Summary
Background
Middle East respiratory syndrome coronavirus (MERS-CoV) is a zoonotic infection causing severe viral pneumonia, with index cases having resided in or recently travelled to the Arabian peninsula, and is a global concern for public health. Limited human-to-human transmission, leading to some case clusters, has been reported. MERS-CoV has been reported in dromedary camels but phenotypic characterisation of such viruses is limited. We aimed to compare MERS-CoV isolates from dromedaries in Saudi Arabia and Egypt with a prototype human MERS-CoV to assess virus replication competence and cell tropism in ex-vivo cultures of human bronchus and lung.
Methods
We characterised MERS-CoV viruses from dromedaries in Saudi Arabia and Egypt and compared them with a human MERS-CoV reference strain. We assessed viral replication kinetics and competence in Vero-E6 cells (rhesus monkey), tissue tropism in cultures of ex-vivo human bronchial and lung tissues, and cytokine and chemokine induction, gene expression, and quantification of viral RNA in Calu-3 cells (human respiratory tract). We used mock-infected tissue as negative controls for ex-vivo experiments and influenza A H5N1 as a positive control for cytokine and chemokine induction experiments in Calu-3 cells.
Findings
We isolated three dromedary strains, two from Saudi Arabia (Dromedary/Al-Hasa-KFU-HKU13/2013 [AH13] and Dromedary/Al-Hasa-KFU-HKU19D/2013 [AH19D]), and one from Egypt (Dromedary/Egypt-NRCE-HKU270/2013 [NRCE-HKU270]). The human and dromedary MERS-CoV strains had similar viral replication competence in Vero-E6 cells and respiratory tropism in ex-vivo cultures of the human respiratory tract, and had similar ability to evade interferon responses in the human-respiratory-tract-derived cell line Calu-3.
Interpretation
The similarity of virus tropism and replication competence of human and dromedary MERS-CoV from the Arabian peninsula, and genetically diverse dromedary viruses from Egypt, in ex-vivo cultures of the human respiratory tract suggests that dromedary viruses from Saudi Arabia and Egypt are probably infectious to human beings. Exposure to zoonotic MERS-CoV is probably occurring in a wider geographical region beyond the Arabian peninsula.
Funding
King Faisal University, Egyptian National Research Centre, Hong Kong Food and Health Bureau, National Institute of Allergy and Infectious Diseases, and European Community Seventh Framework Program.
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is a disease of public health concern globally, with 837 laboratory-confirmed cases and 291 deaths reported to WHO as of July 23, 2014.1 Index-case patients have all resided in or recently travelled to the Arabian peninsula or adjacent countries. Travel-associated cases or secondary transmission arising from such cases have been reported in Europe, North America, Africa, and Asia.2 Some case clusters have been associated with limited human-to-human transmission occurring within family or health-care settings, and the remaining sporadic cases are presumed to be zoonotic in origin.3, 4 A recent increase in reported cases from Saudi Arabia associated with health-care facilities is a particular cause of concern.3
Establishment of the proximate zoonotic source of human infection and modes of transmission from animals to human beings remains crucial for public health. MERS-CoV has been detected in dromedary camels (Camelus dromedarius), in association with and preceding human infection, and in dromedary slaughterhouses and farms.5, 6, 7, 8, 9 Viruses from most of the recent human cases from the Arabian peninsula phylogenetically cluster within clade B, but the earliest human MERS-CoV strains from 2012 are genetically distinct and are denoted as clade A.10 Dromedary MERS-CoV strains detected in the Arabian peninsula in 2012–13 were clade B viruses, phylogenetically related to viruses from human cases from the same regions, but genetically diverse (non-clade A/B) viruses were reported from Egypt.11 Researchers have also reported serological evidence collected between 2009 and 2013 of MERS-CoV infection in dromedary camel serum samples from Tunisia, Nigeria, Ethiopia, and Egypt, suggesting that MERS-CoV or antigenically related viruses might be geographically more widespread than is appreciated at present.6, 12 Findings from two studies of 226 people in Saudi Arabia and 179 people in Egypt working in dromedary abattoirs showed that all remained seronegative for MERS-CoV, suggesting that transmission to humans is uncommon11, 13 and raising the question of whether dromedary MERS-CoV strains have the capacity to infect human beings. Full genome sequences of dromedary viruses suggest that these viruses are similar to viruses in humans.11, 14 There were some aminoacid differences between the human and dromedary viruses, including in the genetic sequence of the spike protein (a major determinant of host specificity), but they did not affect the receptor binding interface with the cell receptor DPP4.11, 14 However, because a single aminoacid change can profoundly affect host range and because the host range determinants of MERS-CoV are not well defined, dromedary viruses need to be phenotypically characterised in a physiologically relevant manner.15 So far, few virus isolates from dromedaries have been available for biological characterisation and direct evidence for replication competence of dromedary MERS-CoV in human respiratory tissues has been lacking. The only biological characterisation of a dromedary MERS-CoV virus so far was the finding of DPP4-receptor-dependent virus replication in Huh-7 cells,16 a transformed human hepatoma cell line physiologically unrelated to the human respiratory tract.
We aimed to compare MERS-CoV isolates from dromedaries in Saudi Arabia and Egypt with the prototype human MERS-CoV EMC strain to assess virus replication competence and cell tropism in ex-vivo cultures of human bronchus and lung.
Methods
Viruses
We used a human MERS-CoV strain EMC (hMERS-CoV) provided by R A M Fouchier (Erasmus University Medical Center, Rotterdam, Netherlands). We prepared the EMC virus stock as previously described.17
Three dromedary camel strains, two from Saudi Arabia (Dromedary/Al-Hasa-KFU-HKU13/2013 [AH13] and Dromedary/Al-Hasa-KFU-HKU19D/2013 [AH19D]) and one from Egypt (Dromedary/Egypt-NRCE-HKU270/2013 [NRCE-HKU270]), were prepared. The dromedary camel MERS-CoV isolates AH13 and AH19D had been isolated in Vero-E6 cells (ATCC, CRL-1586, Manassas, VA, USA) from nasal (AH13) and faecal (AH19D) swabs, collected from dromedaries in Al-Hasa, Saudi Arabia, as previously described.14 The detection of dromedary camel MERS-CoV NRCE-HKU270 from a nasal swab of a dromedary in an abattoir in Egypt has been previously reported.11 For this study, we successfully isolated the virus from this specimen in Vero-E6 cells as previously described.14 Viruses were grown, aliquoted, and stored at −80°C until used. All the dromedary MERS-CoV strains used in this study were passage 3 after isolation in Vero-E6 cells. We had previously compared the full genome sequence of the AH13 virus in the original nasal swab with the Vero-E6 passaged virus used in this experiment. The viruses differed only in two aminoacids: membrane protein T8I, and spike protein S1251F.14
Laboratory procedures
Methods for viral titrations are described in the appendix. The methods we used for ex-vivo organ cultures and infection, quantification of viral RNA and host cytokine and chemokine mRNAs, and quantification of protein and immunohistochemistry have been previously described17 and are also detailed in the appendix.
We did experiments with live MERS-CoV in a biosafety level 3 biocontainment facility at The University of Hong Kong. This study was approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW-13-104).
Viral replication kinetics in Vero-E6 and Calu-3 cells
We cultured Vero-E6 and Calu-3 cells using Dulbecco's modified Eagle's medium. Cells were seeded at 1 × 105 cells per well in 24-well tissue-culture plates and infected with the MERS-CoV strains at a multiplicity of infection of 0·01 or 2 as indicated. After 1 h of virus adsorption at 37°C, we removed the virus inoculum, washed the cells with phosphate-buffered saline (PBS) three times to remove unbound virus, and replenished with fresh culture medium. We measured virus replication kinetics by titrating infectious virus in infected culture supernatants.
Cytokine and chemokine induction, gene expression, and quantification of viral RNA in Calu-3 cells
The human-respiratory-tract-derived cell line Calu-3 (ATCC HTB-55, Manassas, VA, USA) was cultured and infected with virus, as for the Vero-E6 cells. RNA from the infected cells was also collected at 6, 24, and 30 h post infection for analysis of gene expression and quantification of viral RNA. We harvested 1 mL of culture supernatants from Calu-3 cells at 30 h post infection to measure cytokine and chemokine protein concentrations using ELISA (R&D Systems, Minneapolis, MN, USA) and cytometic bead array (BD Bioscience, San Jose, CA, USA), using methods specified by the manufacturer. As a positive control for assessment of cytokine and chemokine induction in Calu-3, we used a highly pathogenic avian influenza H5N1 virus, A/Hong Kong/483/1997. Influenza virus was prepared in Madin-Darby canine kidney cells as previously described.
Thermal inactivation and tissue tropism in ex-vivo cultures of human bronchus and lung
We created thermal inactivation curves of the coronaviruses at 37°C, using culture wells inoculated with virus dilutions in the absence of permissive cells, or with an ex-vivo culture of mouse lung prepared from C57bl/6N mice. The mouse lung culture was prepared similar to the ex-vivo organ culture of human samples. We added 1 mL of virus dilution into 24-well plates with different starting concentrations (104, 103, and 102 TCID50/mL). We collected 130 μL of supernatant at 1, 24, 48, and 72 h post incubation to measure viral titration.
To assess tissue tropism in ex vivo experiments, we infected bronchial and lung tissues with 1 mL of each MERS-CoV virus dilution at a titre of 106 50% tissue culture infective dose per mL (TCID50/mL). After incubation for 1 h at 37°C, we washed the cultures three times with 5 mL of warm PBS to remove unbound virus. Mock-inoculated tissue served as controls (1 mL of Dulbecco's modified Eagle's medium without virus). We collected culture supernatants from the infected cultures at 1, 24, 48, and 72 h post infection and titrated them in parallel for infectious virus using the TCID50 assay. Samples for experiments on the ex-vivo cultures of human bronchus and lung were provided by five independent donors.
Statistical analysis
We assessed the extent of the overall viral replication with area-under-curve analysis using the trapezoid rule. We used the area calculated from 24 to 72 h post infection above the limit of detection of infectious virus titre for this analysis. The results of the four viruses were compared with the non-parametric Friedman test.
We did the experiments with Vero-E6 and Calu-3 cells with three biological replicates of each cell line and calculated mean and standard error of the mean (SEM). We compared differences in log10 transformed viral titres by a two-way ANOVA followed by a Bonferroni multiple-comparison test. We compared the quantitative cytokine and chemokine mRNAs between viruses and over time with a two-way ANOVA followed by a Bonferroni multiple-comparison test. The protein concentrations of the cytokines and chemokines between viruses at 30 h post infection were compared by a one-way ANOVA followed by a Bonferroni multiple-comparison test.
We used p less than 0·05 to indicate statistical significance. Statistical analyses were done with Graphpad Prism version 6.0 for Mac.
Role of the funding source
The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Figure 1 shows the phylogeny of MERS CoV strains used in this study within the global context of MERS-CoV. Only MERS-CoV with full genome sequences were included in this analysis. Most viruses cluster within clade B MERS-CoV, including the dromedary camel virus isolates from Saudi Arabia used in this study (AH13 and AH19D). The prototype human MERS-CoV virus (EMC) is a clade A virus. The dromedary MERS-CoV isolates, NRCE-HKU270 and Dromedary/Egypt-NRCE-HKU205/2013, have a long branch in the phylogenetic tree separating them from clade A and clade B viruses and also from each other, suggesting that they are genetically diverse from previously described MERS-CoV. We used the dromedary MERS-CoV isolate NRCE-HKU270 in this study. The nucleotide and aminoacid similarity between these human and dromedary camel viruses is provided the appendix.
Figure 1.
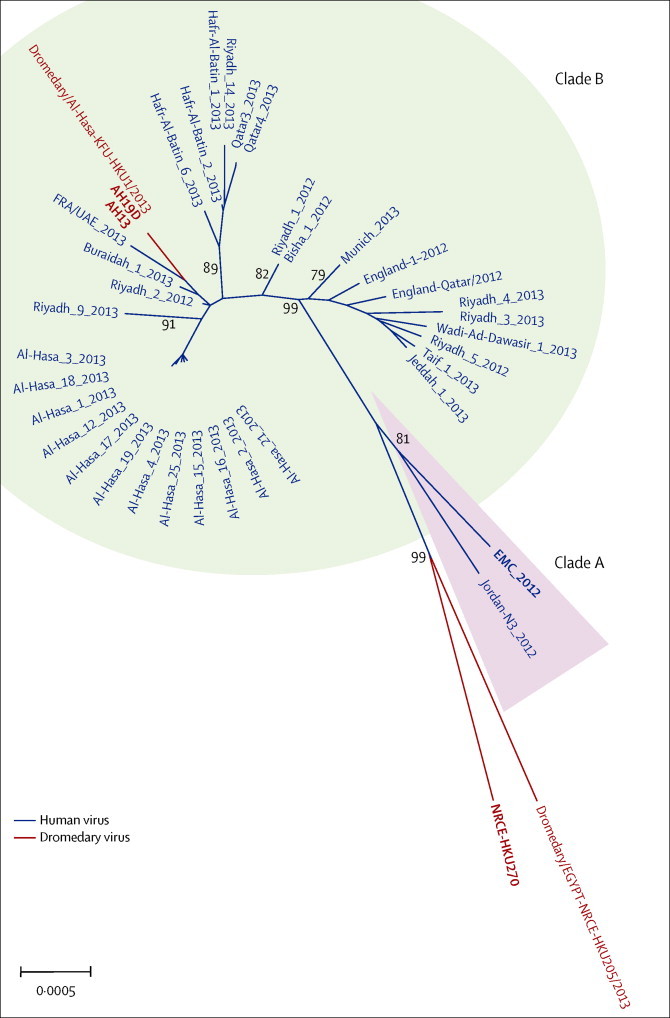
Phylogenetic tree of MERS-CoV full genome sequences
Virus isolates used for this study are in bold. The tree was built by the maximum likelihood method in Mega 6.06 with 500 bootstrap replications. Bootstrap values are shown at the major nodes of the tree. The scale bar shows the mutation rate for the tree. Accession numbers of virus genomes retrieved for this analysis are JX869059, KC776174, KC164505, KC667074, KF192507, KJ156881, KJ156949, KJ156944, KJ556336, KJ156952, KF600613, KF186564, KF600627, KF186567, KF600651, KJ156866, KF600634, KF600632, KF600644, KF186565, KF186566, KF600645, KF600647, KF600630, KF600652, KF745068, KJ156869, KJ650297, KJ650296, KJ650295, KF600628, KF961221, KF961222, KJ156874, KJ156910, KJ156934, KF600612, KF600620, and KJ477102. MERS-CoV=Middle East respiratory syndrome coronavirus. EMC=human MERS-CoV EMC. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. AH19D=MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013.
All four MERS-CoV strains replicated efficiently in Vero-E6 cells at multiplicities of infection of 0·01 and 2 (figure 2 , table ). Egyptian dromedary MERS-CoV NRCE-HKU270 replicated at least as well as did human MERS-CoV EMC, and reached peak titres even faster than human MERS-CoV at 48 h post infection when infected at low multiplicity of infection. Although the two Saudi dromedary MERS-CoV strains (AH13 and AH19D) replicated less efficiently than did human MERS-CoV during the first 24 h post infection, the viral titre of AH13 was similar by 72 h post infection. Replication of dromedary MERS-CoV NRCE-HKU270 was significantly higher than that of the other two dromedary MERS-CoV strains at 24 h and 48 h post infection at multiplicity of infection of 0·01.
Figure 2.
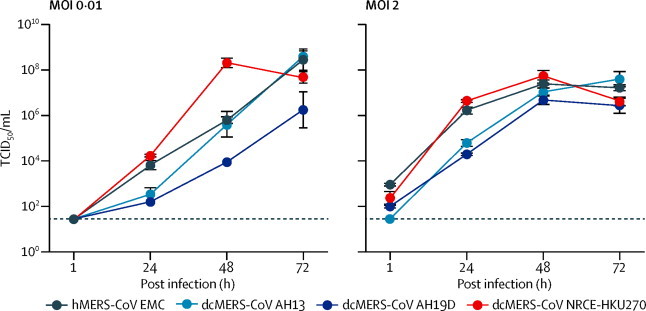
Replication kinetics of the human and camel MERS-CoV strains in Vero-E6 cells
Vero-E6 cells were infected with the indicated viruses at multiplicity of infections of 0·01 and 2 and cultured at 37°C for 72 h. Culture supernatants were harvested at the indicated times, and virus titres were measured by TCID50 assay. Results are presented as individual replicate (mean [SEM] for the three experiments). MOI=multiplicity of infection. TCID50=50% tissue culture infective dose.
Table.
Statistical significance of differences between virus titres
| EMC | AH13 | AH19D | ||
|---|---|---|---|---|
| Multiplicity of infection of 0·01 | ||||
| 24 h | ||||
| AH13 | 0·032* | .. | .. | |
| AH19D | 0·004* | >0·99 | .. | |
| NRCE-HKU270 | >0·99 | 0·002† | 0·0003† | |
| 48 h | ||||
| AH13 | >0·99 | .. | .. | |
| AH19D | 0·0010* | 0·0031* | .. | |
| NRCE-HKU270 | <0·0001† | <0·0001† | <0·0001† | |
| 72 h | ||||
| AH13 | >0·99 | .. | .. | |
| AH19D | <0·0001* | <0·0001* | .. | |
| NRCE-HKU270 | 0·4562 | 0·249 | 0·015* | |
| Multiplicity of infection of 2 | ||||
| 24 h | ||||
| AH13 | <0·0001* | .. | .. | |
| AH19D | <0·0001* | 0·329 | .. | |
| NRCE-HKU270 | 0·639 | <0·0001† | <0·0001† | |
| 48 h | ||||
| AH13 | >0·99 | .. | .. | |
| AH19D | 0·0486* | 0·8686 | .. | |
| NRCE-HKU270 | 0·8686 | 0·0486† | 0·0009† | |
| 72 h | ||||
| AH13 | 0·8686 | .. | .. | |
| AH19D | 0·0209* | 0·0003* | .. | |
| NRCE-HKU270 | 0·1593 | 0·0035* | >0·99 | |
Table shows p values for significance of differences between virus titres at each timepoint. EMC=human MERS-CoV Erasmus Medical Center. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. AH19D=MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013. MERS-CoV=Middle East respiratory syndrome coronavirus.
Reference virus titre (row) significantly lower than the comparator (column).
Reference virus titre (row) significantly higher than the comparator (column).
Immunohistochemistry of ex-vivo cultures of human bronchus and lung infected with human and dromedary MERS-CoV strains suggested that all four viruses could infect these tissues (figure 3 ). Because of their three-dimensional nature, infection in ex-vivo cultures was non-uniform throughout the histological specimen (appendix). Figure 4 shows viral replication kinetics of the viruses in the ex-vivo cultures of human bronchus and lung. In the absence of permissive cells, the thermal inactivation curves of each of these MERS-CoV strains showed virus infectivity largely decreasing to undetectable levels by 24 h post infection (figure 4A). We noted similar thermal inactivation kinetics when we substituted mouse lung tissue (non-permissive to MERS-CoV) for the human ex-vivo tissues (data not shown). Although the extent of virus replication varied from donor to donor, comparison with the thermal inactivation curves provided convincing evidence of virus replication of all four viruses in the ex-vivo human bronchial (figure 4B) and lung (figure 4C) cultures from each donor over the 72 h experiment period. Two replicates of ex-vivo cultures per donor from three donors, infected with the same virus (human MERS-CoV EMC) on different occasions, showed that the change of area-under-curve from the same tissue donor in bronchus ranged between 0 to 1·36-fold and in lung ranged between 0 to 1·46-fold, providing an indication of the range of technical variation in these ex-vivo assays (appendix). By contrast, the difference of area-under-curve for human MERS-CoV EMC between donor 3 and donor 1 was 5·3-fold for the bronchus (area under curve above detection limit 112·8 for donor 1 vs 21·3 for donor 3) and 37·3-fold for the lung ex-vivo cultures (178·8 for donor 1 vs 4·8 for donor 3), suggesting substantial donor-to-donor heterogeneity in virus replication competence (appendix). In view of the patchy nature of the infection, the variation between donors, and lack of adequate statistical power, statistical comparisons between viruses have to be interpreted with caution. Within these limitations, we noted no significant difference in the replication competence of the four viruses in either the bronchus (p=0·151) or the lung (p=0·270) as assessed by the area-under-the-curve using the Friedman test (appendix).
Figure 3.
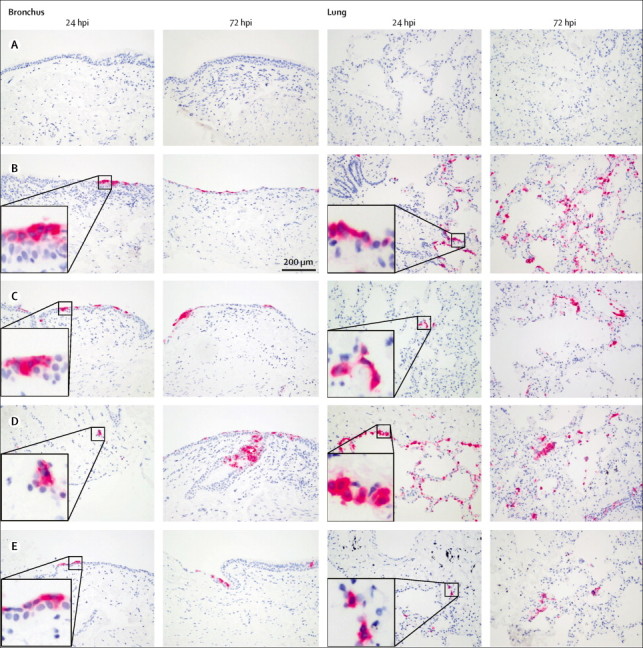
Tissue tropism of human and camel MERS-CoV in human respiratory tract tissues
Figure shows ex-vivo cultures of formalin-fixed paraffin-embedded sections of bronchus and lung. Immunohistochemical staining of MERS-CoV N protein (stained by vector red in pink) in the human bronchus and lung cultures were determined at 24 and 72 h post infection. (A) Mock virus. (B) Human MERS-CoV EMC. (C) MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013 (AH13). (D) MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013 (AH19D). (E) MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013 (NRCE-HKU270). MERS-CoV=Middle East respiratory syndrome coronavirus. hpi=hours post infection.
Figure 4.
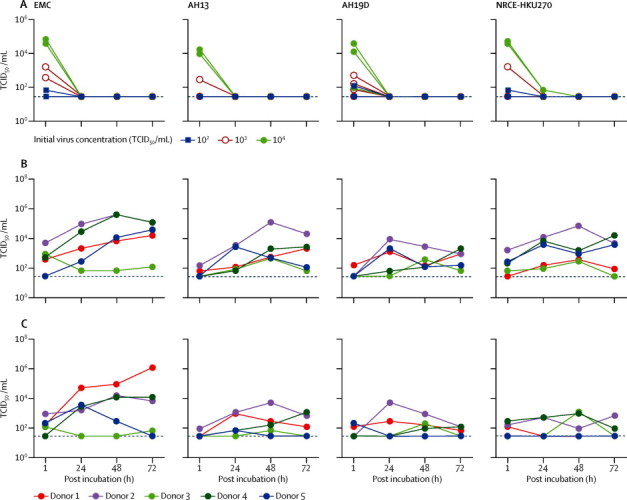
Replication kinetics of human and camel MERS-CoV in ex-vivo cultures of human bronchus and lung
(A) Thermal inactivation of MERS-CoV at 37°C. MERS-CoV strains were all inactivated at 24 h post incubation, irrespective of initial concentration; with the exception of the highest virus input of NRCE-HKU270. Replication curves of MERS-CoV infection of human bronchial (B) and lung (C) tissues, by individual tissue donor. The horizontal dotted line denotes the limit of detection in the TCID50 assay. MERS-CoV=Middle East respiratory syndrome coronavirus. EMC=human MERS-CoV EMC. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. AH19D=MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013. TCID50=50% tissue culture infective dose.
Immunohistochemical analysis of virus-infected bronchial tissues showed that the types of cells infected by human and dromedary MERS-CoV were very similar (figure 5 ). Co-staining of viral antigen with the goblet cell marker MUC5AC (figure 5A) and ciliated cell marker β-tubulin (figure 5B) lent support to the notion that MERS-CoV infects non-ciliated bronchial epithelium other than goblet cells. Co-staining of viral antigen and alveolar epithelial cell marker AE1/3 showed that all four viruses infected alveolar epithelial cells (figure 5C), and co-staining with the alveolar type II epithelial cell marker prosurfactant protein C showed that type II alveolar epithelium was infected (figure 5D). We noted no evidence of infection of alveolar macrophages (figure 5E). All four viruses infected lung endothelium (appendix). Thus, overall, the respiratory tropism of the human and dromedary MERS-CoV was similar.
Figure 5.
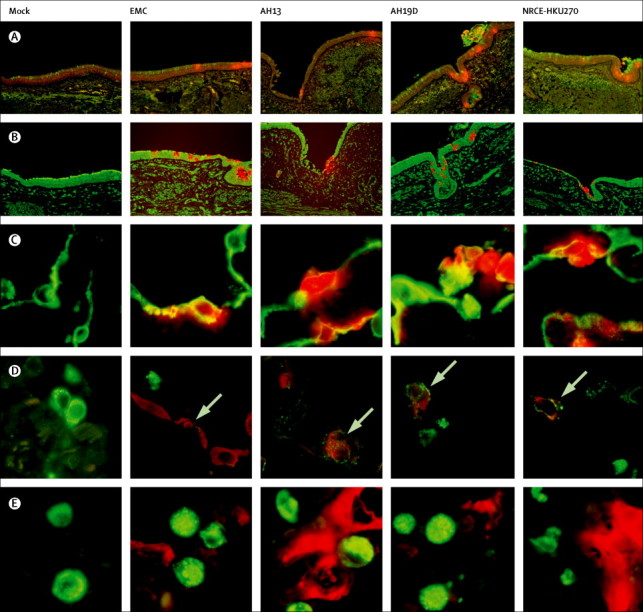
Cellular localisation of human and camel MERS-CoV in bronchus and lung tissue
MERS-CoV was stained with vector red (red) and cell markers conjugated with FITC (green) for detection of MUC5AC (goblet cell marker; A) and β-tubulin (ciliated cell marker; B) in bronchial tissue and human cytokeratin clone AE1/AE3 (epithelial cell marker; C), prosurfactant protein C (type II pneumocyte marker; D) and CD68 (macrophage marker; E), in lung tissue at 72 h post infection. Arrows show cells co-stained with both viral nucleoprotein and prosurfactant C. MERS-CoV=Middle East respiratory syndrome coronavirus. EMC=human MERS-CoV EMC. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. AH19D=MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013.
All three MERS-CoV strains (EMC, AH13, NRCE-HKU270) replicated efficiently in Calu-3 cells (AH19D was not included because it is in the same clade as AH13). To quantify the viral RNA and cytokine and chemokine gene induction and expression, the human-respiratory-tract-derived cell line Calu-3 was cultured and infected with virus as for the Vero-E6 cells. RNA from the infected cells was also collected at 6, 24, and 30 h post infection for analysis of gene expression and quantification of viral RNA. We harvested 1 mL of culture supernatants from Calu-3 cells at 30 h post infection to measure cytokine and chemokine protein concentrations using ELISA (R&D Systems, Minneapolis, MN, USA) and cytometric bead array (BD Bioscience, San Jose, CA, USA), using methods specified by the manufacturer. As a positive control for assessment of cytokine and chemokine induction in Calu-3, we used a highly pathogenic avian influenza H5N1 virus, A/Hong Kong/483/1997. Influenza virus was prepared in Madin-Darby canine kidney cells as previously described.17 Despite efficient replication, none of the MERS-CoV strains induced significant gene expression or protein secretion of tumour necrosis factor α (TNFα), CXCL10, or interferon β (figure 6 ). We used influenza A (H5N1) virus, a potent inducer of interferon β and CXCL10 in epithelial cells, for comparison. We noted evidence of interleukin 6 induction by all three MERS-CoV strains; although AH13 and NRCE-HKU270 induced greater production of interleukin 6 mRNA than the human EMC strain, the human EMC strain induced higher concentrations of interleukin 6 protein than did either dromedary strain (figure 6F).
Figure 6.
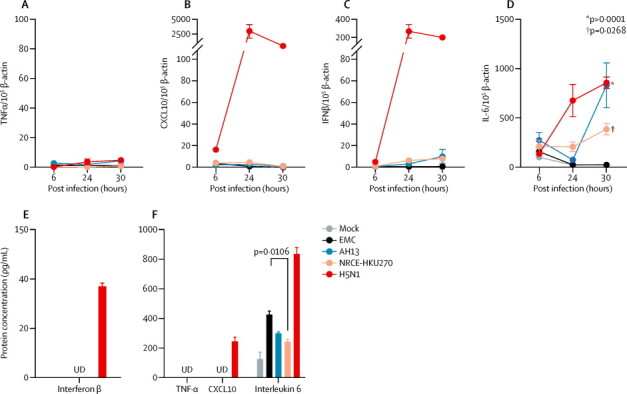
MERS-CoV cytokine responses in Calu-3 cells
Calu-3 cells were infected with MERS-CoV strains and H5N1 at multiplicity of infection of 2 for 1 h at 37°C. Figure shows gene expression of TNFα (A), CXCL10 (B), interferon β (C), and interleukin 6 (D) at 6, 24, and 30 h post infection. Means and SEMs of the mRNA copies expressed per 105 β-actin are shown. The concentration of the cytokine and chemokine in the culture supernatant collected at 30 h post infection were further examined using (E) ELISA and (F) cytometric bead array. The bar chart represents the means and SEMs of the detected protein concentrations. UD represents samples with undetectable concentration, or concentration of the target protein below the detection limit of the ELISA or cytometric bead array kit. p values for significant differences between human and dromedary MERS-CoV are shown. MERS-CoV=Middle East respiratory syndrome coronavirus. EMC=human MERS-CoV EMC. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013. TNFα=tumour necrosis factor α. IL-6=interleukin 6. IFNβ=interferon β. SEM=standard error of the mean.
Discussion
We compared the viral replication competence and respiratory tropism of three MERS-CoV isolates from dromedary camels with the prototype human clade A MERS-CoV isolate EMC (panel ). The two dromedary MERS-CoV isolates from Al-Hasa, AH13 and AH19D, are phylogenetically closely related to recent clade B human viruses from Saudi Arabia.18 As shown by the long-branch in the unrooted phylogenetic tree (figure 1) and genetic similarity tables (appendix), Egyptian dromedary MERS-CoV NRCE-HKU270 and dromedary MERS-CoV NRCE-HKU205 are genetically divergent from all other MERS-CoV detected to date, and are also divergent from each other, suggesting that the global diversity of MERS-CoV is wider than previously appreciated. The lack of a representative human clade B MERS-CoV isolate is a limitation of this study.
Panel. Research in context.
Systematic review
To assess the infection potential of dromedary camel Middle East respiratory syndrome coronavirus (MERS-CoV) strains for humans, genetic analysis should be complemented with phenotypic characterisation in physiologically relevant in-vitro cell cultures. We searched PubMed for reports published up to July 5, 2014, with the terms “MERS coronavirus” and “dromedary” and “culture” or “isolate”. We applied no date or language restrictions. Three papers were retrieved, only two of them related to MERS-CoV isolates from dromedary camels. One other recent paper not yet cited in PubMed was identified through search of the literature. Only one of these studies pertained to the phenotypic characterisation of dromedary MERS-CoV in cell cultures in vitro.16 This study used a transformed human hepatoma cell line, not physiologically relevant to the human respiratory tract.
Interpretation
Our findings show similar virus tropism and replication competence of human and dromedary MERS-CoV in ex-vivo cultures of the human respiratory tract. These findings suggest that dromedary MERS-CoV from the Arabian peninsula, in addition to genetically diverse dromedary viruses from Egypt, can be infectious to human beings. Exposure to zoonotic MERS-CoV might be happening in a wider geographical region beyond the Arabian peninsula. Virological evidence of MERS-CoV needs to be sought in patients with severe unexplained viral pneumonia in Africa in addition to the Middle East.
Comparison of viral genetic data can provide clues, but is not of itself final evidence for the competence of an animal virus to infect human beings. With SARS coronavirus, two aminoacid changes in the spike protein strikingly altered the ability of that virus to transmit efficiently in human beings.15 Because all the crucial determinants of host tropism with MERS-CoV are not fully understood, especially those outside of the spike-receptor binding domain, phenotyping of such viruses provides essential complementary information to the genetic analysis.
The three MERS-CoV isolates from dromedary camels (AH13, AH19D, and NRCE-HKU270) were similar to the human MERS-CoV EMC virus in replication competence in the rhesus monkey cell line Vero-E6. All four viruses replicated in ex-vivo cultures of the human bronchus and lung and infected the same cell types in the bronchus (non-ciliated bronchial epithelium) and lung (alveolar epithelium, including type II alveolar epithelial cells). We noted infection of lung endothelial cells with all four viruses, suggesting that potential for dissemination beyond the respiratory tract was also comparable. Overall, the tropism of human MERS-CoV EMC for the human respiratory tract was similar to that previously reported.17 Phenotypically, human and dromedary MERS-CoV have much the same tropism and replication competence in human respiratory ex-vivo cultures. To our knowledge, this report provides the first comparative data from living human respiratory tissue in situ. Therefore, our study provides new and crucial information to the emerging understanding of a disease with global importance for public health.
The heterogeneity of virus replication between individual donors in our ex-vivo infection experiments might result from differences in host susceptibility, which are physiologically relevant to the reported epidemiology of human MERS and could imply that individual variations might be an important determinant of disease susceptibility and severity. However, this question needs more systematic investigation. Thus, overall, our data support and complement the viral genetic sequence data of the spike proteins of the dromedary viruses, which seem similar to the spike proteins of human MERS-CoV strains,11, 13, 16 and strongly suggest that dromedary MERS-CoV strains have the capacity to infect human beings.
Infection of ex-vivo cultures provided useful information of virus cell tropism in the absence of autopsy data for MERS. Ex-vivo organ cultures provided information about virus tropism at early stages of the infection, which is not usually available in autopsies or from patients with late-stage disease (often after lengthy mechanical ventilation). Thus, our findings emphasise the usefulness of ex-vivo cultures for risk assessment of animal viruses for human health.
MERS-CoV is reported to be a weak inducer of type I and type III interferons.17, 19 Immune evasion mechanisms are relevant to pathogenesis and interspecies transmission, and differences might affect virus replication competence in new hosts. We therefore investigated whether human MERS-CoV and dromedary MERS-CoV differed in their ability to evade eliciting type I interferon and other innate immune responses. Because the multiplicity of infection cannot be accurately controlled in experiments with ex-vivo cultures of the human bronchus and lung, we used Calu-3, an epithelial cell line derived from human lung adenocarcinoma that has been used as a cell line to study viral infections of the human respiratory tract.20 Human MERS-CoV EMC, dromedary MERS-CoV AH13, and dromedary MERS-CoV NRCE-HKU270 replicated efficiently in these cells, although the dromedary MERS-CoV strains replicated significantly more efficiently in these cells (appendix). Despite this efficient virus replication, no virus elicited any TNFα, CXCL10, or interferon β mRNA or protein responses after virus infection (figure 6). By contrast, influenza A H5N1 virus was a potent inducer of interferon β and CXCL10. We noted some induction of interleukin 6 by dromedary MERS-CoV strains AH13 and NRCE-HKU270 and human MERS-CoV at late timepoints post infection (30 h; figure 6E and 6F). Importantly, the human and dromedary MERS-CoV behaved similarly by not eliciting interferon and CXCL10 responses, suggesting that these viruses do not differ in evasion of type I interferon responses.
In view of the high prevalence of infection in dromedaries5, 6, 7, 8, 10 and implied frequent human exposure, the rarity of human infection and disease remains a puzzle and is reminiscent of the epidemiology of avian influenza A H5N1.21 The exact mode of transmission of MERS-CoV from dromedaries to people remains unclear and whether unusual modes of exposure or host susceptibility have a role should be investigated. Serological and virological evidence of MERS-CoV in dromedaries has been reported from countries in east and north Africa outside of the Arabian peninsula,11, 12 but zoonotically acquired MERS has not yet been diagnosed in human beings in the African continent. Our finding that the genetically divergent dromedary MERS-CoV isolated in Egypt is phenotypically similar to human MERS-CoV EMC in explant cultures from human respiratory tissue underscores the possibility that zoonotic MERS might be occurring, unrecognised, in northeast Africa and beyond. At present, many countries only test for MERS-CoV in patients with a recent travel history to the Arabian peninsula, and locally acquired MERS could be missed. Alternatively, differences in cultural practices of camel husbandry, modes of animal contact, and consumption of animal products (eg, fresh camel's milk)22 might account for differences in occurrence of zoonotic disease in different geographical regions.
In conclusion, our findings suggest that dromedary MERS-CoV has competence to infect the human respiratory tract and does not differ phenotypically from human MERS-CoV in this respect. This evidence strengthens the contention that dromedary camels are the proximate source of infection for human beings. Our findings also emphasise the need to consider MERS in the differential diagnosis of viral pneumonia in a much wider geographical region in northeast Africa and beyond.
Acknowledgments
Acknowledgments
We thank Professor R A M Fouchier, Erasmus MC, Netherlands for providing us with the isolate of MERS-CoV strain EMC. We acknowledge Eric H Y Lau for statistical advice and the technical support of Dennis I T Kuok, Hung Sing Li, Kenrie P Y Hui, and Man Chun Cheung at the School of Public Health, and Kevin Fung at the Department of Pathology, The University of Hong Kong. We thank Ralph Baric (University of North Carolina, Chapel Hill, NC, USA) for providing us with the HKU4.2 mouse serum samples. We thank the Deanship of Scientific Research at King Faisal University for their support (Grant No 143011). We acknowledge research funding from Health and Medical Research Fund (Ref: 13121132) from the Research Fund Secretariat, Food and Health Bureau, Hong Kong Special Administrative Region; an internal grant from the National Research Centre, Giza, Egypt; the National Institute of Allergy and Infectious Diseases (NIAID) contract HHSN272201400006C; and a grant from the European Community Seventh Framework Program (FP7/2007-2013) under project European Management Platform for Emerging and Re-emerging Disease entities (grant agreement no. 223498) (EMPERIE).
Contributors
RWYC planned and did the experiments with ex-vivo cultures, analysed the data, and contributed to drafting of the report. MGH, GK, AA, and MAA did the field studies and detection of virus in dromedary specimens, and planned the experiments. DKWC, LLMP, and YG did the genetic sequencing and phylogenetic analysis. KPT did the in-vitro culture and quantitative PCR assays. HYN isolated the MERS-CoV strains. MCWC was involved in study design and obtained research funding. JMN did and interpreted the histology and immunohistochemistry studies. JSMP obtained research funding, planned and coordinated the study, analysed the data, and wrote the report. All authors contributed to the data interpretation, analysis, and provided critical comments on the draft report.
Declaration of interests
We declare no competing interests.
Contributor Information
John M Nicholls, Email: nicholls@pathology.hku.hk.
J S Malik Peiris, Email: malik@hku.hk.
Supplementary Material
References
- 1.WHO Middle East respiratory syndrome coronavirus (MERS-CoV)—update. July 23, 2014. http://www.who.int/csr/don/2014_07_23_mers/en (accessed Aug 12, 2014).
- 2.WHO Middle East respiratory syndrome coronavirus (MERS-CoV) summary and literature update. June 11, 2014. http://www.who.int/csr/disease/coronavirus_infections/MERS-CoV_summary_update_20140611.pdf (accessed July 9, 2014).
- 3.WHO Middle East respiratory syndrome coronavirus (MERS-CoV) 24 April 2014. http://www.who.int/csr/disease/coronavirus_infections/MERS_CoV_RA_20140424.pdf?ua=1 (accessed Aug 23, 2014).
- 4.Cauchemez S, Fraser C, Van Kerkhove MD. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reusken CB, Haagmans BL, Muller MA. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perera RA, Wang P, Gomaa MR. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 7.Haagmans BL, Al Dhahiry SH, Reusken CB. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alagaili AN, Briese T, Mishra N. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5:e00884. doi: 10.1128/mBio.00884-14. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azhar EI, El-Kafrawy SA, Farraj SA. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 10.Cotten M, Watson SJ, Zumla AI. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5:e01062. doi: 10.1128/mBio.01062-13. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu DKW, Poon LLM, Gomaa MM. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis. 2014;20:149–153. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reusken CBEM, Messadi L, Feyisa A. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg Infect Dis. 2014;20:1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aburizaiza AS, Mattes FM, Azhar EI. Investigation of anti-middle East respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia, fall 2012. J Infect Dis. 2014;209:243–246. doi: 10.1093/infdis/jit589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemida MG, Chu DKW, Poon LLM. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes KV. Structural biology: adaptation of SARS coronavirus to humans. Science. 2005;309:1822–1823. doi: 10.1126/science.1118817. [DOI] [PubMed] [Google Scholar]
- 16.Stalin Raj V, Farag EABA, Reusken CBEM. Isolation of MERS coronavirus from dromedary camel, Qatar, 2014. Emerg Infect Dis. 2014;20:1339–1342. doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan RW, Chan MC, Agnihothram S. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briese T, Mishra N, Jain K. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. 2014;5:e01146. doi: 10.1128/mBio.01146-14. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kindler E, Jonsdottir HR, Muth D. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4:e00611. doi: 10.1128/mBio.00611-12. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harcourt JL, Caidi H, Anderson LJ. Evaluation of the Calu-3 cell line as a model of in vitro respiratory syncytial virus infection. J Virol Methods. 2011;174:144–149. doi: 10.1016/j.jviromet.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Doremalen N, Bushmaker T, Karesh WB, Munster VJ. Stability of Middle East respiratory syndrome coronavirus in milk. Emerg Infect Dis. 2014;20:1263–1264. doi: 10.3201/eid2007.140500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


