Abstract
Glioblastoma multiforme (GBM) and other solid malignancies are heterogeneous and contain subpopulations of tumor cells that exhibit stem-like features. Our recent findings point to a de-differentiation mechanism by which reprogramming transcription factors Oct4 and Sox2 drive the stem-like phenotype in glioblastoma, in part by differentially regulating subsets of microRNAs (miRNAs). Currently, the molecular mechanisms by which reprogramming transcription factors and miRNAs coordinate CSC tumor-propagating capacity are unclear. In this study, we identified miR-486–5p as a Sox2-induced miRNA that targets the tumor suppressor genes PTEN and FoxO1 and regulates the GBM stem-like cells. miR-486–5p associated with the GBM stem cell phenotype and Sox2 expression and was directly induced by Sox2 in glioma cell lines and patient-derived neurospheres. Forced expression of miR-486–5p enhanced the self-renewal capacity of GBM neurospheres, and inhibition of endogenous miR-486–5p activated PTEN and FoxO1 and induced cell death by upregulating pro-apoptotic protein BIM via a PTEN-dependent mechanism. Furthermore, delivery of miR-486–5p antagomirs to pre-established orthotopic GBM neurosphere-derived xenografts using advanced nanoparticle formulations reduced tumor sizes in vivo and enhanced the cytotoxic response to ionizing radiation. These results define a previously unrecognized and therapeutically targetable Sox2/miR-486–5p axis that enhances the survival of GBM stem cells by repressing tumor suppressor pathways.
Keywords: miRNAs, CSCs, cancer stem cells, nanomedicine, apoptosis
INTRODUCTION
Neoplastic cells are inherently plastic and reprogramming transcription factors (e.g. Oct4 and Sox2) are commonly over-expressed in cancer and play critical roles in tumor cell fate determination through the process of reprogramming (1). Brain tumors are among the most devastating forms of cancer and glioblastoma (GBM) represents the most aggressive and lethal form of the disease (2). GBM is characterized by small subsets of cells, referred to as glioma stem cells (GSCs), that display stem-like properties including unlimited self-renewal and multi-lineage differentiation (3). These stem-like cells act as critical determinants of GBM resistance to current treatments and are thought to play an important role in tumor recurrence (4). Approaches aimed at targeting mechanisms of GBM cell self-renewal and therapeutic resistance hold great potential for the development of novel therapeutics to treat GBM (4).
Members of the SOX family of transcription factors are potent drivers of somatic cell reprogramming and dysregulation of its members has been implicated in several disease conditions including cancer (5). The SOXB1 sub-class composed of Sox1, 2 and 3, functions to establish and maintain a stem-like state in embryonic stem cells and neural progenitor cells (6). Of these three transcription factors, SOX2 is sufficient to induce cell reprogramming in multiple contexts making it a potent regulator of the stem cell phenotype (7). SOX2 dysregulation alters networks of coding and non-coding genes that affect migration, invasion, growth, survival, and self-renewal capacity of multiple tumor cell types, including GBM (5). SOX2 gene expression is up-regulated in glioma compared to normal brain and silencing of Sox2 results in loss of tumor propagating capacity of GBM stem-like cells (8,9). How stemness-enhancing mechanisms regulated by Sox2 affect the genetic and epigenetic landscape of GBM stem-like cells and which of these changes are amenable to therapeutic intervention remain unclear (5).
Growing evidence highlight microRNAs (miRNAs) as key determinants of cell fate and tumorigenesis (10). These short non-coding RNAs are negative regulators of gene expression that are often dysregulated in cancer with the capacity to act as oncogenes or tumor suppressors (10). Not surprisingly, de-differentiating events driven by reprogramming transcription factors modify non-coding RNA networks and Sox2 has been shown to drive these changes in GBM stem-like cells (11,12). We recently uncovered a novel molecular circuit by which Oct4 and Sox2 co-expression induces glioma cell stemness and tumor propagating capacity by altering DNA methylation and modifying miRNA networks (13). We previously demonstrated that Oct4/Sox2 co-expression induces DNA methylation events that repress stemness-inhibiting miRNAs and that reconstituting these miRNAs potently inhibits GBM cell stemness and tumor growth in vivo (14,15).
The goal of this study is to elucidate how miRNA subsets that are up-regulated by reprogramming transcription factors regulate GBM-propagating stem cells and glioma growth. We present the novel findings that miR-486–5p, a miRNA not previously known to regulate GBM neurospheres, is induced by Sox2 under conditions that promote GBM cell stemness. Exogenous expression of miR-486–5p or Sox2 repressed tumor suppressor genes PTEN and FoxO1 and transgenic miR-486–5p enhanced resistance of GBM neurosphere cells to ionizing radiation (IR) treatment. Additionally, we show that PTEN is the dominant target through which miR-486–5p regulates neurosphere cell survival and that inhibiting miR-486–5p impedes the growth and enhances the radiosensitivity of established GBM xenografts. These findings demonstrate that Sox2 controls GBM cell survival by activating miR-486–5p that inhibits tumor suppressor networks and identifies miR-486–5p inhibition as a potential strategy for treating GBM.
MATERIALS AND METHODS
GBM Neurosphere Culture
GBM-derived neurosphere lines (GBM1A and GBM1B) were originally derived and characterized by Vescovi and colleagues (16). The GBM-KK neurosphere line was derived from a single GBM patient and kindly provided by Dr. Jaroslaw Maciaczyk (University of Freiburg). Low-passage primary neurospheres were derived directly from human GBM clinical specimens and from patient-derived xenografts obtained from pathological GBM specimens obtained during clinically indicated surgeries at Johns Hopkins Hospital using established methods5. The human GBM xenograft line, Mayo39, was originally obtained from the Mayo Clinic (Rochester, MN) (17). All neurospheres were cultured in serum-free medium containing DMEM/F-12 (Invitrogen, Carlsbad, CA, USA), supplemented with 1% BSA, 20 ng/ml epidermal growth factor (EGF) and 10 ng/ml fibroblast growth factor (FGF).
The human embryonic kidney 293FT (HEK293FT) cell line was obtained from the ATCC and was maintained in Dulbecco’s modified Eagle/F12 medium (1:1, vol/vol) supplemented with 10% FBS (Fetal Bovine Serum, Thermo Fisher Scientific Inc, Waltham, MA). The 293FT cells were grown at 37°C in a humidified incubator with 5% CO2. All cell lines used in the study were tested for mycoplasma and were STR profiled. All experiments using transgenic cells were performed within 10 passages of generating the cells lines.
Intra-cranial nano-miR delivery and Tumor formation in vivo
A transcranial cannula was placed so that the tip is in the right caudate/putamen of female athymic nude NCR Nu/Nu mice (8-week old). One week after cannula placement, animals received 1.0x104 GBM1A or Mayo39 tumor propagating cells via the cannula and assigned into different treatment groups in a non-blinded, randomized manner. Using the same cannula, the control cohort received nano-miRs loaded with control miRNA labeled with Dy547 and the experimental group received nano-miRs loaded with the miR-486–5p inhibitor.
Stainless steel guide and dummy cannulas were custom ordered from PlasticsOne (Roanoke, VA). The guide cannula (26 gauge) was designed to have a Decon® mesh under the pedestal and cut 3 mm from the mesh. The guide cannula is capped with a screw-on dummy cannula 6.5 mm long so that a 0.5 mm projection extends past the guide to prevent blockage. Prior to surgical placement of cannulas, mice were anesthetized using a Ketamine (100mg/Kg)/Xylazine (10mg/kg) cocktail and mounted on a stereotactic frame. A rostro-caudal incision was made with a scalpel, the skin is spread apart, the surface of the skull was exposed, and cannulas were placed at coordinates: AP (antero-posterior) 0.0 (0 mm from bregma), L (lateral) 1.8 (1.8 mm right from mid-sagital line).
Lyophilized and resuspended nano-miRs were slowly infused (5μL) into the brains (0.5μL/min with a 2 min wait at the end) twice a week as described for each experiment. At the end of the experiment animals were anesthetized and then sacrificed by perfusion using 4% paraformaldehyde (PFA) according to methods approved by the Animal Use and Care Committee at Johns Hopkins University. All the sectioning and histological analysis was performed in-house. Whole brains were collected and soaked in 4% PFA for 2 days then washed 1X with PBS and soaked in 30% sucrose over-night at 4°C then flash frozen using dry ice. Brains were embedded in Tissue-Tek® O.C.T. Compound (VWR, Radnor, PA) and 20 μm sections were cut using the CryoStat system from Microm (Walldorf, Germany). All tumor sections were analyzed by a neuropahologist in a blinded fashion.
For ionizing radiation experiments, a subset of animals received radiation either alone or in combination with the nano-miR therapy. Radiation was administered starting 8 weeks after tumor cell implantation. Tumor-bearing mice were gently restrained in a 50ml ventilated plastic centrifuge tube encapsulated in lead cylinders to protect normal body parts from radiation. This ensures only the tumor-bearing brain will be irradiated. Animals received 300 cGy (or sham irradiation) once a week for 2 weeks using a collimator 137Cs source. These radiation doses were without adverse side effects.
Tumor growth inhibition was determined by computer-assisted morphometric quantification of tumor area in H&E-stained histologic sections using ImageJ software and volumes calculated using volume = (square root of maximum cross-sectional area)3. Data for all in vivo experiments are shown as the mean tumor area distribution of all animals used in the study. All animal procedures were approved by the Johns Hopkins Institutional Animal Care and Use Committee (Protocol# MO14M307), and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Tumor sections were analyzed by a neuropathologist (H.A.) for histologic features of GBM including necrosis. To measure necrosis, the total areas of viable tumor and necrosis within each tumor were identified by an expert neuropathologist and quantified using ImageJ software from images taken at 10X magnification and represented as (area of necrotic tissue/area of total tumor tissue)*100.
Statistical Analysis
All experiments were performed in triplicates and repeated at least twice in each cell model (N≥6). PRISM GraphPad 7 was used to perform all the statistical analyses presented. Two group comparisons were analyzed for variation and significance using a two-tailed, type 1 t-test and p values lower than 0.05 were considered significant and symbolized by an asterisk in the graphs. One-way or Two-way ANOVA and Tukey post hoc test was used to analyze the relationships when comparing multiple variables, with p values lower than 0.05 considered to be statistically significant. All data shown are representative of means ± S.D. of triplicate results unless otherwise specified.
Additional information about polymer synthesis, primers used for qRT-PCR (Supplemental table S1, S2 and S3), cloning (Supplemental table S4), lentiviral constructs (Supplemental Table S5), antibodies (Supplemental table S6), miRNA array resuls (Supplemental table S7) can be found in the supplemental materials.
RESULTS:
miR-486–5p associates with the GBM stem-cell phenotype and is induced by Sox2.
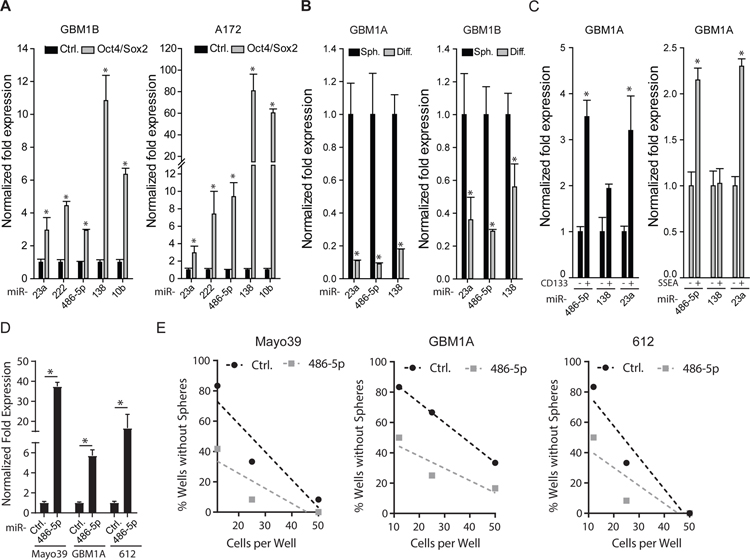
Our previously published miRNA-array screen uncovered ten candidate miRNAs up-regulated (≥2-fold) in human GBM-derived neurospheres following Oct4 and Sox2 co-expression (13). We confirmed by qRT-PCR that Oct4/Sox2 activates the expression of a subset of those precursor (pre-) miRNAs (i.e. miR-23a, miR-10b, miR-138, miR-222, and miR-486–5p) (Fig. 1A). We next examined multiple endpoints to determine if these miRNAs associate with GBM cell stemness. Forced differentiation of GBM neurospheres, a condition that also inhibits Oct4 and Sox2 expression (13), decreased levels of pre-miR-23a, pre-miR-138, and pre-miR-486–5p in multiple neurosphere lines (Fig. 1B). CD133+ and SSEA-1+ cell sub-populations enriched for stem-like cells expressed 2–3 fold higher levels of both pre-miR-23a and pre-miR-486–5p compared to the CD133− or SSEA− cells. We found no significant differential expression of pre-miR-138 in SSEA+ or CD133+ cells (Fig 1C).
Figure 1: miR-486–5p associates with the GBM stem cell phenotype.
(A) qRT-PCR to measure effects of Oct4/Sox2 co-expression on a subset of miRNAs detected by PCR array (ref. 13) to be up-regulated in GBM neurospheres. (B) qRT-PCR analysis shows decreased expression of pre-miR-23a, pre-miR-486–5p, and pre-miR-138 5 days after forced differentiation of GBM neurospheres. (C) GBM neurosphere cells expressing stem cell markers CD133 or SSEA were separated into marker positive and marker negative populations by flow cytometry. qRT-PCR was used to measure expression of selected pre-miRNAs in the different cell subsets. (D) qRT-PCR analysis to measure expression of mature miR-486–5p in 3 different GBM neurosphere isolates. (E) Limiting dilution assay (LDA) in neurosphere isolates transduced with a control lentivirus or a lentivirus expressing miR-486–5p. Statistical significance was calculated using Student’s t-test A, B, C, D and data are presented as means ± S.D. *p *p< 0.05
These associations led us to hypothesize that miR-23a and miR-486–5p act to support the GBM stem-like phenotype, a function consistent with their induction by Oct4/Sox2 expression, up-regulation in SSEA+ and CD133+ cells and repression in response to forced differentiation (13). To test this hypothesis, we forced the expression these miRNAs individually using lenti-viral expression vectors and measured effects on sphere-forming capacity as a surrogate for stem cell self-renewal (Fig. S1). qRT-PCR analysis following lentivirus transduction demonstrated 10–18 fold increase in mature miR-23a, miR-138, and miR-486–5p expression. Only miR-486–5p had the capacity to enhance sphere-formation capacity (Fig. S1). To directly evaluate the effects of miR-486–5p on the GBM stem cell phenotype, three patient-derived neurosphere lines were transduced using lentivirus expressing miR-486–5p precursor miRNA and sphere forming capacity was determined by limiting dilution assay, a quantitative marker of cell stemness and self-renewal. Cells transduced with a scrambled miRNA were used as a negative control. Forced expression of miR-486–5p enhanced self-renewal capacity of all three patient-derived neurosphere lines tested (Fig. 1D and 1E). These results show that Oct4/Sox2 co-expression activates a subset of miRNAs that correlate with the stem-like phenotype of GBM cells and predicts that miR-486–5p functions to regulate GBM cell growth patterns.
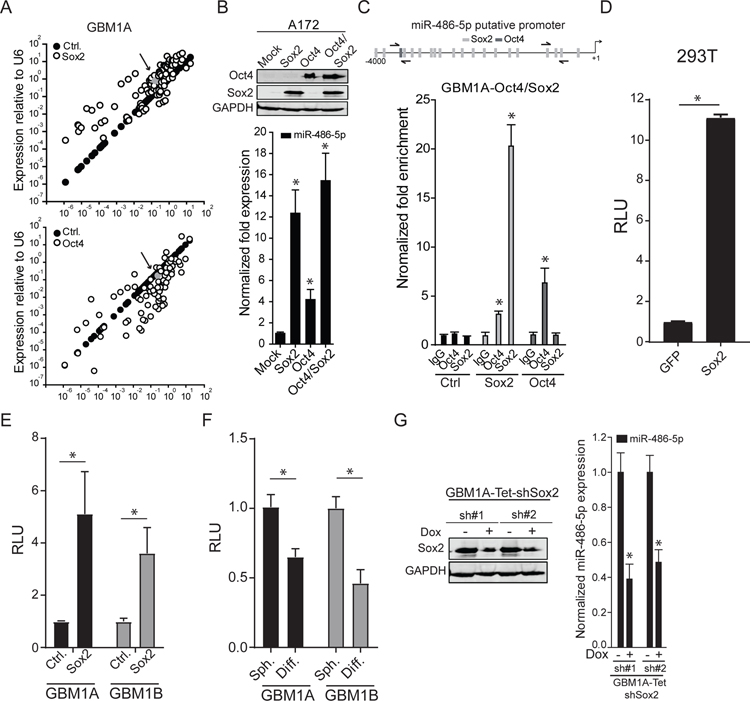
Comparative PCR array data examining changes in miRNA levels in response to expression of either exogenous Oct4 or Sox2 in neurosphere cells suggests that miR-486–5p is preferentially induced by Sox2 alone (Fig. 2A and table S7). To confirm this prediction, we expressed transgenic Oct4, Sox2, or their combination in A172 GBM cells, which have very low expression of endogenous Oct4 and Sox2, and measured expression of mature miR-486–5p using qRT-PCR. Exogenous Sox2, Oct4, or their combination induced mature miR-486–5p expression by 12-, 4-, and 15-fold respectively (Fig. 2B). In silico analysis shows enrichment for Sox2 binding sites in the putative promoter region 5’ from the miR-486–5p transcription start site (Fig. 2C, top panel), suggesting Sox2 directly trans-activates miR-486–5p gene expression in.
Figure 2: miR-486–5p is regulated by SOX2.
(A) Differential miRNA expression was determined in GBM1A neurospheres expressing Oct4 or Sox2 relative to control GBM1A spheres using a Human Brain Cancer miRNA PCR array. Arrow points at grey dot indicating miR-486–5p expression. (B) Western blot showing relative expression of Sox2 and Oct4 (top panel). qRT-PCR to measure expression of mature miR-486–5p 3 days after exogenous expression of Oct4, Sox2, or the combination. (C) Oct4 and Sox2 binding sites on human miR-486–5p promoter, arrows indicate primer sites used for PCR analyses (top panel). DNA purified from chromatin immuno-precipitation was analyzed by qRT-PCR using primer pairs designed to amplify fragments containing Oct4, Sox2, or a control region lacking binding sites for either transcription factor (bottom panels). (D) 293T cells were co-transfected with a luciferase reporter construct spanning the miR-486–5p putative promoter containing the SOX2 binding sites and GFP or SOX2. (E) GBM neurosphere isolates expressing exogenous SOX2 were transfected with the luciferase reporter construct spanning the miR-486–5p putative promoter containing the SOX2 binding sites and luciferase activity was measured 3 days after transfection. (F) GBM1A and GBM1B neurospheres were transfected with luciferase reporter construct covering the miR-486–5p putative promoter containing the SOX2 binding and forced to differentiate. Luciferase activity was measured 3 days after differentiation. (G) Western blot to measure expression of Sox2 (top panel) and qRT-PCR to measure expression of pre-miR-486–5p (bottom panel) 3 days after doxycycline treatment. Student’s t-test was used to calculate statistical significance in panels D,E,F, and G; One-way ANOVA with Tuckey’s post hoc test was used calculate statistical significance in panels B and C. Data are presented as means ± S.D *p< 0.05
Quantitative chromatin immune-precipitation (ChIP) supports this prediction by showing ~20-fold enrichment of Sox2 binding compared with a ~6-fold enrichment of Oct4 binding at the predicted promoter sites (Fig. 2C, bottom panel). Promoter-reporter assays were used to determine if Sox2 transactivates this region of the miR-486–5p promoter. The region of the human miR-486–5p promoter containing the Sox2 binding site identified in our ChIP experiments (Fig. 2C) was cloned into a luciferase reporter cassette and co-transfected into HEK293T in the presence of Sox2 or GFP. Compared with control cells transfected with GFP, Sox2 induced miR-486–5p/luciferase activity ~11 fold (Fig 2D). A similar increase was observed when this same luciferase reporter plasmid was transfected into GBM1A and GBM1B neurospheres stably expressing transgenic Sox2 (Fig. 2E). Using a similar approach, we measured a 40%−50% decrease in luciferase activity from the miR-486–5p luciferase reporter when we forced neurosphere cells to differentiate (Fig. 2F), a condition that decreases endogenous expression of Sox2. To test if Sox2 is required to maintain expression of endogenous miR-486–5p, we generated transgenic neurospheres lines expressing shRNA hairpins specific for Sox2 under the control of a doxycyline-induced promoter (Tet-shSox2). shRNA-mediated knock-down of Sox2 decreased expression of pre-miR-486–5p by 50–60% (Fig. 2G). Taken together these results show that Sox2 plays a significant role in driving miR-486–5p expression.
The Sox2:miR-486–5p axis inhibits tumor suppressor gene expression.
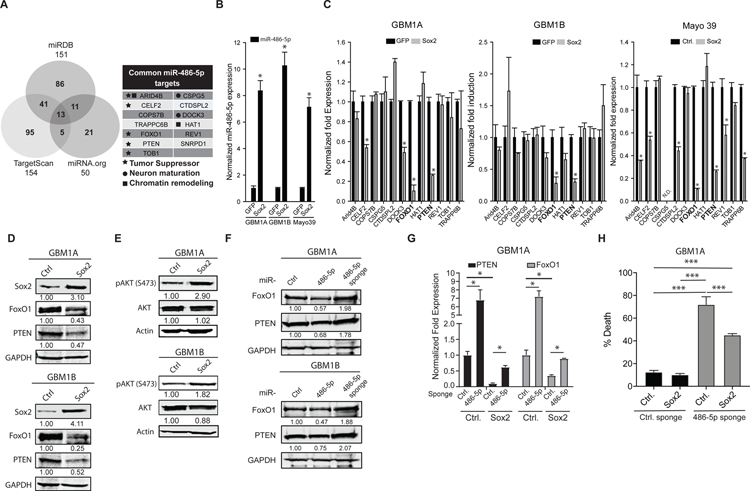
miRNAs function as negative regulators of gene expression and their function is determined by the genes and networks they modulate (18). Combining miR-486–5p targets predicted by 3 different bioinformatics algorithms generated a list of high-confidence gene candidates potentially regulated by the Sox2:miR-486–5p axis in GBM neurospheres (Fig. 3A). We screened neurosphere cells expressing miR-486–5p induced by exogenous Sox2 (Fig. 3B) to identify which of the 13 candidate target genes were likely to function downstream of miR-486–5p. PTEN and FoxO1 were the only candidate targets consistently down-regulated by exogenous Sox2 in multiple GBM cell models (Fig. 3C). Down-regulation of PTEN and FoxO1 by forced Sox2 expression was confirmed using western blot analysis (Fig. 3D and S2A). PTEN inhibits the activation (i.e. phosphorylation) of Akt, a tumor promoter that regulates FoxO1(19). Forced expression of either Sox2 or miR-486–5p increased Akt phosphorylation (pAKT), consistent with the down-regulation of PTEN (Fig 3E and S2B). The seed region for miR-486–5p is highly conserved among several species in the PTEN and FoxO1 3’UTR (Fig. S2C) consistent with their direct targeting by miR-486–5p and functional significance. As predicted, we observed a significant decrease in PTEN and FoxO1 protein and mRNA in response to exogenous miR-486–5p (Fig. 3F and Fig. S2D). Neurospheres were transduced with lentivirus expressing miRNA sponges against miR-486–5p to test if endogenous miR-486–5p regulates PTEN and/or FoxO1 expression. Control lenti-virus expressing a scrambled sequence was used a negative control. miR-486–5p inhibition increased levels of PTEN and FoxO1 in neurosphere lines and primary cells derived from a patient-derived GBM xenograft (Fig. 3G and Fig. S2E). We also observed that inhibiting miR-486–5p partially rescued the repression of PTEN and FOXO1 by exogenous Sox2 (Fig. 3H). Re-expression of PTEN and FOXO1 via miR-486–5p inhibition induced cell death even in the presence of exogenous Sox2, consistent with miR-486–5p functioning as a downstream effector of Sox2. Taken together, these results demonstrate that Sox2 inhibits PTEN and FoxO1 tumor suppressor pathways by directly inducing miR-486–5p as an intermediate.
Figure 3: Sox2:miR-486–5p axis down-regulates tumor suppressor genes in GBM neurospheres.
(A) Venn diagram showing intersection of miR-486–5p target genes using 3 different prediction algorithms (right panel). List of high-confidence miR-486–5p target genes (left panel). (B) qRT-PCR to measure expression of mature miR-486–5p in GBM neurosphere cells expressing transgenic Sox2. (C) qRT-PCR to measure gene levels of high-confidence miR-486–5p target genes in GBM neurospheres expressing transgenic Sox2. Genes highlighted in bold (FOXO1, PTEN) were commonly regulated in 3 distinct GBM neurosphere isolates. (D) Western blot analysis to measure PTEN and FoxO1 protein levels in GBM neurospheres expressing transgenic Sox2. (E) Western blot analysis to measure AKT and pAKT protein levels in GBM neurospheres expressing transgenic Sox2. (F) Western blot analysis to measure PTEN and FoxO1 protein levels in GBM neurospheres 4 days after expressing transgenic miR-486–5p or miR-486–5p inhibition (miR-486–5p sponge). (G) qRT-PCR to measure PTEN and FoxO1 expression in GBM neurospheres expressing transgenic SOX2 after miR-486–5p inhibition. (H) Trypan blue exclusion to measure cell death 4 days after miR-486–5p inhibition in in GBM neurospheres expressing transgenic SOX2. Student’s t-test was used to calculate statistical significance in panels B and C; One-way ANOVA with Tuckey’s post hoc test was used calculate statistical significance in panels H. Data are presented as means ± S.D *p< 0.05
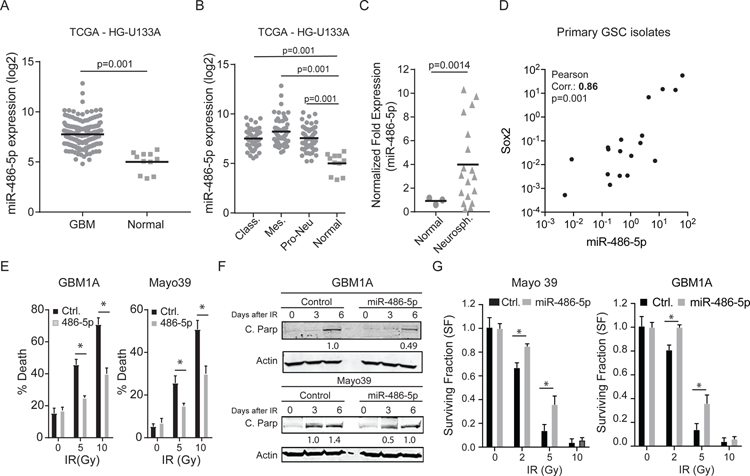
Gene expression profiles from surgical specimens were used to explore the broader potential clinical relevance of the Sox2:miR-486–5p axis. Analysis of TCGA datasets (HG-U133A) reveals high miR-486–5p expression levels in GBM compared to normal brain (Fig. 4A). This same relationship persists in each GBM subtype relative to normal brain (Fig. 4B). Additionally, miR-486–5p expression was found to be up-regulated in a panel of 18 independently isolated primary human GBM-derived neurosphere isolates compared with cells derived from normal human brain (Fig. 4C). Further analysis of these primary neurosphere isolates revealed a significant positive correlation between Sox2 and miR-486–5p expression (Fig. 4D). Thus, miR-486–5p expression was increased in GBM neurospheres, CD133+ and SSEA+ subsets, and directly correlated with Sox2 expression. We also observe a significant negative correlation between Sox2 and PTEN or FoxO1 expression in these clinical datasets (Fig. S3). These observations are further supported by the negative correlation measured between Sox2 and FoxO1 expression in low-passage primary human neurospheres derived directly from human GBM surgical specimens (Fig. S3) consistent with our findings that Sox2 down-regulates PTEN and FoxO1 in vitro. Together, these results are consistent with PTEN and FoxO1 being downstream of a Sox2:miR-486–5p axis in GBM.
Figure 4: miR-486–5p is up-regulated in GBM and enhances resistance to IR treatment.
miR-486–5p expression data was retrieved from the TCGA database using the BETASTASIS portal (http://www.betastasis.com). (A) miR-486–5p levels in normal brain compared to GBM and (B) normal brain compared to GBM subtypes. (C) qRT-PCR to measure miR-486–5p expression in GBM neurospheres lines and primary GBM neurosphere isolates compared to non-tumorigenic glial progenitors (normal) (D) Positive correlation between miR-486–5p and Sox2 expression in primary GBM neurosphere isolates. Pearson coefficient analysis was applied to establish correlations from gene expression data. (E) GBM stem-like cells stably expressing miR-486–5p or a control miRNA (Ctrl.) were treated with increasing doses of IR and cell death was quantified using trypan blue exclusion 3 days after treatment. (F) Western blot to measure cleaved PARP and cleaved Caspase 3 in GBM stem-like cells expressing transgenic miR-486–5p 3 and 6 days after IR (5Gy) treatment. (G) Clonogenic assay in GBM stem-like cells expressing transgenic miR-486–5p after IR treatment. Colonies were counted using Image J script 14 days after treatment. Student’s t-test was used to calculate statistical significance in panels B, C, and D; Two-way ANOVA with Tuckey’s post hoc test was used calculate statistical significance in panels E and G. Data are presented as means ± S.D *p< 0.05
miR-486–5p regulates GBM cell survival.
Our results showing that miR-486–5p inhibits tumor suppressor pathways (Fig. 2) coupled with the clinical associations lead us to hypothesize that miR-486–5p induces glioma resistance to DNA damaging agents. To test this hypothesis we examined the effect of exogenous miR-486–5p on the GBM neurosphere response to ionizing radiation (IR). Cells expressing exogenous miR-486–5p were more resistant to death induced by either 5Gy or 10Gy IR as determined by trypan blue exclusion assay (Fig. 4E). The cytoprotective activity of miR-486–5p was further supported by western blot analysis showing lower levels of cleaved PARP, a well described marker of apoptosis (Fig. 4F). Clonogenic assay for long-term cell survival also showed that exogenous miR-486–5p protects GBM neurospheres from 2Gy and 5Gy radiation toxicity but not 10 Gy (Fig. 4G).
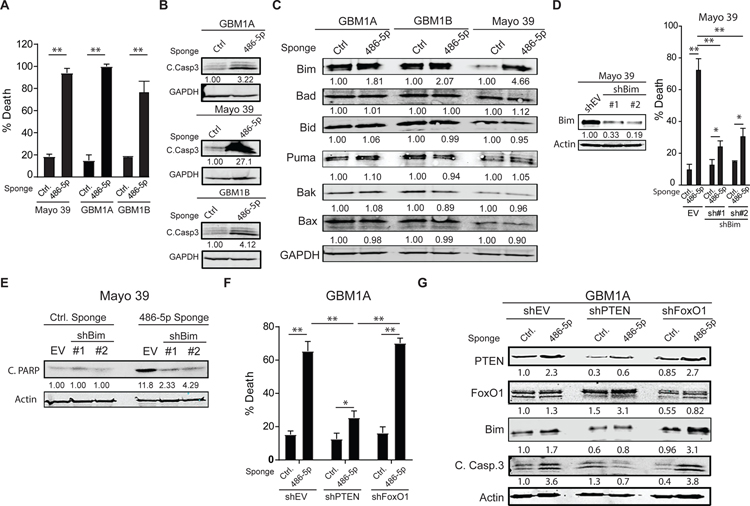
To test the hypothesis that endogenous miR-486–5p functions as a pro-survival factor, GBM-derived neurospheres were transduced with lenti-virus expressing either a scrambled RNA sequence or an anti-sense RNA oligo designed to inhibit miR-486–5p (miR-486–5p sponge). The miR-486–5p sponge efficiently decreased the levels of endogenous pre-miR-486–5p in multiple independent GBM-derived cell models by >80%, as measured by qRT-PCR (Fig. S4). Inhibition of endogenous miR-486–5p triggered apoptosis in these cells as evidenced by an increase in dead cells measured by trypan blue exclusion and PARP cleavage (Fig. 5A and 5B).
Figure 5: miR-486–5p inhibition activates Bim and induces apoptosis in GBM neurospheres.
(A) Equal numbers of GBM1A neurosphere cells transduced with lentiviral constructs expressing a miR-486–5p sponge or a control vector were cultured in neurosphere medium containing EGF/FGF for 14 days and neurosphere numbers (>100µm diameter) were quantified by computer-assisted image analysis. (B) Western blot showing cleaved caspase 3 cleavage 6 days after miR-486–5p inhibition. (C) Western blots showing expression of pro-apoptotic proteins 3 days after miR-486–5p inhibition. (D) Mayo 39 primary GBM cells were transduced with two independent shRNA hairpins targeting BIM. Western blot showing BIM levels following shRNA knock-down (right panel). Cell viability 5 days after miR-485–5p inhibition in cells transduced with shEV or shBim (right panel). (E) Western blot showing PARP cleavage 5 days after miR-485–5p inhibition in cells transduced with shEV or shBim. (F) Cell viability 5 days after miR-485–5p inhibition in cells transduced with shEV, shPTEN, or shFoxO1. (G) Western blot showing expression of PTEN, FoxO1, Bim and cleaved caspase 3 in GBM1A neurospheres after PTEN or FoxO1 knock-down and miR-486–5p inhibition. Student’s t-test was used to calculate statistical significance in panel A; Two-way ANOVA with Tuckey’s post hoc test was used calculate statistical significance in panels D and F. Data are presented as means ± S.D *p< 0.05; **p< 0.01
To further dissect the mechanism of cell death induced by miR-486–5p inhibition we measured expression of multiple members of the Bcl-2 family of proteins that are critical regulators of cell survival (20). Western blot analysis revealed a consistent increase in the BH3-only protein Bim following miR-486–5p inhibition in multiple GBM neurosphere lines (Fig. 5C). Bim is a potent initiator of apoptosis shown to be sufficient to induce cell death in multiple settings (21). To test if Bim up-regulation is responsible for the cell death effects observed after miR-486–5p inhibition, we knocked-down endogenous Bim using 2 independent shRNA hairpins (Fig. 5D, left panel) and then inhibited miR-486–5p as described above. An empty vector (shEV) was used as negative control for shBim. Bim repression rescued cells from the death induced by miR-486–5p inhibition by ~75% as measured by trypan blue exclusion assay (Fig. 5D, right panel) and western blot analysis of PARP cleavage (Fig. 5E).
Both PTEN and FoxO1 have been shown to regulate pro-apoptotic BH3-only protein Bim (22). To determine if either PTEN or FoxO1 plays a dominant role in the capacity of miR-486–5p to regulate Bim and down-stream apoptotic death we examined how PTEN or FoxO1 knock-down effects the response to miR-486–5p inhibition. As before, an empty vector (shEV) was used as negative control. PTEN knock-down was sufficient to impair Bim induction and cell killing in response to miR-486–5p inhibition. In contrast, FoxO1 knock-down had no effect (Fig. 5F and 5G). These observations were confirmed using a second shRNA targeting PTEN (Fig. S5). Together, these results identify miR-486–5p as a critical determinant of cell survival and identify PTEN as the principal functional downstream effector by which miR-486–5p inhibition induces Bim and apoptosis.
Suppression of miR-486–5p in vivo inhibits growth of orthotopic GBM xenograft and sensitizes xenografts to IR treatment.
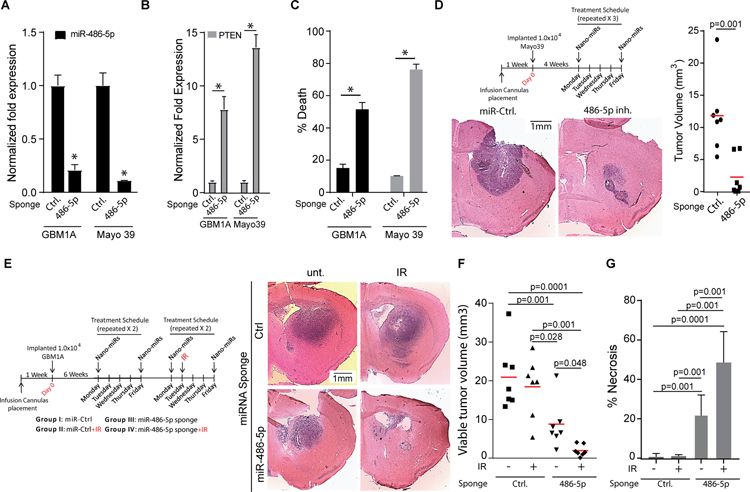
Our in vitro findings predict that in vivo inhibition of miR-486–5p will hinder tumor growth and potentially cooperate with cytotoxic therapeutics. As a first step to evaluate this using a clinically translatable miRNA delivery platform, bioreducible nanoparticles (15) containing a scrambled miRNA sequence or the miR-486–5p sponge were used to transfect GBM neurospheres and primary cells from patient-derived GBM xenografts and pre-miR-486–5p expression was measured using qRT-PCR 3 days after transfection. Nanoparticle-based delivery of miR-486–5p sponge decreased levels of endogenous miR-486–5p by ~80% (Fig. 6A) and simultaneously significantly increased PTEN mRNA levels concurrent with a strong induction of cell death (Fig. 6B and 6C) recapitulating the results using the lenti-viral system. These results show that miR-486–5p sponge retains biological activity and PTEN regulation when delivered to cells using PBAE nanoparticle complexes.
Figure 6: In vivo inhibition of miR-486–5p in pre-established GBM reduces tumor size and sensitizes tumors to IR treatment.
qRT-PCR analysis to quantify expression of miR-486–5p (A) or down-stream targets (B) in GBM1A and Mayo39 cells 3 days after transfection with nano-miRs delivering a non-targeting control miRNA (Ctrl.) or a miR-486–5p sponge. (C) Cell death assay to measure viability of GBM1A and Mayo 39 cells 6 days after transfections with nano-miRs delivering a non-targeting control miRNA (Ctrl.) or a miR-486–5p sponge. (D) Schematic summarizing treatment schedule for in vivo delivery of nano-miRs (top panel). Representative H&E stained brain sections from mice implanted with Mayo 39 cells (N=7 for each group) treated with the indicated nano-miRs. Animals were sacrificed 52 days after cell implantation. (E) Schematic summarizing treatment schedule for in vivo delivery of nano-miRs. (F) Representative H&E stained brain sections from mice implanted with GBM1A neurosphere cells (N=7 for each group) treated with the indicated nano-miRs +/− IR. Animals were sacrificed 70 days after cell implantation. Maximum tumor cross-sectional areas following treatment with nano-miRs representing viable tumor tissue (F) and necrotic tumor tissue (G) were quantified from H&E stained sections using ImageJ software. Student’s t-test was used to calculate statistical significance in panels A-D; One-way ANOVA with Tuckey’s post hoc test was used calculate statistical significance in panels F and G. Data are presented as means ± S.D *p< 0.05
Mice bearing pre-established orthotopic patient-derived GBM xenografts (4 weeks post tumor cell implantation) were treated with nanoparticles containing either scrambled miRNA or miR-486–5p inhibitor by direct intra-tumoral infusion via a trans-cranial cannula twice per week for 3 weeks (Fig. 6D, top panel)(15). Histopathological examination of brains from animals sacrificed 3 days after the last nano-miR treatment (52 days post-cell implantation) showed significantly smaller tumors in response to miR-486–5p antagomir delivery (Fig. 6D). Next, we asked if in vivo miR-486–5p inhibition alters the therapeutic efficacy of ionizing radiation (I.R.), a standard-of-care treatment modality for GBM. For this purpose, GBM derived neurospheres (GBM1A), which generate tumors that closely recapitulate the invasive growth pattern of clinical GBM (16), were implanted and beginning on post-implantation week 6 treated with nano-miRs as described above. A a small number of animals were sacrificed prior to initiating therapies to confirm the presence of tumor (Fig. S6). Two weeks after beginning nanoparticle infusions a subset of animals began IR therapy in combination with either control or anti-miR-486–5p nano-miRs (see Fig. 6E, left panel for treatment schedule). Tumor burden and necrosis were quantified in brains collected 28 days after treatment initiation. Radiation alone had no statistically significant effects on tumor size or tumor-associated necrosis (Fig. 6E–G). Tumor size was significantly decreased in response to the active miRNA sponge and the most profound effects occurred animals treated with miR-486–5p sponge + IR (Fig. 6E and 6F). The decrease in tumor size was accompanied by an increase in tumor tissue necrosis as measured by histopathology (Fig. 6G). These results show that combining in vivo inhibition of miR-486–5p with IR generated a cooperative anti-tumor response.
DISCUSSION:
MicroRNAs (miRNAs) are short non-coding RNAs that control a wide range of biological processes including cell survival, oncogenesis and cell stemness(23). We now understand that miRNAs can function as potent drivers of tumorigenesis and substantial efforts have been devoted to understanding how they are regulated and their downstream effectors to develop efficient ways to replenish tumor suppressor miRNAs and/or inhibit oncomiRs (24). Our results show that Oct4/Sox2 co-expression activates a network of miRNAs that correlate with the stem-like phenotype of GBM cells and predicts that they have oncogenic functions in GBM. In fact, miR-23b is thought to exert oncogenic functions in some cellular contexts by controlling cancer cell metabolism and autophagy (25). MiR-10b-5p has been shown to function as an onco-miR in multiple settings and ex vivo inhibition can repress tumor-initiation capacity (26). miR-138 is dysregulated in multiple cancers and has been found to control many target genes related to proliferation, apoptosis, invasion, and migration (27). Recently, miR-222 has been implicated in controlling pathways that enhance survival of cancer cells to cytotoxic agents (28). However, how these miRNAs regulate the gene networks and phenotypes downstream of Oct4 and Sox2 remains to be elucidated.
miR-486–5p can activate or repress oncogenic pathways in a cell-type specific manner acting as a tumor suppressor in esophageal, colorectal, lung, and liver cancers(29–32) while behaving as an oncomiR in pancreatic, brain, prostate and hematopoietic tumors (33–37). We show that miR-486–5p expression associates with gain of stem cell phenotype in GBM neurospheres (Fig. 1) and is enriched in primary neurosphere isolates compared to glial progenitors (Fig. 4C), conditions where SOX2 is up-regulated (13). Using bioinformatics analysis followed by ChIP-PCR we validated SOX2 binding sites in the miR-486–5p putative promoter and we establish that Sox2 can directly trans-activate miR-486–5p expression in GBM neurospheres using a combination of loss/gain-of-function approach and luciferase reporters (Fig. 2). We also demonstrate that transgenic miR-486–5p enhances GBM neurosphere resistance to IR treatment (Fig. 4) and self-renewal capacity of multiple GBM neurosphere isolates (Fig. 1). Our novel findings in GBM parallel studies showing that miR-486–5p expression is increased in leukemia compared to normal erythroid progenitor cells and transgenic expression enhances self-renewal, growth, survival and drug sensitivity of CML progenitors (33,35). Interestingly, and similar to results in the leukemia models (33), we did not observe a significant change in expression of well-characterized reprogramming transcription factors, stem cell markers, or lineage-specific markers in response to forced miR-486–5p expression. Our results demonstrate that miR-486–5p is a SOX2-regulated oncomiR in GBM neurospheres and suggest that the increase in self-renewal capacity observed in response to transgenic miR-486–5p is not directly related to de-differentiating events but instead related to dysregulation of cell growth and survival mechanisms.
One oncogenic pathway that controls cell growth mechanisms and appears to be commonly affected by SOX2 alterations is the AKT pathway(38,39). AKT activity is controlled by Phosphatase and Tensin homologue (PTEN) and cells that carry inactivating mutations in PTEN gene exhibit constitutively active PI3K:AKT signaling leading to inactivation of FoxO function though a mechanism involving phosphorylation-dependent nuclear exclusion(40). The PTEN:AKT:FoxO axis controls several cellular functions including cell growth, survival and stemness(22,41,42). Interestingly, it has been speculated that miRNA dysregulation plays a significant role in PTEN transcriptional inhibition in gliomas (23). A recent report shows that miR-486–5p drives prostate cancer tumorigenesis by repressing PTEN and FoxO1 (36). Using GBM neurospheres expressing transgenic SOX2, conditions that activate miR-486–5p expression, we also identified PTEN and FoxO1 as miR-486–5p targets (Fig. 3). Furthermore, we show that forced SOX2 or miR-486–5p expression inhibits PTEN and FoxO1 mRNA and protein in addition to increasing AKT phosphorylation (Fig. 3 and S2). Interestingly, we show that PTEN but not FoxO1 knock-down is sufficient to abolish the effects of miR-486–5p inhibition (Fig. 5F and 5G), showing that PTEN is the predominant target responsible for miR-486–5p effects. These results demonstrate the novel findings that SOX2 activates oncogenic signals in GBM, at least in part, by repressing tumor suppressor genes and provides a new mechanism for AKT dysregulation in GBM neurospheres. Interestingly, while TCGA data sets show elevated levels of miR-486–5p GBM compared to non-tumor tissue (Fig. 4) high miR-486–5p levels did not correlate with patient survival when considering all GBM cases (Fig. S7). However, high levels of miR-486–5p, low levels of the miR-486–5p target PTEN and Bim were each found to correlate with worse outcome in patients with proneural GBM (Fig. S7), a subtype characterized by increased Sox2 levels. Interestingly, despite observing an increase in Sox2 gene expression in proneural GBM in the data sets analyzed we did not observe a significant difference in patient survival between high and low expressers. This may be due to the relative low number of patients analyzed resulting in lack of power to achieve statistically significant differences (Fig. S7).
One of the hallmarks of tumor suppressor gene inactivation is inhibition of cell death mechanisms and identifying ways to restore these mechanisms is a promising strategy to treat tumors (22,43). High levels of SOX2 have been shown to inhibit apoptosis and reduce the effects of cancer therapeutics(5), however the exact molecular mechanism involved in controlling this phenotype have been obscure. We show that forced expression of SOX2 represses tumor suppressor genes PTEN and FoxO1, two well-established regulators of apoptosis (19,22), and identified miR-486–5p as a bonafide SOX2-regulated oncomiRs. Additionally, we demonstrate that miR-486–5p inhibition strongly induces apoptosis through activation of pro-apoptotic BH3-only protein Bim (Fig. 5) concurrent with up-regulation of PTEN (Fig. 3G and 3H). Bim is a down-stream effector of PTEN that has been implicated in regulation of cell death mechanisms in CSCs and resistance to GBM therapeutics (44). Also, forced expression of miR-486–5p made GBM neurosphere more resistant to IR (Fig. 4). Consistent with our observations that miR-486–5p acts as a critical determinant of GBM cell survival are reports showing that exosomes enriched with miR-486–5p inhibit apoptosis and reduce ischemic kidney injury by reducing PTEN expression and activating AKT signaling(45). Interestingly, a recent report shows that miR-486–5p is highly enriched in exosomes isolated from GBM patients and can be used as part of a 7 miRNA signature to diagnose GBM preoperatively (46). Also, miR-486–5p plays a key role in maintaining skeletal muscle size by inhibiting PTEN and FoxO1 and activating AKT signaling to promote survival of muscle cells and prevent muscle wasting (47), highlighting how SOX2 dysregulation alters essential physiological pathways leading to enhanced survival. Taken together, these results establish a mechanism in which Sox2 activates miR-486–5p resulting in PTEN and Bim dysregulation that contributes to the resistance to ionizing radiation (Fig. 7).
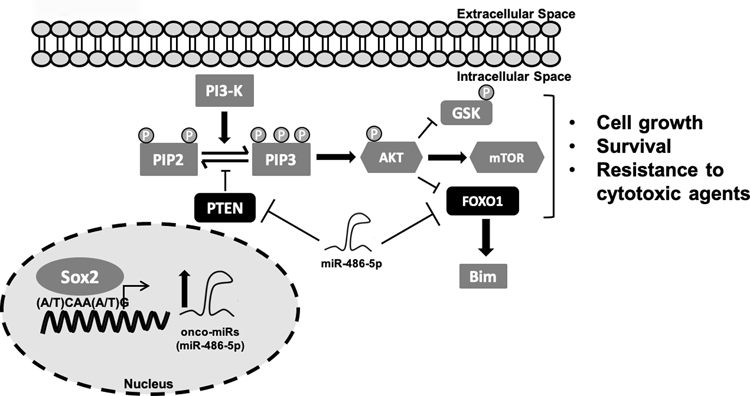
Figure 7:
Sox2 directly binds to the miR-486–5p promoter to activate expression in GBM neurospheres. miR-486–5p regulates expression of PTEN and FoxO1 by directly binding to their respective 3’UTRs and controlling mRNA levels. miR-486–5p activation indirectly contributes to AKT and Bim dysregulation resulting in enhanced cell growth and GBM neurosphere resistance to IR.
Substantial efforts have been devoted to understanding the downstream effectors of reprogramming transcription factors (e.g. SOX2) in an attempt to better understand tumorigenic mechanisms and identify new therapeutic avenues to treat cancer (5). We recently developed self-assembling miRNA-containing polymeric nanoparticles (nano-miRs) that effectively deliver miRNA mimics to human GBM cells in vitro and efficiently distribute through established orthotopic human GBM xenografts (15). Reprogramming transcription factors modulate miRNA networks and we have recently shown that reconstituting miRNA combinations (i.e. miR-148a + miR-296–5p) can have cooperative/synergistic effects leading to the long-term survival and actual cures in pre-established orthotopic GBM xenograft models (15). We expand upon these findings by delivering miR-486–5p antagomirs to pre-established GBM xenografts and show that in vivo miR-486–5p inhibition significantly reduced tumor growth in two different orthotopic GBM models and enhanced the effects of IR therapy (Fig. 6). Although our findings successfully translate the molecular pathway that we rigorously define in vitro to the more complex in vivo setting, they do not completely rule out a role for indirect effects due to the inherent promiscuous nature of miRNAs. Additionally, important future directions of this research is to determine the optimal combinations of miRNAs to “normalize”, through either reconstitution or inhibition, to have a meaningful impact on survival in pre-clinical models as either monotherapy or in combination with current standard-of-care radiation/chemotherapy aimed at producing durable responses by reducing the emergence of more aggressive tumors that commonly result from single agent targeted therapeutics.
In conclusion, SOX2 is an important regulator of tumorigenesis and the cancer stem cell phenotype that controls a vast network of coding and non-coding genes (5,48). Despite our growing understanding of the SOX2 targetome, the targets that specifically regulate the tumor-propagating stem-like phenotype, and the subset of regulated targets amenable to therapeutic intervention remain subject of intense investigation (5). Our current results support a novel mechanism where SOX2 coordinates oncogenic signals in GBM neurospheres by directly trans-activating miR-486–5p which targets tumor suppressor genes PTEN and FoxO1 leading to subsequent inhibition of Bim expression resulting in enhanced cell growth and survival of GBM neurospheres (Fig. 7). Our results demonstrate that miR-486–5p inhibition can be developed as a way to enhance the effects of current GBM standard of care.
Supplementary Material
SIGNIFICANCE:
This study identifies a novel axis that links core transcriptional drivers of cancer cell stemness to miR-486–5p-dependent modulation of tumor suppressor genes that feeds back to regulate glioma stem cell survival.
Acknowledgments
Financial Support:
The authors would like to thank the following organizations for financial support: NIH R25 research fellowship (IK), NSF Graduate Research Fellowship DGE-1232825 (YR), the Bloomberg~Kimmel Institute for Cancer Immunotherapy (JG), Microscopy Core Grant (S10 OD016374) and the United States NIH grants R01NS073611 (JL), R01EB016721 (JG), R01CA195503 (JG).
Footnotes
Conflict of Interest: The authors have no conflict of interest to disclose.
REFERENCES:
- 1.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014;157:580–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, et al. American Brain Tumor Association Adolescent and Young Adult Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-oncology 2016;18 Suppl 1:i1–i50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60 [DOI] [PubMed] [Google Scholar]
- 4.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev 2015;29:1203–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clinical and translational medicine 2014;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell stem cell 2013;12:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julian LM, McDonald AC, Stanford WL. Direct reprogramming with SOX factors: masters of cell fate. Current opinion in genetics & development 2017;46:24–36 [DOI] [PubMed] [Google Scholar]
- 8.Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer genomics & proteomics 2011;8:139–47 [PubMed] [Google Scholar]
- 9.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem cells 2009;27:40–8 [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer 2006;6:259–69 [DOI] [PubMed] [Google Scholar]
- 11.Acanda de la Rocha AM, Lopez-Bertoni H, Guruceaga E, Gonzalez-Huarriz M, Martinez-Velez N, Xipell E, et al. Analysis of SOX2-Regulated Transcriptome in Glioma Stem Cells. PloS one 2016;11:e0163155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC genomics 2011;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cazares H, Eberhart CG, et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015;34:3994–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Bertoni H, Lal B, Michelson N, Guerrero-Cazares H, Quinones-Hinojosa A, Li Y, et al. Epigenetic modulation of a miR-296–5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene 2016 [DOI] [PMC free article] [PubMed]
- 15.Lopez-Bertoni H, Kozielski KL, Rui Y, Lal B, Vaughan H, Wilson DR, et al. Bioreducible Polymeric Nanoparticles Containing Multiplexed Cancer Stem Cell Regulating miRNAs Inhibit Glioblastoma Growth and Prolong Survival. Nano letters 2018 [DOI] [PMC free article] [PubMed]
- 16.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004;64:7011–21 [DOI] [PubMed] [Google Scholar]
- 17.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes, chromosomes & cancer 2004;39:29–36 [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97 [DOI] [PubMed] [Google Scholar]
- 19.Koul D PTEN signaling pathways in glioblastoma. Cancer biology & therapy 2008;7:1321–5 [DOI] [PubMed] [Google Scholar]
- 20.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell death and differentiation 2018;25:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla S, Saxena S, Singh BK, Kakkar P. BH3-only protein BIM: An emerging target in chemotherapy. European journal of cell biology 2017;96:728–38 [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 2011;1813:1978–86 [DOI] [PubMed] [Google Scholar]
- 23.Ames H, Halushka MK, Rodriguez FJ. miRNA Regulation in Gliomas: Usual Suspects in Glial Tumorigenesis and Evolving Clinical Applications. Journal of neuropathology and experimental neurology 2017;76:246–54 [DOI] [PubMed] [Google Scholar]
- 24.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature reviews Drug discovery 2017;16:203–22 [DOI] [PubMed] [Google Scholar]
- 25.Donadelli M, Dando I, Fiorini C, Palmieri M. Regulation of miR-23b expression and its dual role on ROS production and tumour development. Cancer Lett 2014;349:107–13 [DOI] [PubMed] [Google Scholar]
- 26.Sheedy P, Medarova Z. The fundamental role of miR-10b in metastatic cancer. Am J Cancer Res 2018;8:1674–88 [PMC free article] [PubMed] [Google Scholar]
- 27.Sha HH, Wang DD, Chen D, Liu SW, Wang Z, Yan DL, et al. MiR-138: A promising therapeutic target for cancer. Tumour Biol 2017;39:1010428317697575 [DOI] [PubMed] [Google Scholar]
- 28.Dai H, Xu LY, Qian Q, Zhu QW, Chen WX. MicroRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. Biosci Rep 2019;39. [DOI] [PMC free article] [PubMed]
- 29.Lang B, Zhao S. miR-486 functions as a tumor suppressor in esophageal cancer by targeting CDK4/BCAS2. Oncology reports 2018;39:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Li M, Hu Y, Shi N, Yu H, Liu H, et al. miR-486–5p attenuates tumor growth and lymphangiogenesis by targeting neuropilin-2 in colorectal carcinoma. OncoTargets and therapy 2016;9:2865–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ, Lu SD, et al. Direct repression of the oncogene CDK4 by the tumor suppressor miR-486–5p in non-small cell lung cancer. Oncotarget 2016;7:34011–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X, et al. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatology research : the official journal of the Japan Society of Hepatology 2015;45:1312–22 [DOI] [PubMed] [Google Scholar]
- 33.Wang LS, Li L, Li L, Chu S, Shiang KD, Li M, et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 2015;125:1302–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song L, Lin C, Gong H, Wang C, Liu L, Wu J, et al. miR-486 sustains NF-kappaB activity by disrupting multiple NF-kappaB-negative feedback loops. Cell research 2013;23:274–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaham L, Vendramini E, Ge Y, Goren Y, Birger Y, Tijssen MR, et al. MicroRNA-486–5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood 2015;125:1292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Ji C, Guo S, Su X, Zhao X, Zhang S, et al. The miR-486–5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget 2017;8:72835–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mees ST, Mardin WA, Sielker S, Willscher E, Senninger N, Schleicher C, et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Annals of surgical oncology 2009;16:2339–50 [DOI] [PubMed] [Google Scholar]
- 38.Ren C, Ren T, Yang K, Wang S, Bao X, Zhang F, et al. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing’s sarcoma through the PI3K/Akt pathway. Journal of experimental & clinical cancer research : CR 2016;35:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Chen K, Li L, Li R, Zhang J, Ren W. Overexpression of SOX2 is involved in paclitaxel resistance of ovarian cancer via the PI3K/Akt pathway. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 2015;36:9823–8 [DOI] [PubMed] [Google Scholar]
- 40.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology 2012;13:283–96 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nature cell biology 2011;13:1092–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivas S, Gomez-Oro C, Anton IM, Wandosell F. Role of Akt Isoforms Controlling Cancer Stem Cell Survival, Phenotype and Self-Renewal. Biomedicines 2018;6. [DOI] [PMC free article] [PubMed]
- 43.Ma DD, Yang WX. Engineered nanoparticles induce cell apoptosis: potential for cancer therapy. Oncotarget 2016;7:40882–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aroui S, Dardevet L, Najlaoui F, Kammoun M, Laajimi A, Fetoui H, et al. PTEN-regulated AKT/FoxO3a/Bim signaling contributes to Human cell glioblastoma apoptosis by platinum-maurocalcin conjugate. The international journal of biochemistry & cell biology 2016;77:15–22 [DOI] [PubMed] [Google Scholar]
- 45.Vinas JL, Burger D, Zimpelmann J, Haneef R, Knoll W, Campbell P, et al. Transfer of microRNA-486–5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney international 2016;90:1238–50 [DOI] [PubMed] [Google Scholar]
- 46.Saeideh Ebrahimkhani FV, Susannah Hallal, Heng Wei, Maggie Lee, Paul Young, Laveniya Satgunaseelan, Brindha Shivalingam, Catherine Suter, Michael Buckland, View ORCID ProfileKimberley Kaufman. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. bioRxiv 2018 [DOI] [PMC free article] [PubMed]
- 47.Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. The Journal of clinical investigation 2014;124:2651–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song WS, Yang YP, Huang CS, Lu KH, Liu WH, Wu WW, et al. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. Journal of the Chinese Medical Association : JCMA 2016;79:538–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.