Abstract
Loss of podocyte differentiation can cause nephrotic-range proteinuria and Focal and Segmental Glomerulosclerosis (FSGS). As specific therapy is still lacking, FSGS frequently progresses to end-stage renal disease. The exact molecular mechanisms of FSGS and gene expression changes in podocytes are complex and widely unknown as marker changes have mostly been assessed on the glomerular level. To gain a better insight, we isolated podocytes of miR-193a overexpressing mice, which suffer from FSGS due to suppression of the podocyte master regulator Wt1. We characterised the podocytic gene expression changes by RNAseq and identified many novel candidate genes not linked to FSGS so far. This included strong upregulation of the receptor tyrosine kinase EphA6 and a massive dysregulation of circadian genes including the loss of the transcriptional activator Arntl. By comparison with podocyte-specific changes in other FSGS models we found a shared dysregulation of genes associated with the Wnt signaling cascade, while classical podocyte-specific genes appeared widely unaltered. An overlap with gene expression screens from human FSGS patients revealed a strong enrichment in genes associated with extra-cellular matrix (ECM) and metabolism. Our data suggest that FSGS progression might frequently depend on pathways that are often overlooked when considering podocyte homeostasis.
Introduction
Focal and Segmental Glomerulosclerosis (FSGS) is a clinico-pathological syndrome referring to sclerotic lesions in some areas of some kidney glomeruli and nephrotic range proteinuria [1,2]. While corticosteroids or calcineurin inhibitors can ameliorate the disease, FSGS is frequently resistant to treatment and may progress to end stage renal disease (ESRD) [1,3]. The cell type mainly affected and causing FSGS is the podocyte (visceral glomerular epithelial cell). In healthy individuals, interdigitating podocyte foot processes together with the basal lamina and underlying, fenestrated endothelial cells form a sieve-like structure thereby enabling the filtration of blood. A high number of specific genes is necessary to maintain the complex three-dimensional structure of podocytes. The impacts initiating FSGS are heterogeneous and include gene mutations, circulating factors, drugs, viruses and hypertension [1,2]. These very different etiologies lead to a common final path characterised by foot process effacement and loss of podocytes concomitant with nephrotic syndrome, fibrosis and scarring.
Many FSGS-causing mutations have been identified so far [4], but for the majority of patients other causes seem to be responsible and the exact molecular changes are still widely elusive. In order to develop novel therapeutic approaches suitable for a high amount of patients, a better characterisation of the common molecular events in the final path seems to be of high relevance. Several studies addressed gene expression changes in FSGS patients giving insight on the glomerular level, which unfortunately does not allow for a detailed understanding of podocyte-specific events [5–7].
To overcome this limitation, Tobias Huber and coworkers established a protocol to isolate GFP-marked mouse podocytes by FACS sorting and determined the set of expressed genes in healthy podocytes [8]. A similar system was also used by the Potter group to determine specific changes in podocytes, mesangial cells and endothelial cells in Cd2ap-/—and Cd2ap+/-;Fyn-/—driven FSGS [9,10].
In another approach, by translating ribosome affinity purification (TRAP) the group of Benjamin Humphreys has managed to characterize gene expression changes in podocytes upon Actn4 KO [11]. Unexpectedly, in none of these FSGS models, classical podocyte marker genes (e.g. Wt1, Nphs1, Nphs2, Podxl) significantly changed. Therefore, any comparison of gene expression changes during FSGS on the glomerular level runs at high risk of only assessing the changes based on podocyte loss but not the changes within podocytes. These data furthermore raised the question if dysregulation of classical podocyte marker genes in FSGS was generally not very pronounced or just in the models analysed.
Thus, we addressed these issues in the miR-193a FSGS model [12] and tried to identify novel biomarkers and therapeutic targets for FSGS.
Materials and methods
Animal experiments
miR-193a and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (miR-193a) x hNPHS2Cre (podGFP) mice and their handling were described before [8,12]. Genotyping primers were CGATCAGGATGATCTGGACG and CAAGCTCTTCAGCAATATCAC (for miR-193a) and CTCTGCTGCCTCCTGGCTTCT and TCAATGGGCGGGGGTCGT (for Tomato). The two lineages were crossed to obtain miR-193a-overexpressing GFP-positive podocytes (miR-193a x GFP), while their podGFP littermates without transgenic miR-193a construct served as controls (wt x podGFP). miR-193a was induced by 1 mg/ml doxycycline in 5% sucrose, podGFP mice fed with doxycycline solution served as a control. 15 weeks old females were used for this study. Anaesthesia was performed with 100mg/kg Ketamine and 5mg/kg Xylazine i.p. Mice were sacrificed by cervical dislocation. All animal experiments and handling were in accordance with the Austrian law for protection of animals and approved by the animal ethics committee of the Austrian ministry for science and research (66.009/0053-II/3b/2014). AFOG analysis was performed according to a standard protocol. Urinary albumin levels were assessed by ELISA (E90-134, Bethyl Laboratories, Montgomery, TX, USA). Creatinine levels were measured with the Creatinine Assay Kit (STA-378, Cell Biolabs, San Diego, CA, USA).
Human samples
Human data were obtained from the fusion of dysregulated genes found in two independent published studies [5,6]. In short, in the first study [5], all 4 patients suffered from idiopathic FSGS and were female. Urinary protein levels were 4.0, 5.4, 14.7, and 17.0 (g/day) and serum creatinine levels were 0.8, 1.1, 0.9, and 5.3 (mg/dl), respectively. Controls were obtained from ‘normal’ regions of kidneys removed from Wilms’ tumor patients. In the second study [6], FFPE renal biopsy material from 19 patients with idiopathic nephrotic syndrome (edema, proteinuria >3.5 g/day; serum albumin <3.0 g/dl) and 2 without full nephrotic syndrome was used. Controls were renal biopsies that appeared normal by histological and electron microscopic examination obtained from renal biopsies performed for minimal isolated proteinuria or hematuria (seven patients) or tissue from uninvolved portions of a kidney at the time of tumor nephrectomies.
Cell isolation, RNA isolation and qPCR
Podocytes were isolated as described before from 4 animals/group [8]. RNA was isolated with the ReliaPrep RNA kit from Promega according to manufacturer’s instructions. qPCRs for Arntl, Bhlhe41, Cdh11, Dbp, EphA6, Ggt, Mt2, Per2, Per3, Prss23, S100a6, Spns2 and Cyclophilin B (for normalisation) were performed on a CFX96 Real Time System with a C1000 Thermal Cycler (Bio-Rad) using KAPA SYBR FAST from Sigma Aldrich. Primer sequences were from PrimerBank and were TGACCCTCATGGAAGGTTAGAA and GGACATTGCATTGCATGTTGG (Arntl, forward and reverse), TGTGTAAACCCAAAAGGAGCT and TGTTCGGGCAGTAAATCTTTCAG (Bhlhe41), CTGGGTCTGGAACCAATTCTTT and GCCTGAGCCATCAGTGTGTA (Cdh11), GGAAACAGCAAGCCCAAAGAA and CAGCGGCGCAAAAAGACTC (Dbp), TGCGAAGTCCGGGAATTTCTT and GCAACACAACTTGGTTGGAGAC (EphA6), TTCAATGGGACAGAAACCTTGAG and TCCCTGTGTATAAGACCTCCG (Ggt5), GCCTGCAAATGCAAACAATGC and AGCTGCACTTGTCGGAAGC (Mt2), GAAAGCTGTCACCACCATAGAA and AACTCGCACTTCCTTTTCAGG (Per2), AACACGAAGACCGAAACAGAAT and CTCGGCTGGGAAATACTTTTTCA (Per3), GGTGAGTCCCTACACCGTTC and GGCGTCGAAGTCTGCCTTAG (Prss23), ATTGGCTCCAAGCTGCAGG and TCATTGTAGATCAAAGCCAAGG (S100a6), CCATCCTGAGTTTAGGCAACG and GATCACCTTTCTATTGAAGCGGT (Spns2).
RNAseq
RNAseq was performed by the NGS unit of the Vienna Biocenter Core Facilities GmbH (VBCF) (www.viennabiocenter.org/facilities). For RNA preparation we used NEBNext Ultra 2 kit (New England Biolabs; https://international.neb.com/products/e7645-nebnext-ultra-ii-dna-library-prep-kit-for-illumina). The samples were paired end sequenced with a read length of 50 base pairs on an Illumina HiSeq2500 (V4 chemistry) according to the manufacturer’s protocol (RTA 1.18.66.3). RNA-Seq reads were mapped to the reference genome using TopHat [13] and the differential gene expression analysis was calculated by CuffDiff v2.2.1 using blind dispersion method [14].
Statistics
Due to low total RNA yield in podocytes, RNA of 4 mice was pooled for RNAseq making it impossible to calculate p-values of the results for independent mice. For qPCR, results of one of two independent experiments with three wild-type and three miR-193a FSGS mice are shown. Statistical significance was calculated by two-tailed Student’s t-test.
Results
miR-193a suppresses Wt1, a master regulator of podocyte function, thereby causing FSGS [12]. We crossed miR-193a mice with Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J x hNPHS2Cre mice to obtain GFP-tagged podocytes with inducible miR-193a, while GFP-tagged podocytes from littermates without miR-193a construct served as control [8,12]. Irreversible podocyte loss and FSGS were initiated by doxycycline-driven overexpression of miR-193a for 7 weeks, followed by 10 days without doxycycline (Fig 1). This allowed us to focus on transcript changes directly related to FSGS and neglect changes only related to increased miR-193a. Podocytes from FSGS and control mice were isolated by FACS sorting according to an established protocol [8] and submitted to RNAseq analysis.
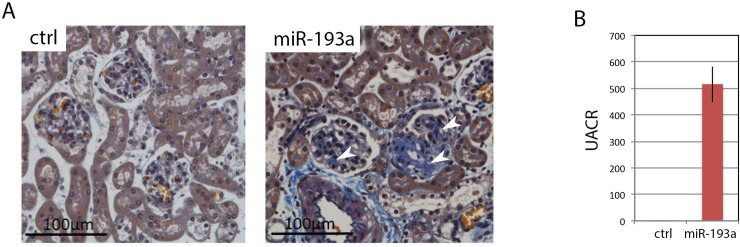
Fig 1. miR-193a-driven FSGS.
A) Histology of miR-193a and control mice 8.5 weeks post FSGS induction. Acid Fuchsin Orange G staining, 400x magnification, scale bars represent 100μm. B) Corresponding UACR of miR-193a and control mice. UACR, urinary albumin:creatinin ratio.
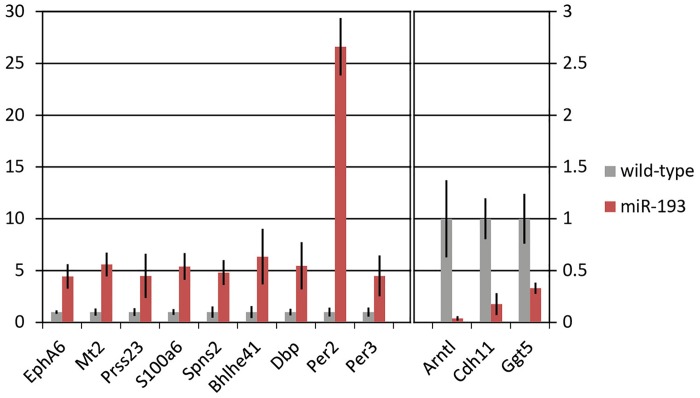
S1 Table depicts the expression changes upon miR-193a-induced FSGS. The strongest upregulation for a protein-coding gene was found for the trans-membrane receptor tyrosine kinase Ephrin receptor A6 (EphA6), which integrates extra-cellular signals and has never been linked to FSGS before. Ephrin receptors can bind to Nck and Src, which both have been implicated in FSGS [15]. Among the strongly upregulated genes were also the metallothioneins Mt1 and Mt2, which can interfere with FSGS and diabetic nephropathy [16,17]. Their upregulation might represent a cell-autonomous mechanism to attenuate FSGS progression. Obscurin (Obscn; Arhgef30; Cytoskeletal Calmodulin And Titin-Interacting RhoGEF), also upregulated and never associated with FSGS, is a modulator of the cytoskeleton and activator of RhoA [18,19] and might therefore be able to induce FSGS [20]. We also found strongly increased levels of Serine Protease 23 (Prss23) and Sphingolipid Transporter 2 (Spns2), which has been associated with kidney fibrosis before [21]. Strikingly, the transcripts of several circadian genes, including Basic Helix-Loop-Helix Family Member E41 (Bhlhe41, Dec2), the Period Circadian Regulators 2 and 3 (Per2, Per3) and D Site Of Albumin Promoter Binding Protein (Dbp) were massively enhanced. In line with this, the protein-coding gene with the strongest downregulation was Aryl Hydrocarbon Receptor Nuclear Translocator Like (Arntl, Bmal, Bhlhe5), which is a circadian master regulator repressed by Per2. Arntl is associated with susceptibility to hypertension and diabetes and can activate the TNF receptor Osteoprotegerin [22–24]. Another strongly downregulated gene was Cadherin 11 (Cdh11), a calcium-dependent mediator of cell adhesion and cytoskeleton and inhibitor of Wnt and Rho activation [25].
Table 1 depicts the 20 strongest up- and downregulated genes identified in the miR-193a model according to RNAseq. We confirmed several of the dysregulated genes by qPCR (Fig 2).
Table 1. The 20 strongest up- and downregulated genes in podocytes during miR193a-driven FSGS compared to wild-type as assessed by RNAseq.
FC, fold change.
| Symbol | FC (log2) | Name |
|---|---|---|
| Epha6 | 8.4 | EPH Receptor A6 |
| Bhlhe41 | 5.8 | Basic Helix-Loop-Helix Family Member E41 |
| Adrb2 | 5.4 | Adrenoceptor Beta 2 |
| Mt2 | 4.7 | Metallothionein 2 |
| Dbp | 4.7 | D-Box Binding PAR BZIP Transcription Factor |
| Ttll7 | 4.5 | Tubulin Tyrosine Ligase Like 7 |
| Abcc3 | 4.5 | ATP Binding Cassette Subfamily C Member 3 |
| Osbpl6 | 4.4 | Oxysterol Binding Protein Like 6 |
| Pnpla7 | 4.1 | Patatin Like Phospholipase Domain Containing 7 |
| A2bp1 | 4.0 | RNA Binding Fox-1 Homolog 1 |
| Pcdh1 | 4.0 | Protocadherin 1 |
| Exdl1 | 3.9 | Exonuclease 3'-5' Domain Containing 1 |
| Obscn | 3.9 | Obscurin, Cytoskeletal Calmodulin And Titin-Interacting RhoGEF |
| Gdnf | 3.8 | Glial Cell Derived Neurotrophic Facto |
| Gng10 | 3.8 | G Protein Subunit Gamma 10 |
| Zbtb16 | 3.8 | Zinc Finger And BTB Domain Containing 16 |
| Pik3ip1 | 3.8 | Phosphoinositide-3-Kinase Interacting Protein 1 |
| Rcan2 | 3.7 | Regulator Of Calcineurin 2 |
| Garnl3 | 3.7 | GTPase Activating Rap/RanGAP Domain Like 3 |
| Per2 | 3.7 | Period Circadian Regulator 2 |
| Arntl | -5.6 | Aryl Hydrocarbon Receptor Nuclear Translocator Like |
| Rtf1 | -5.2 | RTF1 Homolog, Paf1/RNA Polymerase II Complex Component |
| Adra2c | -4.9 | Adrenoceptor Alpha 2C |
| Lmbr1 | -4.0 | Limb Development Membrane Protein 1 |
| Nlrc5 | -4.0 | NLR Family CARD Domain Containing 5 |
| Kcnh3 | -3.9 | Potassium Voltage-Gated Channel Subfamily H Member 3 |
| Ccdc88c | -3.8 | Coiled-Coil Domain Containing 88C |
| Rai2 | -3.7 | Retinoic Acid Induced 2 |
| Amy1 | -3.5 | Amylase Alpha 1A |
| Sp6 | -3.4 | Sp6 Transcription Factor |
| Pim1 | -3.4 | Pim-1 Proto-Oncogene, Serine/Threonine Kinase |
| Ccnjl | -3.4 | Cyclin J Like |
| Spon1 | -3.3 | Spondin 1 |
| Dll1 | -3.2 | Delta Like Canonical Notch Ligand 1 |
| Hoxd9 | -3.2 | Homeobox D9 |
| Cdc7 | -3.1 | Cell Division Cycle 7 |
| Bcl2l11 | -3.0 | Bcl-2-Like Protein 11 |
| Rims1 | -3.0 | Regulating Synaptic Membrane Exocytosis 1 |
| Obsl1 | -2.9 | Obscurin Like Cytoskeletal Adaptor 1 |
| Unc5b | -2.9 | Unc-5 Netrin Receptor B |
Fig 2. Dysregulated genes in podocytes of miR-193a FSGS mice.
Expression changes of selected genes upon miR-193a-driven FSGS in isolated podocytes were assessed by qPCR and normalised to CycB. Results are representative medium values (mean±SEM) of three independent mice of one of two independent experiments. Control was set to 1. Arntl, Aryl Hydrocarbon Receptor Nuclear Translocator Like; Bhlhe41, Basic Helix-Loop-Helix Family Member E41; Cdh11, Cadherin 11; CycB, cyclophilin B; Dbp, D Site Of Albumin Promoter Binding Protein; EphA6, EPH Receptor A6; Ggt5, Gamma-Glutamyltransferase 5; Mt2, Metallothionein 2; Per2, Period Circadian Regulator 2; Per3, Period Circadian Regulator 3; Prss23, Serine Protease 23; S100a6, S100 Calcium Binding Protein A6; Spns2, Sphingolipid Transporter 2; *p < 0.025; **p < 0.005.
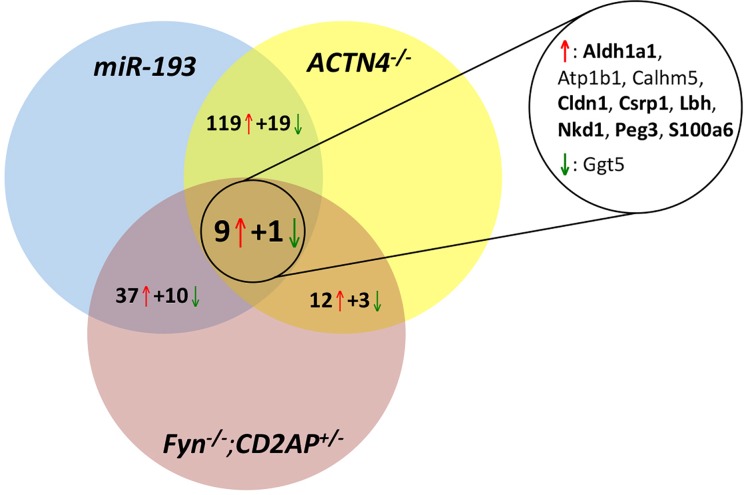
In order to determine which of the detected changes were not only specific for our system, but represented more general changes during FSGS we compared them with data from Actn4-/—and Cd2ap+/-;Fyn-/—driven FSGS [10, 11]. Nine genes were up- and one gene was downregulated in all 3 models (Fig 3). Strikingly, seven (Aldh1a1, Cldn1, Csrp1, Lbh, Nkd1, Peg3, S100a6) of the nine genes upregulated in all three models were associated with the Wnt signaling cascade [26–34] (Fig 3). We found an especially strong overlap between the miR-193a and the Actn4-/- model, suggesting that several of the genes identified by us and Grgic et al. [11] might be of general relevance for FSGS. In line with this, 16 of the top 20 upregulated genes in the Actn4 KO were also strongly increased in our screen (S2 Table) [11].
Fig 3. Dysregulated genes in podocytes in different FSGS models.
An overlap of three independent mRNA expression profiles (miR-193, Actn4-/-, Fyn-/-;Cd2ap+/-,) during FSGS reveals nine commonly upregulated genes (red arrow) and one downregulated gene (green arrow). Seven genes (bold font) are associated with the Wnt signaling pathway. Aldh1a1, Aldehyde Dehydrogenase 1 Family Member A1; Atp1b1, ATPase Na+/K+ Transporting Subunit Beta 1; Calhm5, Calcium Homeostasis Modulator Family Member 5; Cldn1, Claudin 1; Csrp1, Cysteine And Glycine Rich Protein 1; Lbh, LBH Regulator Of Wnt Signaling Pathway; Nkd1, Nkd Inhibitor Of Wnt Signaling Pathway 1; Peg3, Paternally Expressed 3; S100a6, S100 Calcium Binding Protein A6; Ggt5, Gamma-Glutamyltransferase 5.
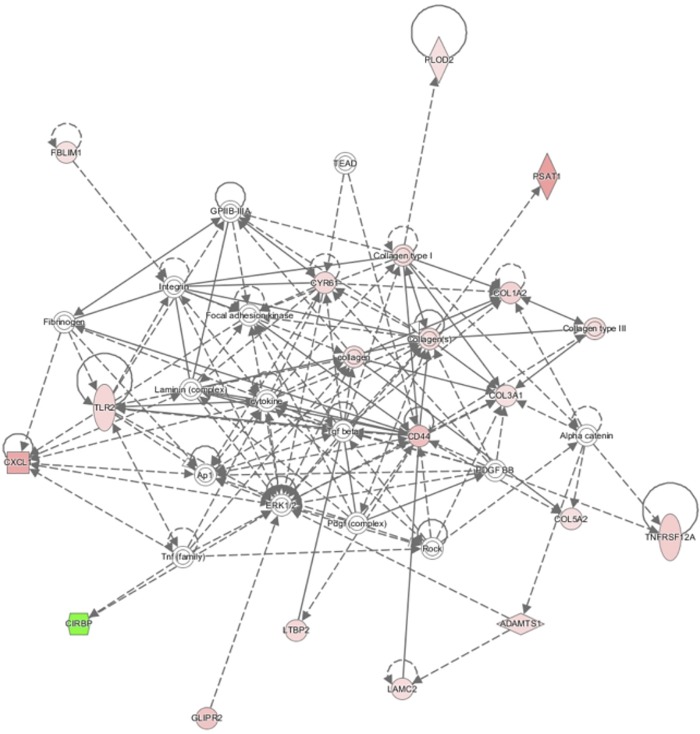
To predict which podocyte-specific changes might occur in human FSGS, we overlapped the at least two-fold dysregulated genes of our screen with the set of at least two-fold dysregulated genes in glomeruli of patients suffering from idiopathic FSGS, as podocytes cannot be isolated from human in bigger quantities [5,6]. While the comparison of genes dysregulated in isolated mouse FSGS podocytes and human FSGS glomeruli bears the risk of missing some changes due to loss of podocytes from the glomeruli during FSGS, we believe that we are still able to define several bona fide podocytic human FSGS genes, especially amongst the upregulated genes. The identified genes included several already associated with FSGS (e.g. Col1a1, Col1a2, Cd24, Cd44, Plau, Umod) and many novel genes not associated with FSGS so far (S3 Table). To narrow down this list even further we overlapped the genes dysregulated in human disease with both the at least two-fold dysregulated genes in the miR-193a model and in the Actn4 KO. This led to the definition of a 35-gene-set which might constitute a core set of podocyte-specific FSGS genes (Table 2). Strikingly, this set contains 17 genes related to ECM modifications, 8 genes closely related to metabolism, and 4 genes related to transmembrane transport. This enrichment in ECM-related genes differs strongly from the most abundant processes in healthy podocytes, namely cytoskeleton regulation and protein transport [8]. Analysis of GO term enrichment also showed collagen fibril organisation/ECM organisation as the top enriched process (Fig 4).
Table 2. An overlap of glomerular changes in human FSGS, and podocytic changes in Actn4-KO- and miR-193a-induced FSGS identifies a core set of 35 commonly dysregulated genes.
| Symbol | Name | context | change |
|---|---|---|---|
| ADAMTS1 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 1 | ECM | up |
| BACE2 | Beta-Secretase 2 | ECM | up |
| CD44 | CD44 Molecule | ECM, cell-cell interactions | up |
| COL1A2 | Collagen Type I Alpha 2 Chain | ECM | up |
| COL3A1 | Collagen Type III Alpha 1 Chain | ECM | up |
| COL5A2 | Collagen Type V Alpha 2 Chain | ECM | up |
| CXCL1 | C-X-C Motif Chemokine Ligand 1 | ECM | up |
| CYR61 | Cellular Communication Network Factor 1 | ECM | up |
| FN1 | Fibronectin 1 | ECM | up |
| GLIPR2 | GLI Pathogenesis Related 2 | ECM | up |
| LAMC2 | Laminin Subunit Gamma 2 | ECM | up |
| LTBP2 | Latent TGF Beta Binding Protein 2 | ECM | up |
| PLOD2 | Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 2 | ECM | up |
| PRSS23 | Serine Protease 23 | ECM | up |
| SLIT3 | Slit Guidance Ligand 3 | ECM | up |
| THBD | Thrombomodulin | ECM | up |
| TNFRSF12A | TNF Receptor Superfamily Member 12A | ECM, inflammation | up |
| ALDH18A1 | Aldehyde Dehydrogenase 18 Family Member A1 | metabolism | up |
| ALDH1A1 | Aldehyde Dehydrogenase 1 Family Member A1 | metabolism | up |
| ASRGL1 | Asparaginase And Isoaspartyl Peptidase 1 | metabolism | up |
| ELOVL7 | ELOVL Fatty Acid Elongase 7 | metabolism | up |
| GUCY1A3 | Guanylate Cyclase 1 Soluble Subunit Alpha 1 | metabolism | up |
| PPP1R3C | Protein Phosphatase 1 Regulatory Subunit 3C | metabolism | up |
| PSAT1 | Phosphoserine Aminotransferase 1 | metabolism | up |
| PTPLAD2 | 3-Hydroxyacyl-CoA Dehydratase 4 | metabolism | up |
| ABCC4 | ATP Binding Cassette Subfamily C Member 4 | transport | up |
| ATP1B1 | ATPase Na+/K+ Transporting Subunit Beta 1 | transport | up |
| SLC1A4 | Solute Carrier Family 1 Member 4 | transport | up |
| SLCO2B1 | Solute Carrier Organic Anion Transporter Family Member 2B1 | transport | up |
| CLDN1 | Claudin 1 | cell contact, slit diaphragm | up |
| FBLIM1 | Filamin Binding LIM Protein 1 | cytoskeleton | up |
| TLR2 | Toll Like Receptor 2 | inflammation | up |
| MAFF | MAF BZIP Transcription Factor F | transcription | up |
| CIRBP | Cold Inducible RNA Binding Protein | survival | down |
| NRIP2 | Nuclear Receptor Interacting Protein 2 | signalling | down |
Fig 4. A core set of FSGS genes.
The network of 35 genes as core set of FSGS identified by Ingenuity Pathway Analysis (Qiagen).
Discussion
In this study we tried to analyse transcriptional changes in podocytes during FSGS in the miR-193a mouse model and identified many genes not associated with FSGS so far. While the RNAseq analysis was performed in pooled samples from four mice per group due to the low yield of podocyte RNA per mouse, we were able to confirm the observed dysregulation for a considerable number of selected genes by qPCR in individual mouse samples (Fig 2) and our data have a good overlap with the data by Grgic et al. [11; S2 Table], which makes us confident that our findings are of relevance.
We observed a massive dysregulation of circadian genes in the miR-193a FSGS model. It is known that circadian genes are crucially involved in the regulation of inflammation, immune response and kidney physiology and disease including glomerular diseases [35,36]. While melatonin has been shown to ameliorate chronic kidney disease via antioxidant effects and modulation of the renin-angiotensin system [37], the detailed interactions between the circadian clock and glomerular diseases are not elucidated and will be a highly interesting field for follow-up studies.
A comparison of our results with published data in other model systems revealed a dysregulation of genes associated with the Wnt cascade as common feature in FSGS. Claudin 1 (Cldn1) and S100 Calcium Binding Protein A6 (S100a6) are β-Catenin targets that can reciprocally activate β-Catenin [26–29]. Enhanced expression of Claudin 1 can induce proteinuria through slit diaphragm destabilisation [38]. Aldehyde Dehydrogenase 1 Family Member A1 (Aldh1a1) and Limb Bud And Heart Development (Lbh) have been shown to be upregulated by β -Catenin and control cell differentiation [30,31]. Aldh1a1 is able to regulate the differentiation and repair process of podocytes during injury via retinoic acid production [39]. Cysteine and glycine-rich protein 1 (Csrp1) and Naked Cuticle Homolog 1 (Nkd1) modulate Wnt signaling and cytoskeletal rearrangements by interaction with Dishevelled [32,33]. The imprinted gene Paternally Expressed 3 (Peg3) is an inhibitor of Wnt signaling [34]. Activation of the Wnt/sβ -Catenin cascade in FSGS patients has also been observed by others [40,41] suggesting that targeting Wnt signalling might be a therapeutic option in FSGS [42].
By overlapping podocyte-specific changes in FSGS models with glomerular data from human patients we found a set of 35 commonly dysregulated genes. Strikingly, this set exhibited a strong enrichment in ECM-associated genes. Of note, it is not the mere loss of functional, differentiated podocytes, but progressive glomerular sclerosis that eventually leads to ESRD. In line with this, several mutations in ECM-associated, FSGS-causing genes have been described (Collagen 4, Laminin B2, the integrins α-4 and β-3, ApoL1, and Cd151) and the circulating factor suPAR is suggested to promote FSGS by modulation of β-3 integrin signaling [4,43]. Therefore, this shared gene set with almost 50% of ECM-associated genes might be of interest for the development of novel therapeutic concepts.
The second big group consisted of 8 genes related to an altered metabolism and might be a consequence of the podocytic attempts to morphologically adjust to loss of podocyte differentiation and number. Stress-related podocyte growth and hypertrophy is a well-known observation during FSGS also receiving a lot of attention recently [44,45].
Of note, only two of the 35 genes were downregulated, while 33 were upregulated. It is a general observation in FSGS profilings, that the majority of genes is upregulated [5,6,10,11], possibly due to gene reactivation during dedifferentiation.
Furthermore, in all three different FSGS models (miR-193a, Actn4 KO, Cd2ap+/-;Fyn-/-) classical podocyte marker genes did not appear strongly changed, with the exception of respective model-specific, disease-causing alterations. This suggests that our current understanding of podocyte biology during FSGS might deserve reconsideration. While these findings might describe the general changes during FSGS relevant for disease progression, we cannot exclude that additional podocyte populations with distinct gene expression signatures exist as FSGS is focal and segmental and injured podocytes are frequently lost before they can be isolated [46]. A thorough analysis of the identified genes and their potential as therapeutic targets should be the focus of follow-up studies.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Tobias Huber, Tillmann Bork and Charlotte Meyer for the podGFP mice and technical support in the podocyte isolation technique.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors have no financial disclosures.
References
- 1.D’Agati VD, Kaskel FJ, Falk RJ. Focal segmental glomerulosclerosis. N Engl J Med. 2011. December 22;365(25):2398–411. 10.1056/NEJMra1106556 [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg AZ, Kopp JB. Focal Segmental Glomerulosclerosis. Clin J Am Soc Nephrol CJASN. 2017. March 7;12(3):502–17. 10.2215/CJN.05960616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurin L-P, Nachman PH, Foster BJ. Calcineurin Inhibitors in the Treatment of Primary Focal Segmental Glomerulosclerosis: A Systematic Review and Meta-analysis of the Literature. Can J Kidney Health Dis. 2017;4:2054358117692559 10.1177/2054358117692559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovric S, Ashraf S, Tan W, Hildebrandt F. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant Off Publ Eur Dial Transpl Assoc—Eur Ren Assoc. 2016. November;31(11):1802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett MR, Czech KA, Arend LJ, Witte DP, Devarajan P, Potter SS. Laser capture microdissection-microarray analysis of focal segmental glomerulosclerosis glomeruli. Nephron Exp Nephrol. 2007;107(1):e30–40. 10.1159/000106775 [DOI] [PubMed] [Google Scholar]
- 6.Hodgin JB, Borczuk AC, Nasr SH, Markowitz GS, Nair V, Martini S, et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am J Pathol. 2010. October;177(4):1674–86. 10.2353/ajpath.2010.090746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong J, Xie J, Ren H, Liu J, Zhang W, Wei C, et al. Comparison of Glomerular Transcriptome Profiles of Adult-Onset Steroid Sensitive Focal Segmental Glomerulosclerosis and Minimal Change Disease. PloS One. 2015;10(11):e0140453 10.1371/journal.pone.0140453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, et al. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013. June;83(6):1052–64. 10.1038/ki.2012.487 [DOI] [PubMed] [Google Scholar]
- 9.Brunskill EW, Potter SS. Pathogenic pathways are activated in each major cell type of the glomerulus in the Cd2ap mutant mouse model of focal segmental glomerulosclerosis. BMC Nephrol. 2015. May 13;16:71 10.1186/s12882-015-0063-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter AS, Drake K, Brunskill EW, Potter SS. A bigenic mouse model of FSGS reveals perturbed pathways in podocytes, mesangial cells and endothelial cells. PloS One. 2019;14(8):e0216261 10.1371/journal.pone.0216261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grgic I, Hofmeister AF, Genovese G, Bernhardy AJ, Sun H, Maarouf OH, et al. Discovery of new glomerular disease-relevant genes by translational profiling of podocytes in vivo. Kidney Int. 2014. December;86(6):1116–29. 10.1038/ki.2014.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebeshuber CA, Kornauth C, Dong L, Sierig R, Seibler J, Reiss M, et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med. 2013. April;19(4):481–7. 10.1038/nm.3142 [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009. May 1;25(9):1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013. January;31(1):46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu T, Shi G, Larose L, Rivera GM, Mayer BJ, Zhou R. Regulation of process retraction and cell migration by EphA3 is mediated by the adaptor protein Nck1. Biochemistry. 2009. July 14;48(27):6369–78. 10.1021/bi900831k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Zheng S, Epstein PN. Metallothionein over-expression in podocytes reduces adriamycin nephrotoxicity. Free Radic Res. 2009. February;43(2):174–82. 10.1080/10715760802657308 [DOI] [PubMed] [Google Scholar]
- 17.Zheng S, Carlson EC, Yang L, Kralik PM, Huang Y, Epstein PN. Podocyte-specific overexpression of the antioxidant metallothionein reduces diabetic nephropathy. J Am Soc Nephrol JASN. 2008. November;19(11):2077–85. 10.1681/ASN.2007080967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009. September;20(17):3905–17. 10.1091/mbc.E08-10-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry NA, Vitolo MI, Martin SS, Kontrogianni-Konstantopoulos A. Loss of the obscurin-RhoGEF downregulates RhoA signaling and increases microtentacle formation and attachment of breast epithelial cells. Oncotarget. 2014. September 30;5(18):8558–68. 10.18632/oncotarget.2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L, Jiang R, Aoudjit L, Jones N, Takano T. Activation of RhoA in podocytes induces focal segmental glomerulosclerosis. J Am Soc Nephrol JASN. 2011. September;22(9):1621–30. 10.1681/ASN.2010111146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard O, Stepanovska B, Starck M, Erhardt M, Römer I, Meyer Zu Heringdorf D, et al. Downregulation of the S1P Transporter Spinster Homology Protein 2 (Spns2) Exerts an Anti-Fibrotic and Anti-Inflammatory Effect in Human Renal Proximal Tubular Epithelial Cells. Int J Mol Sci. 2018. May 17;19(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappa KI, Gazouli M, Anastasiou E, Iliodromiti Z, Antsaklis A, Anagnou NP. The major circadian pacemaker ARNT-like protein-1 (BMAL1) is associated with susceptibility to gestational diabetes mellitus. Diabetes Res Clin Pract. 2013. February;99(2):151–7. 10.1016/j.diabres.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 23.Richards J, Diaz AN, Gumz ML. Clock genes in hypertension: novel insights from rodent models. Blood Press Monit. 2014. October;19(5):249–54. 10.1097/MBP.0000000000000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Yu R, Long Y, Zhao J, Yu S, Tang Q, et al. BMAL1 deficiency promotes skeletal mandibular hypoplasia via OPG downregulation. Cell Prolif. 2018. October;51(5):e12470 10.1111/cpr.12470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Ying J, Li H, Zhang Y, Shu X, Fan Y, et al. The human cadherin 11 is a pro-apoptotic tumor suppressor modulating cell stemness through Wnt/β-catenin signaling and silenced in common carcinomas. Oncogene. 2012. August 23;31(34):3901–12. 10.1038/onc.2011.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhawan P, Singh AB, Deane NG, No Y, Shiou S-R, Schmidt C, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005. July;115(7):1765–76. 10.1172/JCI24543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011. December;141(6):2140–53. 10.1053/j.gastro.2011.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilańczyk E, Graczyk A, Ostrowska H, Kasacka I, Leśniak W, Filipek A. S100A6 is transcriptionally regulated by β-catenin and interacts with a novel target, lamin A/C, in colorectal cancer cells. Cell Calcium. 2012. June;51(6):470–7. 10.1016/j.ceca.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Liu X, Lang H, Zhang S, Luo Y, Zhang J. S100 calcium-binding protein A6 promotes epithelial-mesenchymal transition through β-catenin in pancreatic cancer cell line. PloS One. 2015;10(3):e0121319 10.1371/journal.pone.0121319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, et al. β-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015. April 30;34(18):2297–308. 10.1038/onc.2014.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010. September;30(17):4267–79. 10.1128/MCB.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyasaka KY, Kida YS, Sato T, Minami M, Ogura T. Csrp1 regulates dynamic cell movements of the mesendoderm and cardiac mesoderm through interactions with Dishevelled and Diversin. Proc Natl Acad Sci U S A. 2007. July 3;104(27):11274–9. 10.1073/pnas.0702000104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C, et al. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci U S A. 2001. March 27;98(7):3802–7. 10.1073/pnas.071041898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Yu Y, Yang HW, Agar NYR, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem. 2010. March 12;285(11):8472–80. 10.1074/jbc.M109.069450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hergenhan S, Holtkamp S, Scheiermann C. Molecular Interactions Between Components of the Circadian Clock and the Immune System. J Mol Biol. 2020. January 10; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firsov D, Bonny O. Circadian rhythms and the kidney. Nat Rev Nephrol. 2018;14(10):626–35. 10.1038/s41581-018-0048-9 [DOI] [PubMed] [Google Scholar]
- 37.Ohashi N, Ishigaki S, Isobe S. The pivotal role of melatonin in ameliorating chronic kidney disease by suppression of the renin-angiotensin system in the kidney. Hypertens Res Off J Jpn Soc Hypertens. 2019. June;42(6):761–8. [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Sunq A, Roth RA, Hou J. Inducible Expression of Claudin-1 in Glomerular Podocytes Generates Aberrant Tight Junctions and Proteinuria through Slit Diaphragm Destabilization. J Am Soc Nephrol JASN. 2017. January;28(1):106–17. 10.1681/ASN.2015121324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki A, Ito T, Imai E, Yamato M, Iwatani H, Kawachi H, et al. Retinoids regulate the repairing process of the podocytes in puromycin aminonucleoside-induced nephrotic rats. J Am Soc Nephrol JASN. 2003. April;14(4):981–91. 10.1097/01.asn.0000057857.66268.8f [DOI] [PubMed] [Google Scholar]
- 40.Dai C, Stolz DB, Kiss LP, Monga SP, Holzman LB, Liu Y. Wnt/beta-catenin signaling promotes podocyte dysfunction and albuminuria. J Am Soc Nephrol JASN. 2009. September;20(9):1997–2008. 10.1681/ASN.2009010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato H, Gruenwald A, Suh JH, Miner JH, Barisoni-Thomas L, Taketo MM, et al. Wnt/β-catenin pathway in podocytes integrates cell adhesion, differentiation, and survival. J Biol Chem. 2011. July 22;286(29):26003–15. 10.1074/jbc.M111.223164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He W, Kang YS, Dai C, Liu Y. Blockade of Wnt/β-catenin signaling by paricalcitol ameliorates proteinuria and kidney injury. J Am Soc Nephrol JASN. 2011. January;22(1):90–103. 10.1681/ASN.2009121236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei C, El Hindi S, Li J, Fornoni A, Goes N, Sageshima J, et al. Circulating urokinase receptor as a cause of focal segmental glomerulosclerosis. Nat Med. 2011. July 31;17(8):952–60. 10.1038/nm.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuda A, Chowdhury MA, Venkatareddy MP, Wang SQ, Nishizono R, Suzuki T, et al. Growth-dependent podocyte failure causes glomerulosclerosis. J Am Soc Nephrol JASN. 2012. August;23(8):1351–63. 10.1681/ASN.2012030271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puelles VG, van der Wolde JW, Wanner N, Scheppach MW, Cullen-McEwen LA, Bork T, et al. mTOR-mediated podocyte hypertrophy regulates glomerular integrity in mice and humans. JCI Insight. 2019. 19;4(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou L, Li Y, He W, Zhou D, Tan RJ, Nie J, et al. Mutual antagonism of Wilms’ tumor 1 and β-catenin dictates podocyte health and disease. J Am Soc Nephrol JASN. 2015. March;26(3):677–91. 10.1681/ASN.2013101067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.