Abstract
Objective
Molecular genetic testing for hereditary neuromuscular disorders is increasingly used to identify disease subtypes, determine prevalence, and inform management and prognosis, and although many small disease-specific studies have demonstrated the utility of genetic testing, comprehensive data sets are better positioned to assess the complexity of genetic analysis.
Methods
Using high depth-of-coverage next-generation sequencing (NGS) with simultaneous detection of sequence variants and copy number variants (CNVs), we tested 25,356 unrelated individuals for subsets of 266 genes.
Results
A definitive molecular diagnosis was obtained in 20% of this cohort, with yields ranging from 4% among individuals with congenital myasthenic syndrome to 33% among those with a muscular dystrophy. CNVs accounted for as much as 39% of all clinically significant variants, with 10% of them occurring as rare, private pathogenic variants. Multigene testing successfully addressed differential diagnoses in at least 6% of individuals with positive results. Even for classic disorders like Duchenne muscular dystrophy, at least 49% of clinically significant results were identified through gene panels intended for differential diagnoses rather than through single-gene analysis. Variants of uncertain significance (VUS) were observed in 53% of individuals. Only 0.7% of these variants were later reclassified as clinically significant, most commonly in RYR1, GDAP1, SPAST, and MFN2, providing insight into the types of evidence that support VUS resolution and informing expectations of reclassification rates.
Conclusions
These data provide guidance for clinicians using genetic testing to diagnose neuromuscular disorders and represent one of the largest studies demonstrating the utility of NGS-based testing for these disorders.
Molecular genetic analyses can provide diagnostic clarity for neuromuscular disorders, which demonstrate considerable clinical and genetic heterogeneity.1–4 In some cases, genetically disparate neuromuscular disorders share overlapping pathogenic mechanisms that correlate with and explain the subtleties of the clinical presentations.5–8 Next-generation sequencing (NGS), enabling simultaneous analysis of many genes without significant additional cost has now made genetic testing far more accessible. Studies using NGS in disease-specific contexts have reported high diagnostic yields ranging from 19% for spastic paraplegia to 60% for neuromuscular disorders.9–11 Historically, copy number variant (CNV) analysis has been limited to only a narrow group of genes12 or has not been routinely performed in neuromuscular genetic testing.9,13,14 Recently, the accurate detection of CNVs alongside sequence variants by NGS in a single assay has enabled more comprehensive, more affordable, and faster genetic analysis with a single sample.15
The purpose of this study was to examine a large unselected clinical cohort with neuromuscular disorders to evaluate the diagnostic yield of gene panels with simultaneous sequence and CNV detection. In one of the largest studies of genetic testing for this group of disorders, we also investigated the mutation spectrum, mutation properties, and reclassification of variants of uncertain significance (VUS) to obtain insight into genetic aspects of neuromuscular disorders that should inform clinical diagnosis of affected individuals.
Methods
Gene panel design
Phenotype-specific gene panels were curated by evaluating the strength of evidence in published literature supporting an association between a gene and a disorder, genotype-phenotype correlations, mode(s) of inheritance, and differential diagnoses. This curation led to the development of 3 single-gene tests (DMD, PMP22, and SMN1) and 11 multigene panels, with some overlap in genes to address clinical heterogeneity (table e-1, links.lww.com/NXG/A246).
Next-generation sequencing
NGS-based gene panels (not exome-based) were sequenced at high depth coverage (50× minimum, 350× average) to simultaneously identify single-nucleotide variants (SNVs), short and long indels, exon-level deletions/duplications, or CNVs. Structural rearrangements with breakpoints in coding regions are detected. Validation of bioinformatics methods and the clinical utility of CNV detection in NGS panels have been previously described.15,16 The analysis of SMN1 and SMN2 was conducted using a validated bioinformatics algorithm that gathered sequence reads from both gene copies in a single bin and subsequently used the presence or absence of the c.840C>T variant in exon 7 to disambiguate sequence reads and accurately determine copy number at each locus. The algorithm does not determine the phase of SMN1 gene copies, and thus, silent spinal muscular atrophy (SMA) carriers cannot be detected.
Subjects and reporting criteria
Between October 2014 and April 2019, an unbiased cohort of patients suspected to have a neuromuscular disorder was referred with informed consent for genetic testing (table 1). Only index patients were counted in this study. Clinicians requested testing for all genes on a panel or chose subpanels for narrower clinical indications. NGS testing and clinical variant interpretation was performed as described previously.17 Reports included variants classified as likely pathogenic or pathogenic (LP/P) or VUS; likely benign and benign (LB/B) variants were not reported. A definitive molecular diagnosis included a single LP/P variant in a gene associated with autosomal dominant (AD) or X-linked (XL) inheritance, or 2 variants either in the homozygous or compound heterozygous state in the appropriate phase in genes associated with autosomal recessive (AR) inheritance. LP/P variants were confirmed using the Sanger or PacBio sequencing for sequence variants and exon-focused microarray or NGS-based multiplex ligation-dependent amplification was performed on CNVs larger than 250–500 bp. All variants for this study were collected from Invitae's internal database and annotated based on the guidelines from the Human Genome Variation Society (varnomen.hgvs.org/).
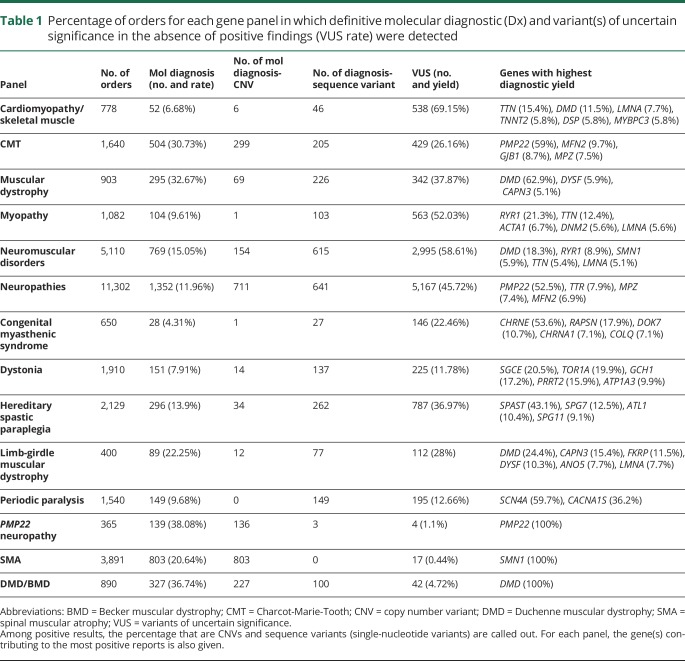
Table 1.
Percentage of orders for each gene panel in which definitive molecular diagnostic (Dx) and variant(s) of uncertain significance in the absence of positive findings (VUS rate) were detected
Data availability
Per institutional review board approval (Western IRB; WIRB #20161796), all reportable variants identified at Invitae were deposited into the ClinVar database.18 The list of genes in each multigene panel is presented in table e-1 (links.lww.com/NXG/A246), and a complete list of variant classifications per gene is listed in table e-2 (links.lww.com/NXG/A247).
Results
We performed a diagnostic genetic testing for 25,356 individuals in aggregate. Because some individuals had multiple tests, a combined total of 32,590 analyses were conducted. The age range of individuals at the time of genetic testing was <1–96 years, with a median of 43 years. Approximately 45% of tested individuals were women. More than 90% of the tests were ordered by neurologists or clinical geneticists, and the remaining were ordered by a wide range of other specialists.
Definitive diagnoses
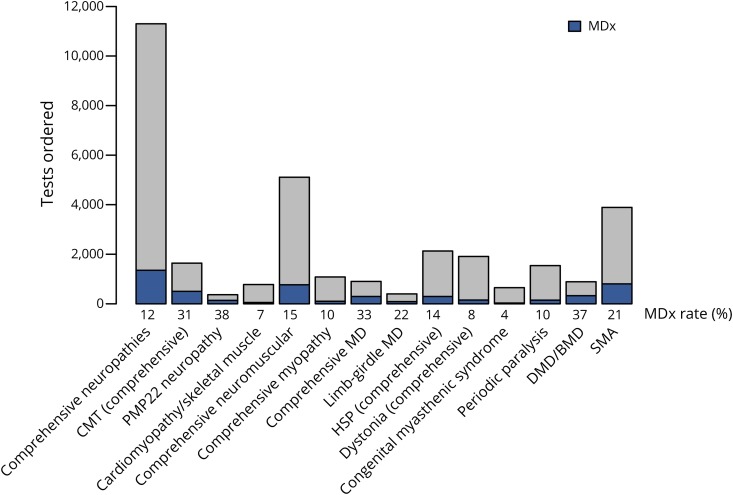
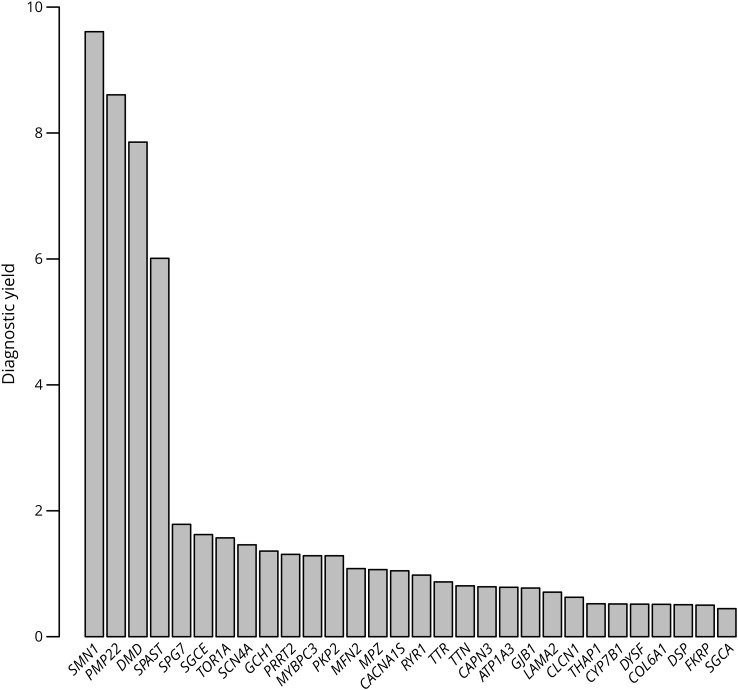
A definitive molecular diagnosis was obtained for 5,055 of the 25,356 individuals, representing a diagnostic yield of 20% overall and a range of 4%–33%, depending on the panel used (figure 1, table 1, table e-2, links.lww.com/NXG/A247). Single-gene tests for Charcot-Marie-Tooth disease type 1A (CMT1A) and Duchenne/Becker muscular dystrophy (DMD/BMD) had 38% and 37% yield, respectively, whereas SMA had a 21% yield. Multigene panels with the highest yield were those for muscular dystrophy, Charcot-Marie-Tooth (CMT), limb-girdle muscular dystrophy, and the comprehensive panel for neuromuscular disorders. Of the 163 genes that provided definitive diagnostic results, PMP22 provided the largest number, followed by SMN1, DMD, MFN2, MPZ, SPAST, TTR, SCN4A, and GJB1 (figure 2, table 1, and table e-1, links.lww.com/NXG/A246). Notably, 17 individuals in this cohort received molecular diagnoses in 2 genes, and one individual received molecular diagnoses in 3. Most of these cases involved pathogenic variants in TTN, MPZ, PMP22, and RYR1. No molecular diagnoses were obtained at all from 103 genes, most of which are associated with extremely rare conditions that have been reported in a handful of individuals and/or in specific ethnic backgrounds.
Figure 1. The number of tests ordered and diagnostic yield by panel.
Diagnostic yield by panel. The percentage of definitive positive results or MDx for each panel is indicated along the X-axis. Total number of orders is indicated along the Y axis. BMD = Becker muscular dystrophy; CMT = Charcot-Marie-Tooth; DMD = Duchenne muscular dystrophy; HSP = hereditary spastic paraplegia; MD = muscular dystrophy; MDx = molecular diagnostic rate; SMA = spinal muscular atrophy.
Figure 2. Diagnostic yield by gene.
The Y axis shows the percentage of positive rate for each gene cumulatively across all panels it was ordered from, with only the top 30 shown from among the 266 total genes included in panels for neuromuscular disorders.
Classification of variants
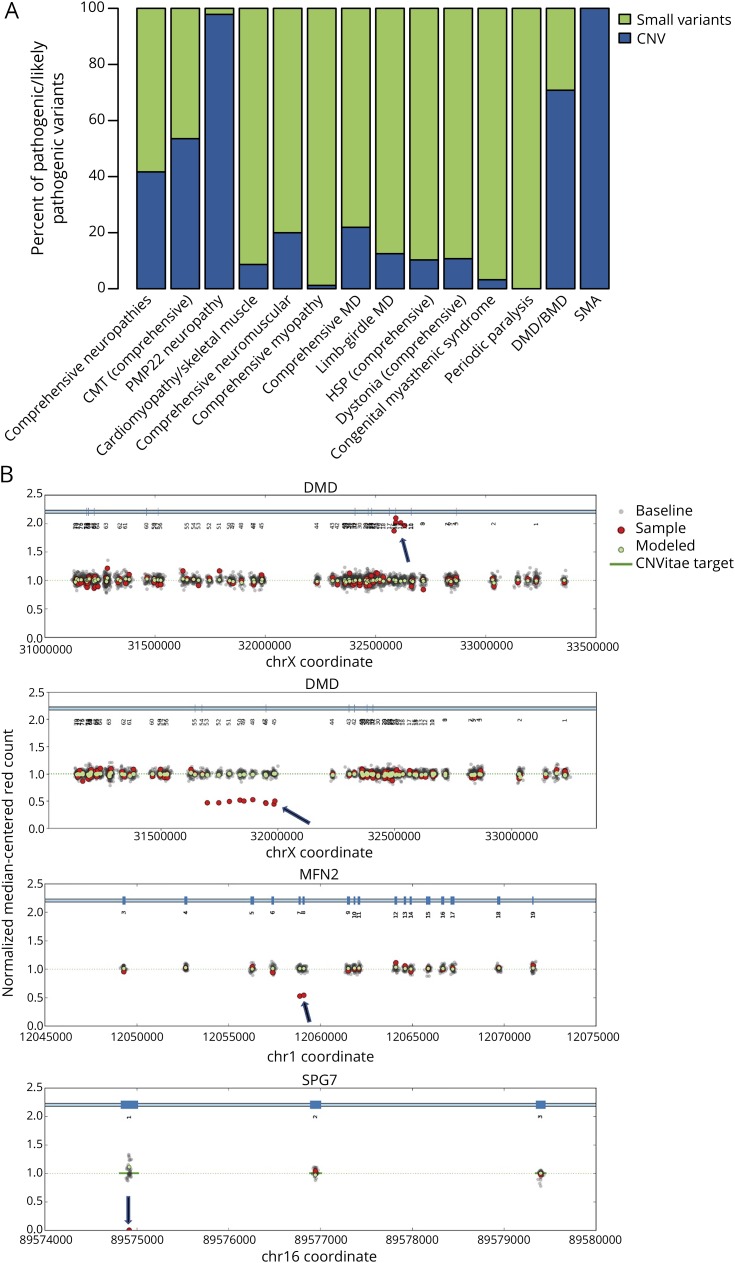
For the 25,356 individuals in this cohort, we reported 33,551 variants classified as LP/P or VUS. Among these variants, 84% were single-nucleotide changes, 6% were indels, and the remaining 10% were intragenic or whole gene CNVs. Notably, 39% of all positive variants in this clinical cohort (7,789 variants) were CNVs, most (93%) of which were in 3 genes—SMN1, PMP22, and DMD. However, it is important that another 77 genes contained 224 clinically significant CNVs, accounting for 7% of all clinically significant CNVs (figure 3, A and B and table 1). Of these, 39 occurred at a low frequency and 113 were novel variants. Of the 3,366 VUS/LP/P CNV reported in aggregate, 75% included loss or gain of a whole gene and the remaining were partial-gene events. However, when restricted to unique CNVs, of which there were 619, only 18% involved a whole gene and the rest were partial-gene events. In other words, many whole gene CNVs were recurrent and often involved SMN1 or PMP22. Furthermore, 1,723 of the 3,051 LP/P CNVs observed in this cohort were in genes associated with AD or XL inheritance and the remaining were in genes associated with AR inheritance. Of the 1,328 LP/P CNVs observed in AR conditions, 30 were observed as heterozygous events in combination with another LP/P non-CNV type of variant and 856 were present in the homozygous state.
Figure 3. Contribution of intragenic CNVs to diagnostic yield by panel.
Contribution of intragenic CNVs to diagnostic yield by panel. (A) The chart shows the percentage of likely pathogenic or pathogenic variants that are sequence variants (green) or intragenic copy number variants (blue) shown by different panels for neuromuscular disorders. Selected cases of intragenic CNVs detected in patients. (B) The baseline (indicated by “1.0”) represents the presence of 2 copies of each gene (1 copy of DMD in males). Gray dots indicate data for each internally included control samples. Green dots represent the calculated median of all control samples. Red dots represent data from the patient sample. Data points on the horizontal axis (chromosomal coordinates) are clustered at targeted exons. Panels from top to bottom show a hemizygous duplication of exons 12-16 of DMD in a male individual, a heterozygous deletion of exons 45-53 of DMD in a female carrier, a heterozygous deletion of exons 7 and 8 of MFN2, and a homozygous deletion of exon 1 of SPG7. BMD = Becker muscular dystrophy; CMT = Charcot-Marie-Tooth; CNV = copy number variant; DMD = Duchenne muscular dystrophy; HSP = hereditary spastic paraplegia; MD = muscular dystrophy; SMA = spinal muscular atrophy.
This cohort revealed several common pathogenic variants that have been previously described. The SMN1 whole gene deletion and the PMP22 whole gene duplication and deletion were the most common pathogenic events in this study. We also observed multiple instances of several other common mutations including 77 instances of GAA c.-32-13T>G, 68 instances of FIG4 c.122T>C (p.Ile41Thr), 63 instances of FKRP c.826C>A (p.Leu276Ile), 63 instances of TTR c.424 G>A (p.Val142Ile), 59 instances of ANO5 c.191dupA (p.Asn64Lysfs*15), 53 instances of SPG7 c.1529C>T (p.Ala510Val), and 48 instances of SH3TC3 c.2860C>T (p.Arg954*).19–22 The recently reported intronic variant in COL6A1 (NM_001848.2:c.930+189C>T) was the most common pathogenic variant affecting a type 6 collagen gene.23,24
Testing patterns for commonly referred disorders
For some neuromuscular disorders, the traditional approach has been to use single-gene tests rather than gene panels. Although this may be useful for individuals who present with overt and characteristic phenotypes, those with milder or atypical clinical presentations may not attract the requisite attention and testing. Even in the case of clearly recognizable disorders, gene panels may be used to curtail a potential diagnostic odyssey or to avoid invasive procedures such as muscle biopsies. Clinicians without extensive training in neuromuscular disorders may also be more comfortable with gene panels if they can address differential diagnoses broadly. DMD and CMT1A are examples of recognizable disorders for which many individuals are diagnosed through multigene panels. In our cohort, the DMD gene provided positive results for 634 male patients. Another 146 female individuals were found to be heterozygous for an LP/P DMD variant; however, inadequate clinical/family history made it impossible to unambiguously categorize the indication for testing as either diagnostic or carrier status. Of the 634 males diagnosed with DMD or BMD, 51% underwent single-gene testing, whereas the remaining underwent multigene testing through a comprehensive muscular dystrophy panel (25%), a limb-girdle muscular dystrophy panel (3%), a comprehensive neuromuscular disorders panel (20%), or a cardiomyopathy and skeletal muscle disease panel (0.5%). Most individuals (61%) with positive results in DMD had pathogenic CNVs, and the remaining had SNVs.
We also observed positive results in PMP22 through both single-gene testing and gene panel testing. Most (83%) of the positive results in PMP22 were represented by the classic whole gene duplication associated with CMT1A. Another 15% were represented by the reciprocal deletion associated with hereditary neuropathy with pressure palsies (HNPP), and the remaining positive results included 22 SNVs and small indels, and one exon 5 deletion. Of the 1,146 individuals who received molecular diagnosis of either CMT1A or HNPP, 62% were identified using the comprehensive neuropathy panel, whereas 26% were identified using the CMT-specific panel and 12% using single-gene analysis. Finally, we observed that individuals referred for genetic testing for SMA received positive diagnostic results most often with single-gene analysis (93% of cases). The remaining obtained positive results through gene panel testing via the comprehensive neuromuscular disorders panel and less often through the comprehensive neuropathies panel.
Rare genetic causes
In more than 6% of individuals for whom a diagnosis of a recognizable condition (e.g., DMD, SMA, or CMT1A) had been established by a referring physician based on clinical features, multigene panel testing revealed a diagnosis in another gene related to the phenotypic spectrum of the disorder. Among 2,501 individuals for whom a clinician received a negative result for a single-gene or small panel and subsequently pursued a larger panel, 200 showed diagnostic results on an expanded panel. In many cases, these results were in genes that are rare contributors to neuromuscular disease. In relation to DMD, PMP22, or SMN1, specifically, when individual analysis of these 3 genes yielded negative results in 339 individuals, a multigene panel provided 57 positive diagnoses that would have otherwise been missed by a traditional single-gene approach.
We also investigated the number of individuals who received a molecular diagnosis that was not consistent with the referral indication, illuminating differential diagnoses that would otherwise go undetected if single-gene or small panels were pursued. For example, among individuals referred with a clinical diagnosis of SMA, most who had a positive molecular diagnostic result indeed had the common deletion of SMN1. However, at least 16 individuals in that group had a molecular diagnosis other than SMN1, with roughly half of these genes associated with muscular dystrophy or myopathies and the other half with neuropathies. Similarly, among males referred with an explicit mention of DMD as a suspected clinical diagnosis and for whom we found a positive molecular diagnostic, three-quarters had a molecular diagnosis in the DMD gene, but the remaining one-quarter had diagnostic variants in other genes. Of interest, 19% of these genes were not in a muscular dystrophy panel but instead in a neuromuscular disease panel. Specifically, at least 3 individuals who were suspected to have DMD were found to have homozygous deletion of SMN1. Other cases were found to have congenital myopathy, cardiomyopathy, congenital myasthenic syndrome, myotonia, or spastic paraplegia.
Variants of uncertain significance
Although NGS provides the advantage of testing many genes at once, it also uncovers variants with meager evidence for pathogenicity that therefore get classified as VUS. We surveyed the VUS in this cohort to determine their distribution and rates of reclassification. In this cohort of 25,356 individuals, the number of VUS ranged from 1 to 13 per person (mean, 1.9; median, 1) and 53% of individuals received reports with at least one VUS (table 1). Among the 25,762 VUS reported in this cohort, there were 17,321 unique variants distributed across 266 genes. At least 16% of individuals with one or more VUS had a co-occurring definitive diagnostic result, indicating that the VUS in these cases were likely unrelated to the presenting clinical phenotype.
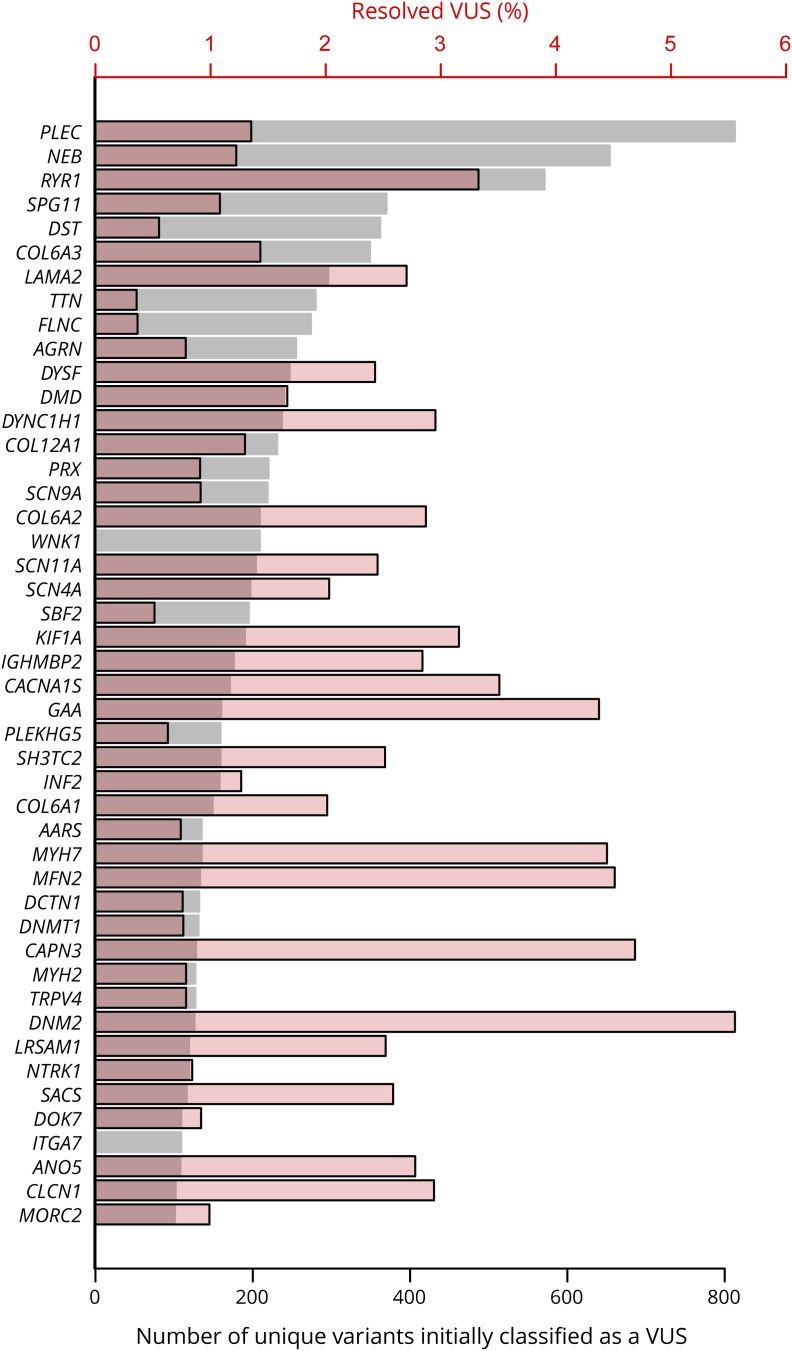
After follow-up studies, 2% of these 17,630 unique variants were reclassified, 44% (or 158 variants) changed to LP/P status, and the remaining 198 variants to LB/B status, and most of the LP/P reclassifications occurred in genes associated with AD disorders and high penetrance. At least 21 genes with over 100 unique VUS had 2% or more of VUS resolved, and one gene (DNM2) had over 5% of VUS resolved (figure 4). Among the additional evidence that supported reclassification of a VUS to a clinically significant LP/P result, de novo occurrence in genes associated with dominant inheritance was the most common contributing factor (39%), followed by case reports in the published literature (34%) and confirmation of trans phase for variants in genes associated with AR inheritance (10%).
Figure 4. Distribution of variants of uncertain significance and rate of reclassification across genes.
Distribution of variants of uncertain significance and rate of reclassification across genes (left axis). The gray bars indicate the number of unique variants (bottom axis) that were classified as variants of uncertain significance in each gene and the red bars indicate the percentage (top axis) that were reclassified to likely pathogenic or pathogenic or to likely benign or benign. Only genes with >100 unique VUS are included. Most de novo events were observed in RYR1, ACTA1, SPAST, and MPZ. Most downgrades of VUS to likely benign or benign classifications occurred in KBTBD13, PLEC, NEB, and LAMA2. VUS = variants of uncertain significance.
Discussion
Our study has demonstrated that NGS-based gene panels with simultaneous sequence and CNV detection can provide a diagnostic yield of 4%–33% in individuals with neuromuscular disease, including muscular dystrophies, SMA, spastic paraplegia, neuropathies, congenital myasthenic syndromes, and congenital myopathies. Although higher diagnostic rates have been published in other studies, they have typically involved small cohorts with homogeneous phenotypes rather than a large unselected cohort, as in this study. This and similar studies have not included repeat expansion/contraction disorders, the diagnosis of which depends largely on nonsequencing methods. Although whole exome sequencing,25–27 and targeted exome sequencing,28 can also be used for diagnosing neuromuscular disorders, their higher cost, incomplete coverage, long turnaround time to results, and limited ability to detect intragenic CNVs make them more appropriate as a second test following a negative panel result.
Most previous studies of the molecular basis of neuromuscular disorders have not routinely included intragenic CNV analysis for all genes. We recently showed that intragenic CNVs are important contributors to pathogenic variant burden in a broad range of hereditary disorders and should be routinely assessed.15 In this cohort, 39% of all positive results included CNVs. Over 80% of unique CNVs in our study included only a few exons, emphasizing the need for high resolution. The majority of pathogenic CNVs were found in PMP22, DMD, and SMN1, as expected. Intragenic CNVs were also identified in 77 other genes for which deletion/duplication analysis is not traditionally performed in a single assay, and these rare CNVs contributed to a molecular diagnosis in 113 cases. Furthermore, identifying CNVs in genes with AR inheritance can be particularly helpful because they can exist as compound heterozygous alleles that may be invisible to traditional sequencing methods. We confirmed 30 compound heterozygous diagnoses involving CNVs in genes such as LAMA2, SPG11, DOK7, and PRKN. CNV analysis is therefore clearly a necessary component of diagnostic genetic testing for inherited neuromuscular conditions to ensure high clinical sensitivity.
Multigene NGS analysis advances the interpretation of heterogeneity for any single clinical disorder and also helps refine differential diagnoses. Panels can also be useful for individuals for whom a single-gene test cannot be confidently selected because of a mild or uncharacteristic phenotype. In illustrating how the challenge of genetic heterogeneity can be overcome, our data show that many individuals who received a molecular diagnosis for a well-recognized disorder, such as DMD or CMT1A, were actually diagnosed through multigene panels rather than single-gene analysis. Moreover, in 2,501 instances in which a clinician received a negative result for a single-gene or small panel test and subsequently pursued testing using a larger panel, a positive diagnostic result was obtained for 200 individuals. Separately, NGS panels also help address differential diagnoses by supporting evaluation of genes for related disorders. Several individuals with clinical suspicion of DMD or CMT1A received positive molecular diagnoses in genes unrelated to their referral indication. For example, 133 individuals suspected to have DMD instead had a molecular diagnosis in a gene unrelated to muscular dystrophy but related to one of the several other types of neuromuscular disorders, including spastic paraplegia, congenital myasthenic syndrome, and SMA.
Our study also provided insight into the complexities of interpreting rare variants observed in the many genes associated with neuromuscular disorders. Investigation of VUS in this cohort showed that most occurred as single heterozygous alleles in genes associated with AR disorders and were therefore less likely to be disease-causing. By contrast, VUS in genes associated with AD disorders and high penetrance have a higher likelihood of being reclassified to a clinically significant status following studies to investigate segregation or de novo status. Of the VUS that were resolved to clinically significant results, 48% were in the genes associated with AD inheritance and were found to be de novo or, alternatively, were in genes associated with AR inheritance for which demonstration of trans phase provided useful evidence toward pathogenicity.
This study has better illuminated the diagnostic yields for various neuromuscular disorders, the contribution of CNVs to these disorders, the importance of addressing genetic heterogeneity and differential diagnoses, and the occurrence and resolution of VUS. As NGS becomes even more accessible to individuals with neuromuscular disorders, these types of studies will provide clinicians the requisite information for understanding the utility of genetic tests, establishing expectations for affected individuals in their course of care, and managing test results.
Glossary
- AD
autosomal dominant
- AR
autosomal recessive
- BMD
Becker muscular dystrophy
- CMT
Charcot-Marie-Tooth
- CMT1A
Charcot-Marie-Tooth disease type 1A
- CNV
copy number variant
- DMD
Duchenne muscular dystrophy
- HNPP
hereditary neuropathy with pressure palsies
- LB/B
likely benign and benign
- LP/P
likely pathogenic or pathogenic
- NGS
next-generation sequencing
- SMA
spinal muscular atrophy
- SNV
single-nucleotide variant
- VUS
variants of uncertain significance
- XL
X-linked
Appendix. Authors
Study funding
No targeted funding reported.
Disclosure
Disclosures available: Neurology.org/NG.
References
- 1.Biancalana V, Laporte J. Diagnostic use of massively parallel sequencing in neuromuscular diseases: towards an integrated diagnosis. J Neuromuscul Dis 2015;2:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohrn MF, Glöckle N, Mulahasanovic L, et al. Frequent genes in rare diseases: panel-based next generation sequencing to disclose causal mutations in hereditary neuropathies. J Neurochem 2017;143:507–522. [DOI] [PubMed] [Google Scholar]
- 3.Nigro V, Savarese M. Next-generation sequencing approaches for the diagnosis of skeletal muscle disorders. Curr Opin Neurol 2016;29:621–627. [DOI] [PubMed] [Google Scholar]
- 4.Reale C, Panteghini C, Carecchio M, Garavaglia B. The relevance of gene panels in movement disorders diagnosis: a lab perspective. Eur J Paediatr Neurol 2018;22:285–291. [DOI] [PubMed] [Google Scholar]
- 5.Bönnemann CG, Wang CH, Quijano-Roy S, et al. Diagnostic approach to the congenital muscular dystrophies. Neuromuscul Disord 2014;24:289–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansone VA. The dystrophic and nondystrophic myotonias. Continuum 2016;22:1889–1915. [DOI] [PubMed] [Google Scholar]
- 7.Boutry M, Morais S, Stevanin G. Update on the genetics of spastic paraplegias. Curr Neurol Neurosci Rep 2019;19:18. [DOI] [PubMed] [Google Scholar]
- 8.Prior R, Van Helleputte L, Benoy V, Van Den Bosch L. Defective axonal transport: a common pathological mechanism in inherited and acquired peripheral neuropathies. Neurobiol Dis 2017;105:300–320. [DOI] [PubMed] [Google Scholar]
- 9.Nallamilli BRR, Chakravorty S, Kesari A, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol 2018;5:1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iqbal Z, Rydning SL, Wedding IM, et al. Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. PLoS One 2017;12:e0174667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian X, Liang WC, Feng Y, et al. Expanding genotype/phenotype of neuromuscular diseases by comprehensive target capture/NGS. Neurol Genet 2015;1:e14 doi: 10.1212/NXG.0000000000000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiVincenzo C, Elzinga CD, Medeiros AC, et al. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med 2014;2:522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stehlíková K, Skálová D, Zídková J, et al. Muscular dystrophies and myopathies: the spectrum of mutated genes in the Czech Republic. Clin Genet 2017;91:463–469. [DOI] [PubMed] [Google Scholar]
- 14.Chae JH, Vasta V, Cho A, et al. Utility of next generation sequencing in genetic diagnosis of early onset neuromuscular disorders. J Med Genet 2015;52:208–216. [DOI] [PubMed] [Google Scholar]
- 15.Truty R, Paul J, Kennemer M, et al. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med 2019;21:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn 2015;17:533–544. [DOI] [PubMed] [Google Scholar]
- 17.Nykamp K, Anderson M, Powers M, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med 2017;19:1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrum MJ, Kattman BL. ClinVar at five years: delivering on the promise. Hum Mutat 2018;39:1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brockington M, Yuva Y, Prandini P, et al. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum Mol Genet 2001;10:2851–2859. [DOI] [PubMed] [Google Scholar]
- 20.Høyer H, Braathen GJ, Busk Ø, et al. Genetic diagnosis of Charcot-Marie-Tooth disease in a population by next-generation sequencing. Biomed Res Int 2014;2014:210401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roxburgh RH, Marquis-Nicholson R, Ashton F, et al. The p.Ala510Val mutation in the SPG7 (paraplegin) gene is the most common mutation causing adult onset neurogenetic disease in patients of British ancestry. J Neurol 2013;260:1286–1294. [DOI] [PubMed] [Google Scholar]
- 22.Bouquet F, Cossée M, Béhin A, et al. Miyoshi-like distal myopathy with mutations in anoctamin 5 gene. Rev Neurol (Paris) 2012;168:135–141. [DOI] [PubMed] [Google Scholar]
- 23.Cummings BB, Marshall JL, Tukiainen T, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolduc V, Foley AR, Solomon-Degefa H, et al. A recurrent COL6A1 pseudoexon insertion causes muscular dystrophy and is effectively targeted by splice-correction therapies. JCI Insight 2019;4:e124403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldrop MA, Pastore M, Schrader R, et al. Diagnostic utility of whole exome sequencing in the neuromuscular clinic. Neuropediatrics 2019;50:96–102. [DOI] [PubMed] [Google Scholar]
- 26.Ghaoui R, Cooper ST, Lek M, et al. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: outcomes and lessons learned. JAMA Neurol 2015;72:1424–1432. [DOI] [PubMed] [Google Scholar]
- 27.Reddy H, Cho K-H, Lek M, et al. The sensitivity of exome sequencing in identifying pathogenic mutations for LGMD in the United States. J Hum Genet 2017;62:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson K, Bertoli M, Phillips L, et al. Detection of variants in dystroglycanopathy-associated genes through the application of targeted whole-exome sequencing analysis to a large cohort of patients with unexplained limb-girdle muscle weakness. Skelet Muscle 2018;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Per institutional review board approval (Western IRB; WIRB #20161796), all reportable variants identified at Invitae were deposited into the ClinVar database.18 The list of genes in each multigene panel is presented in table e-1 (links.lww.com/NXG/A246), and a complete list of variant classifications per gene is listed in table e-2 (links.lww.com/NXG/A247).