Abstract
Background
G protein-coupled estrogen receptor (GPER), or G protein-coupled receptor 30 (GPR30), is reported to mediate non-genomic estrogen signaling. GPR30 associates with breast cancer (BC) outcome and may contribute to tamoxifen resistance. We investigated the expression and prognostic significance of GPR30 in metachronous contralateral breast cancer (CBC) as a model of tamoxifen resistance.
Methods
Total GPR30 expression (GPR30TOT) and plasma membrane-localized GPR30 expression (GPR30PM) were analyzed by immunohistochemistry in primary (BC1; nBC1 = 559) and contralateral BC (BC2; nBC2 = 595), and in lymph node metastases (LGL; nLGL1 = 213; nLGL2 = 196). Death from BC (BCD), including BC death or death after documented distant metastasis, was used as primary end-point.
Results
GPR30PM in BC2 and LGL2 were associated with increased risk of BCD (HRBC2 = 1.7, p = 0.03; HRLGL2 = 2.0; p = 0.02). In BC1 and BC2, GPR30PM associated with estrogen receptor (ER)-negativity (pBC1<0.0001; pBC2<0.0001) and progesterone receptor (PR)-negativity (pBC1 = 0.0007; pBC2<0.0001). The highest GPR30TOT and GPR30PM were observed in triple-negative BC. GPR30PM associated with high Ki67 staining in BC1 (p<0.0001) and BC2 (p<0.0001). GPR30TOT in BC2 did not associate with tamoxifen treatment for BC1. However, BC2 that were diagnosed during tamoxifen treatment were more likely to express GPR30PM than BC2 diagnosed after treatment completion (p = 0.01). Furthermore, a trend was observed that patients with GPR30PM in an ER-positive BC2 had greater benefit from tamoxifen treatment.
Conclusion
PM-localized GPR30 staining is associated with increased risk of BC death when expressed in BC2 and LGL2. Additionally, PM-localized GPR30 correlates with prognostic markers of worse outcome, such as high Ki67 and a triple-negative subtype. Therefore, PM-localized GPR30 may be an interesting new target for therapeutic exploitation. We found no clear evidence that total GPR30 expression is affected by tamoxifen exposure during development of metachronous CBC, or that GPR30 contributes to tamoxifen resistance.
Introduction
Metachronous contralateral breast cancer (CBC) is a second, presumably independent primary tumor (BC2) developed in the contralateral breast after the first breast cancer (BC1). The lifetime risk of a breast cancer (BC) patient developing CBC has been estimated at 2–20%, depending on factors such as family history, prior endocrine treatment, and age at BC1 diagnosis [1–3]. Similar to BC in general, CBC is a heterogenous disease and both disease stage and the molecular characteristics of the tumor is used to assess prognosis and benefit of therapy, where axillary lymph node (LGL) involvement is one of the strongest prognostic factors [4]. At the molecular level, the tumor is generally characterized by the expression of estrogen receptor α (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), as well as the proliferation rate [5]. About 80% of all BC exhibit overexpression of ER, through which the female steroid hormone estrogen acts to stimulate cell growth and proliferation. Therefore, endocrine therapies directed to disrupt ER signaling are central in current BC treatment, acting either by suppressing ER activity, e.g. selective ER modulators or downregulators, or by inhibiting estrogen production, e.g. aromatase inhibitors. The selective ER modulator tamoxifen is one of the most widely prescribed endocrine agents for treatment of ER-positive BC [6]. In the adjuvant setting, 5 years of tamoxifen treatment reduces the 10-year risk of recurrence by almost 50%, and the annual risk of BC mortality by almost one-third [7, 8]. However, not all ER-positive tumors respond to tamoxifen therapy, and resistance may occur de novo or during treatment. Tamoxifen reduces the incidence of CBC, but CBC evolving during tamoxifen treatment is assumed to have intrinsic resistance. Efforts aimed to further understand resistance mechanisms have led to a number of important discoveries, including pathological epigenetic changes or mutations in the ESR1 gene, and interference with other growth stimulatory signaling pathways. These mechanisms subsequently result in augmented receptor activity, ligand-independent growth and transcription, or reduced drug sensitivity [6, 9, 10]. Despite these discoveries, ER remains the only predictive marker for endocrine treatment.
G protein-coupled estrogen receptor (GPER), originally named G protein-coupled receptor 30 (GPR30), is a receptor involved in rapid, non-genomic responses to estrogen [11]. In contrast to the classical ER, which is a soluble receptor residing in the cytoplasm or cell nucleus, GPR30 is a transmembrane receptor reported to be expressed both in the plasma membrane (PM) [12] and in the endoplasmatic reticulum [13]. As an estrogen receptor, GPR30 has caught significant attention in BC research, and the relationship between GPR30 and BC outcome has been addressed in multiple studies. However, results are inconsistent, with the receptor conveying either better [14, 15] or worse prognosis [16, 17], or lacking any prognostic value [18] for BC outcome. Additionally, in vitro studies have shown that GPR30 is pro-apoptotic in the ER-positive BC cell line MCF-7, but proliferative in the ER-negative cell line SkBr3 [19]. Thus, GPR30 may function differently depending on the environment in which it is expressed. Both clinical and pre-clinical studies have shown that subcellular localization is also a factor influencing GPR30 function. Indeed, GPR30 staining specifically located in the PM was found to be a strong prognostic factor for poor prognosis in BC, while the total level of GPR30 staining was not [17]. Consistent with this clinical observation, an in vitro study showed that PM localization of GPR30 is important for receptor stimulation of ERK1/2 activity [20], a cellular signal involved in proliferation and survival. Thus, the biological context of the tumor appears to be critical for GPR30 function in BC, with subcellular localization being a factor of potential importance.
Studies have reported that GPR30 may contribute to tamoxifen resistance [21–24]. Some in vitro data suggest that tamoxifen directly stimulates cell growth via GPR30 [22]. This situation would be of major clinical concern, as many ER-positive BC also express GPR30, and tamoxifen treatment of these BC partly could yield increased cell growth. On the other hand, these observations may also argue that GPR30 is a potential marker to identify BC with poor responsiveness to tamoxifen. GPR30 has been suggested to function as a resistance mechanism to escape tamoxifen responsiveness. However, whether GPR30 expression changes in tumors developing resistance to tamoxifen treatment, has not yet been addressed in a larger cohort of patients.
The aim of this study was to further understand how GPR30 expression relates to BC progression, patient outcome, and previous tamoxifen treatment. To this end, we used a unique retrospective cohort of patients with CBC, either naïve or exposed to tamoxifen following BC1, with paired expression data from primary tumors and lymph node metastases, as a stepwise model of tamoxifen resistance.
Materials and methods
Patient cohort and TMA preparation
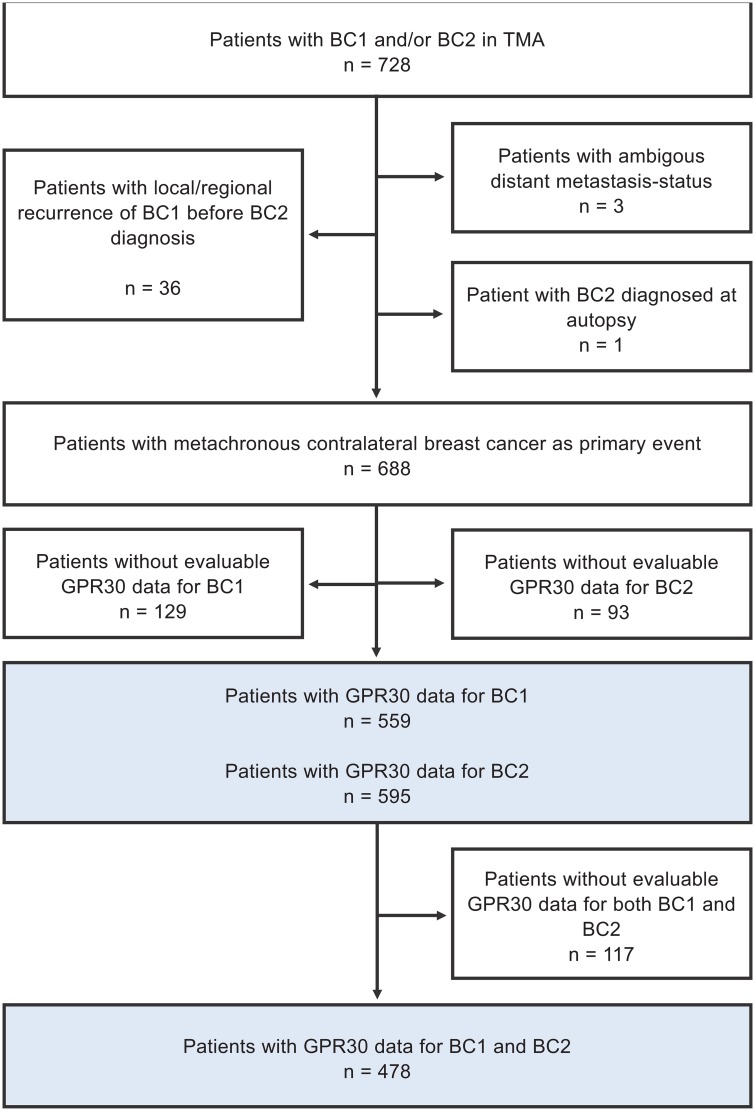
A previously constructed tissue-microarray (TMA) from 728 patients diagnosed with CBC between 1977 and 2007 at 14 hospitals within the Southern Swedish Healthcare Region was used. Details of TMA construction have been previously described [25]. Patient inclusion and number of CBC successfully stained and scored for GPR30 are summarized in Fig 1. Follow-up information was retrieved from patient charts, and cause of death and overall survival data were accessed from the Swedish National Board of Health and Welfare in March 2014. Evaluation of ER, PR, Ki67 and HER2 have been previously described [26, 27].
Fig 1. Flow-chart of inclusion and exclusion for the study cohort.
Cell construction and culture
HFF11 cells, originally constructed from HeLa cells (American Type Culture Collection, ATCC), were kindly provided by K. Kotarsky [28]. HFF11 cells stably expressing the T-Rex system (HeLa TET-On/Off cells), in which the expression of human GPR30 is under the strict control by tetracycline, were constructed according to vendor instructions (ThermoFisher Scientific). MCF7 cells were obtained from ATCC. Cells were confirmed to be mycoplasma-free by the MycoSEQ™ Mycoplasma Detection System (ThermoFisher Scientific). HeLa TET-On/Off cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA), 10% fetal calf serum (FCS; HyClone Laboratories, Logan, UT), and 1% penicillin/streptomycin (Sigma-Aldrich), using blastidine and zeocin as clone selection markers. MCF7 cells were grown in DMEM supplemented with glucose and pyruvate, and with 10% FCS. Both cells were grown in 5% CO2 at 37°C.
GPR30 antibody validation
The specificity of the polyclonal goat GPR30 antibody (AF 5534; R&D Systems) was analyzed in MCF7 human breast cancer cells (American Type Culture Collection), a cell line used extensively to study native GPR30 [19]. Immunoblotting showed that the antibody recognized a receptor species with a molecular mass of 50–55 kDa in MCF7 cells (S1 Fig, panel A), consistent with that described by the vendor (R&D Systems). To ensure that the antibody reactivity was absolutely dependent on GPR30 expression, we immunoblotted HeLa TET-On/Off cells treated with and without 0.1 μM tetracycline. In these cells, the GPR30 antibody recognized three broad receptor species at about 50 kDa, 80 kDa, and 130 kDa following tetracycline treatment, whereas no significant immunoreactivity was observed in the absence of tetracycline treatment (S1 Fig, panel B). These results, and those published by us previously [15, 29], show that GPR30 antibody immunoreactivity is completely dependent on GPR30 expression. Slightly different molecular masses of the recognized receptor species were observed in native MCF7 cells and recombinant HeLa cells. This is common among G protein-coupled receptors (GPCR) and due to variations in posttranslational modifications (e.g. glycosylation, oligomerization, etc.). Confocal immunofluorescence microscopy of MCF7 cells stained live with the GPR30 antibody showed that the antibody detected receptors in the PM in these cells (S1 Fig, panel C). A similar subcellular localization was revealed using M1 FLAG antibodies (Sigma-Aldrich) to detect recombinant human GPR30 tagged at the N terminus with the FLAG epitope transiently expressed in MCF7 cells (S1 Fig, panel D). Thus, the GPR30 antibody is capable of recognizing GPR30 localized in the PM.
IHC staining and scoring of GPR30
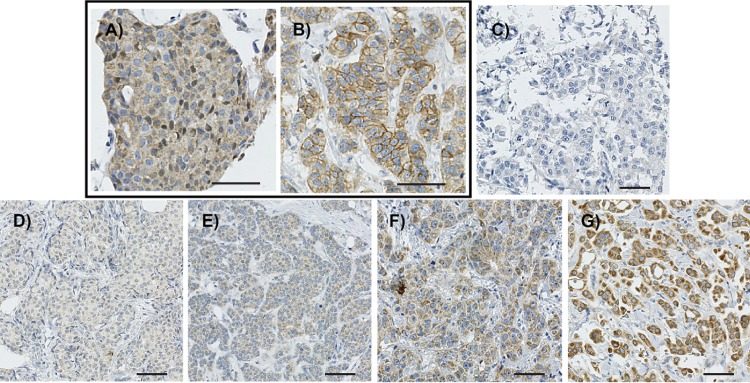
GPR30 expression was monitored by immunohistochemical (IHC) staining with GPR30 antibody (1:50) on 1.0 mm TMA cores. Receptor expression was evaluated as total cellular staining intensity (GPR30TOT; Fig 2A) and PM-specific staining intensity (GPR30PM; Fig 2B). PM-specific staining was defined as a clear increase of immunoreactivity on cell borders, as compared to the cytoplasm. GPR30TOT was scored as overall intensity at 5 levels (Fig 2C–2G; 0 = negative; 1 = very weak; 2 = weak; 3 = moderate; 4 = strong), and GPR30PM at 3 levels (0 = no increase as compared to the cytoplasmic staining; 1 = weak PM-specific staining; and 2 = strong PM-specific staining). Percentage of stained tumor cells was scored, but as the vast majority of tumors had either 0% or >50% stained cells, the intensity was used for further analysis. The staining was visually evaluated by two independent investigators (M.S., K.L.), a well-established widely accepted method to evaluate IHC staining on TMA cores, and the mean score was used, rounding to nearest integer. Due to group sizes, GPR30TOT intensity was combined to two groups of weak (levels 0–2) vs. strong (levels 3–4). GPR30PM+ scores were combined to create a binary variable (levels 0 vs. 1–2).
Fig 2. Representative images of GPR30 staining in BC samples.
A and B, images of GPR30 staining intensity level 3 without plasma-membrane staining (GPR30PM-) (A) and with plasma-membrane staining (GPR30PM+) (B). C-G, images of GPR30 total staining (GPR30TOT) as negative (C), very weak (level 1) (D), weak (level 2) (E), moderate (level 3) (F) and very strong (level 4) (G). Bar, 50 μm.
Statistical analysis
Death from BC (BCD), including BC death or death after documented distant metastasis, was used as primary end-point. Median follow-up time for patients alive at last follow-up was 9.1 years from BC2 diagnosis. Statistical analyses were carried out in RStudio 1.1.442 (RStudio, Boston, MA, USA) using R 3.5.1 [30]. Associations between GPR30 and factors such as patient attributes and clinicopathological markers were evaluated using Pearson’s χ2-test or Mantel-Haenszel χ2-test for trend. GPR30 staining in paired tumors was compared using Wilcoxon matched pairs signed rank test. Cumulative incidence of BCD was calculated with death from other causes as competing risk event using the cmprsk R package [31]. Cox proportional hazards models with Wald test was used to calculate hazard ratios (HR), using the survival R package [32]. Proportional hazards were assessed using Schoenfeldt residuals. As the assumption was not reasonably well met in all the analyses, HRs should be cautiously interpreted as average effects over the follow-up interval, which was restricted to maximum 10 years to reduce the problem of non-proportional hazards.
Ethical approval
All procedures performed in studies involving human participants were approved by the Regional Ethical Review Board of Lund University (LU240-01) and in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Since the study handled saved paraffin material, often several decades old, informed consent was not possible to retrieve from all patients. Nevertheless, all data was analyzed and presented anonymously, and a note was published in the local paper, informing previous BC patients to contact the research group if they did not want their tumor tissue to be used in scientific studies. This procedure was accepted by the Regional Ethical Review Board of Lund University (LU240-01).
Results
GPR30 expression in relation to patient and tumor characteristics
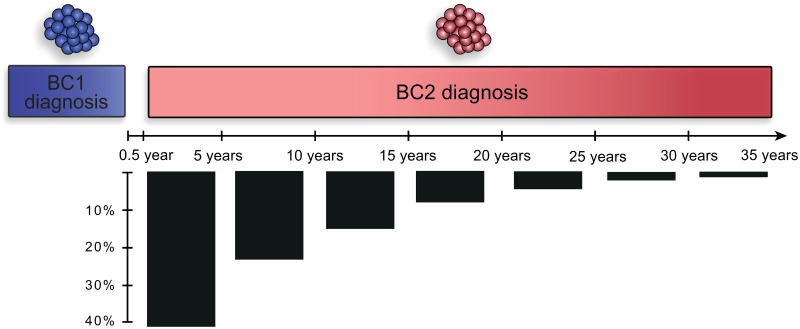
GPR30 expression was assessed in 688 women with metachronous CBC, where BC2 was diagnosed between 6 months and 34.1 years after BC1 diagnosis (Fig 3, median = 6.6 years). Receptor expression was monitored by immunohistochemistry in two variables; receptor staining at the PM (GPR30PM+; Fig 2A and 2B) and total receptor staining intensity (GPR30TOT; Fig 2C–2G). Increasing GPR30TOT was associated with a higher fraction of GPR30PM+ tumors in both BC1 and BC2 (Table 1). As previously observed in three other cohorts [17], GPR30TOT associated with ER and PR expression in a biphasic manner in both BC1 and BC2, where tumors with no or very weak GPR30TOT and tumors with strong GPR30TOT were more likely to be ER-negative and PR-negative, whereas tumors with weak or moderate GPR30TOT were more likely to be ER-positive and PR-positive (Table 1). Interestingly, GPR30PM+ staining associated strongly with ER-negative and PR-negative status in both BC1 and BC2. GPR30PM+ status also strongly associated with high Ki67 staining in both BC1 and BC2 in this cohort. Lastly, both GPR30TOT and GPR30PM correlated with tumor subtype, with strong GPR30TOT and GPR30PM+ status being significantly more prevalent among triple-negative cancers in both BC1 and BC2 (Table 1).
Fig 3. Timeline of CBC in the study cohort.
Timeline showing the distribution of the interval between BC1 and BC2 in the study cohort.
Table 1. Association between total GPR30 intensity score (GPR30TOT), plasma membrane GPR30 (GPR30PM) status, and various clinicopathological variables.
| BC1 n = 559 | GPR30TOT intensity of BC1; n (%) | GPR30PM+ BC1; n (%) | ||||||||
| 0 | 1 | 2 | 3 | 4 | p | No PM GPR30 505 (90) | PM+ GPR30 54 (10) | p | Missing GPR30 data | |
| 46 (8) | 141 (25) | 263 (47) | 103 (18) | 6 (1) | ||||||
| BC1 diagnosis | ||||||||||
| <1977 (n = 122) | 11 (13) | 24 (28) | 36 (42) | 14 (17) | 0 (0) | 0.703 | 74 (87) | 11 (13) | 0.52 | 37 |
| 1977–1986 (n = 210) | 14 (8) | 45 (27) | 79 (47) | 26 (16) | 3 (2) | 149 (89) | 18 (11) | 43 | ||
| 1987–1996 (n = 265) | 15 (7) | 55 (25) | 108 (48) | 43 (19) | 2 (<1) | 0.020a | 204 (92) | 19 (8) | 42 | |
| 1997–2007 (n = 91) | 6 (7) | 17 (20) | 40 (48) | 20 (24) | 1 (1) | 78 (93) | 6 (7) | 7 | ||
| Age at BC1 diagnosis | ||||||||||
| <50 years (n = 189) | 18 (12) | 43 (30) | 63 (43) | 19 (13) | 3 (2) | 0.027 | 127 (87) | 19 (13) | 0.15 | 43 |
| ≥50 years (n = 499) | 28 (7) | 98 (24) | 200 (48) | 84 (20) | 3 (<1) | 0.011a | 378 (92) | 35 (8) | 86 | |
| Interval between tumors | ||||||||||
| <5 years (n = 293) | 21 (8) | 61 (24) | 115 (46) | 51 (20) | 4 (2) | 0.67 | 222 (88) | 30 (12) | 0.14 | 41 |
| ≥5 years (n = 395) | 25 (8) | 80 (26) | 148 (48) | 52 (17) | 2 (<1) | 0.37a | 283 (92) | 24 (8) | 88 | |
| Histological type BC1 | ||||||||||
| Ductal (n = 391) | 21 (6) | 83 (25) | 167 (50) | 58 (17) | 4 (1) | 0.26 | 298 (90) | 35 (10) | 0.01 | 58 |
| Lobular (n = 95) | 11 (14) | 20 (25) | 31 (38) | 19 (24) | 0 (0) | 0.22a | 79 (98) | 2 (2) | 14 | |
| Other (n = 125) | 9 (9) | 25 (26) | 46 (48) | 16 (17) | 0 (0) | 93 (97) | 3 (3) | 29 | ||
| Missing (n = 77) | 5 | 13 | 19 | 10 | 2 | 35 | 14 | 28 | ||
| Tumor subclass | ||||||||||
| Luminal A like (n = 401) | 25 (7) | 95 (25) | 193 (51) | 63 (17) | 0 (0) | <0.0001 | 354 (94) | 22 (6) | <0.0001 | 25 |
| Luminal B like (n = 69) | 2 (3) | 15 (22) | 36 (52) | 16 (23) | 0 (0) | 0.93a | 63 (91) | 6 (9) | 0 | |
| HER2+ luminal (n = 17) | 1 (6) | 2 (12) | 10 (59) | 4 (24) | 0 (0) | 13 (77) | 4 (24) | 0 | ||
| HER2+ non-luminal (n = 13) | 1 (8) | 3 (23) | 8 (62) | 1 (8) | 0 (0) | 11 (85) | 2 (15) | 0 | ||
| Triple-negative (n = 63) | 12 (20) | 19 (31) | 11 (18) | 13 (21) | 6 (10) | 43 (71) | 18 (29) | 2 | ||
| Missing (n = 125) | 5 | 7 | 5 | 6 | 0 | 21 | 2 | 102 | ||
| Node status | ||||||||||
| N0 (n = 416) | 23 (7) | 88 (26) | 157 (46) | 69 (20) | 4 (1) | 0.28 | 305 (89) | 36 (11) | 0.35 | 75 |
| N+ (n = 213) | 17 (9) | 47 (26) | 94 (51) | 25 (14) | 1 (<1) | 0.11a | 170 (92) | 14 (8) | 29 | |
| Missing (n = 59) | 6 | 6 | 12 | 9 | 1 | 30 | 4 | 25 | ||
| Size | ||||||||||
| ≤20 mm (n = 404) | 19 (6) | 79 (24) | 160 (48) | 68 (21) | 5 (1) | 0.047 | 298 (90) | 33 (10) | 0.76 | 73 |
| >20 mm (n = 215) | 23 (12) | 54 (28) | 86 (44) | 30 (16) | 1 (<1) | 0.0033a | 177 (91) | 17 (9) | 21 | |
| Missing (n = 69) | 4 | 8 | 17 | 5 | 0 | 29 | 5 | 35 | ||
| ER status | ||||||||||
| <10% stained (n = 99) | 17 (18) | 29 (31) | 23 (25) | 19 (20) | 6 (6) | <0.0001 | 73 (78) | 21 (22) | <0.0001 | 5 |
| ≥10% stained (n = 494) | 28 (6) | 112 (24) | 240 (52) | 83 (18) | 0 (0) | 0.11a | 431 (93) | 32 (7) | 31 | |
| Missing (n = 95) | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 93 | ||
| PR status | ||||||||||
| <10% stained (n = 165) | 22 (14) | 41 (26) | 56 (36) | 32 (20) | 6 (4) | <0.0001 | 131 (83) | 26 (17) | 0.00070 | 8 |
| ≥10% stained (n = 428) | 23 (6) | 100 (25) | 207 (52) | 70 (18) | 0 (0) | 0.38a | 373 (93) | 27 (7) | 28 | |
| Missing (n = 95) | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 93 | ||
| HER2 staining | ||||||||||
| Negative (n = 544) | 42 (8) | 133 (26) | 241 (47) | 93 (18) | 6 (1) | 0.86 | 469 (91) | 46 (9) | 0.13 | 29 |
| Positive (n = 40) | 3 (7) | 8 (20) | 21 (53) | 8 (20) | 0 (0) | 0.63a | 33 (83) | 7 (17) | 0 | |
| Missing (n = 104) | 1 | 0 | 1 | 2 | 0 | 3 | 1 | 100 | ||
| Ki67 staining | ||||||||||
| <20% stained (n = 356) | 30 (9) | 85 (26) | 160 (48) | 57 (17) | 1 (<1) | 0.19 | 315 (95) | 18 (5) | <0.0001 | 23 |
| ≥20% stained (n = 227) | 15 (7) | 56 (25) | 102 (46) | 44 (20) | 5 (2) | 0.013a | 187 (84) | 35 (16) | 5 | |
| Missing (n = 105) | 1 | 0 | 1 | 2 | 0 | 3 | 1 | 101 | ||
| PM GPR30 status | ||||||||||
| Negative (n = 505) | 46 (9) | 137 (27) | 235 (47) | 87 (17) | 0 (0) | <0.0001 | - | - | - | 0 |
| Positive (n = 54) | 0 (0) | 4 (7) | 28 (52) | 16 (30) | 6 (11) | <0.0001a | - | - | 0 | |
| BC2 n = 595 | GPR30TOT intensity of BC2; n (%) | GPR30PM+ BC2; n (%) | ||||||||
| 0 | 1 | 2 | 3 | 4 | p | No PM GPR30 557 (94) | PM+ GPR30 38 (6) | p | Missing GPR30data | |
| 31 (5) | 151 (25) | 285 (48) | 117 (20) | 11 (2) | ||||||
| CBC diagnosis | ||||||||||
| 1977–1986 (n = 136) | 6 (6) | 27 (25) | 45 (42) | 28 (26) | 1 (<1) | 0.072 | 96 (90) | 11 (10) | 0.18 | 29 |
| 1987–1996 (n = 243) | 11 (5) | 66 (32) | 89 (44) | 33 (16) | 5 (3) | 0.48a | 192 (94) | 12 (6) | 0.101 | 39 |
| 1997–2007 (n = 309) | 14 (5) | 58 (20) | 151 (53) | 56 (20) | 5 (2) | 269 (95) | 15 (5) | 25 | ||
| Age at CBC diagnosis | ||||||||||
| <50 years (n = 67) | 1 (2) | 18 (31) | 26 (45) | 12 (21) | 1 (2) | 0.65 | 53 (91) | 5 (9) | 0.65 | 9 |
| ≥50 years (n = 621) | 30 (6) | 133 (25) | 259 (48) | 105 (20) | 10 (2) | 0.84a | 504 (94) | 33 (6) | 84 | |
| Interval between tumors | ||||||||||
| <5 years (n = 293) | 11 (5) | 64 (26) | 118 (48) | 50 (20) | 4 (2) | 0.95 | 230 (93) | 17 (7) | 0.81 | 46 |
| ≥5 years (n = 395) | 20 (6) | 87 (25) | 167 (48) | 67 (19) | 7 (2) | 0.79a | 327 (94) | 21 (6) | 47 | |
| Histology BC2 | ||||||||||
| Ductal (n = 450) | 24 (6) | 108 (27) | 189 (46) | 82 (20) | 5 (1) | 0.022 | 377 (92) | 31 (8) | 0.031 | 42 |
| Lobular (n = 130) | 2 (2) | 32 (29) | 59 (53) | 17 (15) | 2 (2) | 0.032a | 111 (99) | 1 (1) | 130 | |
| Other (n = 77) | 2 (4) | 8 (15) | 28 (52) | 12 (22) | 4 (7) | 51 (94) | 3 (6) | 77 | ||
| Missing (n = 31) | 3 | 3 | 9 | 6 | 0 | 18 | 3 | 10 | ||
| Tumor subclass | ||||||||||
| Luminal A like (n = 403) | 16 (4) | 88 (23) | 193 (51) | 75 (20) | 4 (1) | <0.0001 | 367 (98) | 9 (2) | <0.0001 | 27 |
| Luminal B like (n = 88) | 4 (5) | 25 (28) | 48 (55) | 11 (13) | 0 (0) | 0.76a | 83 (94) | 5 (6) | 0 | |
| HER2+ luminal (n = 19) | 0 (0) | 5 (26) | 10 (53) | 4 (21) | 0 (0) | 17 (90) | 2 (10) | 0 | ||
| HER2+ non-luminal (n = 10) | 0 (0) | 3 (30) | 3 (30) | 4 (40) | 0 (0) | 9 (90) | 1 (10) | 0 | ||
| Triple-negative (n = 74) | 7 (10) | 19 (27) | 19 (27) | 18 (26) | 7 (10) | 50 (71) | 20 (29) | 4 | ||
| Missing (n = 94) | 4 | 11 | 12 | 5 | 0 | 31 | 1 | 94 | ||
| Node status | ||||||||||
| N0 (n = 371) | 15 (5) | 80 (26) | 155 (50) | 58 (19) | 5 (2) | 0.39 | 290 (93) | 23 (7) | 0.54 | 58 |
| N+ (n = 196) | 13 (7) | 54 (30) | 74 (41) | 38 (21) | 3 (2) | 0.43a | 172 (95) | 10 (5) | 14 | |
| Missing (n = 121) | 3 | 17 | 56 | 21 | 3 | 95 | 5 | 121 | ||
| Size | ||||||||||
| ≤20 mm (n = 481) | 19 (5) | 96 (23) | 199 (48) | 91 (22) | 7 (2) | 0.13 | 387 (94) | 25 (6) | 0.61 | 69 |
| >20 mm (n = 181) | 11 (7) | 53 (31) | 76 (45) | 26 (15) | 4 (2) | 0.027a | 157 (92) | 13 (8) | 11 | |
| Missing (n = 13) | 1 | 2 | 10 | 0 | 0 | 13 | 0 | 13 | ||
| ER status | ||||||||||
| <10% stained (n = 105) | 8 (8) | 30 (30) | 28 (28) | 26 (26) | 7 (7) | <0.0001 | 77 (78) | 22 (22) | <0.0001 | 6 |
| ≥10% stained (n = 524) | 21 (4) | 120 (24) | 256 (52) | 91 (19) | 4 (<1) | 0.47a | 476 (97) | 16 (3) | 32 | |
| Missing (n = 59) | 2 | 1 | 1 | 0 | 0 | 4 | 0 | 55 | ||
| PR status | ||||||||||
| <10% stained (n = 212) | 13 (7) | 49 (25) | 90 (45) | 39 (20) | 9 (5) | 0.0097 | 175 (88) | 25 (12) | <0.0001 | 12 |
| ≥10% stained (n = 412) | 16 (4) | 99 (26) | 192 (50) | 78 (20) | 2 (<1) | 0.62a | 375 (97) | 12 (3) | 25 | |
| Missing (n = 8) | 2 | 3 | 3 | 0 | 0 | 7 | 1 | 56 | ||
| HER2 status | ||||||||||
| Negative (n = 552) | 29 (5) | 139 (25) | 268 (49) | 105 (19) | 11 (2) | 0.24 | 517 (94) | 35 (6) | 0.93 | 32 |
| Positive (n = 37) | 0 (0) | 11 (30) | 15 (40) | 11 (30) | 0 (0) | 0.37a | 34 (92) | 3 (8) | 0 | |
| Missing (n = 67) | 2 | 1 | 2 | 1 | 0 | 6 | 0 | 67 | ||
| Ki67 staining | ||||||||||
| <20% stained (n = 349) | 18 (6) | 73 (23) | 153 (48) | 69 (22) | 6 (2) | 0.40 | 311 (98) | 8 (3) | <0.0001 | 30 |
| ≥20% stained (n = 266) | 10 (4) | 76 (29) | 126 (48) | 47 (18) | 5 (2) | 0.39a | 234 (89) | 30 (11) | 2 | |
| Missing (n = 73) | 3 | 2 | 6 | 1 | 0 | 12 | 0 | 61 | ||
| Radiotherapy BC1 | ||||||||||
| No (n = 257) | 9 (4) | 52 (23) | 119 (52) | 43 (19) | 5 (2) | 0.41 | 218 (96) | 10 (4) | 0.15 | 29 |
| Yes (n = 425) | 22 (6) | 97 (27) | 163 (45) | 74 (20) | 6 (2) | 0.28a | 334 (92) | 28 (8) | 63 | |
| Missing (n = 5) | 0 | 2 | 3 | 0 | 0 | 5 | 0 | 1 | ||
| Chemotherapy for BC1 | ||||||||||
| No (n = 615) | 27 (5) | 135 (25) | 253 (48) | 105 (20) | 10 (2) | 0.98 | 496 (94) | 34 (6) | 1.0 | 85 |
| Yes (n = 66) | 4 (7) | 14 (24) | 29 (49) | 11 (18) | 1 (2) | 0.79a | 55 (93) | 4 (7) | 7 | |
| Missing (n = 7) | 0 | 2 | 3 | 1 | 0 | 6 | 0 | 1 | ||
| Tamoxifen for BC1 | ||||||||||
| All patients | ||||||||||
| No tamoxifen (n = 467) | 25 (5) | 115 (25) | 226 (48) | 92 (20) | 9 (2) | 0.96 | 435 (93) | 32 (7) | 0.57 | 73 |
| Tamoxifen (n = 122) | 6 (5) | 34 (28) | 56 (46) | 24 (20) | 2 (2) | 0.73a | 116 (95) | 6 (5) | 19 | |
| Missing (n = 7) | 0 | 2 | 3 | 1 | 0 | 6 | 0 | 1 | ||
| Tamoxifen treated for BC1 | ||||||||||
| BC2 diagnosis during treatment (n = 60) | 1 (2) | 18 (35) | 19 (37) | 12 (23) | 2 (4) | 0.089 | 46 (89) | 6 (11) | 0.013 | 11 |
| BC2 diagnosis after treatment (n = 81) | 5 (7) | 16 (23) | 37 (53) | 12 (17) | 0 (0) | 70 (100) | 0 (0) | 8 | ||
| Not tamoxifen treated for BC1 | ||||||||||
| BC2<5 years after BC1 (n = 222) | 10 (5) | 45 (24) | 92 (49) | 39 (21) | 2 (1) | 0.83 | 177 (94) | 11 (6) | 0.61 | 34 |
| BC2>5 years after BC1 (n = 318) | 15 (5) | 70 (25) | 134 (48) | 53 (19) | 7 (3) | 0.99a | 258 (93) | 21 (7) | 39 | |
| PM GPR30 status | ||||||||||
| Negative (n = 557) | 31 (6) | 149 (27) | 270 (48) | 103 (19) | 4 (<1) | <0.0001 | ||||
| Positive (n = 38) | 0 (0) | 2 (5) | 15 (40) | 14 (37) | 7 (18) | <0.0001a | ||||
Abbreviations: BC: breast cancer; BC1: the first primary BC; BC2: the second primary BC; CBC: contralateral breast cancer; GPR30: G protein-coupled receptor 30; PM: plasma membrane; HER2: human epidermal growth factor receptor-2; N+/N0: presence or absence of lymph node metastases; ER: estrogen receptor α; PR: progesterone receptor.
Values for p are calculated using Pearson’s χ2-test without continuity correction if otherwise is not stated.
a Calculated using Mantel-Haenszel χ2-test test (χ2-test for trend).
To assess GPR30 expression through tumor progression, GPR30TOT staining was compared between BC1 and BC2, and with their corresponding LGL (Fig 4). A majority of the LGLs had a weaker GPR30TOT compared to the primary BC in both the BC1/LGL1 pair (Fig 4; p<0.0001) and the BC2/LGL2 pairs (p<0.0001).
Fig 4. GPR30 staining in paired tumors.
The change in GPR30 intensity (level 0–4) between paired BC and LGL. In relation to the tumor assumed to have developed earlier, the intensity shift in the second tumor is characterized as decreasing, stable or increasing for each tumor pair respectively. The intensity shift was assessed statistically using Wilcoxon matched pairs signed rank test.
GPR30 expression and risk of BCD in CBC patients
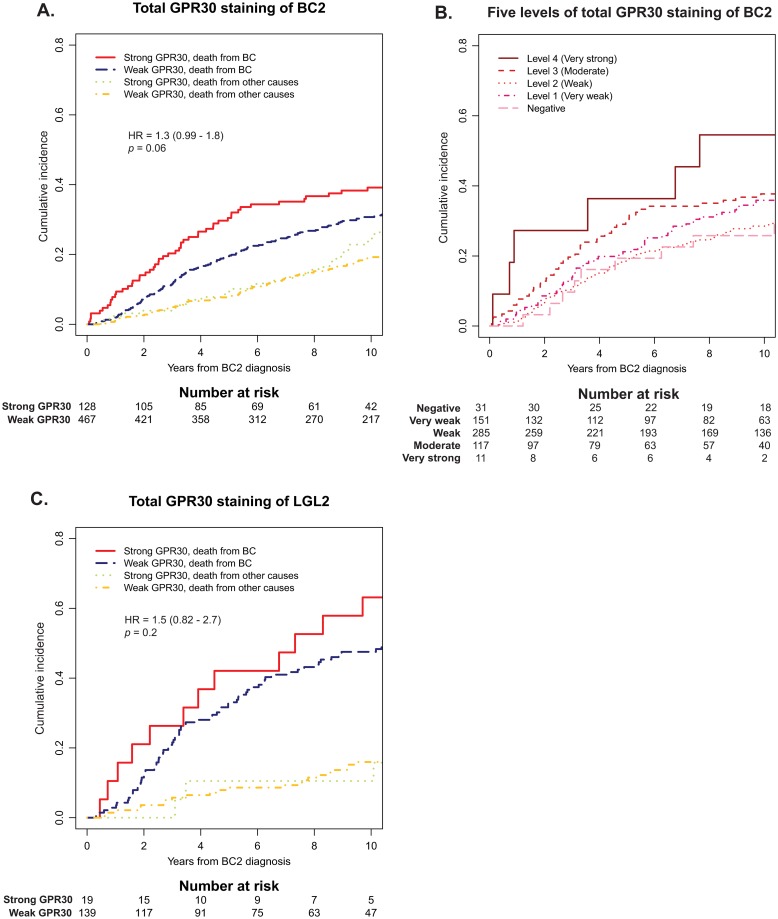
Previous studies by our group showed that characteristics of BC2 have the highest influence on prognosis after development of CBC, although the characteristics of BC1 continue to have some prognostic impact [33]. GPR30PM+ staining in both BC2 and LGL2 was associated with increased risk of BCD (Table 2; Fig 5A and 5B; HRBC2 = 1.7; 95% CI = 1.1–2.7; p = 0.03, and HRLGL2 = 2.0; 95% CI = 1.1–3.4; p = 0.02). A trend was also observed that higher GPR30TOT associated with increased BCD in BC2 and LGL2 (Fig 6A–6C; HRBC2 = 1.3; 95% CI = 0.99–1.8; p = 0.06, and HRLGL2 = 1.5; 95% CI = 0.82–2.7; p = 0.2, respectively). Similar trends were seen when looking at GPR30 expression in BC1.
Table 2. Prognostic effect of total GPR30 staining (GPR30TOT) and PM-specific GPR30 (GPR30PM) staining of BC2 and LGL2 calculated by Cox proportional hazards model with Wald test.
| Unadjusted | Adjusted for BC1a | Adjusted for BC1 and BC2b | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | HR | n | Events | 95% CI | p | HR | n | Events | 95% CI | p | HR | n | Events | 95% CI | p | |
| GPR30TOT | BC2 | 1.3 | 595 | 237 | 0.99–1.8 | 0.06 | 1.2 | 428 | 171 | 0.85–1.8 | 0.3 | 1.4 | 349 | 145 | 0.93–2.1 | 0.11 |
| LGL2 | 1.5 | 151 | 83 | 0.82–2.7 | 0.2 | 1.6 | 119 | 70 | 0.74–3.3 | 0.2 | 2.2 | 110 | 67 | 1.0–4.7 | 0.05 | |
| GPR30PM+ | BC2 | 1.7 | 595 | 237 | 1.1–2.7 | 0.03 | 1.5 | 428 | 171 | 0.87–2.7 | 0.1 | 1.6 | 349 | 145 | 0.89–3.0 | 0.1 |
| LGL2 | 2.0 | 151 | 83 | 1.1–3.4 | 0.02 | 1.4 | 119 | 70 | 0.67–2.8 | 0.4 | 3.0 | 110 | 67 | 1.43–6.1 | 0.004 | |
a Adjusted for the following variables in BC1: GPR30 intensity, tumor size, node status, ER status, HER2 status and ki67 staining.
b Adjusted for factors in a and for the following factors in BC2: size, node status, ER status, HER2 status and Ki67 staining, plus age and calendar interval at BC2 diagnosis and interval between BC1 and BC2.
Fig 5. Cumulative incidence of BCD in relation to PM-specific GPR30 staining (GPR30PM).
A, cumulative incidence of BCD in relation to GPR30PM staining of BC2. B, cumulative incidence of BCD in relation to GPR30PM staining of LGL2. In A and B, cumulative incidences of competing event (death from other cause than BC) are shown for comparison.
Fig 6. Cumulative incidence of BCD in relation to total GPR30 staining (GPR30TOT).
A, cumulative incidence of BCD in relation to GPR30TOT of BC2 in the whole cohort. B, cumulative incidence of BCD in relation to the five GPR30 staining intensity levels of BC2. C, cumulative incidence of BCD in relation to GPR30TOT of LGL2. In A-C, cumulative incidences of competing event (death from other cause than BC) are shown for comparison.
The trend that GPR30 is a negative prognostic marker for BCD remained in multivariate analysis (Table 2), although effect sizes generally decreased. When estimating the prognostic impact of GPR30 in multivariate analyses adjusted for each variable individually, the prognostic effect of GPR30 was found to mainly be affected when adjusting for ER-status (S1 Table). Interestingly, stratified survival analyses based on ER-status showed that both strong GPR30TOT and GPR30PM+ staining in BC2 were graphically associated with increased BCD among patients with ER-positive tumors, but not ER-negative tumors (S2 Fig and S2 Table). However, statistical test for interaction did not show a significant difference in the prognostic value of GPR30 between ER-positive and ER-negative tumors (GPR30TOT pinteraction = 0.7; GPR30PM+ pinteraction = 0.1). To address the combined prognostic information of GPR30 status in both BC1 and BC2, we created combination variables with information from paired BC1 and BC2 for GPR30TOT and GPR30PM respectively. This analysis showed that, although BC2 carried the higher prognostic value, BC1 also added prognostic information (S3 Fig and S3 Table).
GPR30 expression and tamoxifen treatment
To address the GPR30 expression in individual CBC cases, we matched the GPR30 variables of BC1 and BC2 within each patient individually. We observed that GPR30TOT was equally likely to have increased, decreased, or remained the same in BC2 compared to BC1 (Fig 4). Interestingly, the pattern was similar regardless if tamoxifen treatment had been given for BC1 or not (Fig 4). In the whole cohort, there was no association having received tamoxifen treatment for BC1, and score of GPR30TOT or GPR30PM+ in BC2. However, in a subset of patients diagnosed with BC2 during adjuvant tamoxifen treatment for BC1, BC2 were more likely to be GPR30PM+ than in BC2 diagnosed after completed tamoxifen treatment (11% vs 0%, p = 0.01; Table 1).
When stratified for tamoxifen treatment of ER-positive BC2, GPR30PM+ exhibited higher prognostic potential in patients that did not receive tamoxifen for BC2 as compared to patients that did receive tamoxifen. The prognostic potential remained significant in both univariate analysis and multivariate analysis adjusting for attributes of BC1 and BC2 (S2 Table; univariate HR = 2.8, 95% CI = 1.3–6.0, p = 0.01; multivariate HR = 3.8, 95% CI = 1,4–11, p = 0.01). Additionally, when studying the effect of tamoxifen on ER-positive BC2, there was a trend that patients with strong GPR30TOT or GPR30PM+ staining had a greater benefit from tamoxifen treatment than those with lower GPR30TOT or without GPR30PM staining (S4 Fig). However, interaction between GPR30 and tamoxifen could not be confirmed statistically (S2 Table; GPR30TOT pinteraction = 0.4; GPR30PM+ pinteraction = 0.3). Finally, the prognostic effect of GPR30 in BC2 did not seem to be affected by whether tamoxifen had been given for BC1 or not (GPR30TOT pinteraction = 0.2; GPR30PM+ pinteraction = 0.4).
Discussion
GPR30 is a G protein-coupled receptor reported to mediate non-genomic estrogenic signaling and contribute to BC progression and tamoxifen resistance. However, the literature is inconsistent regarding the pathophysiological profile of GPR30 in BC, and the receptor function is still poorly understood. In this study, we sought to clarify the role of GPR30 during BC development and progression with or without tamoxifen exposure. To this end, we used a unique retrospective cohort of CBC, serving as a model of tamoxifen resistance. We show in this material that GPR30 staining is a strong prognostic factor for increased risk of BCD, particularly when expressed in the PM. We also show that the total GPR30 staining generally decreases during tumor progression. Interestingly, we find that total GPR30 staining was unrelated to tamoxifen treatment during tumor development, and we found no clear relationship between the prognostic value of GPR30 and tamoxifen treatment.
Studies have indicated that GPR30 is influenced by the ER modulator tamoxifen, and this has been suggested to contribute to tamoxifen resistance [21–24]. Upregulation of GPR30 expression was observed following tamoxifen treatment in a small BC cohort [21], and GPR30 expression was associated with worse prognosis for BC patients treated with tamoxifen as compared to tamoxifen-naïve patients [24]. In the present study, we assessed the expression of GPR30 in 688 women with metachronous CBC. In 60 of these patients, BC2 tumors developed during ongoing tamoxifen treatment for the first BC, strongly arguing for acquired tamoxifen resistance in these tumors. We hypothesized that if GPR30 contributes to tamoxifen resistance, tumors developed under exposure of tamoxifen would exhibit higher GPR30 expression as a result of selection pressure. In the whole cohort, no difference in total GPR30 staining was observed between the tamoxifen-naïve BC2 and the presumably tamoxifen-resistant BC2. As recent studies suggest that GPR30 may have unique functions when expressed specifically in the PM [12, 20, 34, 35], we also assessed if PM-localization of GPR30 may be involved in resistance mechanisms. In a subgroup analysis, we found that BC2 diagnosed during tamoxifen treatment for BC1 was more likely to express PM-specific GPR30 than BC2 diagnosed after completed treatment. This trend was seen both in ER-positive and ER-negative cases, and could hence not only be explained by a selection of ER-negative BC2 during tamoxifen exposure (ER-negativity associated to GPR30PM+) [17].
To address if GPR30 expression decreases the benefit of tamoxifen, we performed a survival analysis of patients treated with tamoxifen after the diagnosis of an ER-positive BC2, stratified by GPR30 expression. In this material, GPR30 did not associate with worse prognosis. Instead, a trend was noted that among patients not treated with tamoxifen for their ER-positive BC2, PM-localized GPR30 has a higher hazard. Additionally, there was a trend that patients with PM-localized GPR30 staining in an ER-positive BC2 have a greater benefit from tamoxifen treatment. In summary, our data provides no clear evidence that tamoxifen exposure affects the prognostic value of GPR30 in BC2, or that the benefit of tamoxifen depends on GPR30 expression. However, results are not unanimous and the relationship between PM-localized GPR30 and tamoxifen needs to be further addressed in future studies.
We also explored GPR30 through disease progression. The relationship between GPR30 staining in paired BC1 and BC2 appeared to be stochastic, in line with CBC being most often considered an independent primary tumor [36]. On the other hand, LGL-metastases are seeded from, and hence clonally related to, the primary BC. GPR30 staining in the LGL was significantly lower than that observed in the corresponding BC, a pattern observed previously and suggested to reflect a successive downregulation of the receptor during cancer progression [17, 37]. However, a study on samples of normal breast tissue, invasive BC and LGL reported the mean GPR30 expression in normal breast and BC to be equal, but that the expression decreased in the LGL [37]. Unexpectedly given this background, we also show that PM-specific staining in the LGL associates with aggressive tumor characteristics and significantly higher risk of BCD, which would suggest that high GPR30 expression in the LGL is beneficial for tumor cells. Whether lower GPR30 expression is beneficial for tumor cell dissemination to the lymph nodes, or a result of environmental factors in the lymph nodes, is an interesting question for future studies.
A limited amount of data exists regarding the expression of GPR30 in healthy breast tissue. However, one study assessed GPR30 in breast tissue from 12 healthy donors in which all were defined as GPR30 positive, with strong cytoplasmic GPR30 expression in ductal and lobular epithelium, myoepithelium, and stromal fibroblasts, but no expression in smooth muscle or vascular endothelium [16]. In addition, The Human Protein Atlas database reports the level of GPR30 mRNA in normal breast tissue to be at medium level, and protein expression of GPR30 at strong levels in breast myoepithelial cells, but not detected in adipocytes or glandular cells (Data available from v19.3; www.proteinatlas.org [38]). Based on this, it seems unlikely that the mere overexpression of GPR30 could explain the pathological turn the receptor seems to undergo during BC progression. This raises the question if a transforming event changes the localization and function of GPR30, in turn yielding tumor-promoting activity in a minority of BCs. Several GPCR mutations that affect their function have been identified in cancer [39]. However, very little is still known about cancer-related mutations in GPR30. Nevertheless, it has been reported that promoter methylation suppresses GPR30 expression in both BC cell lines and primary BC tissue, and that methylation pattern of GPER1 differ between BC tissue and healthy controls [40, 41]. Among 996 BC samples available in The Cancer Genome Atlas/PanCancer Atlas, genetic alterations in the GPER1 gene are present in less than 2% [42–45]. Hence, it is unlikely that GPER1 mutation causes PM-GPR30 expression, which according to data from this study and earlier is present in around 7–23% of BCs [17]. However, altered methylation may partly explain any BC-specific alterations in GPR30 expression level.
As seen here and previously [17], strong total and PM-specific GPR30 staining associate with ER-negative status. As ER-negativity is a strong marker of poor prognosis, we sought to evaluate if GPR30 adds any prognostic information beyond ER-status. The collinearity between GPR30 and ER is reflected in Cox regression adjusted for only ER (S1 Table), where the effects of both total and PM-specific GPR30 staining are reduced. However, when evaluating the association between GPR30 staining on BC outcome with the cohort stratified for ER status (S3 Fig), we found a trend that both strong total GPR30 and PM-specific GPR30 staining associate with worse prognosis in ER-positive CBC (S2 Fig), suggesting that GPR30 adds prognostic information beyond ER, at least in ER-positive tumors. Even though no prognostic effect of total or PM-specific GPR30 was seen in ER-negative CBC, tests of statistical interaction showed no significant difference in the prognostic effect of GPR30 in the ER-positive and ER-negative groups. Thus, our data suggest that although GPR30 and ER are strongly associated, GPR30 status adds prognostic information beyond ER.
Although associated with the same ligand, ER and GPR30 manifest considerable differences in terms of cellular function. ER is a nuclear receptor; a ligand-activated transcription factor that upon binding estrogen dimerizes and translocates to the nucleus, where it alters expression of target genes [46]. On the other hand, GPR30 is a G protein-coupled receptor (GPCR), and as such an integral membrane protein. In contrast to ER, an active GPCR orchestrates rapid downstream signaling by modifying the activity of several effectors and second messengers [47]. The versatile nature of a GPCR allows it to communicate with a broad signaling network, the profile of which is dependent on the expression profile of the cell. Studies have coupled GPR30 to several signaling events through both G protein-dependent and -independent mechanisms and both in response to estrogen and constitutively. Estrogen was reported to receptor-dependently stimulate increases in intracellular Ca2+ [13, 19], cAMP production [12], and ERK1/2 activity [12, 48–50], the latter through EGFR transactivation [12, 48], and cFos expression [49]. Furthermore, constitutively the receptor was reported to inhibit cAMP production [34] and the Ca2+-pump plasma membrane Ca2+-ATPase 4b [35], and stimulate ERK1/2 activity, the latter through PI3K [20]. Interestingly, many of these effects, including modulation of cAMP production and stimulation of ERK1/2 activity via PI3K and EGFR transactivation, have been found to depend on PM localization of the receptor. In this study, we confirm our previous results that the prognostic potential of GPR30 is more pronounced when the receptor is expressed in the PM [17]. The subcellular distribution of GPR30 is complex, with in vitro studies showing that receptor activity occurs both in the PM [12, 20, 34, 35], endoplasmic reticulum [13], and nucleus [51]. Today, GPR30 function is best described in the PM, which is typical for a GPCR [47], whereas few if any functions have been described in the endoplasmatic reticulum or nucleus.
Recent in vitro data have demonstrated that GPR30-mediated ERK1/2 signaling depends on an amino acid sequence at the receptor intracellular C-terminal end, through which the receptor interacts with scaffold and adaptor proteins localized at the PM [20], and this favors receptor PM localization [20, 34, 35, 52]. Together, these results present a model where intracellular scaffold and adaptor proteins contribute to cell proliferation by retaining GPR30 in the PM, thus spatially positioning the receptor to communicate with the ERK1/2 and EGFR pathways. As both ERK1/2 and EGFR activities are hallmarks of cell proliferation, an intriguing theory is that these interactions contribute to the association of PM-localized GPR30 with high Ki67 and poor BC outcome observed in this CBC material. Therefore, it is well motivated to further study the contribution of scaffold protein interactions to the function of GPR30 in BC pathology, and the potential of PM-localized GPR30 in targeted treatment strategies should be investigated.
In conclusion, this study evaluated the estrogen-responsive receptor GPR30 in a unique and large cohort of CBC with long-term follow-up, serving as a model for tamoxifen exposure and resistance. We conclude that GPR30 has prognostic value in CBC. On the other hand, we find no clear evidence that GPR30 is involved in tamoxifen resistance. GPR30 staining correlates with BC subtype, with the highest total and PM-specific GPR30 observed in triple-negative BC. Additionally, GPR30 is most active in CBC when located in the PM. Thus, PM-localized GPR30 is an interesting candidate for future therapeutic exploitation.
Supporting information
(PDF)
A, MCF7 cell lysates were immunoblotted with goat GPR30 antibody as previously described [20, 29, 34]. B, HeLa TET-On/Off cells were incubated without (-TET) or with 100 ng/ml tetracycline (+TET) for 12 h and then lysed and immunoblotted with GPR30 antibody. C, MCF7 cells were stained live with GPR30 antibody for 30 min and then fixed and stained with Alexa488-labeled anti-goat antibodies (Life Technologies) as previously described [20, 29, 34]. D, MCF7 cells were transiently transfected with a plasmid containing the cDNA of human GPR30 tagged in the N-terminus with the FLAG tag (FLAG-hGPR30), stained live with mouse M1 FLAG antibodies (Sigma-Aldrich) for 30 min, and then fixed and stained with Alexa488-labeled mouse IgG2b antibodies (Life Technologies) as previously described [29, 34]. In C and D, 4',6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining, and fluorescence images were collected using a Nikon Eclipse confocal microscope. The results are representative of experiments performed at least three times. Bar, 10 μm.
(EPS)
Cumulative incidence of competing event (death from other cause than BC) is shown for comparison. HR values were estimated using a cause-specific Cox proportional hazards model, and values of p were calculated using Wald test. A-B, cumulative incidence of BCD in relation to GPR30TOT in CBC patients with ER-positive BC2 (A) or ER-negative BC2 (B). C-D, cumulative incidence of BCD in relation to GPR30PM in CBC patients with ER-positive BC2 (C) or ER-negative BC2 (D).
(PDF)
Presented HR values were estimated using Cox proportional hazards model and p values were calculated using Wald test, where the groups with weak/weak GPR30TOT and PM-/PM- GPR30PM status were used as reference groups. A, cumulative incidence of BCD in relation to GPR30TOT in the tumor pair. B, cumulative incidence of BCD in relation to GPR30PM in the tumor pair.
(PDF)
Cumulative incidence of competing event (death from other cause than BC) is shown for comparison. HR values were estimated using a cause-specific Cox proportional hazards model, and values of p were calculated using Wald test. A-D, cumulative incidence of BCD in relation to tamoxifen treatment of BC2 in CBC patients with weak GPR30 staining (A) or strong GPR30 staining (B), and in patients without PM-specific GPR30 staining (C) and in patients with PM-specific staining (GPR30PM+) (D).
(PDF)
A, multivariate analyses of strong total GPR30 (GPR30TOT) adjusted for each variable separately. B, multivariate analyses of plasma membrane-specific GPR30 (GPR30PM+) adjusted for each variable separately.
(PDF)
Prognostic effect was calculated by cox proportional hazards model with Wald test. Interaction between GPR30 and the stratifying variable (ER or tamoxifen) was assessed using an interaction test. Relationship between GPR30 and risk of death from BC were assessed by Cox regressions.
(PDF)
Prognostic effect was calculated by cox proportional hazards model with Wald test. The groups with weak/weak GPR30TOT and PM-/PM- GPR30PM status were used as reference groups for survival analyses. Relationship between GPR30 and risk of death from BC were assessed by Cox regressions.
(PDF)
Acknowledgments
We thank Kristina Lövgren (K.L.) for excellent technical assistance and BioCare strategic research school for providing an excellent research environment. Results shown in the Discussion section are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
LMFLL: Swedish Cancer Foundation, CAN 2016/423, https://www.cancerfonden.se/; Swedish Research Council, 2016-02427, https://www.vr.se/ SA: Swedish Breast Cancer Association (BRO); Skåne University Hospital foundation; Percy Falks Stiftelse för Forskning Beträffande Prostata- och Bröstcancer; Skåne County Council's Research and Development Foundation; Swedish Governmental Funding of Clinical Research within the National Health Service The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adami HO, Bergstrom R, Hansen J. Age at first primary as a determinant of the incidence of bilateral breast cancer. Cumulative and relative risks in a population-based case-control study. Cancer. 1985;55:643–7. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Thompson W, Semenciw R, Mao Y. Epidemiology of contralateral breast cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:855–61. [PubMed] [Google Scholar]
- 3.Malone KE, Begg CB, Haile RW, Borg A, Concannon P, Tellhed L, et al. Population-based study of the risk of second primary contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2. J Clin Oncol. 2010;28:2404–10. 10.1200/JCO.2009.24.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–7. [DOI] [PubMed] [Google Scholar]
- 5.Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E, et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer. 2017;75:284–98. 10.1016/j.ejca.2017.01.017 [DOI] [PubMed] [Google Scholar]
- 6.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–58. 10.1677/erc.1.00776 [DOI] [PubMed] [Google Scholar]
- 7.Early Breast Cancer Trialists' Collaborative Group (EBCTCG), Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. 10.1016/S0140-6736(11)60993-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- 9.Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun. 2017;8:1865 10.1038/s41467-017-01864-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol. 2016;34:2961–8. 10.1200/JCO.2016.67.3061 [DOI] [PubMed] [Google Scholar]
- 11.Filardo E. J. Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. J Steroid Biochem Mol Biol. 2002;80:231–8. 10.1016/s0960-0760(01)00190-x [DOI] [PubMed] [Google Scholar]
- 12.Thomas P, Pang Y, Filardo EJ, Dong J. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology. 2005;146:624–32. 10.1210/en.2004-1064 [DOI] [PubMed] [Google Scholar]
- 13.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–30. 10.1126/science.1106943 [DOI] [PubMed] [Google Scholar]
- 14.Martin SG, Lebot MN, Sukkarn B, Ball G, Green AR, Rakha EA, et al. Low expression of G protein-coupled oestrogen receptor 1 (GPER) is associated with adverse survival of breast cancer patients. Oncotarget. 2018;9:25946–56. 10.18632/oncotarget.25408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broselid S, Cheng B, Sjöström M, Lövgren K, Klug-De Santiago HL, Belting M, et al. G protein-coupled estrogen receptor is apoptotic and correlates with increased distant disease-free survival of estrogen receptor-positive breast cancer patients. Clin Cancer Res. 2013;19:1681–92. 10.1158/1078-0432.CCR-12-2376 [DOI] [PubMed] [Google Scholar]
- 16.Filardo EJ, Graeber CT, Quinn JA, Resnick MB, Giri D, DeLellis RA, et al. Distribution of GPR30, a seven membrane-spanning estrogen receptor, in primary breast cancer and its association with clinicopathologic determinants of tumor progression. Clin Cancer Res. 2006;12:6359–66. 10.1158/1078-0432.CCR-06-0860 [DOI] [PubMed] [Google Scholar]
- 17.Sjöström M, Hartman L, Grabau D, Fornander T, Malmström P, Nordenskjöld B, et al. Lack of G protein-coupled estrogen receptor (GPER) in the plasma membrane is associated with excellent long-term prognosis in breast cancer. Breast Cancer Res Treat. 2014;145:61–71. 10.1007/s10549-014-2936-4 [DOI] [PubMed] [Google Scholar]
- 18.Kuo WH, Chang LY, Liu DL, Hwa HL, Lin JJ, Lee PH, et al. The interactions between GPR30 and the major biomarkers in infiltrating ductal carcinoma of the breast in an Asian population. Taiwan J Obstet Gynecol. 2007;46:135–45. 10.1016/S1028-4559(07)60007-2 [DOI] [PubMed] [Google Scholar]
- 19.Ariazi EA, Brailoiu E, Yerrum S, Shupp HA, Slifker MJ, Cunliffe HE, et al. The G protein-coupled receptor GPR30 inhibits proliferation of estrogen receptor-positive breast cancer cells. Cancer Res. 2010;70:1184–94. 10.1158/0008-5472.CAN-09-3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez de Valdivia E, Broselid S, Kahn R, Olde B, Leeb-Lundberg LMF. G protein-coupled estrogen receptor 1 (GPER1)/GPR30 increases ERK1/2 activity through PDZ motif-dependent and -independent mechanisms. J Biol Chem. 2017;292:9932–43. 10.1074/jbc.M116.765875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignatov A, Ignatov T, Weissenborn C, Eggemann H, Bischoff J, Semczuk A, et al. G-protein-coupled estrogen receptor GPR30 and tamoxifen resistance in breast cancer. Breast Cancer Res Treat. 2011;128:457–66. 10.1007/s10549-011-1584-1 [DOI] [PubMed] [Google Scholar]
- 22.Ignatov A, Ignatov T, Roessner A, Costa SD, Kalinski T. Role of GPR30 in the mechanisms of tamoxifen resistance in breast cancer MCF-7 cells. Breast Cancer Res Treat. 2010;123:87–96. 10.1007/s10549-009-0624-6 [DOI] [PubMed] [Google Scholar]
- 23.Mo Z, Liu M, Yang F, Luo H, Li Z, Tu G, et al. GPR30 as an initiator of tamoxifen resistance in hormone-dependent breast cancer. Breast Cancer Res. 2013;15:R114 10.1186/bcr3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatov T, Claus M, Nass N, Haybaeck J, Seifert B, Kalinski T, et al. G-protein-coupled estrogen receptor GPER-1 expression in hormone receptor-positive breast cancer is associated with poor benefit of tamoxifen. Breast Cancer Res Treat. 2019;174:121–7. 10.1007/s10549-018-5064-8 [DOI] [PubMed] [Google Scholar]
- 25.Alkner S, Bendahl PO, Fernö M, Manjer J, Rydén L. Prediction of outcome after diagnosis of metachronous contralateral breast cancer. BMC Cancer. 2011;11:114 10.1186/1471-2407-11-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alkner S, Bendahl PO, Ehinger A, Lövgren K, Rydén L, Fernö M. Prior adjuvant tamoxifen treatment in breast cancer is linked to increased AIB1 and HER2 expression in metachronous contralateral breast cancer. PLoS One. 2016;11:e0150977 10.1371/journal.pone.0150977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkner S, Ehinger A, Bendahl PO, Rydén L, Fernö M. Prognosis, stage and oestrogen receptor status of contralateral breast cancer in relation to characteristics of the first tumour, prior endocrine treatment and radiotherapy. Eur J Cancer 2015;51:2304–13. 10.1016/j.ejca.2015.07.016 [DOI] [PubMed] [Google Scholar]
- 28.Kotarsky K, Owman C, Olde B. A chimeric reporter gene allowing for clone selection and high-throughput screening of reporter cell lines expressing G-protein-coupled receptors. Anal Biochem. 2001;288:209–15. 10.1006/abio.2000.4898 [DOI] [PubMed] [Google Scholar]
- 29.Sandén C, Broselid S, Cornmark L, Andersson K, Daszkiewicz-Nilsson J, Mårtensson UE, et al. G protein-coupled estrogen receptor 1/G protein-coupled receptor 30 localizes in the plasma membrane and traffics intracellularly on cytokeratin intermediate filaments. Mol Pharmacol. 2011;79:400–10. 10.1124/mol.110.069500 [DOI] [PubMed] [Google Scholar]
- 30.R: A language and environment for statistical computing. (2018). R Foundation for Statistical Computing, Vienna, Austria., http://www.R-project.org/.
- 31.Gray B (2009) cmprsk: Subdistribution Analysis of Competing Risks. R package version 2.2–7.
- 32.Therneau, T. M. (2015) _A Package for Survival Analysis in S_. version 2.38.
- 33.Alkner S, Ehinger A, Bendahl PO, Rydén L, Fernö M. Prognosis, stage and oestrogen receptor status of contralateral breast cancer in relation to characteristics of the first tumour, prior endocrine treatment and radiotherapy. Eur J Cancer. 2015;51:2304–13. 10.1016/j.ejca.2015.07.016 [DOI] [PubMed] [Google Scholar]
- 34.Broselid S, Berg KA, Chavera TA, Kahn R, Clarke WP, Olde B, et al. G protein-coupled receptor 30 (GPR30) forms a plasma membrane complex with membrane-associated guanylate kinases (MAGUKs) and protein kinase A-anchoring protein 5 (AKAP5) that constitutively inhibits cAMP production. J Biol Chem. 2014;289:22117–27. 10.1074/jbc.M114.566893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran QK, VerMeer M, Burgard MA, Hassan AB, Giles J. Hetero-oligomeric Complex between the G Protein-coupled Estrogen Receptor 1 and the Plasma Membrane Ca2+-ATPase 4b. J Biol Chem. 2015;290:13293–307. 10.1074/jbc.M114.628743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alkner S, Tang MH, Brueffer C, Dahlgren M, Chen Y, Olsson E, et al. Contralateral breast cancer can represent a metastatic spread of the first primary tumor: determination of clonal relationship between contralateral breast cancers using next-generation whole genome sequencing. Breast Cancer Res. 2015;17:102 10.1186/s13058-015-0608-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ignatov T, Weißenborn C, Poehlmann A, Lemke A, Semczuk A, Roessner A, et al. GPER-1 expression decreases during breast cancer tumorigenesis. Cancer Invest. 2013;31:309–15. 10.3109/07357907.2013.789901 [DOI] [PubMed] [Google Scholar]
- 38.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. www.proteinatlas.org/ENSG00000164850-GPER1/tissue/breast. [DOI] [PubMed] [Google Scholar]
- 39.O'Hayre M, Vázquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13:412–24 10.1038/nrc3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weissenborn C, Ignatov T, Nass N, Kalinski T, Dan Costa S, Zenclussen AC, et al. GPER Promoter Methylation Controls GPER Expression in Breast Cancer Patients. Cancer Invest. 2017;35:100–7. 10.1080/07357907.2016.1271886 [DOI] [PubMed] [Google Scholar]
- 41.Weißenborn C, Ignatov T, Ochel HJ, Costa SD, Zenclussen AC, Ignatova Z, et al. GPER functions as a tumor suppressor in triple-negative breast cancer cells. J Cancer Res Clin Oncol. 2014;140:713–23. 10.1007/s00432-014-1620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–19. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. 10.1016/s0092-8674(03)00934-6 [DOI] [PubMed] [Google Scholar]
- 47.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. 10.1038/nrm908 [DOI] [PubMed] [Google Scholar]
- 48.Filardo EJ, Quinn JA, Bland KI, Frackelton AR. Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol Endocrinol. 2000;14:1649–60. 10.1210/mend.14.10.0532 [DOI] [PubMed] [Google Scholar]
- 49.Maggiolini M, Vivacqua A, Fasanella G, Recchia AG, Sisci D, Pezzi V, et al. The G protein-coupled receptor GPR30 mediates c-fos up-regulation by 17beta-estradiol and phytoestrogens in breast cancer cells. J Biol Chem. 2004;279:27008–16. 10.1074/jbc.M403588200 [DOI] [PubMed] [Google Scholar]
- 50.Ding Q, Gros R, Limbird LE, Chorazyczewski J, Feldman RD. Estradiol-mediated ERK phosphorylation and apoptosis in vascular smooth muscle cells requires GPR30. Am J Physiol Cell Physiol. 2009;297:C1178–87. 10.1152/ajpcell.00185.2009 [DOI] [PubMed] [Google Scholar]
- 51.Pupo M, Bodmer A, Berto M, Maggiolini M, Dietrich PY, Picard D. A genetic polymorphism repurposes the G-protein coupled and membrane-associated estrogen receptor GPER to a transcription factor-like molecule promoting paracrine signaling between stroma and breast carcinoma cells. Oncotarget. 2017;8:46728–44. 10.18632/oncotarget.18156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akama KT, Thompson LI, Milner TA, McEwen BS. Post-synaptic density-95 (PSD-95) binding capacity of G-protein-coupled receptor 30 (GPR30), an estrogen receptor that can be identified in hippocampal dendritic spines. J Biol Chem 2013;288:6438–50. 10.1074/jbc.M112.412478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A, MCF7 cell lysates were immunoblotted with goat GPR30 antibody as previously described [20, 29, 34]. B, HeLa TET-On/Off cells were incubated without (-TET) or with 100 ng/ml tetracycline (+TET) for 12 h and then lysed and immunoblotted with GPR30 antibody. C, MCF7 cells were stained live with GPR30 antibody for 30 min and then fixed and stained with Alexa488-labeled anti-goat antibodies (Life Technologies) as previously described [20, 29, 34]. D, MCF7 cells were transiently transfected with a plasmid containing the cDNA of human GPR30 tagged in the N-terminus with the FLAG tag (FLAG-hGPR30), stained live with mouse M1 FLAG antibodies (Sigma-Aldrich) for 30 min, and then fixed and stained with Alexa488-labeled mouse IgG2b antibodies (Life Technologies) as previously described [29, 34]. In C and D, 4',6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining, and fluorescence images were collected using a Nikon Eclipse confocal microscope. The results are representative of experiments performed at least three times. Bar, 10 μm.
(EPS)
Cumulative incidence of competing event (death from other cause than BC) is shown for comparison. HR values were estimated using a cause-specific Cox proportional hazards model, and values of p were calculated using Wald test. A-B, cumulative incidence of BCD in relation to GPR30TOT in CBC patients with ER-positive BC2 (A) or ER-negative BC2 (B). C-D, cumulative incidence of BCD in relation to GPR30PM in CBC patients with ER-positive BC2 (C) or ER-negative BC2 (D).
(PDF)
Presented HR values were estimated using Cox proportional hazards model and p values were calculated using Wald test, where the groups with weak/weak GPR30TOT and PM-/PM- GPR30PM status were used as reference groups. A, cumulative incidence of BCD in relation to GPR30TOT in the tumor pair. B, cumulative incidence of BCD in relation to GPR30PM in the tumor pair.
(PDF)
Cumulative incidence of competing event (death from other cause than BC) is shown for comparison. HR values were estimated using a cause-specific Cox proportional hazards model, and values of p were calculated using Wald test. A-D, cumulative incidence of BCD in relation to tamoxifen treatment of BC2 in CBC patients with weak GPR30 staining (A) or strong GPR30 staining (B), and in patients without PM-specific GPR30 staining (C) and in patients with PM-specific staining (GPR30PM+) (D).
(PDF)
A, multivariate analyses of strong total GPR30 (GPR30TOT) adjusted for each variable separately. B, multivariate analyses of plasma membrane-specific GPR30 (GPR30PM+) adjusted for each variable separately.
(PDF)
Prognostic effect was calculated by cox proportional hazards model with Wald test. Interaction between GPR30 and the stratifying variable (ER or tamoxifen) was assessed using an interaction test. Relationship between GPR30 and risk of death from BC were assessed by Cox regressions.
(PDF)
Prognostic effect was calculated by cox proportional hazards model with Wald test. The groups with weak/weak GPR30TOT and PM-/PM- GPR30PM status were used as reference groups for survival analyses. Relationship between GPR30 and risk of death from BC were assessed by Cox regressions.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.