The deformation of soft nanocapsules during cell binding and endocytosis leads to a lower cellular uptake than the stiff ones.
Abstract
The ability of cells to sense external mechanical cues is essential for their adaptation to the surrounding microenvironment. However, how nanoparticle mechanical properties affect cell-nanoparticle interactions remains largely unknown. Here, we synthesized a library of silica nanocapsules (SNCs) with a wide range of elasticity (Young’s modulus ranging from 560 kPa to 1.18 GPa), demonstrating the impact of SNC elasticity on SNC interactions with cells. Transmission electron microscopy revealed that the stiff SNCs remained spherical during cellular uptake. The soft SNCs, however, were deformed by forces originating from the specific ligand-receptor interaction and membrane wrapping, which reduced their cellular binding and endocytosis rate. This work demonstrates the crucial role of the elasticity of nanoparticles in modulating their macrophage uptake and receptor-mediated cancer cell uptake, which may shed light on the design of drug delivery vectors with higher efficiency.
INTRODUCTION
The perception of mechanical cues is an integral part of cells that influences their performance and adaptation to the surrounding environment (1, 2). Cells are able to perceive forces exerted by substrates or adjacent cells to trigger specific signaling pathways, which, in turn, regulates their biological processes such as cell spreading (3), proliferation (4), and differentiation (5). Current research on the effect of external mechanical information on cellular behaviors mainly focuses on cell-substrate interactions at the macroscale (3–5), cell-cell interactions at the microscale (4, 6), and cell-ligand interactions at single molecular levels (7, 8). However, how cell-nanoparticle (NP) interactions are regulated by NP mechanical properties on the nanoscale remains largely unknown.
Recently, by designing NPs having distinct mechanical properties but otherwise identical, researchers have demonstrated the important role of NP elasticity in regulating their interactions with cells (9–16). However, most of the current studies focus on examining the effects of NP elasticity on nonspecific NP–cell interactions, while the effects of NP elasticity on receptor-mediated cellular interactions remain much less understood despite the fact that specific ligand–receptor interaction has proven to play a critical role in the cellular sensing of external mechanical cues (17). Moreover, the effect of NP mechanical property on their macrophage uptake has important implications, for example, in their blood circulation time. A better understanding of the effect of NP elasticity on these NP-cell interactions as well as the underlying mechanisms will shed light on the future design of better drug delivery vehicles.
Here, we fabricated a library of silica nanocapsules (SNCs) (~150 nm) with Young’s moduli ranging from 0.56 kPa to 1.18 GPa. Their interactions with macrophage cells and cancer cells through nonspecific or receptor-mediated pathways, including cell binding, uptake, and endocytic pathways, were systematically investigated to fundamentally understand the mechanisms underlying their elasticity-dependent NP-cell interactions. Transmission electron microscopy (TEM) study revealed an interesting dynamic shape evolution of the soft SNCs during cell binding and internalization. Single-particle tracking demonstrated the notable difference in internalization kinetics of the soft and stiff SNCs.
RESULTS
Synthesis of SNCs having a broad range of Young’s moduli (560 kPa to 1.18 GPa)
SNCs were synthesized on the basis of a method developed in our laboratory, using a bifunctional peptide SurSi (Ac-MKQLAHSVSRLEHA RKKRKKRKKRKKGGGY-CONH2) (Fig. 1A) (18). The silica precursor, triethoxyvinylsilane (TEVS), allows the formation of very soft SNCs, while tetraethoxysilane (TEOS) gives very stiff SNCs (16). TEOS/TEVS mixtures (0, 5, 10, 20, 40, and 100 mol% of TEOS) were used to tune the elasticity of SNCs. TEM micrographs (Fig. 1A and fig. S2) reveal distinct levels of deformability of different SNCs. Pure TEVS (0% TEOS) generated SNCs with a collapsed structure, whereas TEOS SNCs (100% TEOS) maintained their spherical morphology, indicating a high elasticity (Fig. 1, A and D). The prepared SNCs had hydrodynamic diameters of around 190 nm with small polydispersity index (PDI) values (<0.2) (fig. S1) and zeta (ζ) potentials of around +30 mV. The diameters and shell thickness of the SNCs under TEM were around 150 and 10 nm, respectively.
Fig. 1. Synthesis and characterization of SNCs.
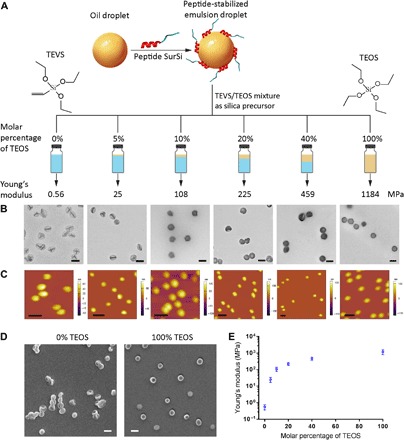
(A) Schematic illustration of the synthesis of SNCs having controllable Young’s moduli. (B) TEM micrographs (scale bars, 200 nm) and (C) AFM height profiles (scale bars, 500 nm) of SNCs. (D) SEM micrographs (scale bars, 200 nm) of SNCs synthesized using 0% (softest) and 100% (stiffest) of TEOS. (E) Young’s moduli of SNCs (values are means ± SD, n = 15).
The mechanical properties of SNCs were characterized using liquid-phase atomic force microscopy (AFM) (Fig. 1C). The Young’s moduli of the SNCs were calculated on the basis of the Hertzian contact model (fig. S3), exhibiting a positive correlation with the molar percentage of TEOS (Fig. 1E). The softest TEVS SNC has a Young’s modulus of 560 kPa, which is comparable to many soft hydrogel NPs, while the stiffest TEOS SNC has a Young’s modulus of 1.18 GPa, representing typical inorganic nanomaterials. The six different SNCs have Young’s moduli of 0.56, 25, 108, 225, 459, and 1184 MPa, respectively, covering an elasticity range much broader than any other previously reported individual NP systems.
Nonspecific and receptor-mediated cell binding and uptake
The SNCs were modified with methoxy-poly(ethylene glycol) (mPEG) (5000 Da) and folate-poly(ethylene glycol) (FA-PEG) (5000 Da) to study the effects of their mechanical properties on nonspecific and specific (receptor-mediated) NP–cell interactions, respectively. After modification and purification, the FA-PEG–modified SNCs (10 mol% FA-PEG with 90 mol% mPEG) remained monodisperse (PDI around 0.1) (Table 1 and fig. S1), with their hydrodynamic sizes rising by 15 nm as a result of PEGylation. The ζ potentials of SNCs reduced from around +30 mV to near neutral (−3 mV). The PEG density of the SNCs (fig. S4 and table S1) was around 0.9 molecules/nm2 (Table 1), which is sufficient for a “brush” conformation that allows effective immune evasion (19). The surface FA density of FA-PEG–modified SNCs was around 0.09 molecules/nm2.
Table 1. Hydrodynamic size, PDI, ζ potential, Young’s modulus, and surface PEG density of FA-PEG–modified SNCs.
Values are means (n = 3) for hydrodynamic diameter, PDI, ζ potential, and Young’s modulus. Coating of FA-PEG–modified SNCs consists of 10% FA-PEG and 90% mPEG (in molar ratio).
|
Molar percentage of TEOS |
0 | 5 | 10 | 20 | 40 | 100 |
| Hydrodynamic diameter (nm) |
202 | 203 | 202 | 205 | 206 | 211 |
| PDI | 0.12 | 0.12 | 0.11 | 0.07 | 0.09 | 0.09 |
| ζ potential (mV) | −3.5 | −3.1 | −3.5 | −3.3 | −3.2 | −2.8 |
| Young’s modulus (MPa) |
0.56 | 25.09 | 108.53 | 224.71 | 459.43 | 1184.46 |
| Surface PEG density (molecules/nm2) |
0.94 | 0.83 | 0.82 | 0.88 | 0.96 | 0.89 |
The impact of SNC elasticity on NP-cell interactions was first demonstrated using the softest (560 kPa) and stiffest (1.18 GPa) SNCs. Three types of NP-cell interactions, including NP–RAW264.7 macrophage, nonspecific NP–SKOV3 cancer cell, and specific (receptor-mediated) NP–SKOV3 cancer cell interactions, were investigated (Fig. 2A). Folic acid modification has been shown to have no significant effect on the RAW264.7 cell uptake of SNCs, while the SKOV3 cells in our laboratory have been proven to have a high level of folate receptor expression that allows a specific ligand-receptor–mediated interaction with FA-PEG–modified SNCs (16). Consistent with our previous study (fig. S5) (16), the stiff SNCs had much higher macrophage uptake (three-time higher) than the soft ones (Fig. 2B). The soft and the stiff PEG-modified SNCs displayed similar nonspecific uptake by SKOV3 cells. In contrast, the stiff FA-PEG–modified SNCs exhibited four-time higher SKOV3 uptake than their soft counterparts. These results raise a fundamental question: Which step in NP uptake is elasticity dependent?
Fig. 2. Effects of SNC elasticity on cellular interactions.
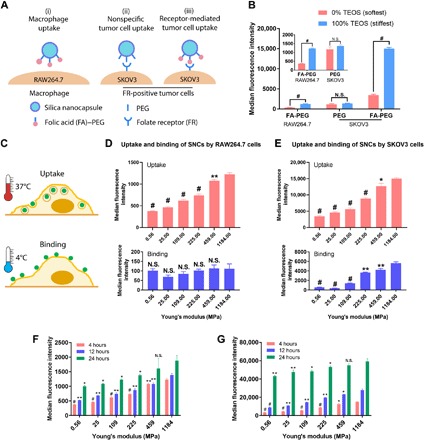
(A) Schematic illustration showing different types of cell-SNC interactions. (B) The uptake of the softest and the stiffest SNCs by RAW264.7 and SKOV3 cells; inset is the magnified graph of the first two groups. (C) The cellular uptake and binding of SNCs were measured at 4° and 37°C, respectively. (D) Four-hour cellular uptake and binding of FA-PEG–modified SNCs in RAW264.7 and (E) SKOV3 cells; values are compared to the SNCs having the highest Young’s modulus. (F) Four-, 12-, and 24-hour cellular uptake of FA-PEG–modified SNCs in RAW264.7 and (G) SKOV3 cells; at every time point, values are compared to the SNCs having the highest Young’s modulus. All values are means ± SD (n = 3, with *P < 0.05, **P < 0.01, and #P < 0.001; N.S., not significant).
NP uptake starts with an initial NP binding onto cell membranes either nonspecifically or through a ligand-receptor recognition, followed by internalization and then trafficking to certain subcellular compartments (20). The cellular binding and uptake of SNCs were investigated by coincubating cells with the FA-PEG–modified SNCs having six different Young’s moduli at 4° and 37°C (Fig. 2C), respectively (20). Figure 2 (D and E) shows that both the macrophage uptake and folate receptor (FR)–mediated SKOV3 cell uptake of the SNCs increased markedly with their Young’s moduli rising from 0.56 to 459 MPa, but less pronouncedly from 459 to 1184 MPa, indicating reduced sensitivity of the cells to very stiff NPs. Similarly, the receptor-mediated binding of the SNCs to SKOV3 cells also increased with their Young’s moduli, exhibiting a positive correlation with their cellular uptake (Fig. 2E). In contrast, the cellular binding of the SNCs to RAW264.7 macrophage cells appeared to be independent of their elasticity (Fig. 2D). The nonspecific cellular binding of PEG-modified SNCs to SKOV3 cells was also elasticity independent (fig. S5). The binding and uptake behaviors suggest that the enhanced cellular binding facilitated receptor-mediated uptake of the stiff SNCs, while the increased macrophage uptake of the stiff SNCs should involve other mechanisms, such as different uptake kinetics or endocytic pathways.
The cellular uptake kinetics of SNCs was studied by incubating SNCs with cells for different time spans (4, 12, and 24 hours) (Fig. 2, F and G). Longer incubation times led to higher SNC uptake by both RAW264.7 and SKOV3 cells, and the differences between the cellular uptake of stiff and soft SNCs became less significant as the incubation time increased. For RAW264.7 cells, the cellular uptake of the softest SNCs was only 31% of that of the stiffest ones at a 4-hour incubation time, but this ratio increased to 38 and 53% at 12 and 24 hours, respectively. Similarly, for SKOV3 cells, this ratio rose from 23% at 4 hours to 32 and 73% at 12 and 24 hours, respectively. Although the uptake of SNCs varies with time, the difference between the macrophage uptake of the softest and the stiffest SNCs at all three time spans was still significant, suggesting that the higher macrophage uptake of stiffer SNCs is not merely a result of the uptake kinetics.
Endocytic pathway–dependent SNC deformation during cellular uptake
Cellular endocytic pathways of the SNCs were investigated using endocytosis inhibitors, including cytochalasin D, nystatin, and chlorpromazine to block their phagocytosis/macropinocytosis, caveolin/lipid raft–mediated, and clathrin-mediated endocytosis, respectively. The FR-mediated uptake of FA-PEG–modified SNCs by SKOV3 cells was mainly through a clathrin-mediated endocytic pathway (Fig. 3A). TEM images show that both the softest and stiffest SNCs adopted a spherical shape when they were not attached to the cell membrane (Fig. 3B and fig. S5). The stiff SNCs remained spherical during either binding to the cell membrane or internalization, whereas the soft SNCs displayed substantial deformations during both binding and internalization (Fig. 3B and fig. S6). As schematically illustrated in Fig. 3C, soft SNCs flattened when they adhered to the cell membrane. Then, the flattened soft SNCs experienced a further morphological change during internalization: from an oblate-like shape to an ellipsoidal shape with its longer axis perpendicular to the cell membrane. Further analysis of the TEM images of soft SNCs shows axis dimension ratios (long axis/short axis) of around 1.37 and 1.27 during cell adhesion and internalization, respectively (fig. S7). Similar shape transformation has been observed in theoretical modeling on the membrane wrapping of fluidic vesicular NPs such as liposomes (21). The deformation of soft SNCs during cellular binding and internationalization has two implications. First, a flattened SNC covers a larger cell surface area and occupies more FRs than its nondeformable stiff counterpart (Fig. 3D), thus leaving less binding sites for other free soft SNCs. Second, the excessive deformation of soft SNCs may increase the energy required for their full wrapping by cell membrane, thus lowering their internalization rate (21–23). Jointly, the reduced binding and internalization of the softer SNCs leads to less cellular uptake as compared to the stiffer SNCs. The different cellular binding of stiff and soft SNCs might also be a result of their different surface ligand orientations. However, the precise effect of NP elasticity on surface ligand orientation and bioavailability remains elusive and warrants further detailed investigations in the future.
Fig. 3. Endocytic pathways and deformations of SNCs during cellular uptake.
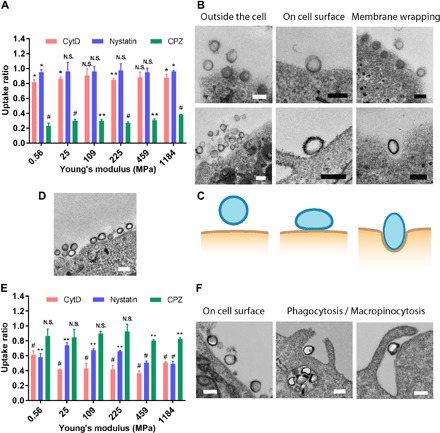
(A) Uptake ratios of the FA-PEG–modified SNCs in SKOV3 cells. (B) TEM micrographs showing the morphological change of the stiffest (top) and softest (bottom) SNCs during receptor-mediated interactions with SKOV3 cells. (C) Schematic illustration showing the deformation of the softest SNCs during receptor-mediated cellular uptake. (D) Considerable proportion of SKOV3 cell surface can be covered by a large number of adhered SNCs. (E) Uptake ratios of the FA-PEG–modified SNCs in RAW264.7 cells. (F) TEM micrographs showing the morphological change of the softest SNCs during interactions with RAW264.7 cells. The uptake ratio represents the uptake of SNCs by cells treated with endocytic inhibitors normalized by the uptake by nontreated ones. Scale bars, 200 nm. All values are means ± SD (n = 3, with *P < 0.05, **P < 0.01, and #P < 0.001; N.S., not significant).
Different from the SKOV3 cells, the RAW264.7 uptake of SNCs mainly relied on phagocytosis/micropinocytosis (Fig. 3E). Unlike their receptor-mediated interactions with SKOV3 cells, the softest SNCs did not flatten on the surface of RAW264.7 cells (Fig. 3F and fig. S8), indicating that there was no apparent force applied on the SNCs. This explains the elasticity-independent cellular binding of SNCs to RAW264.7. However, the softest SNCs did deform during cellular internalization and the protruding pseudopodium structures further proved the phagocytosis/micropinocytosis pathway. It is likely that the deformation of soft SNCs slows their internalization rate, leading to lower macrophage uptake (24). We conducted some theoretical calculation to demonstrate the ability of cells to deform nanocapsules in clathrin-mediated endocytosis and phagocytosis (fig. S9).
The PEG-modified softest SNCs displayed similar nonspecific binding and uptake with their stiffest counterparts in SKOV3 cells (fig. S5), and their endocytosis was not dependent on clathrin, caveolin, or F-actin polymerization (Fig. 4A). Moreover, they did not flatten on the cell membrane during binding and remained spherical during and after internalization (Fig. 4B and fig. S8), confirming that there was minimal deformation of the softest SNCs during the nonspecific NP–cell interaction, thus a similar level of cellular uptake as the stiffest SNCs.
Fig. 4. The endocytic pathway–dependent deformation of SNCs is a result of the associated proteins.
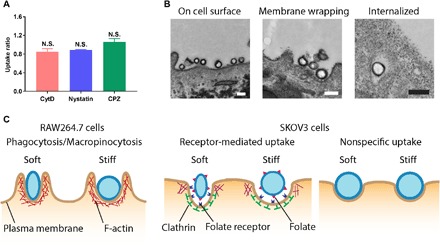
(A) Uptake ratios of the PEG-modified softest SNCs in SKOV3 cells. The uptake ratio represents the uptake of SNCs by cells treated with endocytic inhibitors normalized by the uptake by nontreated ones. (B) TEM micrographs showing the morphological change of the softest PEG-modified SNCs during interactions with SKOV3 cells; scale bars, 200 nm. (C) Schematic illustration showing the proteins associated in the three types of SNC-cell interactions. All values are means ± SD (n = 3).
The above findings demonstrate the important role of SNC morphological change in modulating cellular uptake (Fig. 4C). In active cell interactions such as clathrin-mediated endocytosis and phagocytosis, cell membrane and the associated proteins (e.g., clathrin and cortical actin network) form a composite physical layer to interact with NPs. In these cases, not only the lipid membrane but also the clathrin and cross-linked actin network might matter in the endocytosis. In clathrin-mediated endocytosis and phagocytosis, the softest SNCs deformed owing to the combined force exerted by the cell membrane, underlying protein coating and remodeling actin cytoskeleton. Because the phospholipid bilayer itself exhibits a very low rigidity, it must be the associated membrane-bound proteins that essentially contribute to the increased rigidity of the cell membrane (25). As shown during the protrusion in blebbing cells, the myosin motors exert contractile forces that lead to a compressive loading on the cytoskeletal network (26). Depending on the magnitude of the myosin-mediated contractions, the membrane–cortex layer could be under a smaller tensile or even a compressive tension, which would facilitate the cell uptake especially in the early stage of receptor-mediated endocytosis.
In RAW264.7 cells, the SNCs were internalized mainly through phagocytosis/macropinocytosis that largely depend on the reorganization of actin filaments (27). The recruitment of F-actin increases the cortical rigidity of the microdomain where the particle binds to, thus facilitating the formation of the actin-rich lamellar pseudopods and phagocytosis cups for particle internalization (27). These mechanically stiff structures might cause the deformation of soft SNCs during membrane wrapping and influence their phagocytosis dynamics. In clathrin-mediated endocytosis, clathrin is the main player that assembles at the binding sites of NPs to enforce the curvature of cell membranes (28). F-actin polymerization is also involved in clathrin-mediated endocytosis, especially in conditions of high membrane tensions and/or when the internalized cargos have larger dimensions than the clathrin-coated vesicles (29, 30). The clathrin-coated curvature is known to have a bending rigidity 10 to 20 times higher than that of lipid membranes (28); hence, the soft SNCs can be deformed during the membrane invagination. In contrast, the nonspecific uptake of the PEG-modified SNCs by SKOV3 cells mainly relied on a clathrin- and caveolin-independent pathway, indicating minimal adaptor protein (e.g., F-actin and clathrin) involvement at the microdomains where the membrane wrapping of SNCs occurs. Therefore, the phospholipid bilayer alone is not able to deform even the softest SNCs. In other words, cells cannot distinguish the elasticity difference between the soft and stiff SNCs during their nonspecific interactions.
Internalization rates of soft and stiff SNCs
To further elucidate the effect of NP elasticity on cell uptake dynamics, the cellular internalization of the softest (0.56 MPa) and the stiffest (1184 MPa) SNCs were observed using live-cell imaging and single-particle tracking. The lysosome-labeled SKOV3 cells were coincubated with FA-PEG–modified SNCs at 4°C to allow the cell adhesion of SNCs, followed by increasing temperature to 37°C (time point 0) to initiate SNC internalization. The softest SNCs showed no uptake until 40 min (Fig. 5A), while many stiffest SNCs had already been internalized at 20 min (Fig. 5B). At 60 min, the SKOV3 uptake of the stiffest SNCs was significantly higher than that of the softest ones, which is consistent with the flow cytometry data. Their lysosomal transport was shown as the yellow spots indicating the colocalization of SNCs and lysosomes. The stiffest SNCs displayed a faster lysosomal accumulation as a result of their fast internalization rates. It is worth noting that the internalization of both the soft and stiff SNCs was much slower than their cell binding, suggesting that internalization is the rate-determining step of the entire cell uptake process.
Fig. 5. The internalization kinetics of the softest (0% TEOS, 560 kPa) and the stiffest (100% TEOS, 1.18 GPa) SNCs in receptor-mediated uptake by SKOV3 cells.

(A) Fluorescence live-cell images showing the internalization of the softest and (B) stiffest FA-PEG–modified SNCs by SKOV3 cells and their transport to lysosomes over 60 min; scale bars, 20 μm. Representative time projection images showing the movement of the (C) softest and (D) stiffest SNCs during cellular uptake; tracks of the SNCs are shown in the magnified images of boxed regions. Yellow solid lines indicate the plasma membranes; scale bars, 10 μm. Instantaneous velocity of the (E) softest and (F) stiffest SNCs during stage I and stage II of cellular uptake. MSD analysis of the movement of the (G) softest and (H) stiffest SNCs during stage I and stage II of cellular uptake; insets are magnified graphs. (I) Time taken for the internalization of SNCs. Movements of the SNCs were only recorded for 15 min, as imaging longer than 15 min could lead to fluorescence quenching and focal plane shifting.
The internalization rate of SNCs during receptor-mediated cellular uptake was further investigated using two-dimensional single-particle tracking. The different motion types of SNCs are displayed in the form of time-projection images generated from the time-lapse videos (31). For either the softest (Fig. 5C) or stiffest (Fig. 5D) SNCs, the movements of noninternalized SNCs were confined in a finite area (arrows 2 and 3 in Fig. 5C and arrows 1, 2, and 4 in Fig. 5D), suggesting that they were bound to the cell membrane. The internalized SNCs, in contrast, displayed longer trajectories as marked by arrow 1 in Fig. 5C and arrow 3 in Fig. 5D (see movies S1 and S2). A two-stage movement pattern was exhibited by the SNCs (Fig. 5, E and F): stage I with a low-velocity confined motion (bound to cell membrane), and stage II with a high-speed transport (after being internalized). A mean square displacement (MSD) analysis of the SNC trajectories was conducted to determine their motion types. Particles performing different types of motions (e.g., confined motion, free diffusion, and active transport) exhibit distinct MSD-time curves. Figure 5 (G and H) displays the MSD-time curves of the soft and stiff SNCs, respectively. The red curves correspond to the low-velocity confined motion in stage I, while the green curves exhibit a super-linear MSD-time relationship, indicating directed motion along the microtubules (32).
The time taken for the internalization of SNCs was determined using both velocity and MSD results. The 15 softest SNCs were tracked, and only 1 of them entered the cell at around 14 min, with all the rest of the SNCs remaining on the cell membrane at the end of 15 min (Fig. 5I). In comparison, 6 of the 16 analyzed stiff SNCs entered the cells within 15 min, with three of them entering the cells in the first 5 min. Despite the relatively small sample size, it is clear that SNCs with higher Young’s moduli were internalized more quickly, consistent with the higher cellular uptake of the stiff SNCs.
DISCUSSION
Recently, several theoretical and computational studies have also been performed to investigate the membrane wrapping efficiency of elastic NPs during endocytosis, which has been shown regulated synergistically by NP size, shape, and elasticity (22, 23, 33). However, despite the fact that the effects of NP elasticity on different endocytic uptake mechanisms have been experimentally explored in this work, the theoretical modeling of these uptake mechanisms remains challenging. More thorough and realistic modelings with explicit consideration of the actin network and adaptor proteins underneath the cell membrane are called for. Clathrin- and caveolin-independent endocytosis can be modeled using a membrane wrapping based on the Helfrich-Canham membrane theory (34), suggesting that stiff NPs are energetically more prone to full wrapping than soft ones (21). This elasticity effect on the simple membrane wrapping of an NP gradually becomes less significant as the NP elasticity increases. For isotropic elastic SNCs of a bending rigidity ratio (the NP shell:cell membrane) larger than 5, the stiffness effect can be ignored and the elastic NPs can be approximated as rigid particles (21), which is consistent with our experimental observation that both the softest and stiffest PEG-modified SNCs in nonspecific uptake by SKOV3 cells maintained their spherical shapes (Fig. 4C).
Clathrin-mediated endocytosis not only involves substantial membrane invagination but also entails the protein-induced spontaneous curvature and actin-mediated force on bud formation. Recent theoretical models for clathrin-mediated endocytosis (35–38) are valid for NPs with a radius smaller than that of the self-assembled clathrin scaffold. For relatively large NPs, the compressive mechanical interplay between the engulfed NP and clathrin scaffold that either squeezes the soft NPs and cause substantial NP deformation or expands the relatively small clathrin scaffold needs to be taken into account. In contrast to passive nonspecific endocytosis and less active clathrin endocytosis in which NPs appear to sink into cells, phagocytosis involves the outward actin-mediated membrane protrusion that hugs the phagocytic targets such as NPs, accompanying which cells undergo rapid morphological change with dramatic membrane area variation and active cytoskeletal flow (39, 40). Existing theoretical studies on phagocytosis have only focused on rigid particles. Therefore, future theoretical efforts are needed to provide mechanistic insight into the effects of particle elasticity on phagocytosis of NPs.
In conclusion, we demonstrate that the effect of NP elasticity on cellular uptake is dependent on the type of cell-NP interaction. For macrophage phagocytosis and receptor-mediated endocytosis, the elasticity of NPs significantly affects their cellular uptake, while cancer cells can lose their stiffness sensitivity in nonspecific clathrin- and caveolin-independent endocytosis. The specific ligand–receptor interaction exerts forces on SNCs that deforms the soft SNCs and makes them flattened. The soft SNCs were also deformed during cell membrane wrapping, which is dependent on their endocytic pathway and the associated adaptor proteins. The recruitment of clathrin and F-actin in the clathrin-mediated endocytosis and phagocytosis/macropinocytosis increases the apparent membrane rigidity of the binding sites of SNCs, thus deforming the soft SNCs. In contrast, no adaptor proteins are involved in the clathrin- and caveolin-independent endocytosis; thus, the lipid bilayer alone is not stiff enough to deform even the softest SNCs. Together, the stiff and soft SNCs display different deformation states upon the forces originating from ligand–receptor interactions and cell membrane wrapping, which, in turn, modulates their cellular binding and internalization rates, hence their overall cellular uptake profiles. This study offers fundamental insights into how NPs’ mechanical properties regulate their interactions with cells and highlights the role played by ligand-receptor interactions, which provides guidance for the future design of mechanically engineered NPs for enhanced drug delivery.
MATERIALS AND METHODS
Materials
Peptide SurSi was custom-synthesized by GenScript Corporation (Piscataway, NJ, USA) with a purity ≥95%. Miglyol 812 oil was purchased from Caesar & Loretz GmbH (Hilden, Germany) and passed through heat-activated silica gel (Sigma-Aldrich, Castle Hill, Australia) before use. Water having resistivity >18.2 megohms·cm was obtained from a Milli-Q system (Millipore, Australia) equipped with a 0.22-μm filter. Methoxy-poly(ethylene glycol) succinimidyl carbonate ester (mPEG-NHS, MW 5000) and folate-poly(ethylene glycol) succinimidyl carbonate ester (FA-PEG–NHS, MW 5000) were obtained from Nanocs (New York, NY, USA). DiI and LysoTracker Deep Red stains were purchased from Thermo Fisher Scientific (Scoresby, Australia). Other reagents and chemicals were of analytical grade purchased from Sigma-Aldrich and used as received unless otherwise stated.
Synthesis of SNCs having tunable Young’s moduli
The SNCs were synthesized on the basis of a nanoemulsion- and biosilicification-templated technology developed in our group (18). First, a nanoemulsion template was prepared using peptide SurSi as the surfactant. To do this, a solution of peptide SurSi (400 μM) and ZnCl2 (800 μM) in Hepes buffer (25 mM, pH 7) was prepared. Then, 2940 μl of the peptide solution was mixed with 60 μl of Miglyol 812 oil (2% v/v) and sonicated using a Sonifier 450 ultrasonicator (Branson Ultrasonics, Danbury, CT) to form a nanoemulsion. The nanoemulsion was dialyzed in Hepes buffer (25 mM, pH 7.5) using a dialysis membrane having a cutoff molecular weight of 10 kDa (Thermo Fisher Scientific) for 20 hours at 4°C to remove the excess peptide molecules in the bulk solution. To synthesize oil-core SNCs having tunable Young’s modulus, the dialyzed nanoemulsion was diluted by a factor of 2, and 3 ml of the diluted nanoemulsion was mixed with 80 mM silica precursor, which is a mixture of TEOS and TEVS with the molar percentage of TEOS being 0, 5, 10, 20, 40, and 100. The reaction was conducted under magnetic stirring at room temperature for 30 hours. The prepared nanocapsule suspension was then dialyzed in Hepes buffer (25 mM, pH 7.5) at 4°C for 20 hours to stop the reaction and remove any excess chemicals. To prepare DiI-labeled SNCs, DiI was dissolved in Miglyol 812 oil at a concentration of 500 μg/ml. Then, the SNCs were synthesized following the standard procedure described above.
Surface modification of SNCs with FA-PEG and PEG
Surface of the SNCs was modified with FA-PEG and PEG for specific and nonspecific cellular uptake studies, respectively. Briefly, 3-aminopropyltriethoxysilane (4 mM) was added into the as-prepared nanocapsule suspension and react for 4 hours at room temperature to allow the amination of nanocapsule surfaces. To make FA-PEG–modified SNCs, 7.2 mM mPEG-NHS and 0.8 mM FA-PEG-NHS were added into the suspension, which was then stirred for 16 hours at 4°C. When making PEG-modified SNCs, only 8 mM mPEG-NHS was added into the suspension. The modified nanocapsules were collected by centrifugation twice (21,885g for 5 min) and resuspending in phosphate-buffered saline (PBS).
Characterization of SNCs
Dynamic light scattering
The hydrodynamic sizes and ζ potentials of SNCs were measured by dynamic light scattering using a Malvern Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) at 25°C with a scattering angle of 173°.
Transmission electron microscopy
The morphologies of SNCs were observed by TEM using a JEOL 1010 transmission electron microscope (JEOL, Tokyo, Japan) operated at 100 kV. To prepare samples, 2 μl of SNC suspension was placed on Formvar-coated copper grids (ProSciTech, Townsville, Australia) and air-dried. The deformation of SNCs during cellular uptake was also observed by TEM at 80 kV. To do this, SKOV3 cells were seeded in cell culture petri dish (Nunclon Delta surface, Thermo Fisher Scientific, Australia) at a concentration of 1 × 105 cells/ml and cultured at 37°C for 24 hours. The cells were then coincubated with SNCs (~5 × 1010 nanocapsules/ml) for 12 hours at 37°C to allow the uptake of SNCs. The cells were then washed with PBS and further coincubated with SNCs (~1 × 1010 nanocapsules/ml) for 2 hours at 4°C to allow the binding of SNCs onto cell surface. After this, the cells were incubated at 37°C for 10 min to allow the commencement of internalization of SNCs. The cells were then fixed with glutaraldehyde and OsO4, stained, and dehydrated in a graded acetone series, infiltrated with Durcupan resin and incubated in 60°C for 48 hours. The cell-embedded resin was then sliced into ~50-nm sections for TEM observation.
Scanning electron microscopy
The morphologies (deformation) of SNCs were observed by scanning electron microscopy (SEM) using a JEOL JSM-7001F field emission scanning electron microscope (JEOL, Tokyo, Japan) with a hot (Schottky) electron gun. SNC suspension (20 μl) was placed on a clean coverglass and air-dried. The coverglass was then mounted onto a SEM stub and coated with a thin layer (~10 nm) of iridium using a Quorum Q150T metal coater (Quorum Technologies, Lewes, UK), followed by plasma cleaning using an Evactron 25 plasma cleaner (XEI Scientific, Redwood City, CA, USA).
Atomic force microscopy
The mechanical properties of SNCs were measured by liquid phase AFM using a Cypher atomic force microscope (Asylum Research, Santa Barbara, CA, USA). The SNCs were first dialyzed in Milli-Q water at 4°C for 20 hours to remove salts. Then, 20 μl of SNC suspension was placed onto a freshly cleaved mica surface and air-dried. Before measurements, 100 μl of Milli-Q water was placed on the mica substrate to submerge the SNCs. Because of the electrostatic attraction between the negatively charged mica and the positively charged SNCs, the latter were immobilized during measurements. Cantilevers having a monocrystal silicon tip and a nominal spring constant (kN) of 3.5 N/m (Etalon, NT-MDT, Russia) were used to image all SNCs. The same cantilever was used to conduct force mapping (with a load of 15 nN) to obtain force-indentation curves of the SNCs made by precursors containing 5, 10, 20, 40, and 100% of TEOS. For SNCs made by the precursor containing 0% of TEOS, force mapping (with a load of 3 nN) was conducted using cantilevers having a pyramid tip and a nominal kN of 0.06 N/m (SiNi, BudgetSensors, Bulgaria). For every sample, 15 different SNCs were analyzed. The Young’s modulus maps of the SNCs were obtained by fitting the force-indentation maps using the Hertzian contact model. Before every measurement and imaging, the cantilevers were calibrated on a clean glass slide to ensure that the deviation of kN was within ±10%. AFM data were processed using IGOR Pro software (v. 6.37).
Fluorescence microscopy
All live cell imaging in this study was performed on a Leica TCS SP8 confocal platform (Leica Camera, Wetzlar, Germany) equipped with a 63×, 1.4 numerical aperture oil immersion objective and an Okolab GS35-M stage top incubator (Okolab, Ottaviano, Italy). FluoroBrite Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) was used in the imaging as cell culture medium to reduce the interference of background fluorescence.
Cellular uptake, binding, and endocytic pathway of SNCs
Cell cultures
RAW264.7 (murine macrophage cells) and SKOV3 (human ovarian adenocarcinoma cells) cells were obtained from the American Type Culture Collection and cultured in high-glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco) at 37°C in a humidified atmosphere with 5% CO2.
Cellular binding of SNCs
The cellular binding of FA-PEG–modified SNCs to RAW264.7 and SKOV3 cells was studied on the basis of the method described previously (20). Briefly, RAW264.7 or SKOV3 cells were seeded in 24-well plates (1 × 105 cells per well) and cultured at 37°C for 24 hours. The cells were then cooled in an ice bath and coincubated with DiI-loaded SNCs (at a concentration of around 1.25 × 1010 capsules/ml, see the Supplementary Materials for more details) in an ice bath in a 4°C cold room for 1 hours. The cells were then washed three times with precooled PBS to remove any unbound SNCs. The cells were then resupplied with medium and cultured at 37°C for 3 hours to allow the internalization of bound SNCs. After this, the cells were trypsinized, washed three times, and resuspended in PBS before their fluorescence intensities were measured by flow cytometry using an Accuri C6 flow cytometer (BD Biosciences, North Ryde, Australia). Cells that were not cocultured with SNCs were regarded as blank groups.
Cellular uptake of SNCs
RAW264.7 or SKOV3 cells were seeded in 24-well plates (1 × 105 cells per well) and cultured at 37°C for 24 hours to allow attachment. The cells were then coincubated with DiI-loaded SNCs (at a concentration of around 1.25 × 1010 nanocapsules/ml) at 37°C for 4, 12, or 24 hours. The cells were then trypsinized, washed three times, and resuspended in PBS before their fluorescence intensities were measured by flow cytometry. Cells that were not cocultured with SNCs were regarded as blank groups. To determine the endocytic pathway of SNCs, the cells were cultured in medium containing endocytosis inhibitors: chlorpromazine hydrochloride (20 μg/ml), nystatin (100 U/ml), or cytochalasin D (1 μg/ml) for 2 hours before they were coincubated with DiI-loaded SNCs for another 4 hours (the inhibitors were maintained during the coincubation). Then, the cells were trypsinized, washed three times, and resuspended in PBS, with their fluorescence intensities measured by flow cytometry. Cells that were not cocultured with SNCs were regarded as blank groups.
Observation of SNC lysosomal transportation using live-cell confocal microscopy
SKOV3 cells were seeded in cell imaging dishes equipped with a 170-μm-thick coverglass bottom (Eppendorf, Hamburg, Germany) at a concentration of 4 × 104 cells/ml and were cultured at 37°C for 24 hours. The cells were then cooled down in an ice bath to avoid endocytosis, with the medium removed, leaving a thin fluid film on the cell monolayer. Then, 50 μl of DiI-loaded SNCs (~5 × 1011 nanocapsules/ml) was added to allow a fast binding to the cell surface. One minute after the SNC addition, the cells were washed twice with precooled PBS to remove any unbound SNCs and resupplied with medium. To provide a defined starting point of SNC incubation, 2 min after the cells were mounted to the stage top incubator (at 37°C) was regarded as the time point of zero. Confocal micrographs were acquired every 10 min. To stain lysosomes, the cells were cultured in medium containing 50 nM LysoTracker Deep Red stain 30 min before the addition of SNCs (the stain was maintained during the observation). For dual color detection of SNCs and lysosomes, the cells were excited with alternating 549-nm (DiI) and 647-nm (LysoTracker Deep Red) laser light. Images were analyzed using Leica Application Suite X software (v. 3.0).
Single-particle tracking
SKOV3 cells were seeded in cell imaging dishes equipped with a 170-μm-thick coverglass bottom at a concentration of 4 × 104 cells/ml and were cultured at 37°C for 24 hours. The cells were then cooled down in an ice bath and cultured in medium containing DiI-loaded SNCs (~5 × 108 nanocapsules/ml) for 10 min to allow the binding of SNCs to cell surface. Then, the cells were washed twice with precooled PBS to remove any unbound SNCs and resupplied with medium. To provide a defined starting point of SNC incubation, time point zero was set as 1 min after the cells were mounted to the stage top incubator (at 37°C). Micrograph sequences were acquired by high-speed resonant scanning, with the cells excited at 549-nm laser light for 500 ms per frame. An autofocus system was used to fix the z position of samples during imaging. The overlay images of the green and bright-field channels as well as time projections of the recorded movies were built using ImageJ Fiji software (v. 1.51n). Particle tracking was performed by the TrackMate plugin (31) of ImageJ Fiji, and the tracks were analyzed by MATLAB (R2017b).
Statistical analysis
Statistics were computed using Microsoft Excel 2013. Standard unpaired one-tailed Student’s t test was used to test for statistical significance between groups, with P ≥ 0.05, P < 0.05, P < 0.01, and P < 0.001 denoted as N.S., *, **, and #, respectively. Values for n, P, and the specific statistical test performed for each experiment are included in the appropriate figure legend or in the main text.
Supplementary Material
Acknowledgments
We acknowledge the facilities and the scientific and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, The University of Queensland. This work was performed in part at the Queensland node of the Australian National Fabrication Facility (ANFF-Q), a company established under the National Collaborative Research Infrastructure Strategy to provide nano- and microfabrication facilities for Australia’s researchers. Funding: This research was primarily supported by the Australian Research Council under Discovery Project (DP150100798) and Future Fellowship Project (FT140100726). Y.H. acknowledges scholarships from The University of Queensland and the Chinese Scholarship Council. H.G. acknowledges financial support from the National Science Foundation (grant CMMI-1562904). X.Y. acknowledges support from the National Natural Science Foundation of China (grant no. 11872005). Author contributions: C.-X.Z. designed the project. Y.H. performed most experiments. X.Y. and H.G. performed the theoretical work. D.W. prepared the TEM samples for cells. G.Y. performed the NanoSight measurements. All authors wrote and reviewed the manuscript. Competing interests: A.P.J.M., C.-X.Z., and D.W. are inventors on a patent related to this work filed by The University of Queensland (European Patent Office: EP3046870A1; Australia: AU2014321161B2; United States: US20160193581A1; and Canada: CA2959201A1). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaaz4316/DC1
REFERENCES AND NOTES
- 1.Gillespie P. G., Walker R. G., Molecular basis of mechanosensory transduction. Nature 413, 194–202 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Luo T., Mohan K., Iglesias P. A., Robinson D. N., Molecular mechanisms of cellular mechanosensing. Nat. Mater. 12, 1064–1071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu Y., Brown A. C., Myers D. R., Sakurai Y., Mannino R. G., Tran R., Ahn B., Hardy E. T., Kee M. F., Kumar S., Bao G., Barker T. H., Lam W. A., Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc. Natl. Acad. Sci. U.S.A. 111, 14430–14435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel V., Sheetz M., Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Discher D. E., Janmey P., Wang Y.-L., Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Yap A. S., Duszyc K., Viasnoff V., Mechanosensing and mechanotransduction at cell–cell junctions. Cold Spring Harb. Perspect. Biol. 10, a028761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimatsu M., Mekhdjian A. H., Adhikari A. S., Dunn A. R., Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 13, 3985–3989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Ha T., Defining single molecular forces required to activate integrin and notch signaling. Science 340, 991–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banquy X., Suarez F., Argaw A., Rabanel J.-M., Grutter P., Bouchard J.-F., Hildgen P., Giasson S., Effect of mechanical properties of hydrogel nanoparticles on macrophage cell uptake. Soft Matter 5, 3984–3991 (2009). [Google Scholar]
- 10.Sun J., Zhang L., Wang J., Feng Q., Liu D., Yin Q., Xu D., Wei Y., Ding B., Shi X., Jiang X., Tunable rigidity of (polymeric core)–(lipid shell) nanoparticles for regulated cellular uptake. Adv. Mater. 27, 1402–1407 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Anselmo A. C., Zhang M., Kumar S., Vogus D. R., Menegatti S., Helgeson M. E., Mitragotri S., Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano 9, 3169–3177 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Anselmo A. C., Mitragotri S., Impact of particle elasticity on particle-based drug delivery systems. Adv. Drug Deliv. Rev. 108, 51–67 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Teng Z., Wang C., Tang Y., Li W., Bao L., Zhang X., Su X., Zhang F., Zhang J., Wang S., Zhao D., Lu G., Deformable hollow periodic mesoporous organosilica nanocapsules for significantly improved cellular uptake. J. Am. Chem. Soc. 140, 1385–1393 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Guo P., Liu D., Subramanyam K., Wang B., Yang J., Huang J., Auguste D. T., Moses M. A., Nanoparticle elasticity directs tumor uptake. Nat. Commun. 9, 130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M., Xu L., Tian F., Su Q., Zheng N., Yang Y., Wang J., Wang A., Zhu C., Guo S., Zhang X., Gan Y., Shi X., Gao H., Rapid transport of deformation-tuned nanoparticles across biological hydrogels and cellular barriers. Nat. Commun. 9, 2607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui Y., Wibowo D., Liu Y., Ran R., Wang H.-F., Seth A., Middelberg A. P. J., Zhao C.-X., Understanding the effects of nanocapsular mechanical property on passive and active tumor targeting. ACS Nano 12, 2846–2857 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Chen Y., Ju L., Rushdi M., Ge C., Zhu C., Weaver V. M., Receptor-mediated cell mechanosensing. Mol. Biol. Cell 28, 3134–3155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wibowo D., Zhao C.-X., Middelberg A. P. J., Emulsion-templated silica nanocapsules formed using bio-inspired silicification. Chem. Commun. 50, 11325–11328 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Adumeau L., Genevois C., Roudier L., Schatz C., Couillaud F., Mornet S., Impact of surface grafting density of PEG macromolecules on dually fluorescent silica nanoparticles used for the in vivo imaging of subcutaneous tumors. Biochim. Biophys. Acta. Gen. Subj. 1861, 1587–1596 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Lesniak A., Salvati A., Santos-Martinez M. J., Radomski M. W., Dawson K. A., Åberg C., Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 135, 1438–1444 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Yi X., Shi X., Gao H., Cellular uptake of elastic nanoparticles. Phys. Rev. Lett. 107, 098101 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Yi X., Gao H., Kinetics of receptor-mediated endocytosis of elastic nanoparticles. Nanoscale 9, 454–463 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Shen Z., Ye H., Yi X., Li Y., Membrane wrapping efficiency of elastic nanoparticles during endocytosis: Size and shape matter. ACS Nano 13, 215–228 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Champion J. A., Mitragotri S., Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 103, 4930–4934 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilden J., Krummel M. F., Control of cortical rigidity by the cytoskeleton: Emerging roles for septins. Cytoskeleton 67, 477–486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charras G. T., Yarrow J. C., Horton M. A., Mahadevan L., Mitchison T. J., Non-equilibration of hydrostatic pressure in blebbing cells. Nature 435, 365–369 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pablo R., Veronika M., Dianne C., Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunol. Rev. 256, 222–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saleem M., Morlot S., Hohendahl A., Manzi J., Lenz M., Roux A., A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat. Commun. 6, 6249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulant S., Kural C., Zeeh J.-C., Ubelmann F., Kirchhausen T., Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13, 1124–1131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cureton D. K., Massol R. H., Saffarian S., Kirchhausen T. L., Whelan S. P. J., Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLOS Pathog. 5, e1000394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tinevez J.-Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L., Eliceiri K. W., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Ruthardt N., Lamb D. C., Bräuchle C., Single-particle tracking as a quantitative microscopy-based approach to unravel cell entry mechanisms of viruses and pharmaceutical nanoparticles. Mol. Ther. 19, 1199–1211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Z., Ye H., Li Y., Understanding receptor-mediated endocytosis of elastic nanoparticles through coarse grained molecular dynamic simulation. Phys. Chem. Chem. Phys. 20, 16372–16385 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Helfrich W., Elastic properties of lipid bilayers: Theory and possible experiments. Z. Naturforsch. C 28, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- 35.Liu J., Kaksonen M., Drubin D. G., Oster G., Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl. Acad. Sci. U.S.A. 103, 10277–10282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walani N., Torres J., Agrawal A., Endocytic proteins drive vesicle growth via instability in high membrane tension environment. Proc. Natl. Acad. Sci. U.S.A. 112, E1423–E1432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T., Sknepnek R., Bowick M. J., Schwarz J. M., On the modeling of endocytosis in yeast. Biophys. J. 108, 508–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassinger J. E., Oster G., Drubin D. G., Rangamani P., Design principles for robust vesiculation in clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 114, E1118–E1127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herant M., Heinrich V., Dembo M., Mechanics of neutrophil phagocytosis: Behavior of the cortical tension. J. Cell Sci. 118, 1789–1797 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Herant M., Heinrich V., Dembo M., Mechanics of neutrophil phagocytosis: Experiments and quantitative models. J. Cell Sci. 119, 1903–1913 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Huang X., Chisholm J., Zhuang J., Xiao Y., Duncan G., Chen X., Suk J. S., Hanes J., Protein nanocages that penetrate airway mucus and tumor tissue. Proc. Natl. Acad. Sci. U.S.A. 114, E6595–E6602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi X., Gao H., Cell membrane wrapping of a spherical thin elastic shell. Soft Matter 11, 1107–1115 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Reissner E., Stresses and small displacements of shallow spherical shells, II. J. Math. Phys. 25, 279–300 (1946). [Google Scholar]
- 44.Footer M. J., Kerssemakers J. W. J., Theriot J. A., Dogterom M., Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl. Acad. Sci. U.S.A. 104, 2181–2186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts, P. Walter, Molecular Biology of the Cell (Garland Science, ed. 5, 2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaaz4316/DC1


