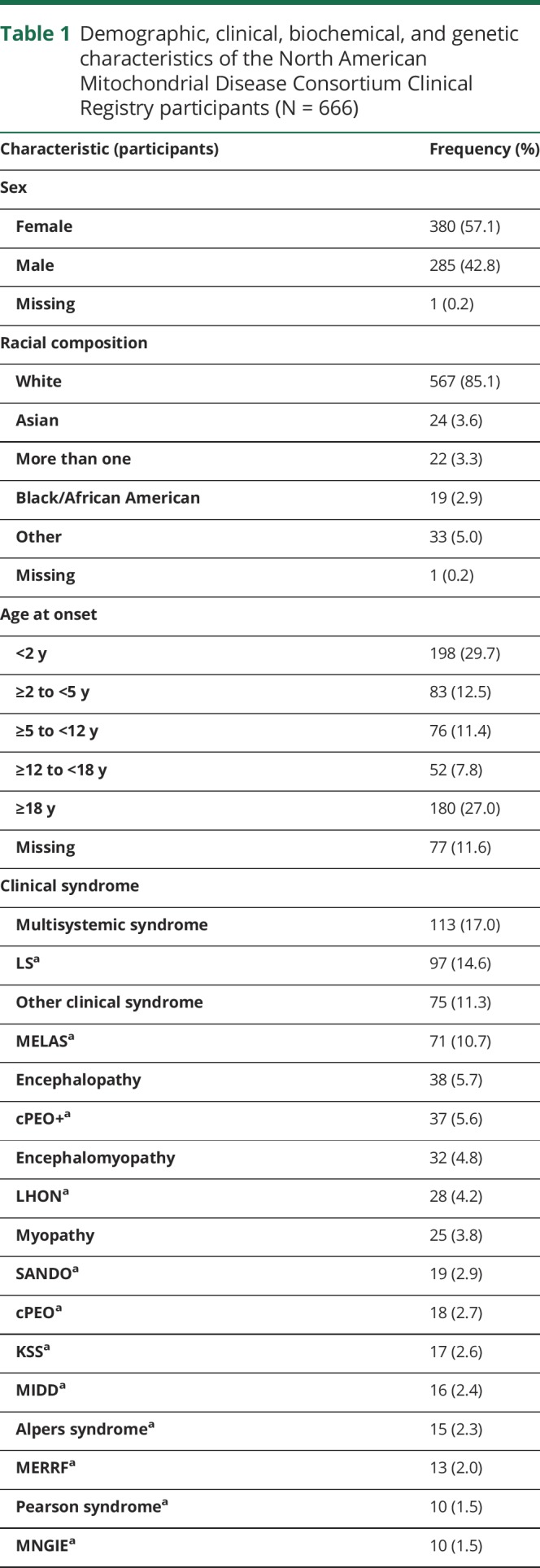
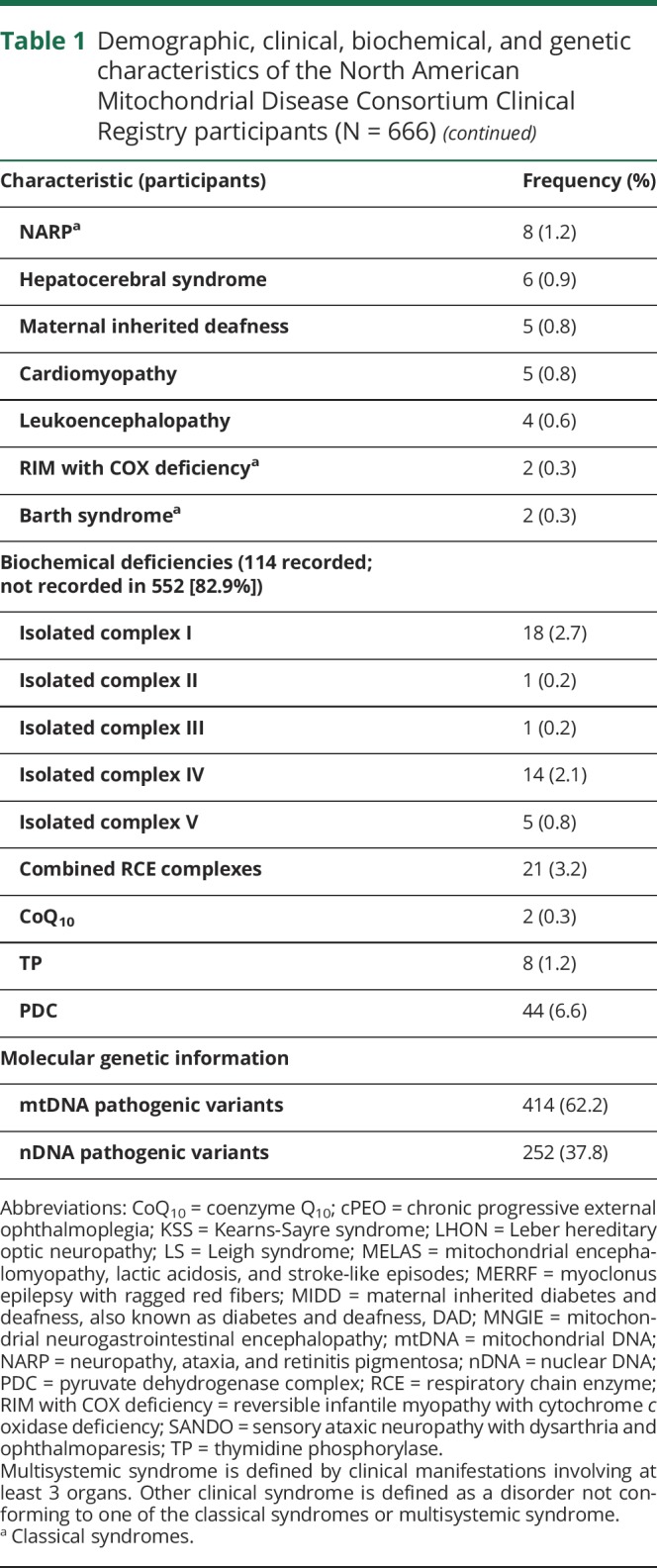
Emanuele Barca
Emanuele Barca, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Yuelin Long
Yuelin Long, MS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Victoria Cooley
Victoria Cooley, MS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Robert Schoenaker
Robert Schoenaker, MD, BS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Valentina Emmanuele
Valentina Emmanuele, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Salvatore DiMauro
Salvatore DiMauro, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Bruce H Cohen
Bruce H Cohen, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Amel Karaa
Amel Karaa, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Georgirene D Vladutiu
Georgirene D Vladutiu, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Richard Haas
Richard Haas, MBBChir
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Johan LK Van Hove
Johan LK Van Hove, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Fernando Scaglia
Fernando Scaglia, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Sumit Parikh
Sumit Parikh, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Jirair K Bedoyan
Jirair K Bedoyan, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Susanne D DeBrosse
Susanne D DeBrosse, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Ralitza H Gavrilova
Ralitza H Gavrilova, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Russell P Saneto
Russell P Saneto, DO, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Gregory M Enns
Gregory M Enns, MBChB
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Peter W Stacpoole
Peter W Stacpoole, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Jaya Ganesh
Jaya Ganesh, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Austin Larson
Austin Larson, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Zarazuela Zolkipli-Cunningham
Zarazuela Zolkipli-Cunningham, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Marni J Falk
Marni J Falk, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Amy C Goldstein
Amy C Goldstein, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Mark Tarnopolsky
Mark Tarnopolsky, MD, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Andrea Gropman
Andrea Gropman, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Kathryn Camp
Kathryn Camp, MS, RD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Danuta Krotoski
Danuta Krotoski, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Kristin Engelstad
Kristin Engelstad, MS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Xiomara Q Rosales
Xiomara Q Rosales, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Joshua Kriger
Joshua Kriger, MS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Johnston Grier
Johnston Grier, MS
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Richard Buchsbaum
Richard Buchsbaum
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
John LP Thompson
John LP Thompson, PhD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,
Michio Hirano
Michio Hirano, MD
1From the Department of Neurology (E.B., V.E., S.D., K.E., X.Q.R., M.H.), Columbia University Medical Center, New York; Department of Biostatistics (Y.L., V.C., J.K., J. Grier, R.B., J.L.P.T.), Mailman School of Public Health, Columbia University, New York; Radboudumc (R.S.), Nijmegen, The Netherlands; Department of Pediatrics (B.H.C.), Northeast Ohio Medical University and Akron Children's Hospital; Genetics Unit (A.K.), Massachusetts General Hospital, Boston; Department of Pediatrics (G.D.V.), State University of New York at Buffalo; Departments of Neurosciences and Pediatrics (R.H.), University of California at San Diego; Department of Pediatrics (J.L.K.V.H., A.L.), University of Colorado School of Medicine, Aurora; Department of Molecular and Human Genetics (F.S.), Baylor College of Medicine, Houston, TX; Texas Children's Hospital (F.S.), Houston; Joint BCM-CUHK Center of Medical Genetics (F.S.), Prince of Wales Hospital, ShaTin, New Territories, Hong Kong; Department of Neurology (S.P.), Cleveland Clinic, OH; Departments of Genetics and Genome Sciences and Pediatrics (J.K.B., S.D.D.), and Center for Human Genetics, University Hospitals Cleveland Medical Center, Case Western Reserve University, OH; Departments of Neurology and Clinical Genomics (R.H.G.), Mayo Clinic, Rochester, MN; Department of Neurology (R.P.S.), University of Washington, Seattle Children's Hospital; Department of Pediatrics (G.M.E.), Stanford University, Palo Alto, CA; Department of Medicine (P.W.S.), University of Florida at Gainesville; Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai (J. Ganesh), New York; Mitochondrial Medicine Frontier Program (Z.Z.-C., M.J.F., A.C.G.), Division of Human Genetics, The Children's Hospital of Philadelphia and University of Pennsylvania Perelman School of Medicine; University of Pennsylvania Perelman School of Medicine (Z.Z.-C.), Philadelphia; Department of Neurology (M.T.), McMasters University, Toronto, Ontario, Canada; Department of Neurology (A.G.), Children's National Health Network, Washington, DC; Office of Dietary Supplements (K.C.), National Institutes of Health, Bethesda, MD; and Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.K.), National Institutes of Health, Bethesda, MD.
1,✉