Abstract
Objective
To describe the clinical and pathologic features of a novel pedigree with heterozygous STUB1 mutation causing SCA48.
Methods
We report a large pedigree of Dutch decent. Clinical and pathologic data were reviewed, and genetic analyses (whole-exome sequencing, whole-genome sequencing, and linkage analysis) were performed on multiple family members.
Results
Patients presented with adult-onset gait disturbance (ataxia or parkinsonism), combined with prominent cognitive decline and behavioral changes. Whole-exome sequencing identified a novel heterozygous frameshift variant c.731_732delGC (p.C244Yfs*24) in STUB1 segregating with the disease. This variant was present in a linkage peak on chromosome 16p13.3. Neuropathologic examination of 3 cases revealed a consistent pattern of ubiquitin/p62-positive neuronal inclusions in the cerebellum, neocortex, and brainstem. In addition, tau pathology was present in 1 case.
Conclusions
This study confirms previous findings of heterozygous STUB1 mutations as the cause of SCA48 and highlights its prominent cognitive involvement, besides cerebellar ataxia and movement disorders as cardinal features. The presence of intranuclear inclusions is a pathologic hallmark of the disease. Future studies will provide more insight into its pathologic heterogeneity.
Spinocerebellar ataxias (SCAs) comprise a heterogeneous group of disorders characterized by progressive cerebellar ataxia in combination with noncerebellar signs, including (extra) pyramidal features, peripheral neuropathy, and cognitive impairment.1 In most cases, prominent loss of cerebellar Purkinje neurons is observed, yet neuropathologic changes can be diverse with degeneration extending to all brain areas. Frequently, intranuclear neuronal inclusions serve as a morphologic marker.2
Currently, over 40 autosomal dominant SCAs have been identified.1 Polyglutamine SCAs caused by trinucleotide CAG repeat expansion represent the most common form. Other genetic causes include repeat expansion in noncoding regions and conventional mutations.3 A large percentage of patients with familial SCA (30%–48%) remain without an identified genetic defect.4 In the past decade, an increasing number of rare causal variants have been recognized in both dominant and recessive forms of ataxia.5 Among these are variants in STUB1 (STIP1 homologous and U-box-containing protein 1; OMIM#607207), the gene encoding the protein C-terminus of Hsp70-interacting protein (CHIP). This molecular co-chaperone plays a prominent role in protein quality control processes and the cellular stress response.6 Recessive STUB1 mutations were identified in families with SCAR16, showing a wide variability of symptoms, including hypogonadism, epilepsy, autonomic dysfunction, and dementia.7,8 More recently, heterozygous variants in STUB1 have been implicated in autosomal dominant SCA (SCA48; OMIM#618093), characterized by a complex phenotype including cognitive/affective symptoms and movement disorders.9–12
In this study, we present a large Dutch SCA48 family associated with a novel heterozygous frameshift mutation in the U-box domain of STUB1 and describe the neuropathologic features of 3 patients in this family.
Methods
Clinical data
Medical records and neuroimaging of 9 affected individuals from 1 large family of Dutch ancestry were collected and reviewed (figure 1 and table 1). Seven of them underwent a single clinical assessment by neurologists from the research group (E.B., J.C.v.S., and L.D.K.) during a local visit. Blood samples were collected from 7 affected and 5 unaffected family members; DNA was extracted using standard procedures.
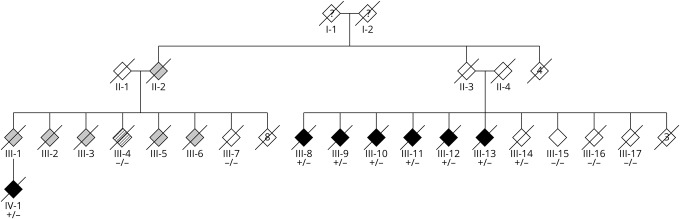
Figure 1. Pedigree of the family.
Filled black symbols represent patients who were personally examined by the researchers. Filled gray symbols are affected individuals based on medical records (III-2 and III-3) or on reports from family members (II-2, III-1, III-5, and III-6). Individuals III-7, III-14, III-15, III-16, and III-17 were clinically unaffected and all deceased >75 years of age. Individual II-3 is an obligate carrier but did not have neurologic symptoms according to the family. Individual III-4 was diagnosed with Alzheimer disease without motor symptoms and without cerebellar atrophy and not considered to have the same phenotype. The numbers inside the symbols represent the number of individuals. Sex is not specified to protect anonymity. Transmission was independent of sex (including male to male transmission). + = mutated allele; – = wild type.
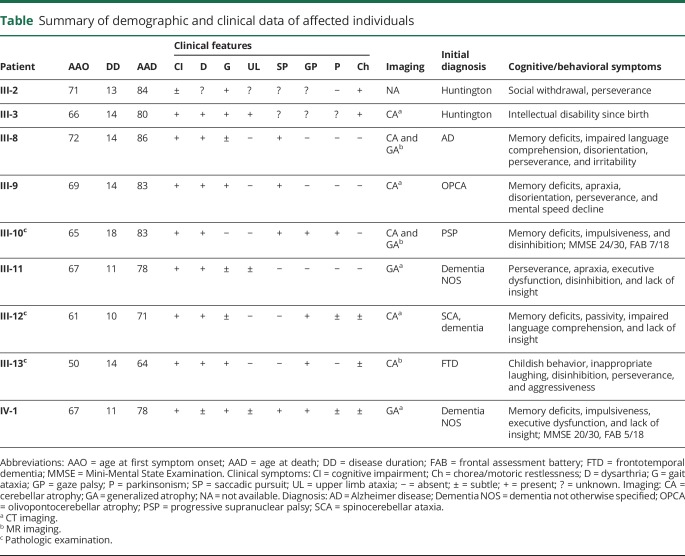
Table.
Summary of demographic and clinical data of affected individuals
Standard protocol approvals, registrations, and patient consents
The study was approved by the Medical Ethical Committee of the Erasmus Medical Center Rotterdam. Brain autopsy was conducted by the Netherlands Brain Bank according to their Legal and Ethical Code of Conduct. Written informed consent was obtained from all participants and/or their legal representatives.
Genetic studies
Linkage analysis
Single nucleotide polymorphism (SNP) array genotyping (Illumina Human OmniExpress-24 v1.0 BeadChip) was performed on 7 patients (III-8, III-9, III-10, III-11, III-12, III-13, and IV-1). SNP call data were adapted by GenomeStudio (Illumina) for linkage analyses using Allegro, implemented in easyLINKAGE Plus.13,14 An affected-only linkage analysis was performed. Mendelian inheritance check was performed with the program PedCheck.15 SNPs showing mendelian inconsistencies were excluded from the calculations. Multipoint linkage analysis was performed with an SNP spacing of 0.3 cM. Logarithm of the odds (LOD) scores were calculated under the assumption of an autosomal dominant disorder.
Whole-exome and genome sequencing
Whole-exome sequencing (WES) was performed on the same 7 patients as those included in the linkage analysis. Whole-genome sequencing (WGS) was performed on 3 of these patients (III-12, III-13, and IV-1). Genomic DNA was fragmented to 150–200 base pairs (bp), end paired, adenylated, and ligated to adapters. The SeqCap capturing kit for Illumina Paired-End Sequencing Library (version 2.0.1; NimbleGen) was used. The captured fragments were purified and sequenced on an Illumina Hiseq2000 platform using 100 bp paired-end reads. WGS was performed by Novogene on an Illumina Hiseq2000 platform using 150 bp paired-end reads. Bioinformatic analysis was performed using an in-house pipeline based on published tools. Briefly, sequence reads were aligned to the human reference genome (hg19) using Burrows-Wheeler Aligner (version 0.7.3a).16 Subsequently, alignment data were processed and recalibrated using Picard (version 1.90) and the Genome Analysis Toolkit (GATK version 2.5.2). SNPs and small indels were called using GATK's HaplotypeCaller and Variant recalibration following best practices.17 The WES and WGS data sets were generated separately. The lists of variants from the WES data were annotated with Annovar18 and filtered to include (1) exonic and splice site variants, (2) nonsynonymous, (3) with rare occurrence in publicly available databases (1000G, Exome Aggregation Consortium and Exome Sequencing Project; minor allele frequency <0.1%), and (4) QD score >5 (quality score normalized by allele depth). Functional predictions by Sorting Intolerant From Tolerant and/or PolyPhen-2 were taken into account. In addition, to detect the possible presence of expanded polyglutamine repeats, short tandem repeats (STRs) were called using WGS data in the chr16p13.3 linkage peak using LobSTR (v3.0.3) according to best practices.19 STRs were filtered for heterozygosity in all 3 carriers. The repeat motif and number of copies per carrier were extracted and analyzed manually. RefSeq gene annotation was added using Annovar (ref; 20601685). The candidate variant was checked by Sanger sequencing (details provided in supplementary file, links.lww.com/NXG/A251).
Neuropathology
Three patients died during follow-up (III-10, III-12, and III-13); brain autopsy was performed by the Netherlands Brain Bank within 4 hours after death. Formalin-fixed (10%) and paraffin-embedded tissue blocks were available for examination. Eight-micrometer sections of all cortical regions, subcortical nuclei, brainstem, and cerebellum underwent routine staining. Immunohistochemistry was performed using the following antibodies: hyperphosphorylated tau (AT8, Innogenetics, Ghent, Belgium; 1:40), beta-amyloid (anti-amyloid, DAKO, Glostrup, Denmark; 1:100, following formic acid pretreatment), alpha-synuclein (anti-synuclein, Zymed Laboratories, San Francisco, CA; undiluted, following formic acid pretreatment), TDP-43 (anti-phospho TDP-43, Cosmo Bio, 1:100 and Proteintech Group, 1:100), ubiquitin (anti-ubiquitin, DAKO, Glostrup, Denmark; 1:500, following 80°C antigen retrieval), p62 (mouse D3 Clone, Santa Cruz, 1:100), 1C2 (mouse 5TF1-1C2-172 Clone, Chemicon, 1:3,200), and STUB1 (rabbit anti-STUB1 Abcam ab2917, 1:100).
Data availability
The deidentified data generated and analyzed in the current study will be available and shared by request to the corresponding author (M.O.M.) from any qualified investigator. These data include deidentified clinical data, raw exome/genome files, microsatellite genotyping, and additional pictures of stainings from the studied material.
Results
Clinical characteristics
The pedigree structure is presented in figure 1, and the main clinical features of 9 affected individuals are summarized in table 1. Individuals II-2, II-3, III-1, III-2, III-3, III-5, and III-6 were deceased before the start of the study, and DNA was not available. Patient III-4 was not included in the genetic analyses (linkage and WES) because the clinical signs did not fully match with the other family members (i.e., isolated cognitive dysfunction suggestive of Alzheimer disease [AD] without ataxia or movement disorders). Mean age at onset of disease in the 9 patients was 65.3 ± 6.6 years. These patients deceased after a mean disease duration of 13.2 ± 2.4 years. The presenting features consisted of slowly progressive gait disturbance (mainly ataxic and frequently described as a waddled gait) combined with prominent cognitive dysfunction and behavioral changes with impaired insight, disinhibition, and perseverations. The cognitive deficits were among the first symptoms in some patients, showing a gradual decline in memory performance and executive functioning. Imaging in 6 of 9 patients showed cerebellar atrophy; 2 had moderate to severe generalized atrophy (for 1 patient, no scan was available). Initial clinical diagnoses by reviewing medical records were olivopontocerebellar atrophy, Huntington disease (without genetic confirmation), unspecified chorea, progressive supranuclear palsy (PSP), AD, frontotemporal dementia (FTD), and unspecified dementia.
Two patients are further described in more detail: patient III-10 presented at age 65 years with a parkinsonian gait, frequent falls, and bradykinesia unresponsive to levodopa. Subsequently, memory problems developed together with behavioral changes and impulsiveness. On neurologic examination, dysarthria, an upward gaze palsy, bilateral bradykinesia, a shuffling gait with freezing, and abnormal postural reflexes were found. Neuroimaging with MRI showed generalized atrophy, including cerebellar volume loss. This patient died 18 years after disease onset at age 83 years.
Patient III-13 presented with progressive behavioral changes from age 50 years, including inappropriate laughing, aggressiveness, and perseverations. Neurologic examination revealed dysarthria, diffuse motor restlessness, and stereotypic movements. Neuropsychological evaluation indicated pronounced frontal symptoms and semantic deficits, but with preservation of episodic memory. On MRI, severe cerebellar and bilateral parietal atrophy was observed. The hippocampus, frontal, and temporal lobes showed no apparent degeneration. This patient died after a disease duration of 14 years.
Genetic analyses
Diagnostic testing excluded mutations in MAPT, C9orf72, PSEN1, PRNP, HTT, DRPLA, FMR1, SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17. Genome-wide linkage analysis using SNP array data did not reveal a significance linkage peak (LOD score >3). However, a few regions showed suggestive linkage, including the region on chromosome 16p13.3 with an LOD score of 2.35. After filtering, WES data of 7 patients revealed 5 candidate variants, all of them in the heterozygous state and located on chromosome 16p13.3. These included 4 nonsynonymous missense variants (AXIN1, IGFALS, PDK1, and THOC6) and 1 frameshift variant (STUB1). Analysis of WGS data of 3 patients using the same filtering steps confirmed these 5 variants; no additional variants were found. In these patients, LobSTR was used on the chromosome 16p13.3 linkage area to identify possible expanded polyglutamine repeats, but none were detected.
Of these 5 candidates, the frameshift variant in STUB1 is the most likely candidate because this gene has recently been linked to both recessive (SCAR16) and dominant ataxia (SCA48). The 2-bp deletion c.731_732delGC (p.C244Yfs*24) is located in exon 6, which encodes the U-box domain together with the last exon. This variant is absent in publicly available databases, and Sorting Intolerant From Tolerant predicts a damaging effect (confidence score 0.86). The mutation was confirmed by Sanger sequencing (figure e-1, links.lww.com/NXG/A251). Subsequently, 5 unaffected relatives were tested (III-7, III-14, III-15, III-16, and III-17), and in 4 of 5, the variant was absent. Individual III-4, who was not considered to have the same phenotype, did not have the STUB1 variant. There was no report of neurologic symptoms in individual II-3, who is an obligate carrier of the STUB1 variant, but died at age 64 years due to a myocardial infarction.
Neuropathology
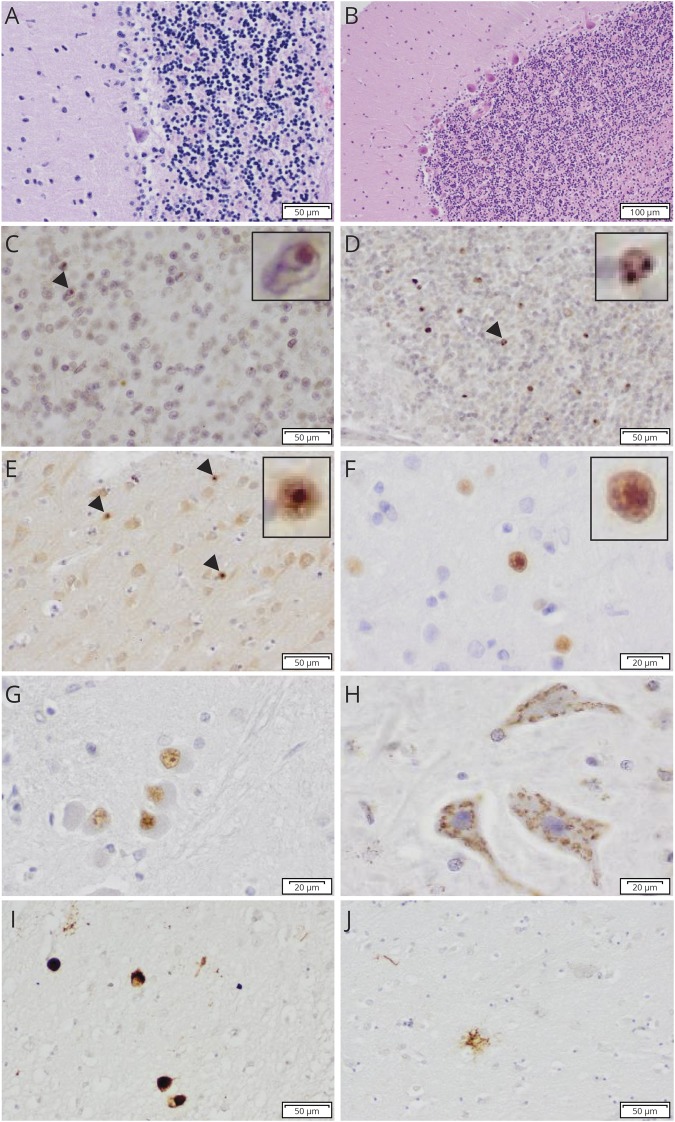
Neuropathologic examination was performed in 3 patients (III-10, III-12, and III-13). Brain weight varied slightly (1,304, 1,223, and 1,090 g, respectively). Macroscopic inspection showed cerebellar atrophy of vermis and hemispheres in all 3 cases, although less profound in III-10. Routine staining on the 3 brains showed no abnormalities in cortical areas, except for focal neuron loss in the parietal and occipital cortices in III-13. Cases III-12 and III-13 contained no apparent hippocampal plaques or tangles (Braak stage 1), whereas a few tangles and neuropil threads were seen in the Ammon horn in III-10 (Braak stage 2). All 3 cases showed nearly complete loss of Purkinje cells with Bergmann gliosis (figure 2A) and severe neuronal loss in the mesencephalon and medulla oblongata. In III-10, the subthalamic nucleus also showed atrophy with severe gliosis. Staining of the cerebellum of III-13 with ubiquitin showed several neuronal intranuclear inclusions (NIIs) in cerebellar granular cells (figure 2C). In all 3 cases, immunohistochemistry with p62 showed similar NII in the cerebellum (figure 2D) and in all cortical areas, most prevalent in posterior regions (figure 2E). NIIs were also present in the substantia nigra and deep pontine nuclei. In the hippocampus, many p62-positive inclusions were found, whereas the dentate gyrus showed punctuate cytoplasmic inclusions (CIs) and occasional diffuse nuclear staining (DNS). Staining with 1C2 antibody in III-13 showed DNS in granular and stellate neurons of the cerebellum and in cortical areas (figure 2F). In the substantia nigra, CIs were observed, and the gyrus cinguli showed CI, DNS, and dystrophic neurites. 1C2 staining also exposed DNS in the inferior olives (figure 2G) and CI in the hypoglossal nucleus with a granular pattern (figure 2H). In III-10, faint 1C2 reactivity was seen in the cerebellum, less prominent than with p62. In cortical regions, similar DNS was seen as in III-13. 1C2 staining was not performed in III-12. Staining with the antibody against CHIP in III-13 showed a diffuse staining of neurons, not different from controls (data not shown). AT8 staining in cases III-12 and III-13 showed few abnormalities, except for a few positive neurons in the temporal neocortex (III-12) and a moderate number of pretangles (III-13). Of interest, AT8 staining in III-10 showed many tufted astrocytes, glial staining, and tangles in the thalamus and subthalamic nucleus (figure 2I). Tau pathology was also present in many other regions (i.e., temporal cortex (figure 2J), hippocampus, caudate, putamen, substantia nigra, locus coeruleus, cerebellar dentate nucleus, and spinal motor neurons). Immunohistochemistry for alpha-synuclein and TDP-43 antibodies in several selected brain regions was negative in all 3 cases.
Figure 2. Histopathologic features of affected cases III-13 (A–H) and III-10 (I and J).
(A and B) Nearly complete loss of Purkinje cells, loss of neurons, and spongiosis in the cerebellar granular layer of the patient (A) compared with the control brain (B) in H/E staining; (C and D) NII in the cerebellar granular layer with ubiquitin (C) and p62 staining (D); (E) NII in the occipital cortex with p62 staining; (F) DNS in the occipital cortex with 1C2 staining; (G) DNS in the inferior olive with 1C2 staining; (H) granular CI in the hypoglossal nucleus with 1C2 staining; (I) AT8-positive neurons, neurites, and glial staining in the thalamic nuclei; (J) AT8-positive tufted astrocyte in the putamen. The arrowheads indicate NII, which are magnified in figure 2C-F. CI = cytoplasmic inclusion; DNS = diffuse nuclear staining; H/E = hematoxylin and eosin staining; NII = neuronal intranuclear inclusion.
Discussion
We present a multigenerational family with late-onset ataxia associated with a novel heterozygous STUB1 mutation, fitting the diagnosis of SCA48. Although the various initial diagnoses of the patients illustrate the clinical heterogeneity of this new SCA subtype, there are some strongly overlapping features. The clinical presentation is dominated by cognitive decline and profound behavioral changes, combined with cerebellar ataxia and movement disorders.
Cognitive impairment has been found in other SCA subtypes, but is usually preceded by cerebellar symptoms.20 Here, it was present in all affected family members and occasionally as the first manifestation. One case was initially diagnosed as FTD, although frontal and temporal atrophy were lacking. Other published SCA48 pedigrees with mutations in close proximity to the current mutation showed a broad range of cognitive symptoms.9–12 Another interesting observation is the presence of other movement disorders besides gait ataxia. Chorea or uncontrolled motor activity was frequently reported, showing resemblance with the chorea described in several other families.10–12 Parkinsonism was also present in 4 of 11 described Italian patients with SCA48.11 Atypical parkinsonism with features resembling PSP was observed in 1 patient. The combination of cognitive and movement disorders in SCA48 also resembles SCA17 and dentatorubral-pallidoluysian atrophy. In both disorders, dementia and chorea are part of the clinical features.1,21
The clinical presentation of SCA48 shows overlap with SCAR16. The latter is more complex with a myriad of phenotypes and seems to constitute a multisystemic disorder usually presenting at an earlier age.7,22 Endocrine abnormalities have been described as a major feature of SCAR16, and hypogonadism was present in patients with SCA48.11 Although not formally tested, endocrine abnormalities were not reported in the medical records of the current pedigree.
The homozygous and compound heterozygous STUB1 mutations in SCAR16, and the recent identification of heterozygous STUB1 mutations in adult-onset familial ataxia, support STUB1 as the most likely candidate gene in this family. The identified frameshift variant is predicted to result in a premature stop codon and is located in the highly conserved U-box domain, similar to previously reported variants causing SCAR16 and SCA48.8,9,11,23 The protein encoded by STUB1, CHIP, regulates several proteins involved in neurodegenerative disorders.6 Through its tetratricopeptide repeat domain, CHIP interacts with chaperones (Hsp70 and Hsp90) and a broad range of co-chaperones. The ubiquitin ligase region (U-box) is responsible for the ubiquitination of misfolded proteins destined for elimination.24 Missense mutations in the U-box domain of CHIP impair the intracellular degradation of ataxin-3 microaggregates by preventing their ubiquitination, thereby accelerating disease progression.25 We hypothesize that a similar pathophysiologic mechanism is likely in SCA48 due to impaired function of CHIP.
Earlier work has shown that mutations in the U-box domain causing SCAR16 destabilize CHIP, supporting a loss-of-function mechanism.26 CHIP null mice and animal models with homozygous missense mutation (p.T246M) exhibited striking similarities with characteristics seen in SCAR16 patients, including neuronal degeneration and ataxic motor disturbances.7,27 Patients with recessive STUB1 mutations had decreased CHIP protein levels in isolated fibroblasts in varying degrees, dependent on specific mutations.27 The U-box domain is especially important, as mutations in this domain have a stronger effect on the overall loss of CHIP function and strongly associate with cognitive dysfunction.28 Indeed, most frameshift mutations identified so far in SCA48 are located in the U-box domain (table e-1, links.lww.com/NXG/A251).9,11 Whether this results in nonsense mediated RNA decay or a truncated protein (with a possible dominant negative effect) remains to be elucidated. Additional functional studies on different mutations in STUB1 are essential to provide more insight why some mutations cause SCA48 in heterozygous state and others SCAR16 in a recessive trait. The co-occurrence of recessive and dominant mutations in the same gene has been described before. For example, mutations in HTRA1 are responsible for cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL), while heterozygous mutations causing a late-onset form of small vessel disease have been reported.29 Likewise, truncating heterozygous progranulin mutations cause FTD, whereas homozygous mutations lead to neuronal ceroid lipofuscinosis.30
In the current study, 2 carriers of the STUB1 mutation did not develop symptoms: 1 unaffected carrier died over age 75 years, and another (obligate) carrier died at age 64 years without neurologic disease. This could indicate incomplete penetrance, or it could reflect the diversity in phenotypical expression, given the large variety in ages at onset in the present family (50–72 years). Therefore, it cannot be excluded that parents of some patients with SCAR16 are at risk of developing SCA48.
It would also be of interest to investigate other genetic factors that could influence disease penetrance and progression. Previous studies have shown that expanded ataxin-2 repeats predispose to other neurodegenerative disorders besides SCA2.31 In line with this, it would be interesting to explore whether repeat length variation in ataxin-3, a known interactor of CHIP, might interfere with the function of CHIP and influence the clinical variability in SCA48. Although we have not found toxic exposures in patients (data not shown), it is worthwhile to study more systematically whether environmental factors can trigger the unfolded protein response and may therefore have an effect on disease onset or progression in STUB1-related disorders.
Neuropathologic changes were present in cerebellar but also neocortical areas, suggesting diffuse involvement of the brain. The almost complete loss of Purkinje cells in the present 3 cases resembles that of most other SCAs.2 Neuropathologic studies of other STUB1 cases are scarce, with a single neuropathologic report of an SCAR16 patient.32 This case showed similar severe Purkinje cell loss and p62-positive NIIs, and CHIP staining revealed nuclear staining in neurons of the deep layers of the frontal cortex. We also observed diffuse staining of neurons with CHIP antibody, but not different from controls (data not shown).
NIIs have been considered the hallmark of disorders with repeat expansions.33 However, several studies have demonstrated the presence of NIIs in the absence of pathologically expanded polyglutamine repeats.34 Moreover, NIIs in the present cases did not show 1C2 positivity, which is typically present in SCA caused by polyglutamine repeat expansion.2 Instead, we did observe a pattern of DNS with 1C2. Granular cytoplasmic and DNS of 1C2 as an earlier stage in aggregate formation have been described in both SCA3 and Huntington disease.35 Other studies have demonstrated that 1C2 recognizes nonexpanded ataxin-3 present in NIIs.36,37 It is possible that recruitment of ataxin-3 into the nucleus is involved in the pathogenesis of STUB1 disease.38 Finally, 1C2 immunoreactivity might be less specific, as it has also been detected in nonrepeat disorder, such as a SCA6 case with 22–24 polyglutamine stretches39 and in healthy controls.40 Future investigations are needed to clarify the exact meaning of the different types of staining patterns on the cellular (dys)function.
Intriguingly, extensive tau pathology was found in 1 patient with clinical similarities to PSP (III-10). This has up till now only been seen in SCA11, a pure progressive cerebellar ataxia.2 Several studies highlight the role of CHIP in tau pathology. It attenuates the pathologic changes associated with tau, playing a primary role in tau ubiquitination and degradation.41 CHIP levels were found to be elevated in AD, suggesting a role in tangle maturation,42 and CHIP depletion in mice was associated with accumulation of hyperphosphorylated tau.43 The discovery of different pathologies within 1 kindred is known to occur also in other (genetic) neurodegenerative diseases.44
The present pedigree along with the recently described families underscores the association between heterozygous STUB1 mutations and SCA48. The combination of familial late-onset gait ataxia, cognitive decline with behavioral changes, and other movement disorders (parkinsonism, chorea, and stereotypic movements) is indicative of SCA48. We show that the pathology of SCA48 is dominated by Purkinje cell loss, p62-positive intranuclear inclusions, and may also include tau pathology. Additional functional studies and reports of newly identified SCA48 and SCAR16 families will provide more insight into the heterogeneous spectrum of STUB1-related disorders.
Acknowledgment
The authors are indebted to all the patients who made this study possible. They also thank Prof. A. Rozemuller from the Netherlands Brain Bank for the neuropathologic examination of the cases.
Glossary
- AD
Alzheimer disease
- CHIP
C-terminus of Hsp70-interacting protein
- CI
cytoplasmic inclusion
- DNS
diffuse nuclear staining
- FTD
frontotemporal dementia
- GATK
Genome Analysis Toolkit
- NII
neuronal intranuclear inclusion
- PSP
progressive supranuclear palsy
- SCA
spinocerebellar ataxia
- STR
short tandem repeat
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
Appendix. Authors
Study funding
This work was financially supported by Prinses Beatrix Fonds (grant number 01-0128) and by the Internationaal Parkinson Fonds (IPF).
Disclosure
Several authors of this publication are members of the European Reference Network for Rare Neurological Diseases - Project ID No 739510. Disclosures available: Neurology.org/NG.
References
- 1.Klockgether T, Mariotti C, Paulson HL. Spinocerebellar ataxia. Nat Rev Dis Primers 2019;5:24. [DOI] [PubMed] [Google Scholar]
- 2.Seidel K, Siswanto S, Brunt ER, den Dunnen W, Korf HW, Rub U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol 2012;124:1–21. [DOI] [PubMed] [Google Scholar]
- 3.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol 2010;9:885–894. [DOI] [PubMed] [Google Scholar]
- 4.Ruano L, Melo C, Silva MC, Coutinho P. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology 2014;42:174–183. [DOI] [PubMed] [Google Scholar]
- 5.Synofzik M, Nemeth AH. Recessive ataxias. Handb Clin Neurol 2018;155:73–89. [DOI] [PubMed] [Google Scholar]
- 6.Dickey CA, Patterson C, Dickson D, Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med 2007;13:32–38. [DOI] [PubMed] [Google Scholar]
- 7.Shi CH, Schisler JC, Rubel CE, et al. Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP. Hum Mol Genet 2014;23:1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Wang J, Li JD, et al. Identification of CHIP as a novel causative gene for autosomal recessive cerebellar ataxia. PLoS One 2013;8:e81884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genis D, Ortega-Cubero S, San Nicolas H, et al. Heterozygous STUB1 mutation causes familial ataxia with cognitive affective syndrome (SCA48). Neurology 2018;91:e1988–e1998. [DOI] [PubMed] [Google Scholar]
- 10.De Michele G, Lieto M, Galatolo D, et al. Spinocerebellar ataxia 48 presenting with ataxia associated with cognitive, psychiatric, and extrapyramidal features: a report of two Italian families. Parkinsonism Relat Disord 2019;65:91–96. [DOI] [PubMed] [Google Scholar]
- 11.Lieto M, Riso V, Galatolo D, et al. The complex phenotype of spinocerebellar ataxia type 48 in eight unrelated Italian families. Eur J Neurol 2020;27:498–505. [DOI] [PubMed] [Google Scholar]
- 12.Palvadeau R, Kaya-Gulec ZE, Simsir G, et al. Cerebellar cognitive-affective syndrome preceding ataxia associated with complex extrapyramidal features in a Turkish SCA48 family. Neurogenetics 2020;21:51–58. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann K, Lindner TH. easyLINKAGE-plus—automated linkage analyses using large-scale SNP data. Bioinformatics 2005;21:3565–3567. [DOI] [PubMed] [Google Scholar]
- 14.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000;25:12–13. [DOI] [PubMed] [Google Scholar]
- 15.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gymrek M, Golan D, Rosset S, Erlich Y. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res 2012;22:1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burk K. Cognition in hereditary ataxia. Cerebellum 2007;6:280–286. [DOI] [PubMed] [Google Scholar]
- 21.Tsuji S. Dentatorubral-pallidoluysian atrophy. Handb Clin Neurol 2012;103:587–594. [DOI] [PubMed] [Google Scholar]
- 22.Hayer SN, Deconinck T, Bender B, et al. STUB1/CHIP mutations cause Gordon Holmes syndrome as part of a widespread multisystemic neurodegeneration: evidence from four novel mutations. Orphanet J Rare Dis 2017;12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Synofzik M, Schule R, Schulze M, et al. Phenotype and frequency of STUB1 mutations: next-generation screenings in caucasian ataxia and spastic paraplegia cohorts. Orphanet J Rare Dis 2014;9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murata S, Chiba T, Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol 2003;35:572–578. [DOI] [PubMed] [Google Scholar]
- 25.Williams AJ, Knutson TM, Colomer Gould VF, Paulson HL. In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis. Neurobiol Dis 2009;33:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanack AJ, Newsom OJ, Scaglione KM. Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP. J Biol Chem 2018;293:2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi CH, Rubel C, Soss SE, et al. Disrupted structure and aberrant function of CHIP mediates the loss of motor and cognitive function in preclinical models of SCAR16. PLoS Genet 2018;14:e1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madrigal SC, McNeil Z, Sanchez-Hodge R, et al. Changes in protein function underlie the disease spectrum in patients with CHIP mutations. J Biol Chem 2019;294:19236–19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favaretto S, Margoni M, Salviati L, Pianese L, Manara R, Baracchini C. A new Italian family with HTRA1 mutation associated with autosomal-dominant variant of CARASIL: are we pointing towards a disease spectrum? J Neurol Sci 2019;396:108–111. [DOI] [PubMed] [Google Scholar]
- 30.Ward ME, Chen R, Huang HY, et al. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med 2017;9:eaah5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross OA, Rutherford NJ, Baker M, et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet 2011;20:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bettencourt C, de Yebenes JG, Lopez-Sendon JL, et al. Clinical and neuropathological features of spastic ataxia in a Spanish family with novel compound heterozygous mutations in STUB1. Cerebellum 2015;14:378–381. [DOI] [PubMed] [Google Scholar]
- 33.Yamada M, Sato T, Tsuji S, Takahashi H. CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol 2008;115:71–86. [DOI] [PubMed] [Google Scholar]
- 34.Fujigasaki H, Uchihara T, Koyano S, et al. Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado-Joseph disease brains. Exp Neurol 2000;165:248–256. [DOI] [PubMed] [Google Scholar]
- 35.Seidel K, Siswanto S, Fredrich M, et al. Polyglutamine aggregation in Huntington's disease and spinocerebellar ataxia type 3: similar mechanisms in aggregate formation. Neuropathol Appl Neurobiol 2016;42:153–166. [DOI] [PubMed] [Google Scholar]
- 36.Perez MK, Paulson HL, Pittman RN. Ataxin-3 with an altered conformation that exposes the polyglutamine domain is associated with the nuclear matrix. Hum Mol Genet 1999;8:2377–2385. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi J, Tanaka J, Arai K, et al. Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J Neuropathol Exp Neurol 2001;60:369–376. [DOI] [PubMed] [Google Scholar]
- 38.Scaglione KM, Zavodszky E, Todi SV, et al. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol Cell 2011;43:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishikawa K, Owada K, Ishida K, et al. Cytoplasmic and nuclear polyglutamine aggregates in SCA6 Purkinje cells. Neurology 2001;56:1753–1756. [DOI] [PubMed] [Google Scholar]
- 40.Herndon ES, Hladik CL, Shang P, Burns DK, Raisanen J, White CL III. Neuroanatomic profile of polyglutamine immunoreactivity in Huntington disease brains. J Neuropathol Exp Neurol 2009;68:250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saidi LJ, Polydoro M, Kay KR, et al. Carboxy terminus heat shock protein 70 interacting protein reduces tau-associated degenerative changes. J Alzheimers Dis 2015;44:937–947. [DOI] [PubMed] [Google Scholar]
- 42.Sahara N, Murayama M, Mizoroki T, et al. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem 2005;94:1254–1263. [DOI] [PubMed] [Google Scholar]
- 43.Dickey CA, Yue M, Lin WL, et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J Neurosci 2006;26:6985–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santpere G, Ferrer I. LRRK2 and neurodegeneration. Acta Neuropathol 2009;117:227–246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The deidentified data generated and analyzed in the current study will be available and shared by request to the corresponding author (M.O.M.) from any qualified investigator. These data include deidentified clinical data, raw exome/genome files, microsatellite genotyping, and additional pictures of stainings from the studied material.