Abstract
Enzymes of the chalcone synthase (CHS) family participate in the synthesis of multiple secondary metabolites in plants, fungi and bacteria. CHS showed a significant correlation with the accumulation patterns of anthocyanin. The peel color, which is primarily determined by the content of anthocyanin, is an economically important trait for eggplants that is affected by heat stress. A total of 7 CHS (SmCHS1-7) putative genes were identified in a genome-wide analysis of eggplants (S. melongena L.). The SmCHS genes were distributed on 7 scaffolds and were classified into 3 clusters. Phylogenetic relationship analysis showed that 73 CHS genes from 7 Solanaceae species were classified into 10 groups. SmCHS5, SmCHS6 and SmCHS7 were continuously down-regulated under 38°C and 45°C treatment, while SmCHS4 was up-regulated under 38°C but showed little change at 45°C in peel. Expression profiles of key anthocyanin biosynthesis gene families showed that the PAL, 4CL and AN11 genes were primarily expressed in all five tissues. The CHI, F3H, F3’5’H, DFR, 3GT and bHLH1 genes were expressed in flower and peel. Under heat stress, the expression level of 52 key genes were reduced. In contrast, the expression patterns of eight key genes similar to SmCHS4 were up-regulated at a treatment of 38°C for 3 hour. Comparative analysis of putative CHS protein evolutionary relationships, cis-regulatory elements, and regulatory networks indicated that SmCHS gene family has a conserved gene structure and functional diversification. SmCHS showed two or more expression patterns, these results of this study may facilitate further research to understand the regulatory mechanism governing peel color in eggplants.
Introduction
Eggplant (S. melongena L.) is one of the most important thermophilic vegetables produced in many tropical and temperate regions around the world. The optimum growth temperature for eggplant is between 22 and 30°C. After subjected to high temperature treatment, eggplants may exhibit to stagnation of growth, abortion of flower buds, and pollen viability rate and fruit set decrease, and the peel’s color will turn light when the temperature is over 35°C. High temperature severely reduces the yield and affects the appearance quality of eggplant. However, the molecular mechanism of heat stress response in eggplants has not been thoroughly elucidated.
Anthocyanins are plant secondary metabolites and are among the most abundant natural pigments, that are responsible for the characteristic colors in flowers, fruits and vegetables plant tissues. The anthocyanin biosynthesis pathway has been studied in numerous plant species and most of the genes involved in this process have been identified. Moreover, anthocyanins play an important role in plant survival under stressful environmental conditions. High temperatures are known to reduce anthocyanin accumulation and have discoloration effects in many plant tissues, causing drastic effects in colored flowers [1, 2], and affecting the skin of such fruits as grape berries, apples and eggplant [3–7].
It is well known that CHS is the gatekeeper of the anthocyanin pathway [8]. Enzymes of chalcone synthase (CHS) are member of the plants-specific type III polyketide synthase (PKS) [9, 10], family and catalyze the first committed step of the branch of the phenylpropanoid pathway, which leads to the synthesis of flavonoids [11, 12]. Flavonoids are well known as a group of plant secondary metabolites that comprise several different classes of compounds, such as chalcones, flavones, flavonol isoflavones and anthocyanins. Flavonoids have a wide variety of biological functions in flower pigmentation, protection against UV radiation, pathogen defense, auxin transport and pollen fertility [13–15]. CHS also showed a significant correlation with synthesis of flavonoid compounds during heat stress defense. Heat stress responsive element in bread wheat (Triticum aestivum L.) has been found in the promoter of Chs-D1 gene [16]. High-temperature stress had a large impact on the expression of CHS7, CHS8 in both seeds and pods of Soybean [17]. The transcript levels of CHS decreased in apple peel and rose flower after heat treatment [1, 4]. In cork oak, CHS gene expression exhibited an increase under 45°C, but showed a decreased expression at 55°C [18]. The emergence of CHSV and CHSVII is important for the development of fungal heat stress tolerance and pathogenicity in pathogenic fungi. [19]. In addition, CHS (Sme2.5_00283.1_g00002.1) was up-regulated, and the other two CHS gene members were down-regulated under heat stress in peel of eggplant [7].
The product of the CHS reaction is a pivotal precursor for a large array of secondary metabolites derived from malonyl-CoA and p-coumaroyl-CoA. CHS exists as homodimeric iterative PKS (monomer size of 42–45 kDa) with two independent active sites that catalyze a series of decarboxylation, condensation, and cyclization reactions [10, 20]. Member of the CHS superfamily share high similarity in their amino acid sequence, which contains the structurally conserved catalytic center consisting of four residues, Cys-His-Asn-Phe, and most of the genes contain two exons and one intron [21]. However, the CHS gene family has not been characterized in eggplants to date.
In the current study, all SmCHS family members were identified in eggplant. A comprehensive analysis of members was performed, including gene structures, the biochemical characteristics of putative CHS protein, promoter cis-elements, phylogenetic relationships among members in other relative species, and their expression profiles in various organs/tissues under high temperature stress. The findings of the present study may facilitate functional studies on eggplant SmCHS family genes.
Materials and methods
Plant materials and RNA extraction
The eggplant cultivar ‘Tewangda’ is a cold-tolerant cultivar with blackish purple skin. This cultivar grows vigorously and has good fruit setting. The fruit size was about 27.6 cm in length, 5.4 cm transverse diameter and a 209 g single fruit weight on average. ‘Tewangda’ fruits were grown at the same growth stage and were randomly selected. These plants were grown 144 days after sowing, and then placed inside incubators set at 27°C (CK), 38°C or 45°C for 3 or 6 h (three plants per treatment). For each treatment, the tissue samples of root, stem, leaf, flower and peel were obtained and immediately frozen in liquid nitrogen and stored at -80°C for RNA extraction and other analyses. All plant materials examined in this study were obtained from Shanghai Academy of Agricultural Sciences. Total RNA was extracted from each tissue sample using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s protocol. The extracted total RNA was stored at -80°C. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Identification of the CHS family members in the eggplant genome
The whole protein sequence of Solanum melongena L. (eggplant) were obtained from the Eggplant Genome DataBase (http://eggplant.kazusa.or.jp) [22], and those of Solanum tuberosum L. (potato, http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml) [23], Solanum lycopersicum (tomato, https://solgenomics.net/organism/Solanum_lycopersicum/genome) [24], Solanum penellii (wild tomato, https://www.plabipd.de/project_spenn/start.ep) [25], Capsicum annuum L. (pepper, http://peppergenome.snu.ac.kr) [26], Petunia axillaris (https://solgenomics.net/organism/Petunia_axillaris/genome) [27], Petunia inflate (https://solgenomics.net/organism/Petunia_inflata/genome) [27], and Nicotiana tabacum (common tobacco, https://www.ncbi.nlm.nih.gov/nuccore/AYMY00000000) [28]. The profiles of CHS (PF00195 and PF02797) were downloaded from the Pfam protein family database (http://pfam.xfam.org/), and these profile sequences were used as queries to perform BLASTP searches against the protein sequence data of all the species mentioned above with a maximum E-value of 1×10−3, respectively [29]. To further verify the exact copy number of CHS and remove redundant sequences, the Pfam database and Genome websites were also searched using “chalcone synthase” as keywords. All CHS sequences were submitted to EXPASy (https://web.expasy.org/protparam/) to calculate the number of amino acids, molecular weights and theoretical isoelectric points (pI).
Structural characterization
The locations and intron numbers of CHS were acquired through the genome website. All of the acquired protein sequences were first aligned by ClustalX software with the default parameters [30]. An unrooted maximum-likelihood phylogenetic tree was constructed using MEGA6 software with a Bootstarp value of 1000 times [31]. The MEME program (Version 5.0.5, http://meme-suite.org/tools/meme) was used to identify the conserved motif of the CHS sequences with the following parameters: any number of repetitions, maximum of 10 misfits and optimum motif width of 6–200 amino acid residues. The WoLF PSORT program was used to predict the subcellular localization information of CHS proteins (https://www.genscript.com/wolf-psort.html) [32].
Analysis of cis–acting elements in SmCHS
The upstream sequences (2 kb) of the SmCHS coding sequences in eggplant were retrieved from the genome sequence and then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify regulatory elements [33].
Phylogenetic analysis of CHS genes
The full-length protein sequences of all eight species in Solanaceae were used for phylogenetic analysis. All of the protein sequences were first aligned by ClustalX software with the default parameters [30]. The phylogenetic tree was generated with MEGA6 software with a bootstrap test of 1000 times. The final tree was viewed and modified in Evolview software [34]. The CHS genes were classified into different groups according to the topology of the phylogenetic tree.
Expression analysis of antyocyanin biosynthetic genes and construction of the mRNA regulatory network
The RNA-seq results were obtained by our lab [35]. Gene expression level was estimated from mean FPKM (fragments per kilobase of exon model per million reads mapped) values for each treatment, and showed the expression patterns in heatmap.Significant differentially expressed genes (fold change ≥ 2 and p-value ≤ 0.05) were used to calculate the Pearson correlation coefficient between CHS genes and other genes. The TBtools program was used to elucidate the Gene Ontology (GO) functional classification for the mRNAs with correlation coefficients greater than 0.9 [36]. The top 5 regulatory mRNAs annotated by GO enrichment for the genes associated with anthocyanin biosynthesis were collected to construct the regulatory network. The network was visualized using Cytoscape [37].
qRT-PCR analysis
Expression level of anthocyanin biosynthesis genes, including phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarateCoA ligase (4CL), CHS, chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′5′-hydroxylase (F3′5′H), dihydroflavonol4-reductase (DFR), anthocyanidin synthase (ANS), anthocyanidin 3-O-glucosyltransferase (3GT), Anthocyanins11 (AN11), and most transcription factors, such as myeloblastosis (MYB), basic helix-loop-helix (bHLH) and metadata authority description schema(MADS1), were analyzed. First-strand cDNA was synthesized from 1 μg total RNA from 5 tissues (root, stem, leaf, flower and peel) using a Prime Script RT Reagent Kit (Takara, Dalian, China). The qRT-PCR reactions were performed in 96-well plates using the ABI 7500 fast Real-Time PCR system (Applied Biosystems, USA) with the QuantiFast SYBR Green PCR Kit (Qiagen, Duesseldorf, Germany). The qRT-PCR parameters were as follows: 95°C for 5 min, then 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. The relative mRNA expression levels were calculated using the 2-ΔΔCT method [38]. PGK(JX154676) was used as an internal control to normalize the data. For each sample, three biological repeats were performed, the relative expression levels were calculated using the standard curve and normalized by the control’s expression, the results were display by heatmap. The primer sequences are listed in S3 Table.
Results
Identification of CHS genes and sequence analysis in Solanaceae species
A total of 7 CHS (SmCHS1-7) genes in eggplant were identified after being verified by protein sequence analysis and BlAST search using the eggplant genome annotation database (S1A Table). The length of SmCHS protein ranged from 327 to 396 amino acids (Table 1, S2 Table). The PKS type III active sites of the enzymes and Phe215 connected with CoA binding are conserved among all SmCHS (S1 Fig). In addition, 66 CHS genes were characterized from 7 other Solanaceae species. The subfamily numbers of CHS genes ranged from 6 (Solanum penellii) to 13 (Petunia axillaris) (Table 1, S1B–S1H Table). The length for the other 7 Solanaceae species (Solanum tuberosum L., Solanum lycopersicum, Solanum penellii, Capsicum annuum L., Petunia axillaris, Petunia inflate and Nicotiana tabacum) proteins ranged from 156 to 431 amino acids (S1A–S1G Table). The average number of amino acids was calculated, and all the amino acids of each species are arranged in same order to form a data set. The correlation coefficients among the above data set were all greater than 0.99. This finding suggests that CHS genes are conserved in Solanaceae species.
Table 1. Features of SmCHS genes identified in eggplant.
| Gene Name | Gene ID | Number of amino acids |
|---|---|---|
| SmCHS1 | Sme2.5_01077.1_g00016.1 | 333 |
| SmCHS2 | Sme2.5_02154.1_g00001.1 | 389 |
| SmCHS3 | Sme2.5_13923.1_g00001.1 | 389 |
| SmCHS4 | Sme2.5_00283.1_g00002.1 | 392 |
| SmCHS5 | Sme2.5_01039.1_g00002.1 | 327 |
| SmCHS6 | Sme2.5_00346.1_g00019.1 | 396 |
| SmCHS7 | Sme2.5_05261.1_g00004.1 | 383 |
Structure and conserved motif analysis of SmCHS
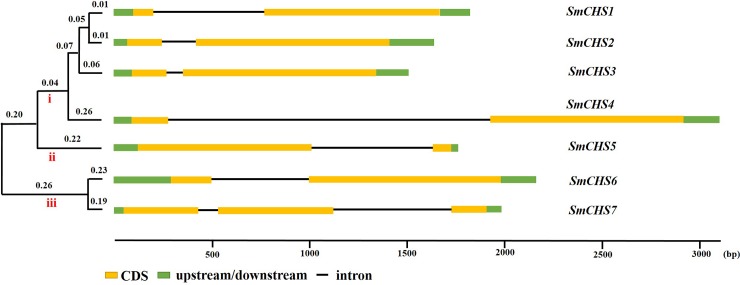
The 7 SmCHS genes were distributed on 7 scaffolds. To better understand the evolution of SmCHS genes, an unrooted maximum-likelihood tree was constructed based on the 7 SmCHS protein sequences, and the SmCHS were classified into 3 clusters (i, ii and iii) (Fig 1). Among the SmCHS genes, only one SmCHS7 had three exons, and the others had two exons (Fig 1) based on information available from the genome annotation. These results suggest the potential diversity of the biological functions of the SmCHS genes in eggplants, previous studies also have similar conclusion [39, 40].
Fig 1. Phylogenetic relationship and gene structure analysis of SmCHS genes.
The phylogenetic tree was on the left of the figure and showed that the SmCHS genes were classified into three clusters (i, ii and iii). The exon/intron organization of SmCHS is shown on the right of the figure. For SmCHS gene organization, yellow boxes represent exons, black lines represent introns, and green boxes indicate upstream/downstream regions. The lengths of the exons and introns are drawn to scale.
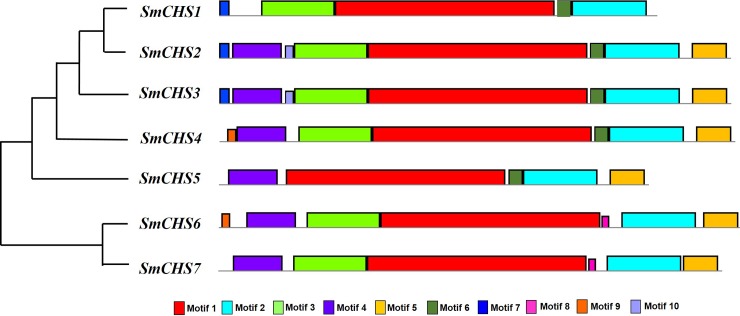
To understand the functional diversification of SmCHS, the conserved motifs of these 7 protein sequences were identified by the MEME program, and 10 conserved motifs were detected in eggplant (Fig 2, Table 2). The Chal_sti_synt_C domain and Chal_sti_synt_N domain were included in motifs 1 and motifs 2, respectively. For all 7 eggplant SmCHS proteins, Motif 1 and motif 2 exist in all of them, motif 3 is only absent in SmCHS5, and motif 4 and motif 5 are only absent in SmCHS1. The N-terminal domain (PF00195) of the CHS protein contained motif 1 and the combination of motifs 3, 4, 6, 7 and 9. The C-terminal domain (PF02797) of the CHS protein contained motif 2 and the combination of motifs 5, 8 and 10. Therefore, the motif configuration of the SmCHS reflects the conservation and diversity of the CHS family. To further investigate the subcellular localization information of SmCHS proteins, the WoLF PSORT program was used to predict the localization of SmCHS protein [31]. SmCHS7 was predicted to localize in the nucleus, whereas SmCHS4 and SmCHS6 were predicted to localize in the chloroplast. The others SmCHS proteins were predicted to localize in the cytoplasm. The different compositions of the domains and subcellular localization may indicate functional diversity.
Fig 2. Motifs conserved across all CHS proteins in eggplant.
Ten conserved motifs are indicated in differently colored boxes.
Table 2. List of the putative motifs of CHS proteins.
| Motif | Length (amino acids) | Best possible match |
|---|---|---|
| Motif 1 | 167 | IKEWGQPKSKITHLVFCTTSGVDMPGADYQLTKLLGLRPSVKRFMMYQQGCFAGGTVLRLAKDLAENNKGARVLVVCSEITAVGFRGPSETHPDSLVGQA |
| Motif 2 | 57 | DWNSJFWIAHPGGPAILDQVELKLGLKPEKLRATRQVLSDYGNMSSACVLFILDEMR |
| Motif 3 | 56 | RLCDKSMIKKRYMHLTEEILKENPNLCEYMAPSLDARQDIVVVEVPKLGKEAAQKA |
| Motif 4 | 38 | QRAEGPATILAIGTATPSNCVDQSTYPDYYFRITNSEH |
| Motif 5 | 27 | TTGEGLDWGVLLGFGPGLTIETIVLHS |
| Motif 6 | 11 | LIEAFEPLGIS |
| Motif 7 | 8 | MVTVEEVR |
| Motif 8 | 6 | FCEKLI |
| Motif 9 | 7 | QNIGKVN |
| Motif 10 | 7 | ELKEKFK |
Stress-related cis-elements in SmCHS promoters
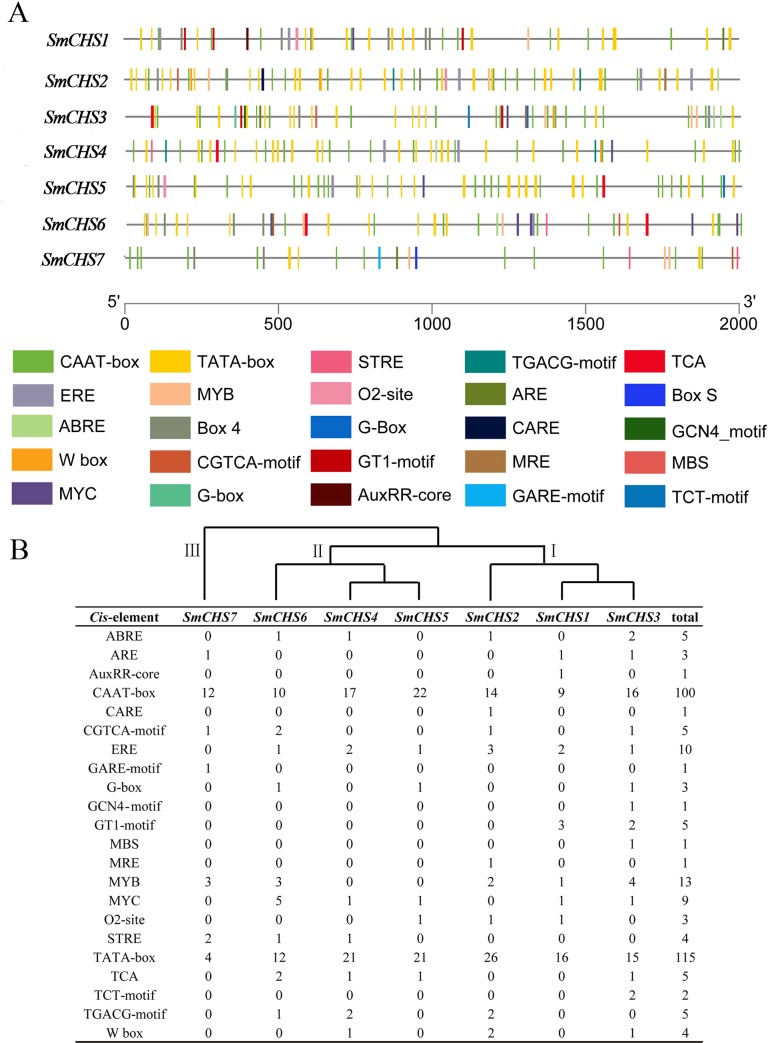
To further study the potential regulatory mechanisms of SmCHS during abiotic stress responses, the 2 kb upstream sequences from the translation start sites of SmCHS were used to identify the cis-elements (Fig 3A). The results showed that all SmCHS had common upstream promoter elements, including TATA-box and CAAT-box, which occurred more than 100 times; therefore, these sequences were presumed to be the promoter sequences (Fig 3B). The elicitor response element (ERE) and myeloblastosis binding cis-elements (MYB) occurred more than 10 times in the SmCHS upstream sequences. Research has shown that an increase in CHS activity causes a high accumulation of flavonoids that inhibits polar auxin transport [8, 41, 42]. Two cis-acting elements (ABRE, involved in abscisic acid responsiveness; AuxRR, involved in auxin responsiveness) were found in the upstream regions. MYB and myelocytomatosis (MYC) binding sites have also been identified, which may greatly influence plant stress tolerance. Cluster analysis of cis-element number showed that 7 SmCHS genes were divided into 3 groups (Ⅰ, Ⅱ, Ⅲ), and SmCHS1, SmCHS2 and SmCHS3 had similar regulatory pattern (Fig 3A). Five cis-elements (CARE, GCN4-motif, GT1-motif, MRE and TCT-motif) exist only in group Ⅰ, GARE-motif only exist in group Ⅲ. STRE exist in group Ⅱ and Ⅲ. These results showed that SmCHS is activated by a wide range of environmental and developmental stimuli, and there are many complex means of regulating SmCHS activity in eggplants.
Fig 3. Cis-elements in CHS family gene promoters.
(A) Frequency of cis-element occurrence in upstream sequences. (B) Predicted cis-elements in CHS gene promoters. The scale bar indicates the length of promoters.
Phylogenetic analysis of CHS genes in Solanaceae
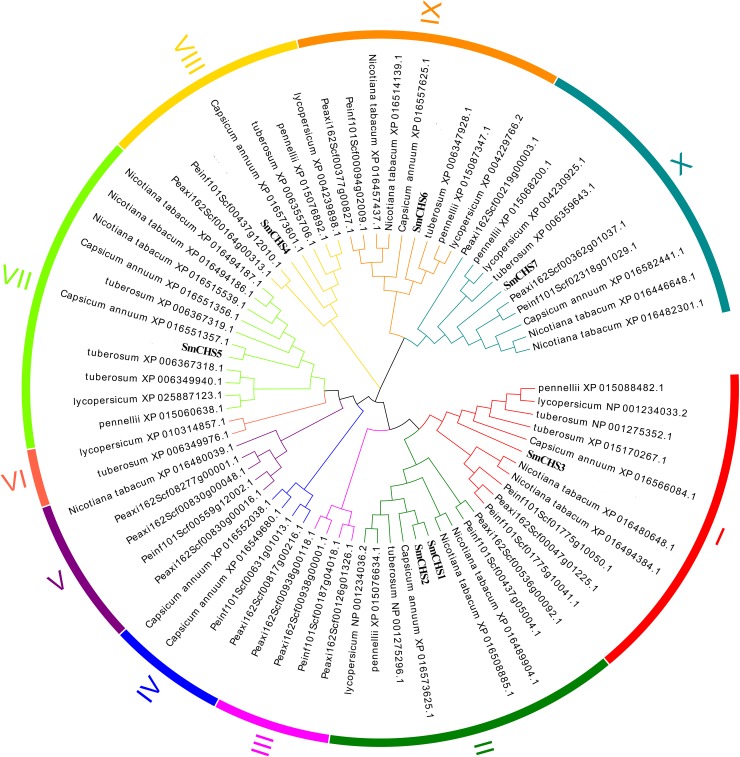
To analyze the evolutionary relationships of CHS genes in Solanaceae, an unrooted phylogenetic tree was constructed using full-length amino acid sequences. All 73 CHS genes were classified into 10 groups (Fig 4, Table 3), and the number of CHS gene groups ranged from two to eleven. The 7 SmCHS were categorized into 6 groups (groups Ⅰ, Ⅱ, Ⅶ, Ⅷ, Ⅸ and Ⅹ), and group Ⅱ contained SmCHS1 and SmCHS2. Groups Ⅰ, Ⅱ, Ⅸ and Ⅹ exist in all eight species, and groups Ⅲ, Ⅳ and Ⅴ were absent in Solanum melongena L., Solanum penellii, Solanum lycopersicum and Solanum tuberosum L.. The group Ⅵ is absent in Capsicum annuum L., Nicotiana tabacum, Petunia inflate and Petunia axillaries (Table 3). The Ⅷ, Ⅸ and Ⅹ groups are distinguished from other groups mainly depends on the position 1–164 amino acids, GroupsⅠ, Ⅱ and Ⅲ are relatively conservative at the position 260–360 amino acids, in which the other groups are very diverse(S1 Fig). These results suggested that the CHS were conserved, but small variations existed among the eight species in Solanaceae and showed that SmCHS1, SmCHS2 and SmCHS3 were more conserved than SmCHS4 according to the phylogenetic tree.
Fig 4. Phylogenetic tree of CHS genes in Solanaceae species.
The colored region is associated with 10 groups of proteins (Group Ⅰ to Ⅹ).
Table 3. Distribution of CHS genes in the phylogenetic tree.
| Plant Pecies | Number | Phylogenetic Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ⅰ | Ⅱ | Ⅲ | Ⅳ | Ⅴ | Ⅵ | Ⅶ | Ⅷ | Ⅸ | Ⅹ | ||
| Solanum melongena L. | 7 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Solanum penellii | 6 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Solanum lycopersicum | 7 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| Solanum tuberosum L. | 10 | 2 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 1 |
| Capsicum annuum L. | 9 | 1 | 1 | 0 | 2 | 0 | 0 | 2 | 1 | 1 | 1 |
| Nicotiana tabacum | 12 | 2 | 2 | 0 | 0 | 1 | 0 | 3 | 0 | 2 | 2 |
| Petunia inflate | 9 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| Petunia axillaris | 13 | 1 | 1 | 3 | 1 | 3 | 0 | 0 | 1 | 1 | 2 |
Expression profile of key anthocyanin biosynthesis genes in eggplants under heat stress
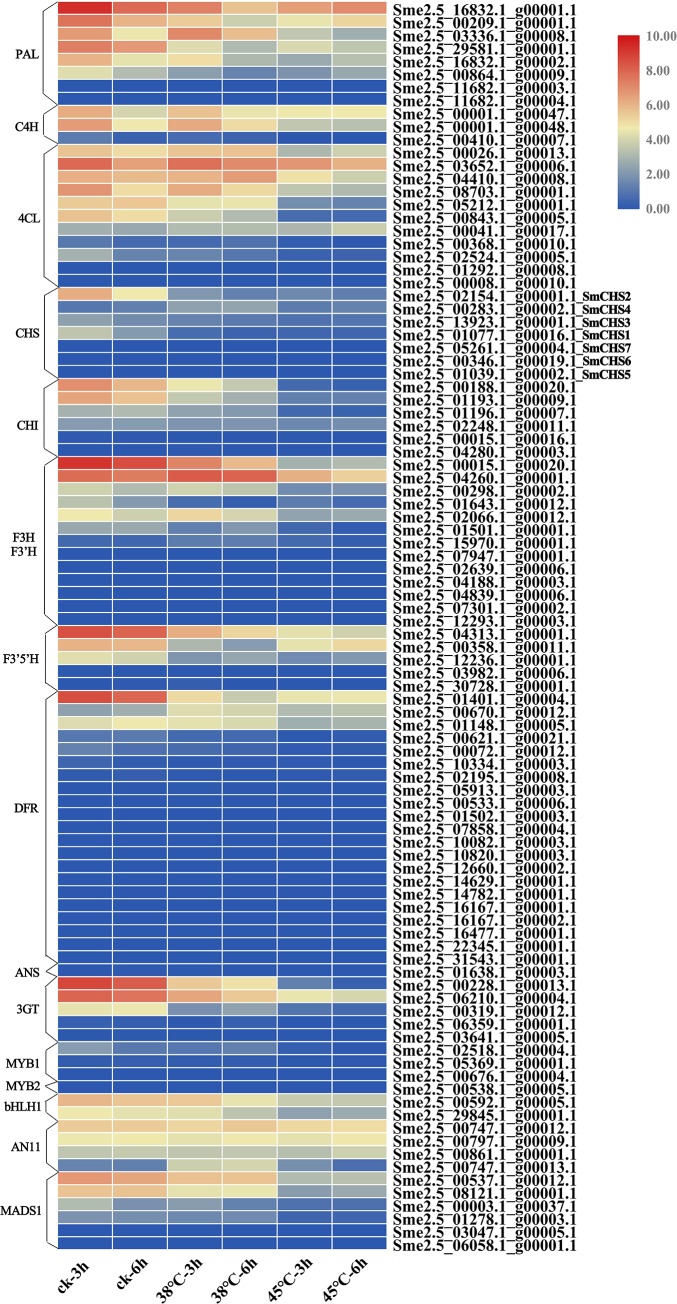
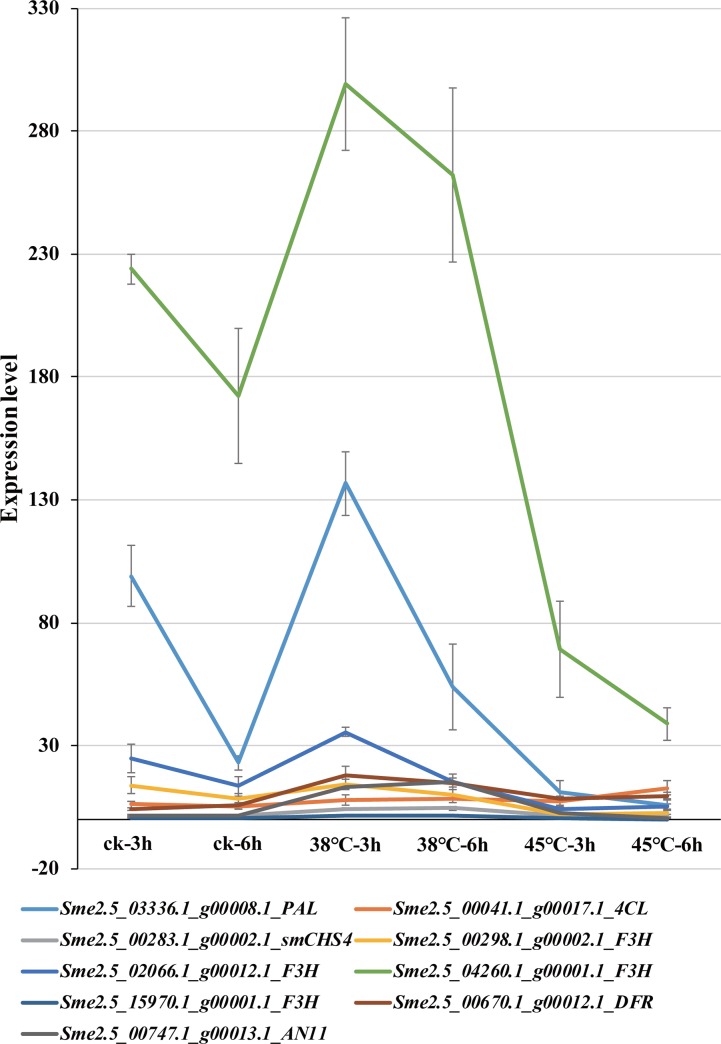
Using the RNA-seq data of eggplant peel, a heatmap of 96 key anthocyanin biosynthesis genes (PAL, C4H, 4CL, CHS, CHI, F3H/F3’H, F3’5’H, DFR, ANS, 3GT, MYB1, MYB2, bHLH1, AN11, MADS1) was established under heat stress (Fig 5). The expression of anthocyanidin synthase (ANS) and MYB2 was not identified during this sampling period (the undetected genes were also color-coded for 0 in Fig 5). For seven SmCHS genes, expression of three genes(SmCHS5, SmCHS6, and SmCHS7) were not identified, and the other four SmCHS genes were divided into two groups according to their expression patterns. Three of those four SmCHS genes (SmCHS1, SmCHS2, and SmCHS3) were continuously down-regulated under 38°C and 45°C treatment compared with the CK. However, SmCHS4 was up-regulated under 38°C, but showed little change under 45°C in peel. These phenomena have also been observed in some other key gene families associated with anthocyanin biosynthesis. According to the RNA-seq results of 96 anthocyanin biosynthesis key genes in eggplant peel, SmCHS4 showed the highest expression level at the 38°C-3h along with eight other genes (Sme2.5_03336.1_g00008.1_PAL, Sme2.5_00041.1_g00017.1_4CL, Sme2.5_00283.1_g00002.1_smCHS4, Sme2.5_00298.1_g00002.1_F3H, Sme2.5_02066.1_g00012.1_F3H, Sme2.5_04260.1_g00001.1_F3H, Sme2.5_15970.1_g00001.1_F3H, Sme2.5_00670.1_g00012.1_DFR, Sme2.5_00747.1_g00013.1_AN11) (Fig 6). In particular, Sme2.5_03336.1_g00008.1_PAL expression level under 38°C doubled but was down-regulated at 45°C compared with CK; Sme2.5_00670.1_g00012.1_DFR, Sme2.5_00747.1_g00013.1_AN11 expression level increased 3–4 fold and 7–10 fold under 38°C, respectively. These results suggest that these genes may have tissue-specific or functionally differentiation.
Fig 5. Heatmap of 96 key anthocyanin biosynthesis genes expression level in eggplants peel under heat stress.
The color box from blue to red indicate an increased expression level.
Fig 6. Expression profiles of SmCHS4 and eight anthocyanin biosynthesis genes in response to heat stress.
These genes have the highest expression level at 38°C-3h in eggplant peel. The error bars represent the standard error of the means of three biological replicates.
mRNA regulatory network associated with anthocyanin biosynthesis in eggplant
Pearson correlation coefficient was calculated between all the mRNAs and anthocyanin biosynthesis related genes, the mRNAs with correlation coefficient greater than 0.9 are considered to be genes that are co-expressed with the anthocyanin biosynthesis genes. A total of 4928 mRNA correlation coefficients were more than 0.9, and all of these mRNAs were functionally categorized in the GO database. The top 20 GO enrichment results of biological processes are shown in Table 4. The genes involved in the regulation of biological processes (GO:0050789), regulation of cellular metabolic processes (GO:0031323) and regulation of gene expression (GO:0010468) were collected and filtered to construct a regulatory network. In totally, 67 anthocyanin biosynthesis key genes and 146 regulatory mRNAs were included in this regulatory network (S2 Fig). These GO enrichment results suggest that the anthocyanin biosynthesis pathway may be regulated by a wide range of environmental and developmental stimuli.
Table 4. Top 20 GO enrichment results of biological processes.
| GO term | GO ID | P value |
|---|---|---|
| cellular biosynthetic process | GO:0044249 | 0 |
| cellular nitrogen compound biosynthetic process | GO:0044271 | 0 |
| cellular response to chemical stimulus | GO:0070887 | 0 |
| cellular response to stress | GO:0033554 | 0 |
| regulation of biological process | GO:0050789 | 0 |
| regulation of cellular macromolecule biosynthetic process | GO:2000112 | 0 |
| developmental process | GO:0032502 | 0 |
| regulation of RNA biosynthetic process | GO:2001141 | 0 |
| regulation of cellular metabolic process | GO:0031323 | 0 |
| cellular component organization | GO:0016043 | 0 |
| response to organic substance | GO:0010033 | 1.11E-16 |
| protein metabolic process | GO:0019538 | 1.11E-16 |
| regulation of nitrogen compound metabolic process | GO:0051171 | 1.11E-16 |
| regulation of gene expression | GO:0010468 | 1.11E-16 |
| response to stimulus | GO:0050896 | 1.11E-16 |
| regulation of nucleobase-containing compound metabolic process | GO:0019219 | 1.11E-16 |
| response to chemical | GO:0042221 | 1.11E-16 |
| cell communication | GO:0007154 | 1.11E-16 |
| response to stress | GO:0006950 | 1.11E-16 |
| oxoacid metabolic process | GO:0043436 | 2.22E-16 |
Expression pattern of anthocyanin biosynthesis key genes in different tissues under heat stress
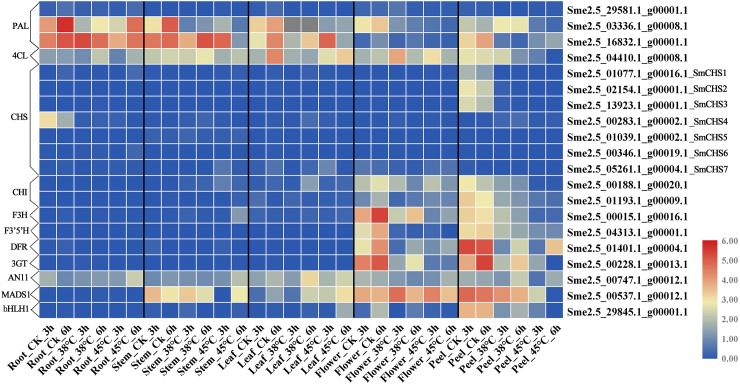
Using the qRT-PCR data, a heatmap of 20 key anthocyanin biosynthesis genes was established in different tissues under heat stress (Fig 7). The qRT-PCR results showed a high consistency with the RNA-seq data, which suggested that the RNA-seq data were credible. Most of the CHS genes were expressed in peel and were expressed at low levels in other tissues. The PAL, 4CL and AN11 genes were mainly expressed in all five tissues. The CHI, F3H, F3’5’H, DFR, 3GT and bHLH1 genes were expressed in flower and peel. MADS1 was expressed in stems, leaves, flowers and peels. Under heat stress, cluster i (cluster show in Fig 2) was continuously downregulation, cluster ii was up-regulated 4 times under 38°C compared with CK in peel, and cluster iii was not detected in most eggplant tissues.
Fig 7. Expression profiles of 20 key anthocyanin biosynthesis genes in different tissues.
Discussion
It is well-known that the CHS gene family plays a significant role in the growth and development of plants. In many species, multigene families of CHS have been identified. For example, six CHS genes have been described in turnip [43]. In maize, 14 complete CHS genes have been identified [39]. A total of 27 CHS genes were found in rice [40]. These studies showed that CHS members were divided into two or more subclasses according to phylogenetic analysis. Generally, genes grouped into the same subclasses shared similar evolutionary features, and obtained the same expression pattern. In our study, the identified sequences showed a high level of coding sequence similarity (above 90%). The SmCHS were classified into three clusters based on the results of the maximum-likelihood tree. At 35°C, previous studies showed that SmCHS1 and SmCHS3 (Sme2.5_01077.1_g00016.1, Sme2.5_13923.1_g00001.1) were down-regulated in peels of eggplant [7], which is in keeping with our results, other two clusters CHS genes show different expression patterns. These results suggest the functional diversification of SmCHS.
Flavonoids have numerous functions and contribute to pigments, signaling molecules, and protectants against biotic and abiotic stresses. The flavonoid biosynthetic pathway is one of the most intensively investigated pathways for applied biological and genetic processes, as well as for understanding gene regulation, characterizing transposable elements and producing of agronomically stress-tolerant plants and natural dietary antioxidants. Biosynthesis of anthocyanins responds to environmental stress factors, such as light, nutrient depletion, and temperature change. The peel color determined by the content of anthocyanin is a majority economically important trait for eggplant, and this color is modulated by the genes in the flavonoid biosynthesis pathway. Compared with other tissues, SmMYB1 and all anthocyanin biosynthetic key genes (SmCHS, SmCHI, SmF3H, SmDFR) except SmPAL were dramatically up-regulated in the fruit skin of the purple cultivar [44]. The full length cDNA of SmCHS, SmCHI, SmF3’5’H, and SmDFR were isolated from eggplants by Jiang (2016) [45]. These genes have the highest expression levels in peels except for SmF3H, which was detected in stems [45]. The expression profiles of these key gene families under heat stress were investigated in our study. ‘These anthocyanin biosynthesis key genes (PAL, 4CL, AN11, CHI, F3H, F3’5’H, DFR, 3GT and bHLH1) show tissue specific expression, suggesting that these genes respond at the late stage of the anthocyanin pathway and directly regulate the color of fruit skin and flower.
Heat stress reduced the anthocyanin content and the enzyme activities of CHS, DFR, ANS and 3GT/UFGT in eggplant peel and strengthened the activity of PAL [35, 46]. When the temperature exceeds 35°C, the eggplant will be dehydrated and shrink, and the peel’s color will lighten. CHS is a key enzyme of the flavonoid biosynthesis pathway. Most of the genes associated with flavonoid biosynthesis were down-regulated under heat stress. In this study, the genes of the flavonoid biosynthesis pathway showed tissue-specificity, and genes expressed in different phases and tended to change over time (Fig 7). Under heat stress, SmCHS4 and some anthocyanin biosynthesis related genes show different expression profiles at 38°C-3h (Fig 6), suggest that these co-up-regulated genes contribute to protect the eggplant at beginning of heat stress defense. In addition, 52 gene expression levels were reduced under heat stress, which was similar to Lv’s (2019) results [7], while 35 gene expression levels were not identified. These results suggest that some key anthocyanin biosynthesis genes help to protect the eggplant from damage to heat stress. Moreover, these gene families exhibited two or more expression patterns and performed multiple genetic functions to regulate anthocyanin content. Combined with regulatory networks, it is possible to further understand the regulatory mechanism of peel color in eggplants.
Conclusions
In this study, a genome-wide analysis of the SmCHS gene family in eggplants was performed. The CHS protein biochemical characteristics, phylogenetic relationships, gene structures, cis-regulatory elements, regulatory network and functional predictions of the smCHS gene family members were examined. The SmCHS gene family has conserved gene structure and functional diversification. CHS plays important roles in the anthocyanin biosynthesis pathway, exhibits two or more expression patterns and executes multiple functions to regulate anthocyanin content in eggplant peels under heat stress. The result of this study may contribute to the production of heat-resistant eggplant for further research on the functions, regulation and evolution of the CHS family.
Supporting information
(XLSX)
(XLSX)
(DOCX)
Color bars on the left represent the 10 groups in Fig 4. Active site residues are highlighted in yellow, malony-CoA binding sites are highlighted in blue and other conserved sequence are shown in green.
(JPG)
The pink labels represent the CHS gene family.
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
In addition, the authors have declared this work was supported by the Agricultural Committee Basic Project (Shanghai Agricultural word (2015) No 6-2-3), the National Key Technology R&D Program during the 13th Five-Year Plan Period (2017YFD0101904) and the China Agriculture Research System (Grant No. CARS-25). And the funding bodies supporting this work did not play a role in the design of the study and the collection, analysis, and interpretation of data or in the composition of the manuscript.
References
- 1.Dela G, Or E, Ovadia R, Nissim-Levi A, Weiss D, Oren-Shamir M. Changes in anthocyanin concentration and composition in 'Jaguar' rose flowers due to transient high-temperature conditions. Plant Sci. 2003; 164(3): 333–340. 10.1016/S0168-9452(02)00417-X. [DOI] [Google Scholar]
- 2.Nozaki K, Takamura T, Fukai S. Effects of high temperature on flower colour and anthocyanin content in pink flower genotypes of greenhouse chrysanthemum (Chrysanthemum morifolium Ramat.). J Hortic Sci Biotechnol. 2006; 81(4): 728–734. 10.1080/14620316.2006.11512130. [DOI] [Google Scholar]
- 3.Mori K, Goto-Yamamoto N, Kitayama M, Hashizume K. Loss of anthocyanins in red-wine grape under high temperature. J Exp Bot. 2007; 58(8): 1935–1945. 10.1093/jxb/erm055 . [DOI] [PubMed] [Google Scholar]
- 4.Lin-Wang K, Micheletti D, Palmer J, Volz R, Lozano L, Espley R, et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011; 34(7): 1176–1190. 10.1111/j.1365-3040.2011.02316.x . [DOI] [PubMed] [Google Scholar]
- 5.Movahed N, Pastore C, Cellini A, Allegro G, Valentini G, Zenoni S, et al. The grapevine VviPrx31 peroxidase as a candidate gene involved in anthocyanin degradation in ripening berries under high temperature. J Plant Res. 2016; 129(3): 513–526. 10.1007/s10265-016-0786-3 . [DOI] [PubMed] [Google Scholar]
- 6.Pastore C, Dal Santo S, Zenoni S, Movahed N, Allegro G, Valentini G, et al. Whole plant temperature manipulation affects flavonoid metabolism and the transcriptome of grapevine berries. Front Plant Sci. 2017; 8: 929 10.3389/fpls.2017.00929 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv LL, Feng XF, Li W, Li K. High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome. 2019; 62(7): 503–512. 10.1139/gen-2019-0021 . [DOI] [PubMed] [Google Scholar]
- 8.Dao T, Linthorst H, Verpoorte R. Chalcone synthase and its functions in plant resistance. Phytochem Rev. 2011; 10(3): 397 10.1007/s11101-011-9211-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schröder J. A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci. 1997; 2(10): 373–378. 10.1016/S1360-1385(97)01104-7. [DOI] [Google Scholar]
- 10.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003; 20(1): 79–110. 10.1039/b100917f . [DOI] [PubMed] [Google Scholar]
- 11.Winkel-Shirley B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001; 127(4): 1399–1404. 10.1104/pp.127.4.1399 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang Y, Shen G, Wu W, Liu X, Lin J, Tan F, et al. Characterization and expression of chalcone synthase gene from Ginkgo biloba. Plant Sci. 2005; 168(6): 1525–1531. 10.4238/2014.April.30.6 [DOI] [Google Scholar]
- 13.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr Opin plant Bio. 2002; 5(3): 218–223. 10.1016/s1369-5266(02)00256-x . [DOI] [PubMed] [Google Scholar]
- 14.Buer CS, Imin N, Djordjevic MA. Flavonoids: new roles for old molecules. J Integr plant biol. 2010; 52(1): 98–111. 10.1111/j.1744-7909.2010.00905.x . [DOI] [PubMed] [Google Scholar]
- 15.Falcone FM, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012; 3: 222 10.3389/fpls.2012.00222 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glagoleva AY, Ivanisenko NV, Khlestkina EK. Organization and evolution of the chalcone synthase gene family in bread wheat and relative species. BMC Genet. 2019; 20(1): 30 10.1186/s12863-019-0727-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chennupati P, Seguin P, Chamoun R, Jabaji S. Effects of high-temperature stress on soybean isoflavone concentration and expression of key genes involved in isoflavone synthesis. J AgricFood Chem. 2012; 60(51): 12421–12427. 10.1021/jf3036319 . [DOI] [PubMed] [Google Scholar]
- 18.Correia B, Rodriguez JL, Valledor L, Almeida T, Santos C, Cañal MJ, et al. Analysis of the expression of putative heat-stress related genes in relation to thermotolerance of cork oak. J Plant Physiol. 2014; 171(6): 399–406. 10.1016/j.jplph.2013.12.004 . [DOI] [PubMed] [Google Scholar]
- 19.Liu R, Xu C, Zhang Q, Wang S, Fang W. Evolution of the chitin synthase gene family correlates with fungal morphogenesis and adaption to ecological niches. Sci Rep. 2017; 7: 44527 10.1038/srep44527 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tropf S, Kärcher B, Schröder G, Schröder J. Reaction Mechanisms of Homodimeric Plant Polyketide Synthases (Stilbene and Chalcone Synthase). A single active site for the condensing reaction is sufficient for synthesis of stilbenes, chalcones, and 6'-deoxychalcones. J Biol Chem. 1995; 270(14): 7922–7928. 10.1074/jbc.270.14.7922 . [DOI] [PubMed] [Google Scholar]
- 21.Ferrer J-L, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Mol Biol. 1999; 6(8): 775 10.1038/11553 . [DOI] [PubMed] [Google Scholar]
- 22.Hideki H, Kenta S, Koji M, Tsukasa N, Satomi N, Akio O, et al. Draft genome sequence of eggplant (Solanum melongena L.): the representative Solanum species indigenous to the old world. DNA Res. 2014; 21(6): 649–660. 10.1093/dnares/dsu027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium PGS. Genome sequence and analysis of the tuber crop potato. Nature. 2011; 475(7355): 189 10.1038/nature10158 . [DOI] [PubMed] [Google Scholar]
- 24.Consortium TTG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012; 485(7400): 635 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt MH-W, Vogel A, Denton AK, Istace B, Wormit A, van de Geest H, et al. De novo assembly of a new Solanum pennellii accession using nanopore sequencing. Plant Cell. 2017; 29(10): 2336–2348. 10.1105/tpc.17.00521 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014; 46(3): 270 10.1038/ng.2877 . [DOI] [PubMed] [Google Scholar]
- 27.Bombarely A, Moser M, Amrad A, Bao M, Bapaume L, Barry CS, et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat Plants. 2016; 2(6): 16074 10.1038/nplants.2016.74 . [DOI] [PubMed] [Google Scholar]
- 28.Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 2014; 5: 3833 10.1038/ncomms4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41(W1): W29–W33. 10.1093/nar/gkt282 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997; 25(24): 4876–4882. 10.1093/nar/25.24.4876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30(12): 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007; 35(suppl_2): W585–W587. 10.1093/nar/gkm259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002; 30(1): 325–327. 10.1093/nar/30.1.325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian B, Gao S, Lercher MJ, Hu S, Chen W-H. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019. 10.1093/nar/gkz357 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhang A, Wu X, Zhu Z, Yang Z, Zhu Y, et al. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. 2019; 19(1): 1–13. 10.1186/s12870-018-1600-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, Xia R, Chen H, He Y. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018: 289660 10.1101/289660. [DOI] [Google Scholar]
- 37.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010; 27(3): 431–432. 10.1093/bioinformatics/btq675 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001; 25(4): 402–408. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 39.Han Y, Ding T, Su B, Jiang H. Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int J Mol Sci. 2016; 17(2): 161 10.3390/ijms17020161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Y, Cao Y, Jiang H, Ding T. Genome-wide dissection of the chalcone synthase gene family in Oryza sativa. Mol Breeding. 2017; 37(10): 119 10.1007/s11032-017-0721-x. [DOI] [Google Scholar]
- 41.Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988; 241(4863): 346–349. 10.1126/science.241.4863.346 . [DOI] [PubMed] [Google Scholar]
- 42.Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, et al. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001; 126(2): 524–535. 10.1104/pp.126.2.524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou B, Wang Y, Zhan Y, Li Y, Kawabata S. Chalcone synthase family genes have redundant roles in anthocyanin biosynthesis and in response to blue/UV A light in turnip (Brassica rapa; Brassicaceae). AM J Bot. 2013; 100(12): 2458–2467. 10.3732/ajb.1300305 . [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Hu Z, Chu G, Huang C, Tian S, Zhao Z, et al. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J Agr Food Chem. 2014; 62(13): 2906–2912. 10.1021/jf404574c . [DOI] [PubMed] [Google Scholar]
- 45.Jiang M, Liu Y, Ren L, Lian H, Chen H. Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol Plant. 2016; 38(7): 163 10.1007/s11738-016-2172-0 [DOI] [Google Scholar]
- 46.Wu X, Zhang A, Zhu Z, Yao J, Zha D, Li X. Effects of High ÿtemperature Stress on Active Oxygen Metabolism, Anthocyanin Content and Its Main Synthases in Eggplant Peel. Acta Ag Jiangxi. 2018; 30(06): 1–5. https://doi.org/10.19386j.cnki.jxnyxb.2018.06.01. [Google Scholar]