Abstract
The complete mitochondrial genome (mitogenome) of Epicauta impressicornis Pic (Coleoptera: Meloidae) was determined. The circular genome is 15,713-bp long, and encodes 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region (CR). The 13 PCGs start with the typical ATN codon and terminate with the typical stop codon TAA (ND2, ND4L, ND6, ATP6, ATP8, and CYTB), TAG (ND1 and ND3), and T- (COX1, COX2, COX3, ND4, and ND5). The two rRNA genes (rrn12S and rrn16S) are encoded on the minority strand. All tRNAs genes except trnS1 (AGN) are predicted to fold into the typical cloverleaf structure. The longest overlap (10 bp) is observed between ATP8 and ATP6. CR mainly harbors a conserved poly-T stretch (15 bp), a short repeat unit (17 bp), some universal microsatellite-like repeats, and a canonical poly-A tail. Phylogenetic analysis using Bayesian inferences and maximum likelihood based on nucleotide and corresponding amino acid sequences of the 13 PCGs showed that E. impressicornis is closely related to E. chinensis, this relationship is and supported within Cucujiformia belonging to Meloidae (Tenebrionoidea). Our results further confirmed the monophyly of Tenebrionoidea, Lymexyloidea, Curculionoidea, Chrysomeloidea, Cucujoidea, Coccinelloidea, and Cleroidea within Cucujiformia, and revealed the sister relationships of (Cleroidea + Coccinelloidea), (Lymexyloidea + Tenebrionoidea), and ((Chrysomeloidea + Cucujoidea) + Curculionoidea). We believe that the complete mitogenome of E. impressicornis will contribute to further studies on molecular bases for the classification and phylogeny of Meloidae or even Cucujiformia.
Keywords: medicinal insect, mitogenome, meloidae, molecular phylogeny
As a traditional material in Chinese medicine, blister beetles have widely been used to treat various human diseases (e.g., edema, warts, molluscum, furuncles, hemorrhoids, and most types of cancers) because of the beetles’ secretion of cantharidin which is an active toxic sesquiterpenoid (Carrel et al. 1993, Nikbakhtzadeh et al. 2007, National Pharmacopoeia Editorial Board 2010, Kim et al. 2013, Prasad and Verma 2013, Verma and Prasad 2013, Vakharia et al. 2018). Thus, blister beetles are of great medicinal value (National Pharmacopoeia Editorial Board 2010, Wang et al. 2010). The blister beetle Epicauta impressicornis Pic, which belongs to the family Meloidae, has attracted attention in research due to its importance in both pharmacology and agriculture (Pinto 1999, Bologna and Pinto 2002, Bologna et al. 2008, Wang et al. 2010, Du et al. 2016). Beetles of the genus Epicauta (one of many genera in the Meloidae family) show several interesting biological characteristics, such as hypermetamorphosis, adult clustering, and larval semi-parasitism (feeding on grasshopper eggs) with adult herbivory (feeding mainly on the flowers and leaves) (Pinto 1999; Bologna et al. 2008; Shintani et al. 2011; Terao et al. 2012, 2015; Mu and Chen 2014, 2015; Shintani et al. 2017; Liu et al. 2018, 2019). In addition, the larvae and adults show beneficial and harmful effects in agriculture, respectively; larvae can be used for the biological control of grasshoppers, while adults are important pests of crops.
In recent years, the molecular phylogenies of several insect families have been studied in detail; however, there have been few studies on the molecular phylogeny of Meloidae species. Moreover, many studies of E. impressicornis have mainly focused on morphological taxonomy (Bologna and Pinto 2002, Bologna et al. 2008, Giulio et al. 2014, Pan and Bologna 2014, Liu et al. 2016, Liu et al. 2019), ecology and behavior (Shintani et al. 2011;Terao et al. 2012, 2015; Mu and Chen 2014, 2015; Shintani et al. 2017; Liu et al. 2018), molecular biology (Du et al. 2016, 2017; Jie et al. 2016; Wu et al. 2018a, b), and cantharidin characteristics (Jiang et al. 2019). Nevertheless, in the previous molecular phylogenetic studies of this family, Bologna et al. (2005, 2008) analyzed the phylogeny and evolution of Meloidae species in Europe using three gene fragments [COI, 16S ribosomal RNA (rRNA), and ITS2] and Du et al. (2017) studied the phylogenetics of several Meloidae species based on comparative genomics.
In studies of molecular phylogeny of insects, mitochondrial genomes (mitogenomes) play an important role because of their rapid, comprehensive, and evolutionarily low levels of maternal inheritance and recombination (gene rearrangements), which are important for species identification, comparative genomics, population genetics, phylogenetic reconstruction, and evolutionary biology (Zhang and Hewitt 1997; Cameron 2014; Du et al. 2016, 2017). Mitochondrial DNA is a highly compact and a covalently closed circular double-stranded DNA molecule (~14–20 kb); it typically contains 37 genes, including 13 protein-coding genes (PCGs) [ATPase subunits 6 and 8 (ATP6 and ATP8), cytochrome c oxidase subunits I–III (COX1–COX3), cytochrome B (CYTB), and NADH dehydrogenase subunits 1–6 and 4 l (ND1–6 and ND4L)] involved in oxidative phosphorylation and electron transport, two rRNAs, 22 transfer RNAs (tRNAs), and a noncoding control region (CR) of variable length comprising fundamental regulatory elements for transcription and replication (Wolstenholme 1992, Boore 1999). As of May 2019, only nine complete Meloidae mitogenomes had been published in GenBank (namely those of E. aptera Kaszab, E. chinensis Laporte, E. gorhami Marseul, E. tibialis Waterhouse, Hycleus cichorii (Linnaeus), H. marcipoli Pan & Bologna, H. phaleratus (Pallas), Lytta caraganae (Pallas), and Mylabris aulica (Menetries) [Coleoptera: Meloidae]) (Du et al. 2016, 2017; Jie et al. 2016; Wu et al. 2018b). An incomplete mitogenomes of H. chodschenticas (Ballion) (Coleoptera: Meloidae), with many individual genes and incomplete gene fragments, has also been reported (Du et al. 2017). To date, the mitogenome of the pharmacologically and agriculturally important E. impressicornis has not been published. Currently, the general taxonomy of Chinese Meloidae is relatively disorganized due to simplistic species descriptions, lack of detail regarding morphological characteristics, and absence of molecular phylogenetic evidence from Epicauta species. Consequently, medicinal blister beetles are often misidentified in China, which seriously hinders the further development and use of these beetles as medicinal resources (Wang et al. 2010).
In the present study, we sequenced and annotated the complete mitogenome of E. impressicornis. Furthermore, we constructed phylogenetic trees based on the complete mitogenomes of 53 species of the beetle infraorder Cucujiformia, and five species of the infraorder Scarabaeiformia (as outgroups). The results of this study will contribute to understanding based on the characteristics of the E. impressicornis mitogenome, help determine the phylogenetic position of Meloidae, and reveal the phylogenetic relationships of members of Cucujiformia within Coleoptera, and facilitate future studies in all these areas.
Materials and Methods
Sample Collection, DNA Extraction, and Sequencing
Adults of E. impressicornis were collected from Luokun Village (106.58° E, 25.42° N), Luodian, Guizhou Province, China, in May 2018. All specimens were initially identified using morphological characteristics (Liu et al. 2016) and were immediately preserved in absolute ethanol and placed in a refrigerator at −20°C for 1 d in the Institute of Entomology of Guizhou University, before being transferred and stored in a refrigerator at −80°C in fresh absolute ethanol. Total genomic DNA was extracted from the samples using Takara Genome DNA Extraction Kits according to the manufacturer’s instructions (Sangon Biotech, Shanghai, China), and DNA quality was evaluated using 1% agarose gel electrophoresis. The complete mitogenomes were sequenced at OriGene (Beijing, China).
Sequence Assembly and Genome Annotation
Raw data were retrieved and qualified using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/), CLC Genomics Workbench 10.0 (CLC bio, Arhus, Denmark), Cutadapt (http://cutadapt.readthedocs.org/), and Pilon (Walker et al. 2014) using the default settings. Clean data were compared against reference mitochondrial genomes from GenBank (specifically E. aptera, E. chinensis, E. gorhami, and E. tibialis). Mitogenomes were reconstructed using Minimap2 (Li et al. 2018) and Nanopolish (https://github.com/jts/nanopolish) with the default parameters. Annotation of the mitochondrial genomes and comparative analyses were performed using GeSeq (Tillich et al. 2017) and Geneious R10 (Biomatters, Auckland, New Zealand), with BLAST against and related species from GenBank as the reference (Altschul et al. 1990, Kearse et al. 2012). Composition skewness was calculated according to the formulas AT-skew = [A − T] / [A+T] and GC-skew = [G − C] / [G + C] (Perna and Kocher 1995). Nucleotide composition and relative synonymous codon usage (RSCU) were analyzed and calculated using MEGA 7.0 (Kumar et al. 2016). Tandem repeats in the control region (CR) were predicted using the Tandem Repeats Finder program (http://tandem.bu.edu/trf/trf.basic.submit.html). The mitogenome sequence of E. impressicornis was deposited into GenBank under accession number MN082609. Transfer RNAs (tRNA) and secondary structures were identified and predicted using MITOS Web Server (Bernt et al. 2013). The tRNAs were visually inspected and compared with reference mitogenomes.
Phylogenetic Analysis
Phylogenetic analysis included sequences from the mitochondrial genomes of 58 species of Coleoptera, including 53 species of Cucujiformia, and five species of Scarabaeiformia, that were used as outgroups (Supp Table 1 [online only]). Nucleotide and amino acid sequences of 13 mitochondrial genes were aligned using Clustal X 1.83 (Thompson et al. 2017) with the default settings, and the resulting alignments were concatenated into two sets of sequences using SequenceMatrix 1.8 (Vaidya et al. 2011). The nucleotide matrix and amino acid matrix were used for phylogenetic analysis using two methods: Bayesian inference using MrBayes 3.2 (Ronquist et al. 2012) and maximum likelihood using IQ-TREE 1.6.2 (Nguyen et al. 2015). The best-fitting model (GTR + I + G) for the nucleotide sequences was selected using the Akaike information criterion (AIC) in jModeltest 2.1.10 (Darriba et al. 2012). MtArt + I + G + F was found to be the best-fitting model for the amino acid sequences using ProtTest 3.4 (Abascal et al. 2005). In the BI analysis, two runs of 10,000,000 generations were conducted for each matrix. Each set was sampled every 1,000 generations, with a burn-in of 25%. In the ML analysis, the node support values were assessed with 1,000 bootstrap resampling replicates. The resulting phylogenetic trees were visualized in FigTree 1.4.0 (Mousavi et al. 2014).
Results and Discussion
Mitogenome Organization and Base Composition
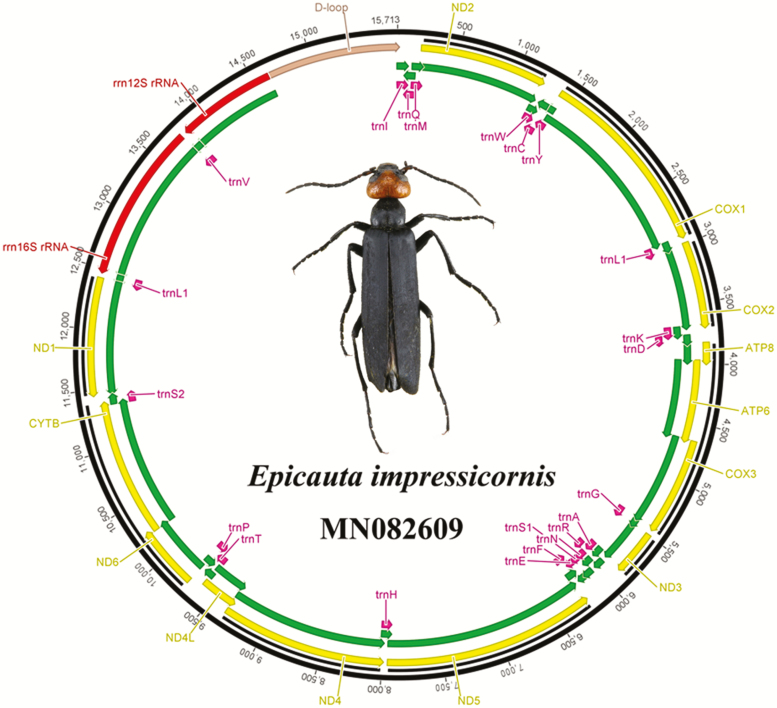
The complete mitogenome of E. impressicornis is 15,713 bp in length and is a typical closed circular DNA molecule. The length of this mitogenome lies between 15,645 bp of E. aptera (Jie et al. 2016) and the 15,717 bp of E. chinensis (Du et al. 2016). The E. impressicornis mitogenome is composed of 37 typical mitochondrial genes including 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a CR. This composition is similar to that observed in many insect mitogenomes: 23 genes (9 PCGs and 14 tRNAs) are encoded on the majority strand, and 4 PCGs, 8 tRNAs, and 2 rRNAs are encoded on the minority strand (Fig. 1, Table 1). The mitogenome gene order and orientation of E. impressicornis were the same those of other previously sequenced Meloidae and were also identical to ancestral arthropod mitogenomes (Bogdanowicz et al. 2000, Li et al. 2016). Specifically, the mitogenome contains 19 overlapped genes (49 bp), and the longest overlap (10 bp) is identical to that reported in other Meloidae species (Du et al. 2016, 2017), with the exception of E. aptera (Jie et al. 2016). Most of the gene overlaps appear in tRNA genes, which may be explained by the lower evolutionary constraints of these genes (Sheffield et al. 2008). Intergenic spacers in the E. impressicornis mitogenome have seven regions ranging from 1 to 17 bp (a total of 45 bp), with the longest region detected between trnS2 (UCN) and ND1 (Table 1). This particular intergenic spacer is similar to that previously reported in other Meloidae species (Du et al. 2016, 2017) and is common in to most Coleopteran mitogenomes (Sheffield et al. 2008, Yuan et al. 2016). Moreover, this intergenic spacer is apparently the basis for transcription termination site recognition using the transcriptional machinery (Sheffield et al. 2008). The nucleotide composition of the E. impressicornis mitogenome is as follows: A = 36.90%, T = 34.00%, G = 11.30%, and C = 17.90%. The A+T and G+C contents are 70.90 and 29.10%, respectively. The AT-skew of the whole mitogenome is positive (0.04)and the GC-skew is negative (−0.23) (Supp Table 2 [online only]); this finding indicates a strand compositional bias characterized by a strong excess of C over G nucleotides and a slight excess of A over T nucleotides, which also corresponds well to the A + T bias generally observed in Meloidae mitogenomes (Du et al. 2017).
Fig. 1.
Gene map of the mitogenome of E. impressicornis. Gene abbreviations are as follows: COX1–3, cytochrome c oxidase subunits 1–3; ATP6 and ATP8, ATP synthase subunits 6 and 8; CYTB, cytochrome B; ND1–6 and ND4L, NADH dehydrogenase subunits 1–6 and 4 L. In addition, trnS1, trnS2, trnL1, and trnL2 denote codons tRNA-Ser (AGN), tRNA-Ser (UCN), tRNA-Leu (CUN), and tRNA-Leu (UUR), respectively.
Table 1.
Summary of the mitogenome of E. impressicornis
| Gene | Strand | Location | Size (bp) | Anticodon | Start codon | Stop codon | Intergenic nucleotides |
|---|---|---|---|---|---|---|---|
| trnI | + | 1–66 | 66 | GAT | |||
| trnQ | − | 63–132 | 70 | TTG | 4(–) | ||
| trnM | + | 132–200 | 69 | CAT | 1(–) | ||
| ND2 | + | 201–1214 | 1,014 | ATA | TAA | ||
| trnW | + | 1,213–1,280 | 68 | TCA | 2(–) | ||
| trnC | − | 1,280–1,343 | 64 | GCA | 1(–) | ||
| trnY | − | 1,347–1,410 | 64 | GTA | 3(+) | ||
| COX1 | + | 1,403–2,945 | 1,543 | ATT | T | 8(–) | |
| trnL2(UUR) | + | 2,946–3,010 | 65 | TAA | |||
| COX2 | + | 3,011–3,698 | 688 | ATA | T | ||
| trnK | + | 3,698–3,770 | 73 | CTT | 1(–) | ||
| trnD | + | 3,769–3,833 | 65 | GTC | 2(–) | ||
| ATP8 | + | 3,843–3,995 | 153 | ATA | TAA | 9 | |
| ATP6 | + | 3,986–4,657 | 672 | ATG | TAA | 10(–) | |
| COX3 | + | 4,657–5,437 | 781 | ATG | T | 1(–) | |
| trnG | + | 5,438–5,501 | 64 | TCC | |||
| ND3 | + | 5,502–5,855 | 354 | ATT | TAG | ||
| trnA | + | 5,854–5,918 | 65 | TGC | 2(–) | ||
| trnR | + | 5,918–5,981 | 64 | TCG | 1(–) | ||
| trnN | + | 5,981–6,045 | 65 | GTT | 1(–) | ||
| trnS1(AGN) | + | 6,046–6,102 | 57 | TCT | |||
| trnE | + | 6,103–6,164 | 62 | TTC | |||
| trnF | − | 6,163–6,226 | 64 | GAA | 2(–) | ||
| ND5 | − | 6,227–7,937 | 1,711 | ATT | T | ||
| trnH | − | 7,937–8,002 | 66 | GTG | 1(–) | ||
| ND4 | − | 8,002–9,334 | 1,333 | ATG | T | 1(–) | |
| ND4L | − | 9,328–9,615 | 288 | ATG | TAA | 7(–) | |
| trnT | + | 9,618–9,680 | 63 | TGT | 2(+) | ||
| trnP | − | 9,680–9,745 | 66 | TGG | 1(–) | ||
| ND6 | + | 9,747–10,238 | 492 | ATT | TAA | 1(+) | |
| CYTB | + | 10,238–11,377 | 1,140 | ATG | TAA | 1(–) | |
| trnS2(UCN) | + | 11,376–11,443 | 68 | TGA | 2(–) | ||
| ND1 | − | 11,461–12,411 | 951 | ATT | TAG | 17(+) | |
| trnL1(CUN) | − | 12,412–12,475 | 64 | TAG | |||
| rrn16S | − | 12,482–13,747 | 1,266 | 6(+) | |||
| trnV | − | 13,755–13,823 | 69 | TAC | 7(+) | ||
| rrn12S | − | 13,824–14,613 | 790 | ||||
| Control region (CR) | 14,614–15,713 | 1,100 |
In the column for intergenic length, a positive sign indicates the interval in base pairs between genes, while the negative sign indicates overlapping base pairs between genes.
Protein-Coding Genes
In the mitogenome of E. impressicornis, the 13 typical PCGs (11,115 bp in length) commonly found in other reported Meloidea species (Du et al. 2016, 2017) and previously described in the Introduction were found, i.e., the seven NADH genes, two ATPase genes, and four cytochrome dehydrogenase genes (Fig. 1, Supp Table 2 [online only]), all of which are commonly found in other reported Meloidea species (Du et al. 2016, 2017). Nine of the 13 PCGs are encoded on the majority strand (COX1, COX2, COX3, CYTB, ND2, ND3, ND6, ATP6, and ATP8) and are on the minority strand (ND1, ND4, ND4L, and ND5) (Fig. 1, Table 1). All 13 PCGs start with a typical ATN start codon. The conventional termination codons TAA and TAG are assigned to six PCGs (ND2, ND4L, ND6, ATP6, ATP8, and CYTB) and two PCGs (ND1 and ND3), respectively, whereas COX1, COX2, COX3, ND4, and ND5 genes show T- as the incomplete stop codon (Table 1). Similar situations have been observed in many other insect species (Boore et al. 2001, Kim et al. 2006, Du et al. 2016), incomplete stop codons may emerge as functional stop codons via polyadenylation processes and polycistronic transcription cleavage; moreover, this may function to minimize gene overlaps and intergenic spacers (Ojala et al. 1981).
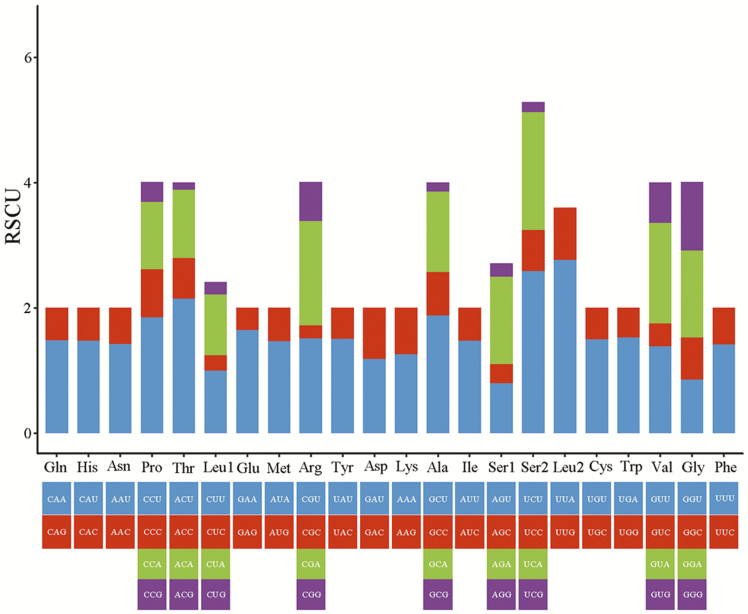
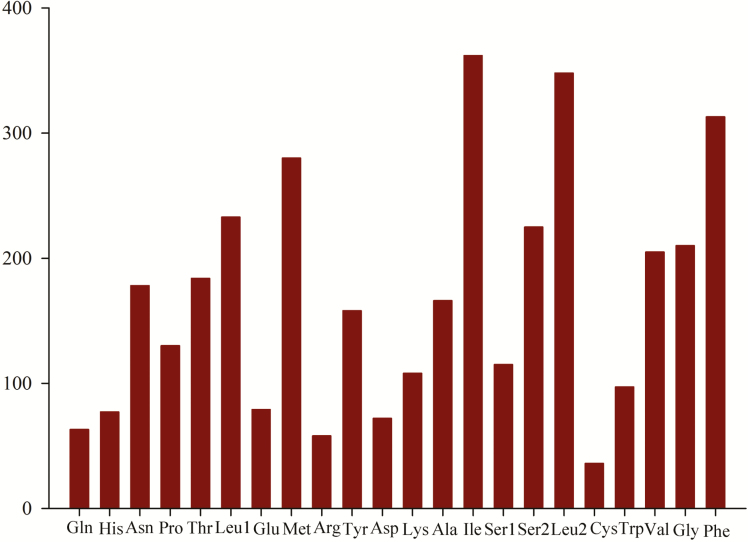
The results of RSCU analysis for the 13 PCGs, comprising 3,705 codons excluding start and stop codons, in the E. impressicornis mitogenome are presented in Fig. 2 and Supp Table 3 [online only]. In addition, codon distribution analysis (Fig. 3, Supp Table 3 [online only]) demonstrated that UUU (222 incidences), UUA (268), AUU (268), and AUA (206) were the most frequently used codon families (they occurred >200 times and accounted for 26.02% of the codons), followed by AAU(127) and UAU (119). All these codons consist of A or T nucleotides, which is indicative of the biased usage of A and T nucleotides in E. impressicornis PCGs. Many other Coleopteran species have a similar pattern of codon usage in their mitogenomes (Kim et al. 2009, Du et al. 2016, Wang et al. 2019). Nine codons in the 13 PCGs (UCG, ACG, GCG, UAA*, UAG*, UGC, CGC, CGG, and AGG) do not exceed 10 instances. Furthermore, the AT and GC-skews), of these PCGs were negative (Supp Tables 2 and 3 [online only]).
Fig. 2.
Relative synonymous codon usage (RSCU) in the mitogenome of E. impressicornis.
Fig. 3.
Amino acid composition of the mitogenome of E. impressicornis.
Transfer and Ribosomal RNAs
The typical set of 22 tRNA genes identified within the E. impressicornis mitogenome, which showed a total length of 1,441 bp, varied from 57 bp (trnS1) to 73 bp (trnK) in length (Table 1 and Supp Table 2 [online only])). The positions of tRNAs in the mitogenome are usually conserved across insect species (Du et al. 2016, 2017; Wang et al. 2019). In E. impressicornis, these genes had an A+T content of 72.90% and a positive AT (0.04) and GC (0.18) skews (Supp Table 2 [online only]). Fourteen tRNA genes are encoded on the majority strand and eight on the minority strand. Anticodons of all of the tRNA genes were identical to those found in other Meloidae mitogenomes (Du et al. 2016, 2017; Jie et al. 2016). Most of the identified tRNA genes had a typical cloverleaf secondary structure, with the exception of trnS1 (AGN), which had lost the dihydrouridine (DHU) arm (Supp Fig. 1 [online only]). Similarly, in many non-Meloidae insect mitogenomes, trnSer (AGN) lacks the DHU arm and cannot fold into the emblematic cloverleaf structure; thus, this is a common phenomenon (Wolstenholme 1992; Cameron 2014; Du et al. 2016, 2017; Jie et al. 2016; Wang et al. 2019). However, increasing variation is usually accompanied more compensatory base changes in stems, which results in tRNAs that are more or less unconserved (Du et al. 2017).
Two rRNA genes were discovered on the minority strand (rrn12S and rrn16S) of the E. impressicornis mitogenome; these genes are typically observed in other Meloidae species and in many other insect mitogenomes (Du et al. 2016, 2017; Wang et al. 2019). In total, these rRNA genes were 2,056 bp in length: rrn16S (1,266 bp in length) was found to be situated between trnL1 (CUN) and trnV, whereas rrn12S (790 bp in length) was located between trnV and the CR (Table 1). The gene rrn16S was assumed to supplement the blanks between trnL1 (CUN) and trnV (Fig. 1). The terminal region of the rRNA genes may have extended to the boundaries of flanking genes because it was difficult to accurately resolve by DNA sequence alone (Boore 2001, 2006). The boundary between rrn12S and the putative CR can be determined by an alignment with homologous sequences in other Meloidae insects (Boore and Brown 2000; Jie et al. 2016; Du et al. 2016, 2017). For rRNAs in E. impressicornis, the AT-skew was negative (−0.06) and the GC-skew was positive (0.35), with a high A+T content in the mitogenome (74.60%) (Supp Table 2 [online only]). The locations of rrn12S and rrn16S in the E. impressicornis mitogenome each correspond to those in the mitogenomes of other known Meloidae species (Supp Table 4 [online only]); this finding demonstrates that rRNAs are conserved in Meloidae and possess sites that might have played an important role in phylogenetic evolution (Du et al. 2016, 2017; Jie et al. 2016).
Control Region
The mitochondrial CR is involved in the initiation and regulation of replication and transcription in insects (Zhang et al. 1995, Zhang and Hewitt 1997, Fernandez-Silva et al. 2003). The length of the CR in the E. impressicornis mitogenome, which was 1,100 bp, lies between the lengths of L. caraganae (1,015 bp) and M. aulica (1,139 bp) (Supp Table 4 [online only]). It was found to be located between rrn12S and trnI. With 884 bp, it exhibited the highest A+T content (80.36%) of the entire of E. impressicornis mitogenome, which is in agreement with other Meloidae mitogenomes. The overall AT-skew and the GC-skew in the CR were 0.04 and −0.18, respectively (Fig. 1, Supp Table 2 [online only]). Since high A+T content limits the utility of genes as molecular markers, it is possible that the PCGs might be more variable than the CR (Zhang and Hewitt 1997, Du et al. 2016). A conserved poly-T stretch (15 bp) was found in the CR of the E. impressicornis mitogenome, and it was comparable to that in other Meloidae mitogenomes (Du et al. 2016, Jie et al. 2016); this has been reported to function as a possible recognition site for the origin of the DNA replication minor strand (Zhang and Hewitt 1997, Andrews et al. 1999, Saito et al. 2005, Kim et al. 2009). A tandem repeat sequence was identified centrally in the E. impressicornis CR with the consensus pattern ATTAATAATGATTATAT (17 bp), which was repeated by base insertions/deletions/mismatches. Given that other published Meloidae mitogenomes do not have tandem repeats, this feature might provide a useful molecular marker for E. impressicornis in relation to Meloidae species identification (except in phylogeographic and population genetics studies) (Zhang and Hewitt 1997; Du et al. 2016, 2017). Microsatellite-like elements such as (AT)3 and (AT)8 also appeared in the CR between the tandem repeat sequence and trnI. In addition, as in other Meloidae mitogenomes (Du et al. 2016), poly-A was identified at the end of the CR (Supp Fig. 2 [online only]).
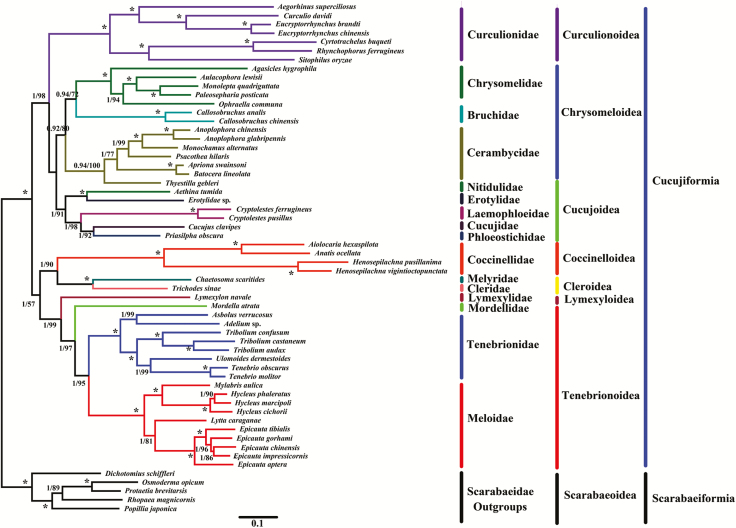
Phylogenetic Analysis
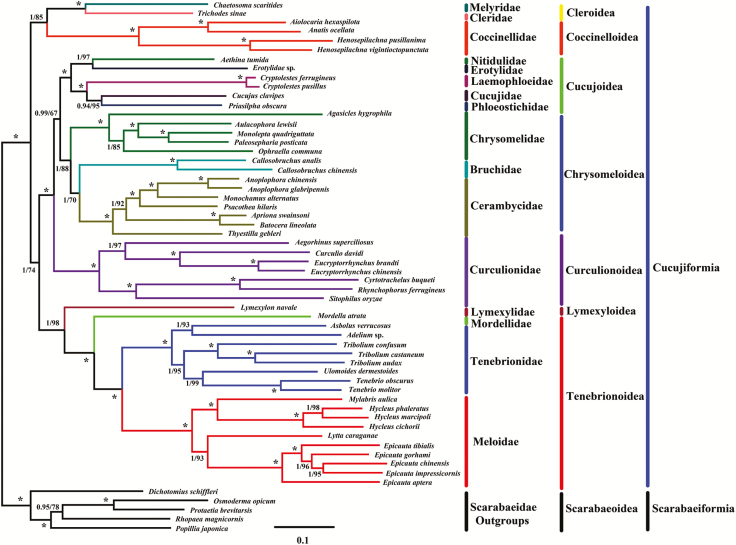
In the present study, phylogenetic analyses were based on the nucleotide and amino acid sequences of the 13 PCGs from the complete mitogenomes of 53 species of Cucujiformia (Figs. 4 and 5). Previous phylogenetic studies have shown that different datasets and inference methods affect the topological structure of Cucujiformia (Beutel and Haas 2000, Timmermans et al. 2015, Yuan et al. 2016). Thus, similarities and differences exist in different phylogenetic trees.
Fig. 4.
Phylogeny of E. impressicornis. The phylogenetic tree was inferred from the nucleotide sequences of 13 PCGs in the mitogenome using Bayesian Inferences (BI) and Maximum Likelihood (ML) methods. Numbers on branches indicate BI posterior probability (PP, left) and ML bootstrap support (BS, right). An asterisk indicates PP =1.0 and BS = 100.
Fig. 5.
Phylogeny of E. impressicornis. The phylogenetic tree was inferred from the amino acid sequences of the 13 PCGs in mitogenome using Bayesian Inferences (BI) and Maximum Likelihood (ML) methods. Numbers on branches indicate BI posterior probability (PP, left) and ML bootstrap support (BS, right). An asterisk indicates PP =1.0 and BS = 100.
Here, BI trees provided significantly higher support values than ML trees in the same dataset, especially at branches that involved the superfamily-level relationships of the 13 PCG nucleotide sequences at the support values (PP/BS = 0.99/67) of (Chrysomeloidea + Cucujoidea) (Fig. 4) and the 13 PCGs amino acid sequences at the support values (PP/BS = 1/57) of (Coccinelloidea + Cleroidea) + (Lymexyloidea + Tenebrionoidea) (Fig. 5). However, the lack of resolution at some nodes affected the tree topology; these results might have been due to limited taxon sampling, which is one of the most important determinants of accurate phylogenetic inference, particularly in species-rich lineages (Zwick and Hillis 2002, Rosenberg 2007).
Unsurprisingly, E. impressicornis clustered with the genus Epicauta in Meloidae. In particular, E. impressicornis had a close relationship with E. chinensis. Similar to previous findings (Du et al. 2017), phylogenetic analyses among representative members of Meloidae resulted in two major clades: Epicauta and Lytta formed one clade (a sister group) while Hylceu and Mylabris formed the other. At the family level, Meloidae, Tenebrionidae, Mordellidae, and Lymexylidae formed one clade; Cleridae and Melyridae formed a sister group; while Cerambycidae, Chrysomelidae, and Bruchidae formed the final clade. These results were congruent with previous phylogenetic studies using mitogenomes (Timmermans et al. 2015, Yuan et al. 2016).
At the superfamily level, the phylogenetic relationships in Cucujiformia were somewhat ambiguous. The phylogenetic topologies of Coccinelloidea, Cleroidea, Curculionoidea, Chrysomeloidea, Cucujoidea, Lymexyloidea, and Tenebrionoidea were well-established and formed their respective clades. Three sister groups were identified, (Cleroidea + Coccinelloidea), (Lymexyloidea + Tenebrionoidea), and ((Chrysomeloidea + Cucujoidea) + Curculionoidea), which were consistent with previous phylogenetic research using the mitogenomes (Timmermans et al. 2010, Gunter et al. 2014, Timmermans et al. 2015, Yuan et al. 2016). However, the position of (Coccinellidea + Cleroidea) was not stable. For example, in the nucleotide sequence analysis, (Coccinelloidea + Cleroidea) and (Lymexyloidea + Tenebrionoidea) collectively constituted a clade (Fig. 4). However, in amino acid sequence analysis (Coccinelloidea + Cleroidea) and ((Lymexyloidea + Tenebrionoidea) + ((Chrysomeloidea + Cucujoidea) + Curculionoidea)) comprised a clade (Fig. 5). Thus, the phylogenetic relationships in Cucujiformia were distinctive in the nucleotide sequence analysis (Fig. 4) and the amino acid sequence analysis (Fig. 5). Although this finding was inconsistent with previous mitogenomes-based phylogenetic research (Timmermans et al. 2015, Yuan et al. 2016), (Lymexyloidea + Tenebrionoidea) and ((Cucujoidea + Chrysomeloidea) + Curculionoidea) could be well clustered into a sole branch. As previously reported in mitogenomic studies, there are some well-supported populations and some inconsistent relationships among major lineages in Cucujiformia. Cucujiformia is a diverse clade and contains the highest abundance of polyphaga species in Coleoptera (Sheffield et al. 2009, Bocak et al. 2014, Timmermans et al. 2015, Du et al. 2016, Li et al. 2016). Consequently, it remains a challenge to construct a well-supported Cucujiformia system.
At present, a successful analysis of the phylogenetic relationships in Cucujiformia based on either morphological or molecular characteristic does not exist. Indeed, many phylogenetic studies of Coleoptera have failed to provide strong support or to identify and infer phylogenetic relationships within Cucujiformia (Timmermans et al. 2015, Yuan et al. 2016). The phylogenetic relationships reported in the present study differed from those reported in previous phylogenetic studies; this might be due to differences in datasets (Marvaldi et al. 2008, Kim et al. 2009, Coates 2014) and/or the inconsistency among different mitochondrial regions detected in diverse species (Meiklejohn et al. 2014). Nevertheless, our study will be useful for providing a molecular basis for the classification and phylogeny of Cucujiformia. Considering the diversity of Cucujiformia and the limitations of the current molecular information, the accurate phylogeny within Cucujiformia will require more molecular markers (either from mitochondrial data or nuclear segments) and additional samples.
Supplementary Material
Acknowledgments
We sincerely appreciate Guo-yong Li (Guiyang University) and Zhao Pan (Hebei University) for their help in sample collection and identification, respectively. This research was supported by National Natural Science Foundation of China (81460576), the Program of Science and Technology Innovation Talents Team, Guizhou Province, China (20144001), and the Program of Excellent Innovation Talents, Guizhou Province, China (20154021).
References Cited
- Abascal F., Zardoya R., and Posada D.. . 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J.. . 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews R. M., Kubacka I., Chinnery P. F., Lightowlers R. N., Turnbull D. M., and Howell N.. . 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23: 147. [DOI] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., and Stadler P. F.. . 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69: 313–319. [DOI] [PubMed] [Google Scholar]
- Beutel R., and Haas F.. . 2000. Phylogenetic relationships of the suborders of Coleoptera (Insecta). Cladistics 16: 103–141. [DOI] [PubMed] [Google Scholar]
- Bocak L., Barton C., Crampton-Platt A., Chesters D., Ahrens D., and Vogler A. P.. . 2014. Building the Coleoptera tree-of-life for >8000 species: composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 39: 97–110. [Google Scholar]
- Bogdanowicz S. M., Schaefer P. W., and Harrison R. G.. . 2000. Mitochondrial DNA variation among worldwide populations of gypsy moths, Lymantria dispar. Mol. Phylogenet. Evol. 15: 487–495. [DOI] [PubMed] [Google Scholar]
- Bologna M. A., and Pinto J. D.. . 2002. The Old World genera of Meloidae (Coleoptera): a key and synopsis. J Nat. Hist. 36: 2013–2102. [Google Scholar]
- Bologna M. A., D’Inzillo B., Cervelli M., Oliverio M., and Mariottini P.. . 2005. Molecular phylogenetic studies of the Mylabrini blister beetles (Coleoptera, Meloidae). Mol. Phylogenet. Evol. 37: 306–311. [DOI] [PubMed] [Google Scholar]
- Bologna M. A., Oliverio M., Pitzalis M., and Mariottini P.. . 2008. Phylogeny and evolutionary history of the blister beetles (Coleoptera, Meloidae). Mol. Phylogenet. Evol. 48: 679–693. [DOI] [PubMed] [Google Scholar]
- Boore J. L. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol. Phylogenet. Evol. 18: 1413–1416 [DOI] [PubMed] [Google Scholar]
- Boore J. L. 2006. The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda). BMC Genomics. 7: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L., and Brown W. M.. . 2000. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 17: 87–106. [DOI] [PubMed] [Google Scholar]
- Cameron S. L. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 59: 95–117. [DOI] [PubMed] [Google Scholar]
- Carrel J. E., McCairel M. H., Slagle A. J., Doom J. P., Brill J., and McCormick J. P.. . 1993. Cantharidin production in a blister beetle. Experientia. 49: 171–174. [DOI] [PubMed] [Google Scholar]
- Coates B. S. 2014. Assembly and annotation of full mitochondrial genomes for the corn rootworm species Diabrotica virgifera virgifera and Diabrotica barberi (Insecta: Coleoptera: Chrysomelidae) using Next Generation Sequence data. Gene 542: 190e197. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., and Posada D.. . 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., He S., Song X., Liao Q., Zhang X., and Yue B.. . 2016. The complete mitochondrial genome of Epicauta chinensis (Coleoptera: Meloidae) and phylogenetic analysis among Coleopteran insects. Gene. 578: 274–280. [DOI] [PubMed] [Google Scholar]
- Du C., Zhang L., Lu T., Ma J., Zeng C., Yue B., and Zhang X.. . 2017. Mitochondrial genomes of blister beetles (Coleoptera, Meloidae) and two large intergenic spacers in Hycleus genera. BMC Genomics. 18: 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Silva P., Enriquez J. A., and Montoya J.. . 2003. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 88: 41–56. [DOI] [PubMed] [Google Scholar]
- Giulio A. D., Carosi M., Khodaparast R., and Bologna M. A.. . 2014. Morphology of a new blister beetle (Coleoptera, Meloidae) larval type challenges the evolutionary trends of phoresy-related characters in the genus Meloe. Entomologia 2: 69–79. [Google Scholar]
- Gunter N. L., Levkaničová Z., Weir T. H., Ślipiński A., Cameron S. L., and Bocak L.. . 2014. Towards a phylogeny of the Tenebrionoidea (Coleoptera). Mol. Phylogenet. Evol. 79: 305–312. [DOI] [PubMed] [Google Scholar]
- Jiang M., Lü S. M., Qi Z. Y., and Zhang Y. L.. . 2019. Characterized cantharidin distribution and related gene expression patterns in tissues of blister beetles, Epicauta chinensis. Insect Sci. 26: 240–250. [DOI] [PubMed] [Google Scholar]
- Jie H., Lei M. Y., Li P. M., Feng X. L., Zeng D. J., Zhao G. J., Zhu J. B., Zhang C. L., Yu M., Huang Y., . et al. 2016. The complete nucleotide sequence of the mitochondrial genome of Epicauta aptera Kaszab. Mitochondrial DNA B 1: 489–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., . et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Lee E. M., Seol K. Y., Yun E. Y., Lee Y. B., Hwang J. S., and Jin B. R.. . 2006. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol. Biol. 15: 217–225. [DOI] [PubMed] [Google Scholar]
- Kim K. G., Hong M. Y., Kim M. J., Im H. H., Kim M. I., Bae C. H., Seo S. J., Lee S. H., and Kim I.. . 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol. Cells. 27: 429–441. [DOI] [PubMed] [Google Scholar]
- Kim Y. M., Ku M. J., Son Y. J., Yun J. M., Kim S. H., and Lee S. Y.. . 2013. Anti-metastatic effect of cantharidin in A549 human lung cancer cells. Arch. Pharm. Res. 36: 479–484. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., and Tamura K.. . 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34: 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Ou J., Wei Z. M., Li Y. X., and Tian Y. F.. . 2016. The mitogenomes of three beetles (Coleoptera: Polyphaga: Cucujiformia): new gene rearrangement and phylogeny. Biochem. Syst. Ecol. 69: 101–107. [Google Scholar]
- Liu Y. Y., Pan Z., and Chen X. S.. . 2016. Comparative morphological study on five medicinal Epicauta species from Guizhou Province. Sichuan J. Zool. 35: 84–92. [Google Scholar]
- Liu Y. Y., Li G. Y., Yang L., Chi H., and Chen X. S.. . 2018. Demography and mass rearing of the medicinal blister beetle Epicauta impressicornis (Pic) (Coleoptera: Meloidae) at different temperatures. J. Econ. Entomol. 111: 2364–2374. [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Li G. Y., Yang L., and Chen X. S.. . 2019. Observation of fine structure of antennal sensilla in sexually dimorphic antennae of the blister beetle Epicauta impressicornis (Coleoptera: Meloidae) adults. Acta Entomol. Sin. 62: 442–452. [Google Scholar]
- Marvaldi A. E., Duckett C. N., Kjer K. M., and Gillespie J. J.. . 2008. Structural alignment of 18S and 28S rDNA sequences provides insights into phylogeny of Phytophaga (Coleoptera: Curculionoidea and Chrysomeloidea). Zool. Scr. 38: 63e77. [Google Scholar]
- Meiklejohn K. A., Danielson M. J., Faircloth B. C., Glenn T. C., Braun E. L., and Kimball R. T.. . 2014. Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol. Phylogenet. Evol. 78: 314e323. [DOI] [PubMed] [Google Scholar]
- Mousavi S. A., Österman J., Wahlberg N., Nesme X., Lavire C., Vial L., Paulin L., de Lajudie P., and Lindström K.. . 2014. Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst. Appl. Microbiol. 37: 208–215. [DOI] [PubMed] [Google Scholar]
- Mu Y. L., and Chen X. S.. . 2014. Reproductive habit of Epicauta impressicornis. Guizhou Agricul. Sci. 42: 49–52. [Google Scholar]
- Mu Y. L., and Chen X. S.. . 2015. Studying the relationship between feeding and growing development of Epicauta impressicornis Pic larva. J. Environ. Entomol.37: 407–411. [Google Scholar]
- National Pharmacopoeia Editorial Board. 2010. The pharmacopoeia of the people’s Republic of China, Vol. 1 China Medical Science and Technology Press, Beijing, China. [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A., and Minh B. Q.. . 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikbakhtzadeh M. R., Dettner K., Boland W., Gäde G., and Dötterl S.. . 2007. Intraspecific transfer of cantharidin within selected members of the family Meloidae (Insecta: Coleoptera). J. Insect Physiol. 53: 890–899. [DOI] [PubMed] [Google Scholar]
- Ojala D., Montoya J., and Attardi G.. . 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290: 470–474. [DOI] [PubMed] [Google Scholar]
- Pan Z., and Bologna M. A.. . 2014. Taxonomy, bionomics and faunistics of the nominate subgenus of Mylabris Fabricius, 1775, with the description of five new species (Coleoptera: Meloidae: Mylabrini). Zootaxa. 3806: 1, 3–1,78. [DOI] [PubMed] [Google Scholar]
- Perna N. T., and Kocher T. D.. . 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41: 353–358. [DOI] [PubMed] [Google Scholar]
- Pinto J. D. 1999. The new world genera of Meloidae (Coleoptera): a key and synopsis. J. Nat. Hist. 33: 569–620. [Google Scholar]
- Prasad S. B., and Verma A. K.. . 2013. Cantharidin-mediated ultrastructural and biochemical changes in mitochondria lead to apoptosis and necrosis in murine Dalton’s lymphoma. Microsc. Microanal. 19: 1377–1394. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., and Huelsenbeck J. P.. . 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A. 2007. Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution. 61: 317–323. [DOI] [PubMed] [Google Scholar]
- Saito S., Tamura K., and Aotsuka T.. . 2005. Replication origin of mitochondrial DNA in insects. Genetics. 171: 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield N. C., Song H., Cameron S. L., and Whiting M. F.. . 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 25: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield N. C., Song H., Cameron S. L., and Whiting M. F.. . 2009. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 58: 381–394. [DOI] [PubMed] [Google Scholar]
- Shintani Y., Hirose Y., and Terao M.. . 2011. Effects of temperature, photoperiod and soil humidity on induction of pseudopupal diapauses in the bean blister beetle, Epicauta gorhami. Physiol. Entomol. 36: 14–20. [DOI] [PubMed] [Google Scholar]
- Shintani Y., Terao M., and Tanaka S.. . 2017. Adaptive significance of precocious pupation in the bean blister beetle, Epicauta gorhami (Coleoptera: Meloidae), a hypermetamorphic insect. J. Insect Physiol. 99: 107–112. [DOI] [PubMed] [Google Scholar]
- Terao M., Hirose Y., and Shintani Y.. . 2012. Effects of temperature and photoperiod on termination of pseudopupal diapause in the bean blister beetle, Epicauta gorhami. J. Insect Physiol. 58: 737–742. [DOI] [PubMed] [Google Scholar]
- Terao M., Hirose Y., and Shintani Y.. . 2015. Food-availability dependent premature metamorphosis in the bean blister beetle, Epicauta gorhami (Coleoptera: Meloidae), a hypermetamorphic insect that feeds on grasshopper eggs in the larval stage. Entomol. Sci. 18: 85–93. [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., and Higgins D. G.. . 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 173–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E. S., Fischer A., Bock R., and Greiner S.. . 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45: W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M. J., Dodsworth S., Culverwell C. L., Bocak L., Ahrens D., Littlewood D. T., Pons J., and Vogler A. P.. . 2010. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 38: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M. J. T. N., Barton C., Haran J., Ahrens D., Culverwell C. L., Ollikainen A., Dodsworth S., Foster P. G., Bocak L., and Vogler A. P.. . 2015. Family-level sampling of mitochondrial genomes in coleoptera, compositional heterogeneity and phylogenetics. Genome Biol. Evol. 8: 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G., Lohman D. J., and Meier R.. . 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. [DOI] [PubMed] [Google Scholar]
- Vakharia P. P., Chopra R., Silverberg N. B., and Silverberg J. I.. . 2018. Efficacy and safety of topical cantharidin treatment for molluscum contagiosum and warts: a systematic review. Am. J. Clin. Dermatol. 19: 791–803. [DOI] [PubMed] [Google Scholar]
- Verma A. K., and Prasad S. B.. . 2013. Changes in glutathione, oxidative stress and mitochondrial membrane potential in apoptosis involving the anticancer activity of cantharidin isolated from redheaded blister beetles, Epicauta hirticornis. Anticancer. Agents Med. Chem. 13: 1096–1114. [DOI] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C. A., Zeng Q., Wortman J., Young S. K., . et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. Plos One. 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. P., Pan Z., and Ren G. D.. . 2010. The Chinese genera of Meloidae (Coleoptera: Tenebrionoidea). Entomotaxonomia 32: 43–52. [Google Scholar]
- Wang Q. Q., and Guang H. T.. . 2019. The mitochondrial genomes of two walnut pests, Gastrolina depressa depressa and G. depressa thoracica (Coleoptera: Chrysomelidae), and phylogenetic analyses. PeerJ 6: e4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme D. R. 1992. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141: 173–216. [DOI] [PubMed] [Google Scholar]
- Wu Y. M., Li J., and Chen X. S.. . 2018a. Draft genomes of two blister beetles (genus: Hycleus) harvested for the potent blistering agent cantharidin. Gigascience 7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. M., Liu Y. Y., and Chen X. S.. . 2018b. The complete mitochondrial genomes of Hycleus cichorii and Hycleus phaleratus (Coleoptera: Meloidae). Mitochondrial DNA Part B 3: 159–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M. L., Zhang Q. L., Zhang L., Guo Z. L., Liu Y. J., Shen Y. Y., and Shao R.. . 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol. Phylogenet. Evol. 104: 99–111. [DOI] [PubMed] [Google Scholar]
- Zhang D. X., and Hewitt G. M.. . 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 25: 99–120. [Google Scholar]
- Zhang D. X., Szymura J. M., and Hewitt G. M.. . 1995. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 40: 382–391. [DOI] [PubMed] [Google Scholar]
- Zwick D. J., and Hillis D. M.. . 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 51: 588–598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.