Abstract
Recent advances in J-difference-edited proton magnetic resonance spectroscopy (1H MRS) data acquisition and processing have led to the development of Hadamard Encoding and Reconstruction of MEGA-Edited Spectroscopy (HERMES) techniques, which enable the simultaneous measurement of ɣ-aminobutyric acid (GABA), the primary inhibitory amino acid neurotransmitter in the central nervous system, and of glutathione (GSH), the most abundant antioxidant in living tissue, at the commonly available magnetic field strength of 3 Tesla. However, the reproducibility of brain levels of GABA and GSH measured across multiple scans in human subjects using HERMES remains to be established. In the present study, twelve healthy volunteers completed two consecutive HERMES scans of the dorsal anterior cingulate cortex (dACC) to assess the test-retest reproducibility of the technique for GABA and GSH measurements at TE=80ms. Eleven of the twelve participants additionally completed two consecutive MEGA-PRESS scans at TE = 120 ms, with editing pulses configured for GSH acquisition, to compare the reliability of GSH in the same voxel measured using the standard MEGA-PRESS at TE=120ms. The primary findings of study were that, 1) the coefficient of variation (CV) of measuring GABA with HERMES was 16.7%, which is in agreement with the reliability we previously reported for measuring GABA using MEGA-PRESS; and 2) the reliability of measuring GSH with MEGA-PRESS at TE=120ms was more than twice as high as that for measuring the antioxidant with HERMES at TE=80ms (CV=7.3% vs. 19.0% respectively). These findings suggest that HERMES and MEGA-PRESS offer similar reliabilities for measuring GABA, while MEGA-PRESS at TE=120ms is more reliable for measuring GSH relative to HERMES at TE=80ms.
Keywords: GABA, GSH, j-difference editing, reproducibility, MEGA-PRESS, HERMES
1. INTRODUCTION
Proton magnetic resonance spectroscopy (1H MRS) of the brain has commonly been restricted to the reliable quantification of a small number of metabolites (i.e., N-acetylaspartate, choline-containing compounds, creatine-containing compounds, glutamate + glutamine [Glx], and myo-inositol) due to the challenges inherent to measuring low-concentration metabolites containing J-coupled protons [1]. Over the past decade or so, advances in spectral editing acquisition and processing techniques, primarily involving the J-difference editing sequence, “MEGA-PRESS” [2], have enabled the reliable measurement of additional metabolites of vital interest to the neurosciences at the common magnetic field strength of 3 Tesla, including ɣ-aminobutyric acid (GABA) and glutathione (GSH), the brain’s primary inhibitory neurotransmitter and antioxidant, respectively [3].
Although J-difference editing techniques have proved valuable to advancing the goals of neuroscientific research, they have remained relatively inefficient in practice, providing reliable measurement of a single metabolite from a relatively lengthy (e.g., 10-min) acquisition. Recent efforts to improve the efficiency of J-difference acquisition methods have enabled editing of two or more metabolites (e.g., GABA and GSH) within a single acquisition by applying editing pulses according to a Hadamard encoding scheme (Hadamard Encoding and Reconstruction of MEGA-edited Spectroscopy, HERMES, [4]); initial simulation, phantom, and human evaluations of the HERMES sequence have supported the validity of this approach [5]. Nonetheless, given that the echo time (TE) for successful J-difference editing depends on the J-evolution of the spin system under investigation, selection of a single TE to investigate differentially coupled systems necessarily represents a compromise. Given that J-difference editing has commonly been used to measure GABA levels, initial evaluation of HERMES has used a TE commonly used for GABA measurement (i.e., TE = 80 ms, [6]). It is likely that GSH-only editing is better performed at a longer TE, although the editing benefits of longer TE are mitigated by the relaxation losses in vivo [7].
In the present study, twelve healthy volunteers completed two consecutive 1H MRS scans of the dorsal anterior cingulate cortex (dACC), separated by removal from the scanner (interval: 5–10 min), to test the hypotheses that HERMES at TE = 80 ms would produce GABA reproducibility estimates comparable to those previously reported in investigations of MEGA-PRESS at TE = 68–80 ms, and that MEGA-PRESS at TE=120 ms would result in improved reproducibility of GSH estimates relative to HERMES at TE=80 ms.
2. MATERIALS AND METHODS
Scans were conducted on a 2.89 Tesla SIEMENS Prisma MR system using a 32-channel head coil (Erlangen, Germany), under a local Institutional Review Board-exempt quality assurance protocol. J-difference editing was performed using the universal editing sequence, with editing pulses separated by TE/2 [8]. Twelve healthy volunteers (9 female, mean ± s.d. age = 25 ± 2.5) completed two consecutive TE=80ms HERMES scans (NEX = 256, each) of the dACC (3.0 × 2.5 × 2.5 cm3, see Figure 1a for sample voxel positioning). Eleven of these volunteers (8 female, mean ± s.d. age = 25 ± 2.4) additionally completed two consecutive TE-120 ms MEGA-PRESS scans (NEX = 256, each) of the dACC. These scans were acquired in a fixed order, with each TE=120 ms scan following a preceding TE=80 ms scan. Each individual acquisition was immediately followed by a matched water-unsuppressed acquisition (NEX = 16, each) for phase and eddy-current correction as well as for concentration referencing. Common sequence parameters included TR = 2000 ms, 16-step phase cycling, spectral width = 2.5 kHz, and 2048 complex data points. Each set of acquisitions began with a T1-weighted MPRAGE scan for voxel positioning and subsequent segmentation and correction of partial volume effects [9], and was separated by a brief (5–10-min) break during which participants were removed from the scanner and then repositioned. All 1H MRS analyses were conducted on raw (TWIX) data files using Gannet (version 3.1) within the MATLAB environment [10], with frequency and phase correction applied prior to fitting [11]. Co-registration of each individual’s voxel mask to their MPRAGE, followed by within-voxel tissue segmentation, was performed in SPM12 [12]. The between-scan reliability of each metabolite by sequence combination was represented by within-subject CVs, with within-subject standard deviations calculated according to [13]. Bland-Altman plots were created to visualize the agreement between scans 1 and 2 for subjects’ metabolite measurements [14]. These are shown as the mean between scans 1 and 2 and the percentage difference between the scans.
Figure 1.
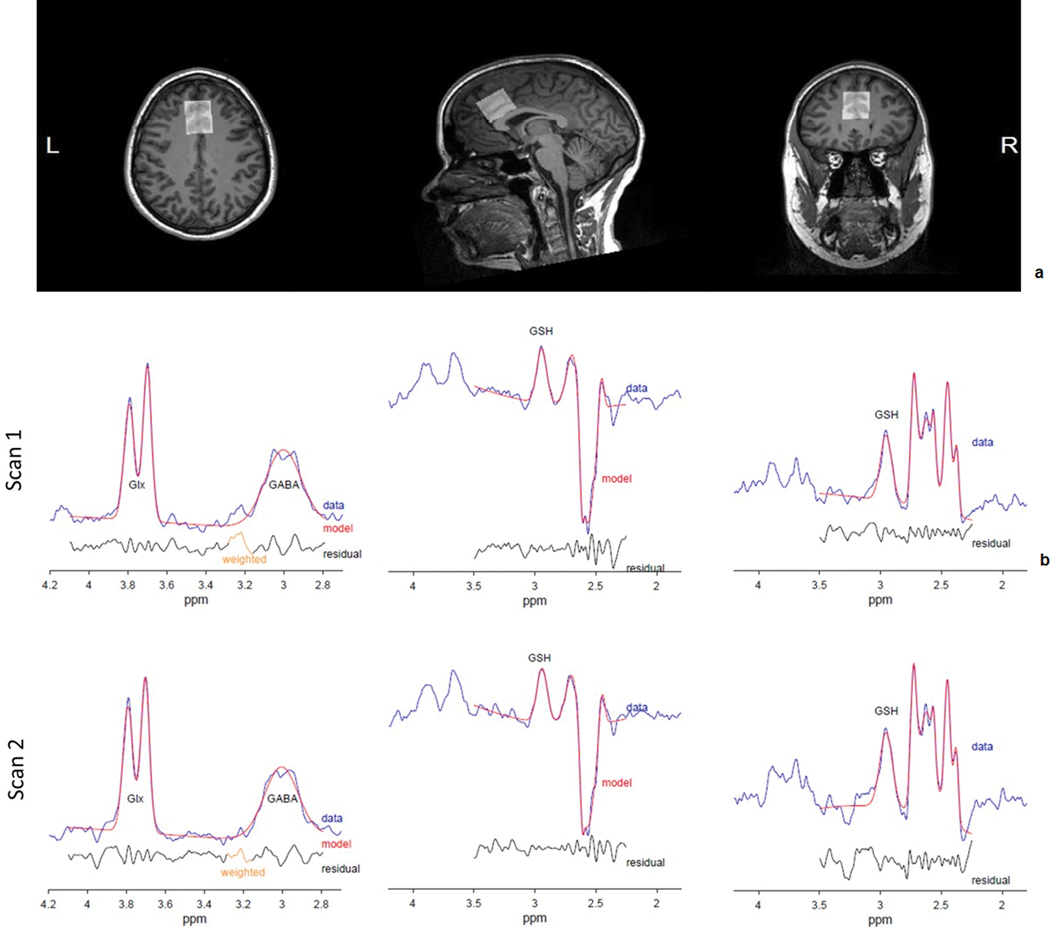
a) Sample dorsal Anterior Cingulate Cortex (dACC) voxel placement, and b) Sample fitted spectra from TE = 80 ms HERMES (Glx, GABA: left; GSH: middle) and TE = 120 ms MEGA-PRESS (GSH: right) by scan (1: top, 2: bottom).
3. RESULTS
Water linewidths were similar across scan sets (TE = 80 ms, scan 1: 8.29 ± 0.49 Hz, scan 2: 8.29 ± 0.38 Hz, t = 0.00, p = 1.00; TE = 120 ms, scan 1: 7.98 ± 0.51 Hz, scan 2: 8.10 ± 0.69 Hz, t = –1.22, p = 0.25). Furthermore, gray matter tissue fractions between scan sets (mean ± s.d., 1: 67.5 ± 3.3%, 2: 67.0 ± 3.3%) were very highly intercorrelated (r = 0.95, p < 0.001), consistent with reliable between-scan-set voxel placement. Figure 1b shows example difference spectra with corresponding model fits from one subject. Table 1 contains summary information regarding metabolite levels and fit errors at each scan, along with between-scan, within-subject %CVs.
Table 1.
Metabolite levels / model fit errors by scan, and between-scan (within-subject) %CV from HERMES / MEGA-PRESS
| Scan 1 | Scan 2 | ||||
|---|---|---|---|---|---|
| Level (I.U.) Mean (SD) | Fit Error (%) Mean (SD) | Level (I.U.) Mean (SD) | Fit Error (%) Mean (SD) | %CV | |
| HERMES (TE = 80 ms) | |||||
| GABA | 3.77 (0.58) | 8.83 (2.27) | 3.91 (0.76) | 7.72 (1.85) | 16.74 |
| Glx | 14.58 (1.48) | 3.59 (0.85) | 14.05 (1.19) | 3.42 (0.79) | 6.36 |
| GSH | 1.90 (0.52) | 7.29 (2.16) | 2.02 (0.47) | 6.36 (2.59) | 19.04 |
| MEGA-PRESS (TE=120 ms) | |||||
| GSH | 3.91 (0.48) | 8.51 (1.69) | 3.83 (0.39) | 8.77 (1.86) | 7.25 |
Note: TE = echo time, I.U. = Institutional Units, CV = Coefficient of Variation, Glx = glutamate+glutamine, GSH = glutathione.
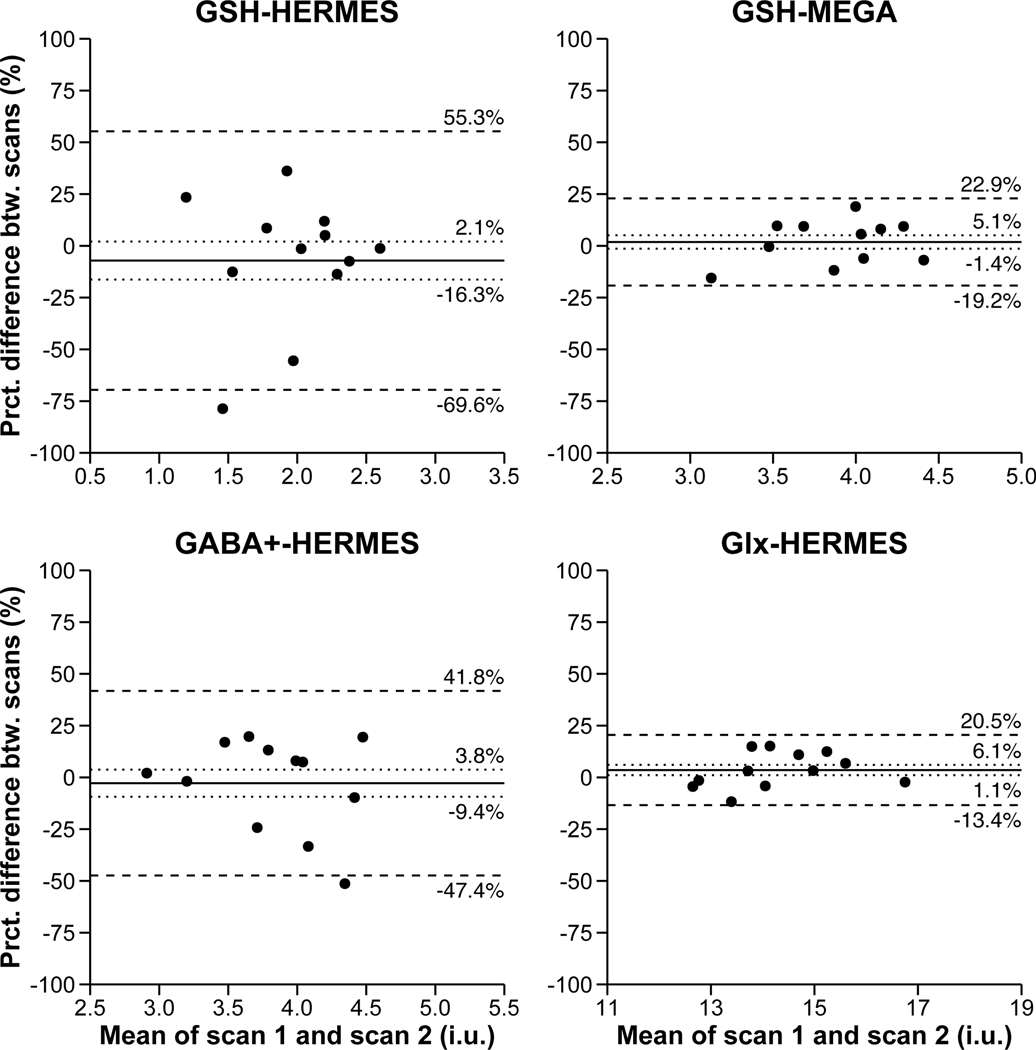
HERMES (TE = 80 ms) produced GABA level estimates with an average CV of 16.7%. GSH estimates acquired at TE = 80 ms via HERMES produced between-scan reliability coefficients that were more than 2.5x the size of those acquired at TE = 120 ms via MEGA-PRESS (CV = 19.0% vs. 7.3%). Although not the focus of the present study, HERMES (TE = 80 ms) produced edited glutamate+glutamine (Glx) level estimates with an average between-scan reliability coefficient (i.e., CV) of 6.4%. Please see Figure 2 for Bland-Altman plots representing the agreement between scans 1 and 2 for subjects’ metabolite measurements.
Figure 2.
Bland-Altman plots showing the agreement between scans 1 and 2 for GSH as measured by HERMES, GSH as measured by MEGA-PRESS, GABA+, and Glx. The solid line is the mean percentage difference, the dotted lines are the 95% confidence intervals of the mean percentage difference, and the dashed lines are the limits of agreement (mean +/− 1.96 standard deviations). The confidence intervals and limits of agreements are also given by each line.
4. DISCUSSION
The primary findings from the present study were that, 1) the HERMES acquisition sequence produced GABA levels with between-scan reliability estimates in line with those reported for MEGA-PRESS editing of GABA [15, 16], and 2) a MEGA-PRESS sequence with TE = 120 ms, optimized for GSH acquisition, produced GSH levels with between-scan reliability estimates that were far (i.e., >2.5x) lower relative to those produced by HERMES editing of GSH with TE = 80 ms. Together, these results lead us to the conclusion that, a) if the primary interest is in measuring GABA for a given study, using the HERMES sequence with TE = 80 ms would be a good choice, as it provides equivalent measurement of GABA relative to standard TE = 68 ms MEGA-PRESS along with measurement of GSH, the brain’s primary antioxidant and a metabolite of growing interest to neuroscience [17]; b) however, if the primary interest is in measuring GSH for a given study, using a standard MEGA-PRESS sequence with TE = 120 ms would be a better sequence choice, as it provides far more robust measurement of GSH, but at the expense of simultaneously measuring GABA. Of course, a HERMES sequence could easily be configured, by simply changing the default sequence-TE, to acquire both GSH and GABA at TE = 120 ms, however, such a sequence would be unlikely to produce robust measurement of GABA levels [18].
Contextualizing the observed GSH reliability estimates in the extant literature is difficult, as very few relevant studies have been published. Nonetheless, at least two studies have shown GSH detection using TE = 120 ms to yield higher SNR than using TE = 68 ms [7, 19]. A recent study suggested that J-difference editing of GSH achieves a more reliable detection of GSH with concentrations in the physiological range than (non-edited) PRESS at short TE [20]. GSH reliability estimates from the present study, using TE = 120 ms, were comparable to reports of those obtained with 2D J-resolved PRESS (i.e., mean CV = 7.5%, [21]. Similarly, a recent study demonstrated linearity and excellent reproducibility (mean CV = 5.4%) between actual and observed GSH levels using a very low TE (i.e., 6.5 ms) “phase-rotated” STEAM sequence, but found that, unlike MEGA-PRESS, STEAM detected positive GSH levels when GSH was completely absent [22]. In contrast to the finding of the present study, they also found in that study that the reproducibility of TE = 120 ms, MEGA-edited GSH (mean CV = 13.5%) was substantially worse than that of STEAM (M CV = 5.4%), as well as that of TE = 120ms, MEGA-edited GSH in the present study (M CV = 6.6%), despite similar study methods. However, the sequence used in that work did not adjust the editing pulse timing for changes in TE, as required for optimal editing [1]. In sum, the results from the present study suggest that J-difference editing of GSH using TE = 120 ms may be the best available sequence for measuring GSH, as it provides reliability estimates very similar to low TE STEAM and PRESS sequences, but, unlike those sequences, correctly detects the absence of GSH when no GSH is present. This latter consideration is likely important to the field of neuropsychiatry, where the disorders of interest are characterized by low brain GSH levels and where 1H MRS-guided evaluation of treatments would need to be able to sensitively detect changes in GSH levels in the very low to normal range [17].
The findings of the present study should also be contextualized relative to its limitations. A more direct comparison between TE = 80 ms HERMES and TE = 120 ms HERMES would have arguably provided a more ideal basis for comparison of GSH levels acquired at different TEs. However, given that GABA cannot be reliably measured at TE = 120 ms, we chose to compare two J-difference editing sequences that would at least be potentially useful to researchers. Alternatively, we could have compared TE = 80 ms MEGA-PRESS and TE = 120 ms MEGA-PRESS, both with selective refocusing pulses configured to edit GSH. However, given that TE = 80 ms MEGA-PRESS would not be used to measure GSH in practice, unless that TE was chosen as a compromise to measure multiple metabolites (as with HERMES), we again chose to compare two sequences that would be used by applied researchers. Importantly, and the reason why HERMES is a useful acquisition sequence is because (by design), the ON editing pulses relevant to GABA in HERMES do not significantly affect the edited GSH signal [5].
These limitations notwithstanding, the present study demonstrated that the HERMES sequence produced GABA estimates with similar reliability to published studies of MEGA-PRESS, along with GSH estimates with reasonable reproducibility, and that J-difference editing of GSH with TEs of 120 ms versus 80 ms produced GSH estimates with >2.5x better reliability. These findings may suggest that TE 120 ms J-difference editing (e.g., MEGA-PRESS) should be used to measure GSH, if GSH is of primary importance to the research question at hand, but that HERMES provides excellent measurement of GABA along with reasonable measurement of GSH, useful particularly if measuring GSH is of secondary importance to the question at hand.
ACKNOWLEDGMENTS
The authors would like to thank James Coatsworth, Shannon Powers, and Scott Henderson for their invaluable assistance in coordinating and executing data acquisition for the present study.
Funding Sources: This study applies tools developed under NIH R01 EB016089, R01 EB023693 and P41 EB015909; RAEE also receives salary support from these grants.
Footnotes
Declarations of Interest: none.
REFERENCES
- [1].de Graaf R. In Vivo NMR Spectroscopy: Principles and Techniques. 2nd ed.: Wiley; 2013. [Google Scholar]
- [2].Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR in biomedicine 1998;11(6):266–72. [DOI] [PubMed] [Google Scholar]
- [3].Harris AD, Saleh MG, Edden RA. Edited (1) H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magnetic resonance in medicine 2017;77(4):1377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magnetic resonance in medicine 2016;76(1):11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Saleh MG, Oeltzschner G, Chan KL, Puts NAJ, Mikkelsen M, Schar M, et al. Simultaneous edited MRS of GABA and glutathione. NeuroImage 2016;142:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Edden RA, Puts NA, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magnetic resonance in medicine 2012;68(3):657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chan KL, Puts NA, Snoussi K, Harris AD, Barker PB, Edden RA. Echo time optimization for J-difference editing of glutathione at 3T. Magnetic resonance in medicine 2017;77(2):498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saleh MG, Rimbault D, Mikkelsen M, Oeltzschner G, Wang AM, Jiang D, et al. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. NeuroImage 2019;189:425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, et al. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magnetic resonance in medicine 2006;55(6):1219–26. [DOI] [PubMed] [Google Scholar]
- [10].Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. Journal of magnetic resonance imaging : JMRI 2014;40(6):1445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mikkelsen M, Saleh MG, Near J, Chan KL, Gong T, Harris AD, et al. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magnetic resonance in medicine 2018;80(1):21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Harris AD, Puts NA, Edden RA. Tissue correction for GABA-edited MRS: Considerations of voxel composition, tissue segmentation, and tissue relaxations. Journal of magnetic resonance imaging : JMRI 2015;42(5):1431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Synek V. Evaluation of the standard deviation from duplicate results. Accreditation and Quality Assurance 2008;13:335–7. [Google Scholar]
- [14].Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1(8476):307–10. [PubMed] [Google Scholar]
- [15].Mikkelsen M, Singh KD, Sumner P, Evans CJ. Comparison of the repeatability of GABA-edited magnetic resonance spectroscopy with and without macromolecule suppression. Magnetic resonance in medicine 2016;75(3):946–53. [DOI] [PubMed] [Google Scholar]
- [16].Geramita M, van der Veen JW, Barnett AS, Savostyanova AA, Shen J, Weinberger DR, et al. Reproducibility of prefrontal gamma-aminobutyric acid measurments with J-edited spectroscopy. NMR in Biomedicine 2011;24:1089–98. [DOI] [PubMed] [Google Scholar]
- [17].Rae CD, Williams SR. Glutathione in the human brain: Review of its roles and measurement by magnetic resonance spectroscopy. Analytical biochemistry 2017;529:127–43. [DOI] [PubMed] [Google Scholar]
- [18].Edden RAE, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T2 in vivo with J-difference editing: Application to GABA at 3 tesla. Journal of Magnetic Resonance Imaging 2011;35:229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].An L, Zhang Y, Thomasson DM, Latour LL, Baker EH, Shen J, et al. Measurement of glutathione in normal volunteers and stroke patients at 3T using J-difference spectroscopy with minimized subtraction errors. Journal of magnetic resonance imaging : JMRI 2009;30(2):263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sanaei Nezhad F, Anton A, Parkes LM, Deakin B, Williams SR. Quantification of glutathione in the human brain by MR spectroscopy at 3 Tesla: Comparison of PRESS and MEGA-PRESS. Magnetic resonance in medicine 2017;78(4):1257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Prescot AP, Renshaw PF. Two-dimensional J-resolved proton MR spectroscopy and prior knowledge fitting (ProFit) in the frontal and parietal lobes of healthy volunteers: assessment of metabolite discrimination and general reproducibility. Journal of magnetic resonance imaging : JMRI 2013;37(3):642–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wijtenburg SA, Near J, Korenic SA, Gaston FE, Chen H, Mikkelsen M, et al. Comparing the reproducibility of commonly used magnetic resonance spectroscopy techniques to quantify cerebral glutathione. Journal of magnetic resonance imaging : JMRI 2019;49(1):176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]