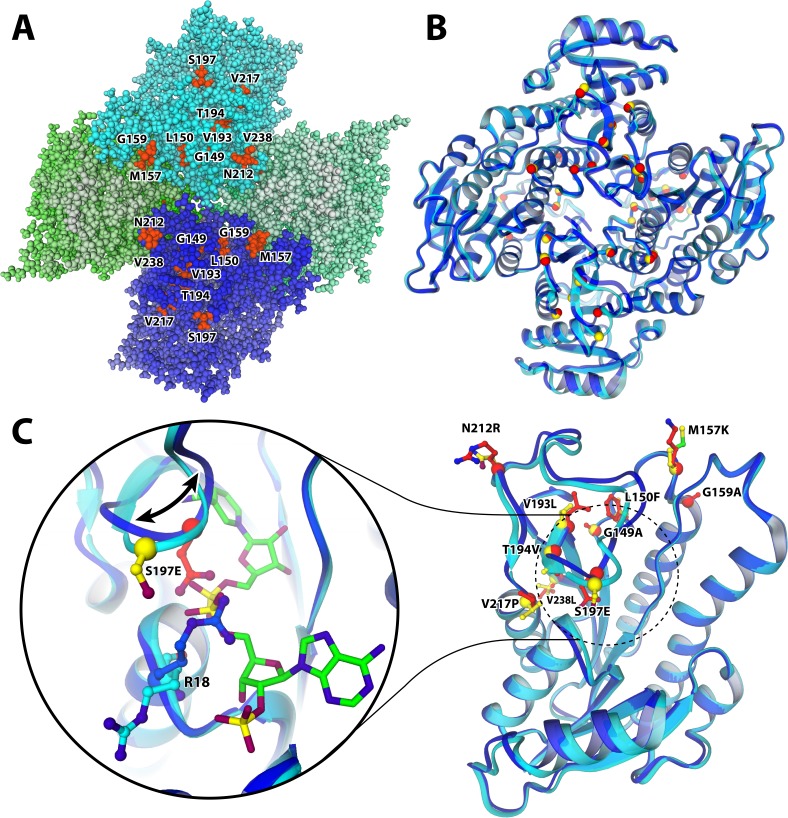
Figure 4. Structure of the M9 mutant of ADHA with mutated resides highlighted.
(A and B) quaternary structure of M9. The tetramer is organized such that the N-termini are on the outside (on the edge of the top-down view of A and B), whereas the C-termini all point inwards; which is where most and the most stabilizing mutations were found. (A) M9 structure with all atoms represented as balls. The four monomers are shaded in various colours, highlighting the particular clustering of the observed stabilizing mutations. (B) The structure as ribbon model, superimposing the mutant (blue ribbon, red spheres indicate mutated residue) and the wild type (cyan ribbon, yellow spheres). (C) Colour scheme as in B. The loop (196-214) that is dislocated as a result of the S197E mutation, compared to the structure of wild-type ADHA. The shift is accompanied by a flip of R18 into the NADP-binding pocket, likely due to an electrostatic attraction from the mutant glutamate. As a result, the cofactor (green carbons) is only bound in the wild type, while absent from the mutant structure.