:23–38. doi: [DOI] [PubMed] [Google Scholar]
- 10. Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci. 1992;102(Pt 2):341–351. [DOI] [PubMed] [Google Scholar]
- 11. Chen Q, Shou P, Zheng C, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–1139. doi: 10.1038/cdd.2015.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kokabu S, Lowery JW, Jimi E. Cell fate and differentiation of bone marrow mesenchymal stem cells. Stem Cells Int. 2016;2016:3753581. doi: 10.1155/2016/3753581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verma S, Rajaratnam JH, Denton J, Hoyland JA, Byers RJ. Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J Clin Pathol. 2002;55:693–698. doi: 10.1136/jcp.55.9.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021 [DOI] [PubMed] [Google Scholar]
- 15. Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocr Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033 [DOI] [PubMed] [Google Scholar]
- 16. Manolagas SC, Jilka RL. Bone marrow, cytokines, and bone remodeling. Emerging insights into the pathophysiology of osteoporosis. N Engl J Med. 1995;332:305–311. doi: 10.1056/NEJM199502023320506 [DOI] [PubMed] [Google Scholar]
- 17. Franceschi C, Bonafè M, Valensin S, et al. Inflammaging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x [DOI] [PubMed] [Google Scholar]
- 18. Florez H, Troen BR. Fat and inflammaging: a dual path to unfitness in elderly people? J Am Geriatr Soc. 2008;56:558–560. doi: 10.1111/j.1532-5415.2007.01584.x [DOI] [PubMed] [Google Scholar]
- 19. Cao J, Ou G, Yang N, et al. Impact of targeted PPARγ disruption on bone remodeling. Mol Cell Endocrinol. 2015;410:27–34. doi: 10.1016/j.mce.2015.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H, Zhang A, Kohan DE, Nelson RD, Gonzalez FJ, Yang T. Collecting duct-specific deletion of peroxisome proliferator-activated receptor γ blocks thiazolidinedione-induced fluid retention. Proc Natl Acad Sci USA. 2005;102:9406–9411. doi: 10.1073/pnas.0501744102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu F, Woitge HW, Braut A, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–653. doi: 10.1387/ijdb.041816fl [DOI] [PubMed] [Google Scholar]
- 22. Yu K, Xu J, Liu Z, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
- 23. De Langhe SP, Carraro G, Tefft D, et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS One. 2008;3:e1516. doi: 10.1371/journal.pone.0001516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akiyama TE, Sakai S, Lambert G, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol Cell Biol. 2002;22:2607–2619. doi: 10.1128/mcb.22.8.2607-2619.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang W, Ou G, Hamrick M, et al. Age-related changes in the osteogenic differentiation potential of mouse bone marrow stromal cells. J Bone Miner Res. 2008;23:1118–1128. doi: 10.1359/jbmr.080304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang N, Baban B, Isales CM, Shi XM. Crosstalk between bone marrow-derived mesenchymal stem cells and regulatory T cells through a glucocorticoid-induced leucine zipper/developmental endothelial locus-1-dependent mechanism. FASEB J. 2015;29:3954–3963. doi: 10.1096/fj.15-273664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie D, Cheng H, Hamrick M, et al. Glucose-dependent insulinotropic polypeptide receptor knockout mice have altered bone turnover. Bone. 2005;37:759–769. doi: 10.1016/j.bone.2005.06.021 [DOI] [PubMed] [Google Scholar]
- 28.Pan G, Cao J, Yang N, et al. Role of glucocorticoid-induced leucine zipper (GILZ) in bone acquisition. J Biol Chem. 2014;289:19373–19382. doi: 10.1074/jbc.M113.535237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao JJ, Gregoire BR, Shen CL. A high-fat diet decreases bone mass in growing mice with systemic chronic inflammation induced by low-dose, slow-release lipopolysaccharide pellets. J Nutr. 2017;147:1909–1916. doi: 10.3945/jn.117.248302 [DOI] [PubMed] [Google Scholar]
- 30. Parfitt AM, Drezner MK, Glorieux FH, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617 [DOI] [PubMed] [Google Scholar]
- 31. Zhang W, Yang N, Shi XM. Regulation of mesenchymal stem cell osteogenic differentiation by glucocorticoid-induced leucine zipper (GILZ). J Biol Chem. 2008;283:4723–4729. doi: 10.1074/jbc.M704147200 [DOI] [PubMed] [Google Scholar]
- 32.He F, Wilson A, Desvergne B. Bone Marrow Adiposity Affects Osteoclastogenesis by Modulating the Bone Marrow Niche. In: ASBMR 2011 Annual Meeting Abstracts. September 16-20, 2011, San Diego. Washington, DC: JBMR; 2011.
- 33.Almeida M, Palmieri M, Kim H, et al. PPARγ in cells of the mesenchymal lineage is dispensable for the age-dependent decline of bone mass and hematopoietic changes in the appendicular skeleton. In: ASBMR 2018 Annual Meeting Abstracts. September 28–October 1, 2018, Montreal. Washington, DC: JBMR; 2018. [Google Scholar]
- 34. Sun H, Kim JK, Mortensen R, Mutyaba LP, Hankenson KD, Krebsbach PH. Osteoblast-targeted suppression of PPARγ increases osteogenesis through activation of mTOR signaling. Stem Cells. 2013;31:2183–2192. doi: 10.1002/stem.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duque G, Li W, Vidal C, Bermeo S, Rivas D, Henderson J. Pharmacological inhibition of PPARγ increases osteoblastogenesis and bone mass in male C57BL/6 mice. J Bone Miner Res. 2013;28:639–648. doi: 10.1002/jbmr.1782 [DOI] [PubMed] [Google Scholar]
- 36. Botolin S, McCabe LR. Inhibition of PPARγ prevents type I diabetic bone marrow adiposity but not bone loss. J Cell Physiol. 2006;209:967–976. doi: 10.1002/jcp.20804 [DOI] [PubMed] [Google Scholar]
- 37. Yu WH, Li FG, Chen XY, et al. PPARγ suppression inhibits adipogenesis but does not promote osteogenesis of human mesenchymal stem cells. Int J Biochem Cell Biol. 2012;44:377–384. doi: 10.1016/j.biocel.2011.11.013 [DOI] [PubMed] [Google Scholar]
- 38. Cock TA, Back J, Elefteriou F, et al. Enhanced bone formation in lipodystrophic PPARgammahyp/hyp mice relocates haematopoiesis to the spleen. EMBO Rep. 2004;5:1007–1012. doi: 10.1038/sj.embor.7400254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang F, Mullican SE, DiSpirito JR, Peed LC, Lazar MA. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc Natl Acad Sci USA. 2013;110:18656–18661. doi: 10.1073/pnas.1314863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duque G, Huang DC, Macoritto M, et al. Autocrine regulation of interferon gamma in mesenchymal stem cells plays a role in early osteoblastogenesis. Stem Cells. 2009;27:550–558. doi: 10.1634/stemcells.2008-0886 [DOI] [PubMed] [Google Scholar]
- 41. Duque G, Huang DC, Dion N, et al. Interferon-gamma plays a role in bone formation in vivo and rescues osteoporosis in ovariectomized mice. J Bone Mineral Res. 2011;26:1472–1483. doi: 10.1002/jbmr.350 [DOI] [PubMed] [Google Scholar]
- 42. Dormady SP, Bashayan O, Dougherty R, Zhang XM, Basch RS. Immortalized multipotential mesenchymal cells and the hematopoietic microenvironment. J Hematother Stem Cell Res. 2001;10:125–140. doi: 10.1089/152581601750098372 [DOI] [PubMed] [Google Scholar]
- 43. Ruiz C, Pérez E, García-Martínez O, Díaz-Rodríguez L, Arroyo-Morales M, Reyes-Botella C. Expression of cytokines IL-4, IL-12, IL-15, IL-18, and IFNgamma and modulation by different growth factors in cultured human osteoblast-like cells. J Bone Miner Metab. 2007;25:286–292. doi: 10.1007/s00774-007-0767-7 [DOI] [PubMed] [Google Scholar]
- 44. Su Y, Shen X, Chen J, Isales CM, Zhao J, Shi XM. Differentially expressed genes in PPARγ-deficient MSCs. Mol Cell Endocrinol. 2018;471:97–104. doi: 10.1016/j.mce.2017.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y, Zhu X, McLntee FL, et al. Interferon regulatory factor-1 mediates PPARgamma-induced apoptosis in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2004;24:257–263. doi: 10.1161/01.ATV.0000109170.43400.2f [DOI] [PubMed] [Google Scholar]
- 46. Cunard R, Eto Y, Muljadi JT, Glass CK, Kelly CJ, Ricote M. Repression of IFN-gamma expression by peroxisome proliferator-activated receptor gamma. J Immunol. 2004;172:7530–7536. doi: 10.4049/jimmunol.172.12.7530 [DOI] [PubMed] [Google Scholar]
- 47. Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARγ and PPARδ negatively regulate specific subsets of lipopolysaccharide and IFN-γ target genes in macrophages. Proc Natl Acad Sci USA. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spiegelman BM PPARgamma in monocytes: less pain, any gain? Cell. 1998;93:153–155. doi: 10.1016/s0092-8674(00)81567-6 [DOI] [PubMed] [Google Scholar]
- 49. Jiang C, Ting AT, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184 [DOI] [PubMed] [Google Scholar]
- 50. Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52. doi: 10.1038/83336 [DOI] [PubMed] [Google Scholar]
- 51. Matthews L, Berry A, Tersigni M, D’Acquisto F, Ianaro A, Ray D. Thiazolidinediones are partial agonists for the glucocorticoid receptor. Endocrinology. 2009;150:75–86. doi: 10.1210/en.2008-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.